Abstract
The mechanism of action of the antiviral compound 3(2H)-isoflavene against Sabin type 2 poliovirus has been studied, and interference with virus uncoating was demonstrated. Isolation and sequencing of drug-resistant variants revealed single amino acid substitutions (I194M or D131V) in the VP1 capsid protein. While M194 is located in a hydrophobic pocket and should partially fill the space occupied by the isoflavene ring, V131 is exposed on the VP1 surface, forming a contact with VP4. The D131V mutation most likely induces local conformational changes in VP1 and/or VP4 that affect viral flexibility. Two dependent variants, N53S of VP1 and K58E of VP4, both located on the inner surface of the capsid, near the threefold axis of symmetry, were also selected. Both mutations affected viral stability, allowing the transition to 135S particles in the absence of drug, without the involvement of the viral receptor.
Enteroviruses, a genus of the family Picornaviridae, which includes polioviruses, echoviruses, group A and B coxsackieviruses, and numbered enteroviruses (48), have been implicated in a large variety of human diseases, ranging from mild illnesses to severe clinical diseases such as myocarditis, meningitis, encephalitis, and paralysis (31, 37).
Polioviruses, the prototype of the enterovirus group, have a positive-sense single-stranded RNA genome packaged inside a protein capsid. The genome codes for 11 individual polypeptides, initially synthesized as a large precursor and subsequently processed into mature polypeptides by virus-encoded proteases. The icosahedral capsid contains 60 copies of each of the VP1, VP2, and VP3 proteins, which interact with an additional smaller polypeptide, VP4, situated on the inner capsid surface (42). The first step in the poliovirus replication cycle is the attachment of the viral particle to its receptor on the cell surface (32). The availability of the three-dimensional structures of poliovirus and other members of the picornavirus family, alone or in complex with their receptors, has greatly enhanced the understanding of the receptor binding and the early steps of poliovirus replication at the atomic level (14, 22, 36, 45). The poliovirus receptor binding site has been mapped to the bottom of a canyon around the icosahedral fivefold axis formed by VP1, VP2, and VP3 molecules (14). The canyon is also the site of entry into an internal hydrophobic pocket normally occupied by an uncharacterized sphingosine-like molecule (pocket factor), the precise function of which is still unknown (11, 21, 27).
Cell surface attachment is followed by conformational changes of the viral capsid leading to uncoating of the viral genome. The loss of the VP4 protein and the extrusion of the VP1 amino terminus generate an altered A particle with a reduced sedimentation coefficient and increased hydrophobicity (14). It is still unknown whether uncoating of poliovirus RNA occurs concomitantly with penetration or subsequently in the cytosol. Successful completion of this process is a prerequisite for the initiation of the replication cycle (10, 21), thus making receptor binding and virus uncoating important targets for antiviral therapy, as demonstrated by the extensive literature describing antipicornavirus drugs (17, 44, 49, 56).
Human rhinovirus (HRV) and poliovirus infections can be prevented by uncoating inhibitors that either block virus-receptor interactions (major-group HRVs) or prevent the low pH-induced conformational change necessary for uncoating (minor-group HRVs) (2, 13, 17, 25, 38, 39, 56). Among these inhibitors, the drug pleconaril (Viropharma, Exton, Pa.), a novel molecule with broad antipicornavirus activity, has been used for the treatment of the common cold in humans (19) and more recently to cure enterovirus infections and poliovirus type 2 (PV2) and PV3 excretion in immunodeficient subjects (28, 30, 40, 43, 46, 47).
4′,6-Dichloroflavan, a natural compound extracted from plants, is a potent inhibitor of replication of several rhinovirus serotypes and prevents viral uncoating (1, 52). A synthetic derivative of this molecule is 3(2H)-isoflavene, which has been reported to be effective not only against rhinovirus but also against a broader spectrum of picornaviruses (4, 16, 51).
Previous biological characterization of the Sabin PV2 and 3(2H)-isoflavene drug-resistant and -dependent variants selected after multiple passages suggested that the compound may act by interfering with viral replication at a phase between uncoating and viral RNA synthesis (9, 15).
In the present study, we report on the molecular characterization of resistant and dependent viral variants capable of either bypassing the 3(2H)-isoflavene-induced block of viral replication or growing only in the presence of the compound. Differently from previous investigations, these variants were selected by a single-step passage in cell culture, thus allowing the identification of single mutations that correlate with the associated phenotype. Our results demonstrate that 3(2H)-isoflavene exerts its action during the uncoating step and exclude an effect of 3(2H)-isoflavene on viral RNA synthesis. Analysis of mutations conferring drug resistance within the structural context of the poliovirus capsid also provides strong evidence that 3(2H)-isoflavene inserts into the well-known hydrophobic pocket, thereby blocking the uncoating process.
The binding and uncoating of drug-dependent variants were unaffected by the drug: the mutations conferring dependence on the compound for infectivity were identified and found to be important for the stability of the derived viruses.
MATERIALS AND METHODS
Compounds.
3(2H)-Isoflavene was synthesized as described previously (4, 9). A stock solution in ethanol (1 mg/ml) was stored at 4°C before dilution to a final concentration of 20 μM with tissue culture medium. WIN 51711 (5-[7-[4-(4,5-dihydro-2-oxazolyl)phenoxyl]heptyl]-3-methylisoxazole) was obtained from the Sterling-Winthrop Research Institute (Rensselaer, N.Y.) and was used as a control at a final concentration of 1.5 μg/ml of medium (4.4 μM) in dimethyl sulfoxide (13, 56).
Cells and viruses.
HeLa R19 cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 200 U of penicillin per ml, and 200 μg of streptomycin per ml (growth medium). PV2 Sabin (ATCC VR-1003) was used as the reference strain. A PV2 viral stock was produced after transfection of HeLa cells with viral RNA transcribed from pBR322 plasmid p2/T7/SAB2 (41). This preparation was used for selection of resistant and dependent variants. Confluent cells were infected with PV2 Sabin at a multiplicity of infection (MOI) of 5 PFU per cell in DMEM with 2% FCS (maintenance medium). After virus adsorption for 1 h at 37°C, fresh maintenance medium was added and the cells were incubated at 37°C. When the viral cytopathic effect was complete, the cells were harvested and titration of virus was performed by plaque assay (15).
Selection of resistant and dependent variants.
PV2-resistant and -dependent variants were selected in HeLa cell monolayers by plaquing serial dilutions (10−1 to 10−5 PFU) of viral stock and including 20 μM 3(2H)-isoflavene in the overlay medium during growth, which corresponds to approximately 12.0 times the 50% effective concentration (1.7 μM) (16). Several single well-developed viral plaques were randomly picked from each plate. Differentiation between drug-resistant and drug-dependent variants was achieved by comparing the titers in the presence of the compound and those in the absence of the compound. The selected plaques were than subjected to at least two further cycles of plaque purification in the presence of the compound. Before the experiments were run, stock viruses were treated with chloroform to remove the compound (52).
Sequence determination.
Sequencing of the entire genomes of viral variants and PV2 Sabin was carried out as described previously (12). After heat denaturation, genomic RNA was subjected to reverse transcription and cDNA amplification with a panel of synthetic oligonucleotides as primers. The amplified products were purified and sequenced with a Dye Deoxy Terminator cycle sequencing kit (Applied Biosystems, Perkin-Elmer, Foster City, Calif.) and the same primers used for amplification. The sequences obtained were aligned with the sequence of the reference PV2 Sabin strain (41, 53).
Construction of recombinant variants.
Variant viral RNA fragments containing the selected mutations were reverse transcribed, amplified by PCR, and subcloned into plasmid p2/T7/SAB2 (41). To obtain the A919G mutation, a fragment spanning from nucleotides 162 to 1340 of the D6/01 viral genome was generated and restricted with MluI-SacI; the A2639G mutation was obtained after reverse transcription-PCR amplification of the region from nucleotides 1201 to 2840 of D7/01 RNA and digestion with SacI-BssHII; the fragments from nucleotides 2700 to 4640 of R1/01 and R2/01, which contain the mutations A2873G and A3063G, were amplified by PCR and restricted with BssHII-BstEII and BssHII-SnaBI, respectively. Nucleotide changes were confirmed by sequencing the recombinant plasmids. Viruses were obtained by transfection of HeLa cells with RNAs transcribed from the plasmids.
RNA transfection.
RNA transfection was performed as described previously (9, 15), with minor modifications. Briefly, HeLa cells at 60 to 70% confluence were transfected with viral RNAs of PV2 Sabin and recombinant variants in the presence of 0.5 mg of DEAE-dextran (molecular weight, 500,000) per ml in phosphate-buffered saline (PBS) with calcium and magnesium ions. Following incubation for 30 min at room temperature in the presence or absence of the drug, the inoculum was removed and maintenance medium was added. After 6 h at 37°C, the cells were frozen-thawed three times and centrifuged at low speed to remove cell debris. Titers were determined by plaque assay in the absence of drug for PV2 Sabin and resistant variants and in the presence of 3(2H)-isoflavene for the dependent variant.
Viral RNA synthesis.
HeLa cell monolayers in 60-mm-diameter dishes were incubated for 30 min at 37°C with PV2 Sabin and viral variants at MOIs of 10 or were transfected with RNA extracted from purified viruses. After a single step of viral replication (6 h postinfection) in the presence or absence of drug (20 μM), the cells were treated with TRIzol reagent (Invitrogen, Glasgow, United Kingdom) and total RNA was extracted by following the instructions of the manufacturer. Dot blot analysis was performed with 5 to 10 μg of RNA as described previously (54). Filters were hybridized with a 32P-labeled probe whose sequence spanned from nucleotides 1500 to 1940 of the PV2 genome sequence. Following autoradiography, RNA was quantified with Quantity One software (Bio-Rad Laboratories, Hercules, Calif.).
Heat inactivation.
Experiments of heat inactivation of viral infectivity were performed as described previously (16). Briefly, viral suspensions diluted in PBS (5 × 108 PFU/ml) were incubated overnight at 4°C with or without 3(2H)-isoflavene (20 μM) or with WIN 51711 (4.4 μM) and for 30 min at temperatures of 37, 42, and 48°C. After the mixtures were cooled on ice to stop the reaction, they were diluted and the titers were determined by plaque assay in the presence or absence of the compound.
Virus labeling and purification.
Viruses were radiolabeled with [35S]methionine (1,000 Ci/mmol; ICN) as described previously (9) and purified by CsCl2 gradient centrifugation. The titers of the purified viruses were determined by plaque assay.
Binding assay.
Six-well dishes containing HeLa cell (8 × 105) monolayers were precooled on ice for 10 min, washed with maintenance medium, and then incubated for 1 h at 4°C with 200,000 cpm of [35S]methionine-labeled virions in the presence or absence of compounds [20 μM 3(2H)-isoflavene, 4.4 μM WIN51711]. The cells were washed three times with ice-cold PBS to remove unbound virus and lysed in 0.2 ml of 0.2% Nonidet P-40 in PBS. Cell-associated radioactivity was determined by scintillation counting. The assay was performed in triplicate.
Sucrose gradient analysis.
HeLa cell monolayers in 60-mm-diameter dishes were chilled for 30 min on ice and infected with 500,000 cpm of [35S]methionine-labeled viruses in the presence or absence of 3(2H)-isoflavene or WIN 51711. Adsorption was allowed to proceed for 60 min on ice. The cells were washed three times with ice-cold PBS to remove unbound particles, and 1 ml of DMEM supplemented with 2% FCS with or without the drugs was added. The cells were incubated at 37°C for 0, 30, or 60 min. The monolayers were washed twice with ice-cold PBS, lysed with 0.8 ml of lysis buffer (0.01 M Tris [pH 7.2], 0.15 M NaCl, 0.1% sodium dodecyl sulfate, 1% deoxycholate, 1% Triton X-100), and frozen at −80°C. The lysates were thawed, clarified by centrifugation at 10,000 × g for 5 min, and layered onto 15 to 30% sucrose gradients prepared in PBS. Gradients were centrifuged at 4°C in a Beckman SW41 rotor at 40,000 rpm for 1.5 h. Fractions were collected from the top, and the radioactivity of each fraction was determined in a scintillation counter.
Thermostability assay for the dependent variant.
Variant D6/01 at an MOI of 10 PFU was bound to HeLa cells for 1 h at 4°C in the absence of the drug. Cells were washed and incubated at different temperatures (25, 29, 33, and 37°C) for 6 h in the presence (20 μM) or absence of the compound. Viral titers were determined by plaque assay in the presence of the compound. The drug-plating index was expressed as the ratio of the viral titer obtained in the presence of the compound to that obtained in the absence of the compound.
Modeling and structural analysis.
Substitutions on individual residues were analyzed in the structural context of the PV2 (strain Lansing) crystal structure (Protein Data Bank [PDB] entry 1EAH) (3, 26) with Insight II modeling software (Accelrys, San Diego, Calif.). The few amino acid sequence differences (no insertions or deletions) that existed between the Sabin strain and the Lansing strain VP1 to VP4 capsid proteins could readily be modeled into the 1EAH template structure without consequences regarding the local or overall protein conformation. Modeling of the 3(2H)-isoflavene compound into the pocket was performed manually by using PDB entry 1EAH and related structures of poliovirus-inhibitor complexes as a guide. Icosahedral assemblies were constructed by using the BIOMT transformations provided in the 1EAH PDB file. Pictures were generated with WebLab software (Accelrys).
RESULTS
Selection of 3(2H)-isoflavene-resistant and -dependent variants.
Drug-dependent and -resistant variants were isolated by selection of primary plaques developed during a single passage in HeLa cells in the presence of 20 μM 3(2H)-isoflavene. Viral stocks from seven randomly selected PV2 variants were propagated in the presence of drug, and titers were determined in the presence or absence of the compound. Five of the seven variants showed a resistant phenotype, growing at the same titer and exhibiting a similar plaque shape independent of the presence of compound, with a drug-plating index of approximately 1. The remaining two variants behaved as drug-dependent variants, requiring the presence of the compound to yield high-titer viral progeny (drug-plating index, >104).
Amino acid substitutions in 3(2H)-isoflavene-resistant and -dependent variants.
Full-length genome sequencing of the new viral variants revealed that a single amino acid substitution was associated with each phenotype (Table 1). Four of the five drug-resistant variants (R2/01 to R5/01) carried an identical mutation, an isoleucine-to-methionine change at residue 194 of the VP1 capsid protein; in the remaining variant (R1/01), aspartic acid was replaced by valine at residue 131 of VP1. The amino acid changes of the drug-dependent phenotype were located either in VP4 (variant D6/01), with lysine 58 replaced by glutamic acid, or in VP1 (variant D7/01), with asparagine 53 replaced by serine.
TABLE 1.
Nucleotide and amino acid changes in PV2 Sabin and 3(2H)-isoflavene-resistant and -dependent variants
| Variant(s) | Phenotype | Nucleotide change | Amino acid changea |
|---|---|---|---|
| R1/01 | Resistant | A2873T | Asp to Val (1131) |
| R2/01 to R5/01 | Resistant | A3063G | Ile to Met (1194) |
| D6/01 | Dependent | A919G | Lys to Glu (4058) |
| D7/01 | Dependent | A2639G | Asn to Ser (1053) |
The first digit of each number in parentheses is the number of the viral capsid protein, and the next three digits give the number of the amino acid in that protein.
Recombinant variants carrying the respective single nucleotide changes were constructed by site-specific mutagenesis, confirming that the amino acid changes identified were both necessary and sufficient to generate the observed phenotypes.
Computer-assisted modeling of 3(2H)-isoflavene-PV2 interaction and mechanism of drug action.
Although no structural data are available for the PV2 Sabin strain, the high amino acid sequence conservation of the VP1 to VP4 capsid proteins among polioviruses (for example, PV2 Sabin and PV2 Lansing [PDB entry 1EAH]) (26) allowed us to construct very accurate structural models and to address the mechanisms of drug binding. By placing 3(2H)-isoflavene at the bottom of the known hydrophobic pocket in VP1, a mechanism of action can be supposed for the four identical Ile194Met resistance mutations (in R2/01 to R5/01) (Fig. 1A): addition of a methyl group at the end of the amino acid 194 side chain would affect drug binding inside the hydrophobic pocket by steric hindrance. An analogous mechanism which confers resistance to WIN 51711, a known inhibitor of poliovirus uncoating, was reported for mutation Ile192Phe in PV3 Sabin (34).
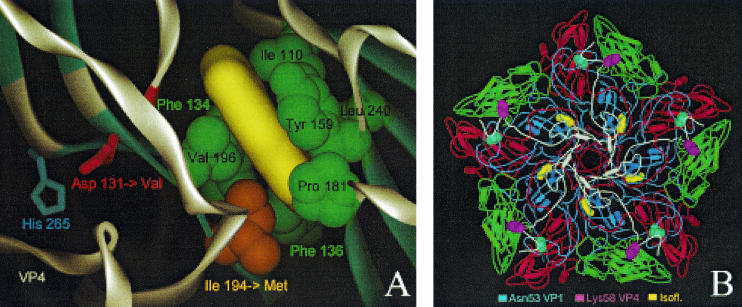
FIG. 1.
Localization of drug-resistant and -dependent mutations and modeling of drug binding. (A) Close-up of the 3(2H)-isoflavene binding site near VP1 Ile194. The 3(2H)-isoflavene ligand (yellow) is modeled into the bottom of the hydrophobic pocket formed by VP1 Ile110, Phe134, Phe136, Tyr159, Pro181, Ile194, Val196, and Leu240. Mutation of Ile194 into a longer Met side chain (indicated by an orange arrow) partially fills the bottom of the pocket, abolishing efficient binding of the isoflavene. Also shown is VP1 Asp131 (red), a second mutation (Asp→Val) that confers drug resistance. Asp131 forms contacts with VP4 (through His265) and the backbone (NH of Tyr198) of one of the beta strands that is part of the pocket wall. In particular, loss of the latter contact could induce a local conformational change of the Val196-Pro197-Tyr198 backbone segment (highlighted in red), leading to a change in shape and, possibly, also leading to a change in the accessibility of the pocket. (B) The locations of the two mutations resulting in a drug-dependent phenotype, VP1 Asn53 to Ser and VP4 Lys58 to Glu, are shown in the context of a VP1 to VP4 pentamer seen from inside the capsid (VP1, blue; VP2, green; VP3, red; VP4, white). Both residues lie on the inner surface of the capsid and are far from the isoflavene-binding pocket (yellow).
Because the mutation is located immediately outside the hydrophobic pocket, the mutation of Asp131 to valine can abolish both contact to VP4 (mediated by the VP1 His265 side chain) and contact to the backbone (mediated by the NH of Tyr198) of the neighboring β strand that is part of the pocket wall (Fig. 1A). The absence of the latter contact and the rearrangement of the hydrophobic Val131 side chain could induce a local conformational change of the segment Val196-Pro197-Tyr198 that affects the pocket shape and/or that increases the flexibility of VP1. The latter effect has been postulated to confer drug resistance by the mutation Asp129Val found in the WIN 51711-resistant PV3 Sabin variant (34).
While the model allowed us to derive an acceptable explanation for drug resistance, conclusions regarding the underlying mechanism for the opposite phenotype, drug dependence, associated with VP1 Asn53Ser and VP4 Lys58Glu mutations, were much harder to draw. Both residues are situated on the inner surface of the capsid (Fig. 1B), far from the drug-binding pocket, with VP4 Lys58 completely exposed and VP1 Asn53 making contacts with other VP1 and VP3 residues. These contacts would be weakened or abolished if asparagine was replaced by serine. Similar mutations, including an identical VP1 Asn51Ser change, have been observed in WIN 51711-dependent PV3 Sabin variants (34), confirming that this internal region of the capsid is a hot spot for drug-dependent mutations.
Although the modeling analysis and the location of the mutations within VP1 strongly suggested that 3(2H)-isoflavene interferes with the viral cell entry and/or uncoating process, we also performed a more detailed biological characterization of one resistant variant (R2/01) and one dependent variant (D6/01).
Effect of 3(2H)-isoflavene on viral RNA synthesis.
To exclude a possible direct interaction of the compound with viral RNA synthesis, HeLa cells were transfected with purified RNAs of the VP2 Sabin, R2/01, or D6/01 variant in the presence or absence of drug. The cells were lysed 6 h after transfection. Total RNAs were extracted and adsorbed on filter paper, and RNA synthesis was measured by dot blot hybridization, as described in Materials and Methods (Fig. 2A). Viral infection with the PV2 Sabin strain or variants was monitored in parallel (Fig. 2B). HeLa cells were infected with the three viruses at MOIs of 10 at 37°C in the presence or absence of the compound. Infection was stopped at 6 h to avoid a possible inhibitory effect of the drug on newly synthesized viral particles.
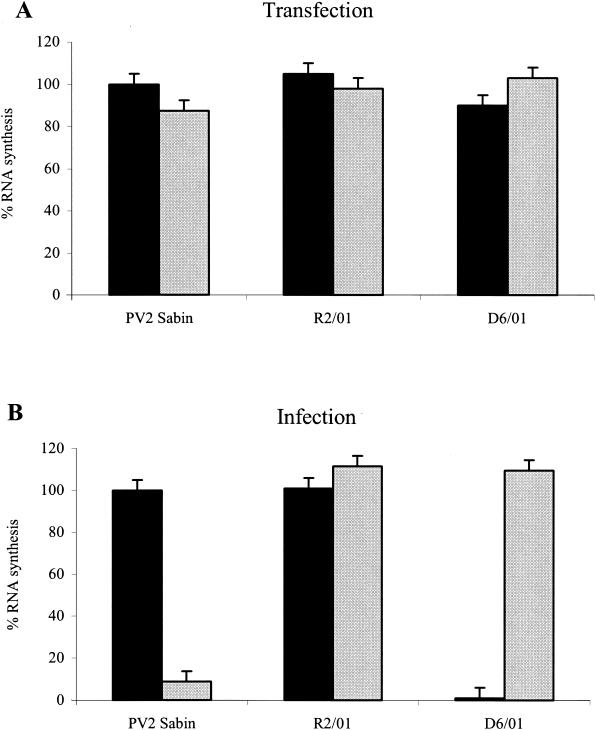
FIG. 2.
Effect of 3(2H)-isoflavene on RNA synthesis of PV2 Sabin, R2/01, and D6/01 variants. (A) HeLa cells monolayers at 60% confluency were transfected with purified viral RNA from PV2 Sabin and viral variants in the presence of 0.5 mg of DEAE-dextran per ml. Following incubation for 6 h in the presence or absence of the drug (20 μM), RNAs were extracted, adsorbed on filter paper (10 μg), and hybridized with a probe (specific for PV2 nucleotides 1500 to 1940). After autoradiography, RNAs were quantified as described in Materials and Methods. (B) HeLa cell monolayers were infected with viral variants and the reference Sabin PV2 strain at MOIs of 10 and incubated at 37°C in the absence or presence of the antiviral compound (20 μM). After a single step of viral replication (6 h postinfection), total RNAs were extracted and adsorbed (5 μg) on filter paper. Hybridization was performed as described for panel A. Solid bars, without antiviral compound; shaded bars, with antiviral compound.
As shown in Fig. 2A, 3(2H)-isoflavene had no effect on RNA synthesis of PV2 Sabin or the R2/01 and D6/01 variants when it was added after RNA transfection. On the contrary, the presence of the compound during infection led to a 95% decrease in the level of PV2 Sabin RNA synthesis (Fig. 2B). As expected, the D6/01 variant underwent efficient genome replication only in the presence of 3(2H)-isoflavene, whereas the drug did not interfere with RNA synthesis of the resistant variant (Fig. 2B).
Effect of 3(2H)-isoflavene on thermal inactivation.
To investigate whether 3(2H)-isoflavene could bind to PV2 Sabin or R2/01 variants and protect them from heat inactivation, the viruses were first incubated overnight at 4°C and then at 37, 42, and 48°C for 30 min in the presence or absence of the compound. Viral titers were determined by plaque assay. The capsid binding compound WIN 51711 was included as a control. As shown in Fig. 3A, PV2 Sabin was stable up to 42°C in the absence of drug, and a significant (approximately 3 log10) decrease in viral titer was observed only when the virus was incubated at 48°C. At this temperature, the presence of 3(2H)-isoflavene completely protected PV2 Sabin from thermal inactivation with an efficiency comparable to that of WIN 51711. At 48°C, variant R2/01 was sensitive to heat inactivation in either the presence or the absence of the compound (Fig. 3B). The reduction in viral titer of approximately 4 log10 PFU/ml, slightly higher than that observed for PV2 Sabin, strongly indicated that the drug did not bind to the capsid of this variant. The R1/01 variant showed a heat inactivation profile identical to that of R2/01 (data not shown).
FIG. 3.
Effect of 3(2H)-isoflavene on thermal inactivation of PV2 Sabin and R2/01. Each virus (5 × 108 PFU) was incubated overnight at 4°C in the presence or absence of antiviral compounds. Following 30 min of incubation at the indicated temperatures, the viruses were chilled on ice and titers were determined by plaque assay (the data represent the means of three separate experiments). Solid bars, without antiviral compounds; shaded bars, with WIN51711; open bars, with 3(2H)-isoflavene.
Effect of 3(2H)-isoflavene on binding and uncoating.
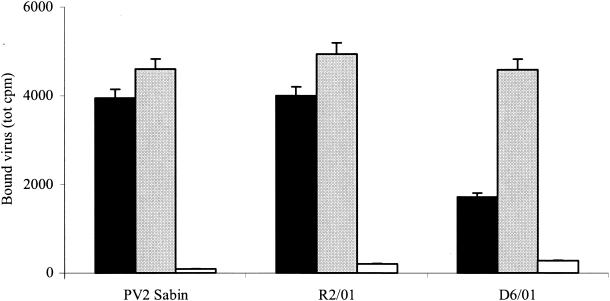
To determine whether 3(2H)-isoflavene could interfere with binding of PV2 Sabin during infection, we measured the amount of cell-associated radiolabeled virus after 1 h of adsorption to HeLa cells at 4°C in the presence or absence of the compound. The resistant and dependent variants were also assayed. As shown in Fig. 4, PV2 Sabin and R2/01 bound to the cells to the same extent in the presence or absence of the drug, indicating that 3(2H)-isoflavene did not affect virus adsorption to cells. The D6/01 variant showed a binding capacity similar to that of PV2 Sabin in the presence of drug. Conversely, binding of this variant decreased (2.7-fold) in the absence of drug. This result is more likely due to the instability of the D6/01 variant rather than to a decrease in binding capacity: this is better demonstrated by the thermolability experiments described below. Similar results have been also reported for PV3 WIN 51711-dependent variants (33).
FIG. 4.
Effect of 3(2H)-isoflavene on binding of PV2 Sabin, R2/01, and D6/01 variants. HeLa cell monolayers were infected with 200,000 cpm of [35S]methionine-labeled purified viruses in the presence or absence of the drug (20 μM). After 1 h of incubation at 4°C, the cells were washed with PBS and lysed with 0.2% Nonidet P-40, and the cell-bound radioactivity was measured in a beta-scintillation counter. A polyclonal antibody against PV2 Sabin was used as the virus binding inhibitor. The experiment was repeated three times. Solid bars, without 3(2H)-isoflavene; shaded bars, with 3(2H)-isoflavene; open bars, with anti-PV2 antibody.
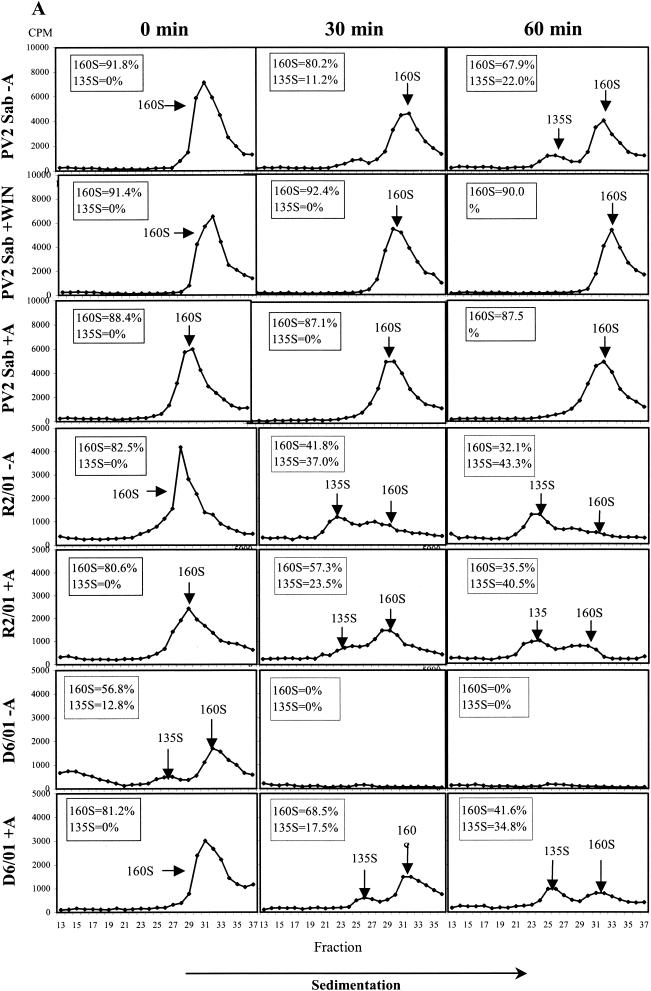
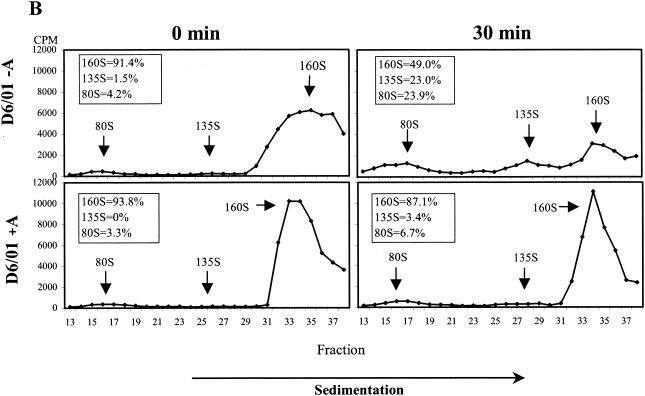
The effect of the compound on viral uncoating was studied by analyzing the pattern of viral particle and subviral particle accumulation during HeLa cell infection with PV2 Sabin. The R2/01 and D6/01 variants were also included in the study. Binding of radiolabeled purified viruses was synchronized by 1 h of incubation at 4°C; after removal of unbound viruses, the cells were kept at 37°C for 0, 30, and 60 min in the presence or absence of compound and then lysed. The conversion from 160S native virions into 135S particles was measured after sucrose gradient centrifugation of cell-associated radiolabeled viruses. WIN 51711 was included as a control (13, 56). As shown in Fig. 5A, 3(2H)-isoflavene completely inhibited the formation of PV2 Sabin 135S particles, proving to be as effective as WIN 51711. The PV2 subviral particles released in the medium were to a large extent composed of 160S particles and a very small amount of 135S particles, which appeared after 30 to 60 min at 37°C (data not shown). In the case of the R2/01 variant, uncoating was unaffected by the drug, and the kinetics of conversion from 160S to 135S and 80S particles were faster compared to those for PV2 Sabin. In the presence of drug, the D6/01 variant showed a normal uncoating process, with the kinetics comparable to those for variant R2/01. In the absence of drug, D6/01 160S virions were converted to 135S and 80S particles even at time zero after binding, indicating the marked instability of this virus (Fig. 5A).
FIG. 5.
Effect of 3(2H)-isoflavene on sucrose gradient sedimentation profiles of PV2 Sabin (Sab), R2/01, and D6/01 variants. (A) Binding of purified [35S]methionine-labeled viruses to HeLa cells was synchronized by 1 h of incubation at 4°C in the presence or absence of the compound (20 μM). After removal of unbound virus, cells were incubated in the presence or absence of the compound at 37°C for the indicated times and then lysed. Cell-associated radiolabeled viral particles were centrifuged through a 15 to 30% sucrose gradient. Fractions were collected, and radioactivity was measured in a beta-scintillation counter. The percentages of mature 160S and 135S viral particles are indicated. WIN 51711 was used in the assay as an inhibitor of uncoating. (B) [35S]methionine-labeled variant D6/01 virions were incubated with or without compound at 37°C without cells for the indicated times. Virus-associated radioactivity was analyzed on a 15 to 30% sucrose gradient, as specified above. The percentages of the 160S, 135S, and 80S viral particles are indicated.
The instability of the dependent variant was also confirmed by experiments in which radiolabeled D6/01 particles were incubated at 37°C with or without drug in the absence of cells. Analysis by sucrose gradient centrifugation showed that, in the absence of drug, mature 160S viral particles were rapidly converted into the thermodecay products of 135S and 80S (Fig. 5B). Conversely, the addition of the compound restored the sedimentation profile characteristic of PV2 Sabin (Fig. 5B). Similar experiments could not be performed with the D7/01 variant due to the extreme instability of this virus in the absence of the compound, which did not enable the purification of 160S viral particles (data not shown).
Thermolability of the drug-dependent variant.
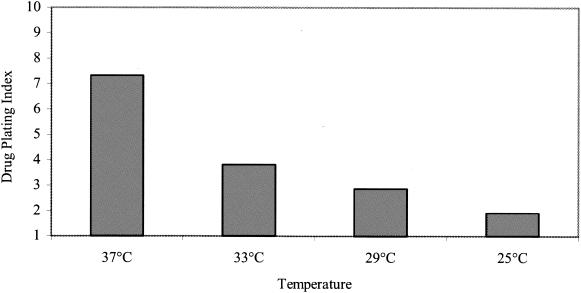
In an attempt to prevent the spontaneous degradation of the D6/01 variant in the absence of drug at 37°C, further experiments were carried out at lower incubation temperatures. After virus adsorption to HeLa cells for 1 h at 4°C, the cells were incubated for 6 h at 25, 29, 33, or 37°C in the presence or absence of the compound. Viral titers were determined by plaque assay in the presence of the compound. As shown in Fig. 6, the stability of the viral particles was gradually recovered by lowering the growth temperature. At 25°C, the titers in the absence or presence of drug were found to be closer (drug-plating index, approximately 2).
FIG. 6.
Thermostability of the dependent variant. HeLa cells were incubated with the D6/01 variant in the absence of the drug for 1 h at 4°C. After infection for 6 h in the presence or absence of the drug at the indicated temperatures, the cells were lysed and viral titers were determined by plaque assay in the presence of the compound. The drug-plating index is expressed as the ratio between the plaque titer in the presence of the drug and that in the absence of the drug.
DISCUSSION
Even though successful vaccination with live oral vaccine has pushed poliomyelitis to the verge of global eradication (6, 23), the discovery of prolonged poliovirus excretion by immunodeficient individuals (24, 29) has raised increasing concern in the World Health Organization. These individuals could in fact represent reservoirs for both wild-type and vaccine-derived poliovirus strains after eradication. Indeed, at least three outbreaks of poliomyelitis caused by highly mutated vaccine strains have been reported so far (7, 8, 24). The only available therapy for these patients is immunoglobulin substitution therapy, which, however, has not proved to be sufficient for virus clearance from the intestine for some long-term poliovirus excretors. Recently, the new compound pleconaril (VP 63843; Viropharma) has been used for this purpose, proving effective against PV2, PV3, and other enteroviruses but not against PV1 (30). Treatment of these patients thus requires the development of new specific therapies active against poliovirus infections and, possibly, also other important picornavirus infections against which vaccines are not yet available (18, 55).
Soon after cell entry, native infectious poliovirus particles undergo a conformational change induced by the cellular receptor (PVR), resulting in the formation of subviral A particles which sediment at 135S and which still contain genomic viral RNA. Release of the RNA from the protein shell creates the subviral B particles, which sediment at 80S. During this process the virus presumably penetrates into the cytosol by breaking the endosomal membrane (21). Understanding of the relevant events during early viral infection has been greatly improved by the availability of specific inhibitors and the analysis of resistant and dependent variants (33-35).
3(2H)-isoflavene, a derivative of 4′,6-dichloroflavan, specifically inhibits poliovirus type 2 replication in HeLa cells at a concentration of 20 μM. At this concentration, viral protein synthesis and virus-induced shutoff of cellular protein synthesis are inhibited (15). The results presented in this report show that the compound does not affect binding of poliovirus to the cellular receptor and efficiently blocks uncoating of Sabin PV2, proving that it is as effective as WIN 51711. Genovese et al. (15) found evidence for a possible effect of 3(2H)-isoflavene on viral RNA synthesis. Most likely, this observation was caused by the long incubation time in the presence of the antiviral agent prior to virus recovery from transfected cells (18 h). In the present study, a shorter incubation period (6 h) discriminated the effect of the drug on the uncoating of newly synthesized viral particles from apparent interference with transcription from input RNA.
To identify the compound binding site, drug-resistant variants were isolated and sequenced and the amino acid substitutions responsible for the observed phenotype were localized. We demonstrated that single amino acid substitutions (Ile194Met or Asp131Val) in the VP1 capsid protein are responsible for the resistant phenotype, and site-directed mutagenesis confirmed that they independently confer drug resistance.
The major determinant of 3(2H)-isoflavene resistance, VP1 Met194, is located in the well-known VP1 hydrophobic pocket, with a second resistance mutation, Asp131Val, being close by. The pocket is normally occupied by sphingosine-like molecules (pocket factor), including palmitic and myristic acids (50) of unknown function, and has been shown to represent the binding site for a variety of distinct hydrophobic compounds (11). Given their vicinity to the pocket, both mutations could affect drug binding by changing the pocket properties: by steric hindrance for Ile194Met or through local conformational changes in the case of Asp131Val. This hypothesis was also supported by the inability of the drug to protect the resistant variants against thermal inactivation or inhibit the conversion of mature R2/01 160S particles into 135S particles, indicating a complete loss or a significant reduction of virus-drug binding.
The mutations found in 3(2H)-isoflavene-resistant and -dependent viruses map into regions that coincide with regions harboring analogous mutations for PV3 Sabin WIN 51711 resistance variants (34), with two mutations being identical and one occurring at the identical position. At a first glance this might be expected, given the high amino acid sequence identity between the two viruses. However, the observed mutations are induced by two structurally unrelated compounds, 3(2H)-isoflavene and WIN 51711, which show very similar antiviral effects on early virus-cell interactions: prevention of uncoating without interference with host cell functions. More importantly, these overlapping spectra of mutations clustered in two distinct separate regions, suggesting that these mutations do not occur in a random fashion. Apparently, the structural transformations that the virus undergoes during its life cycle are only compatible with a limited repertoire of possible modifications that the virus can use to counterbalance the inhibitory effects of drugs.
The sedimentation profile of the drug-dependent variant after binding to cells at 4°C for 1 h in the absence of drug indicated a rapid conversion to 135S and 80S particles. This capacity to initiate uncoating at temperatures lower than those required for the wild-type virus suggests that the observed loss of infectivity at 37°C in the drug-dependent variant results from thermolability. This was indeed confirmed by experiments in which the D6/01 variant was incubated at 37°C for 30 min in the absence of compound and cell substrate. This condition was sufficient to rapidly convert 160S mature particles into thermodecay products sedimenting at 135S and 80S. The addition of the drug completely restored the stability of this variant.
In the absence of the compound, virus stability was gradually recovered when infection was carried out at lower temperatures that reached a maximum at 25°C. In fact, at 4°C a detectable amount of the D6/01 variant also bound to cells in the absence of the compound, and at 25°C almost all virus penetrated into the cells and could be rescued after a single step of growth.
While 3(2H)-isoflavene is apparently not critical for the attachment or uncoating of the dependent variant, during infection it promotes stabilization of the thermolabile variants long enough to allow them to survive during the interval between release of the progeny virus in the extracellular medium and infection of new cells. Similar findings were observed for PV3 WIN 51711-dependent variants that were also protected against thermal inactivation (5, 20, 33, 44). The enhanced uncoating shown in the case of R2/01 and D6/01 in the presence of drug may possibly reflect a faster process of decapsidation, which is accounted for by the corresponding amino acid changes that for the latter are most likely also related to the increased instability of the capsid.
In summary, we have shown that 3(2H)-isoflavene acts as a potent inhibitor of PV2 uncoating and targets the VP1 protein. Elucidation of the underlying mechanism of action provides additional information about the process by which the capsid triggers the conversion of native virus into 135S particles and, in particular, the involvement of the VP1 protein in this process. The possible use in vivo of 3(2H)-isoflavene or other halogen-substituted derivatives of 4′,6-dichloroflavan available in our laboratories is under investigation in animal models of poliovirus infection.
In addition to the effects of 3(2H)-isoflavene on PV2 Sabin reported herein, the drug also inhibits PV3 and PV1 replication (the latter is inhibited to a lower extent, however) and coxsackievirus type A9 replication (data not shown). Modification of the compound could therefore enhance its capability to interact with and inhibit a larger number of viruses belonging to the family Picornaviridae. Although picornavirus infections cause significant morbidity and even mortality, no vaccines are available at present, with the exception of vaccines against poliovirus and hepatitis A virus. The development and possible exploitation of drugs for therapeutic use are therefore needed, in addition to the design of effective and safe vaccines.
Acknowledgments
WIN 51711 was kindly provided by the Sterling-Winthrop Research Institute.
This work was partially supported by a grant from the Ministry of Health of Italy (to L.F.), “Mechanism of action of antiviral compounds on enterovirus replication and therapeutic use in animal model” (2001-2002), art. 12 D.L. 502/92.
REFERENCES
- 1.Bauer, D. J., J. W. T. Selway, J. F. Batchelor, M. Tisdale, I. C. Caldwell, and D. A. B. Young. 1981. 4′,6-Dichloroflavan (BW683C), a new anti-rhinovirus compound. Nature 292:369-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, N., E. Prchla, M. Schwab, D. Blaas, and R. Fuchs. 1999. Human rhinovirus HRV14 uncoats from early endosomes in the presence of bafilomycin. FEBS Lett. 463:175-178. [DOI] [PubMed] [Google Scholar]
- 3.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissing, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burali, C., N. Desideri, M. L. Stein, C. Conti, and N. Orsi. 1987. Synthesis and anti-rhinovirus activity of halogen-substituted isoflavenes and isoflavanes. Eur. J. Med. Chem. 22:119-123. [Google Scholar]
- 5.Caliguiri, L. A., J. J. McSharry, and G. W. Lawrence. 1980. Effect of arildone on modifications of poliovirus in vitro. Virology 105:86-93. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Progress toward global eradication of poliomyelitis, 2001. Morb. Mortal. Wkly. Rep. 51:253-256. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2002. Acute flaccid paralysis associated with circulating vaccine-derived poliovirus: Philippines 2001. JAMA 287:311. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2002. Public health dispatch: poliomyelitis, Madagascar, 2002. Morb. Mortal. Wkly. Rep. 51:622. [Google Scholar]
- 9.Conti, C., D. Genovese, R. Santoro, M. L. Stein, N. Orsi, and L. Fiore. 1990. Activities and mechanisms of action of halogen-substituted flavonoids against poliovirus type 2 infection in vitro. Antimicrob. Agents Chemother. 34:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danthi, P., M. Tosteson, Q.-H. Li, and M. Chow. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiore, L., B. Ridolfi, D. Genovese, G. Buttinelli, S. Lucioli, A. Lahm, and F. M. Ruggeri. 1997. Poliovirus Sabin type 1 neutralization epitopes recognized by immunoglobulin A monoclonal antibodies. J. Virol. 71:6905-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, M. P., M. J. Otto, and M. A. McKinlay. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51711, a new antiviral drug. Antimicrob. Agents Chemother. 30:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genovese, D., S. Catone, M. E. Farah, A. Gambacorta, and L. Fiore. 1999. Isolation and biological characterization of 3(2H)-isoflavene-resistant and -dependent poliovirus type 2 Sabin variants. J. Gen. Virol. 80:157-167. [DOI] [PubMed] [Google Scholar]
- 16.Genovese, D., C. Conti, P. Tomao, N. Desideri, M. L. Stein, S. Catone, and L. Fiore. 1995. Effect of chloro-, cyano-, and amidino-substituted flavonoids on enterovirus infection in vitro. Antivir. Res. 27:123-136. [DOI] [PubMed] [Google Scholar]
- 17.Grant, R. A., C. N. Hiremath, D. J. Filman, R. Syed, K. Andries, and J. M. Hogle. 1994. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr. Biol. 4:784-797. [DOI] [PubMed] [Google Scholar]
- 18.Halliday, E., J. Winkelstein, and A. D. Webster. 2003. Enteroviral infections in primary immunodeficiency (PID): a survey of morbidity and mortality. J. Infect. 46:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Hayden, F. G., D. T. Herrington, T. L. Coats, K. Kim, E. C. Cooper, S. A. Villano, S. Liu, S. Hudson, D. C. Pevear, M. Collett, M. McKinlay, and Pleconaril Respiratory Infection Study Group. 2003. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin. Infect. Dis. 36:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz, B. A., R. R. Rueckert, D. A. Shepard, F. J. Dutko, M. A. McKinlay, M. Fancher, M. G., Rossmann, J. Badger, and T. J. Smith. 1989. Genetic and molecular analyses of spontaneous variants of human rhinovirus 14 that are resistant to an antiviral compound. J. Virol. 63:2476-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 23.Hull, H. F. 2001. Progress towards global polio eradication. Dev. Biol. 105:3-7. [PubMed] [Google Scholar]
- 24.Kew, O. M., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, et al. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 25.Kim, K. H., P. Willingmann, Z. X. Gong, M. J. Kremer, M. S. Chapman, I. Minor, M. A. Oliveira, M. G. Rossmann, K. Andries, G. D. Diana, et al. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206-227. [DOI] [PubMed] [Google Scholar]
- 26.Lentz, K. N., A. D. Smith, S. C. Geisler, S. Cox, P., Buontempo, A. Skelton, J. DeMartino, E. Rozhon, J. Schwartz, V. Girijavallabhan, J. O'Connell, and E. Arnold. 1997. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973, comparison of the structural and biological properties of three poliovirus serotypes. Structure 15:961-978. [DOI] [PubMed] [Google Scholar]
- 27.Macadam, A. J., C. Arnold, J. Howlett, A. John, S. Marsden, et al. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccines. Virology 172:408-414. [DOI] [PubMed] [Google Scholar]
- 28.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, J., E. Samoilovich, G. Dunn, A. Lackenby, E. Feldman, A. Heath, E. Svirchevskaya, G. Cooper, M. Yermalovich, and P. D. Minor. 2002. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J. Virol. 76:10921-10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinlay, M. A. 2001. Recent advances in the treatment of rhinovirus infections. Curr. Opin. Pharmacol. 1:477-481. [DOI] [PubMed] [Google Scholar]
- 31.Melnick, J. L. 1996. Current status of poliovirus infections. Clin. Microbiol. Rev. 9:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 33.Mosser, A. G., and R. Rueckert. 1993. WIN-51711-dependent variants of poliovirus type 3: evidence that virions decay after release from cells unless drug is present. J. Virol. 67:1246-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosser, A. G., J. Y. Sgro, and R. R. Rueckert. 1994. Distribution of drug resistance mutations in type 3 poliovirus identifies three regions involved in uncoating functions. J. Virol. 68:8193-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosser, A. G., D. A. Shepard, and R. R. Rueckert. 1994. Use of drug-resistance variants to identify functional regions in picornavirus capsid proteins. Arch. Virol. 9:111-119. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira, M. A., R. Zhao, W. M. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, et al. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 37.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, p. 723-775. In B. N. Fields, D. Knipe, and P. Howley (ed.), Virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 38.Perchla, E., E. Kuechler, D. Blaas, and R. Fuchs. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol. 68:3713-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pevear, D. C., M. J. Fancher, P. J. Felock, M. G. Rossmann, M. S. Miller, G. Diana, A. M. Treasurywala, M. A. McKinlay, and F. J. Dutko. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pevear, D. C., T. M. Tull, M. E. Seipel, and J. M. Groake. 1999. Activity of pleconaril against enteroviruses. Antimicrob. Agents Chemother 43:2109-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard, S. R., G. Dunn, N. Cammack, P. D. Minor, and J. Almond. 1989. Nucleotide sequence of a neurovirulent variant of the type 2 oral poliovirus vaccine. J. Virol. 63:4949-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In B. N. Fields, D. Knipe, and P. Howley (ed.), Virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Rogers, J. M., J. D. Diana, and M. A. McKinlay. 1999. Pleconaril. A broad spectrum antipicornaviral agent. Adv. Exp. Med. Biol. 458:69-76. [PubMed] [Google Scholar]
- 44.Rombaut, B., K. Andries, and A. Boeye. 1991. A comparison of WIN 51711 and R 78206 as stabilizers of poliovirus virions and procapsids. J. Gen. Virol. 72:2153-2157. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, et al. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 46.Rotbart, H. A. 1999. Antiviral therapy for enteroviral infections. Pediatr. Infect. Dis. J. 18:632-633. [DOI] [PubMed] [Google Scholar]
- 47.Rotbart, H. A., A. D. Webster, and the Pleconaril Treatment Registry Group. 2001. Treatment of potentially life-threatining enterovirus infections with pleconaril. Clin. Infect. Dis. 32:228-235. [DOI] [PubMed] [Google Scholar]
- 48.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 723-775. In B. N. Fields, D. Knipe, and P. Howley (ed.), Virology, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 49.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 50.Smyth, M., T. Pettitt, A. Symonds, and J. Martin. 2003. Identification of the pocket factors in a picornavirus. Arch. Virol. 148:1225-1233. [DOI] [PubMed] [Google Scholar]
- 51.Superti, F., L. Seganti, N. Orsi, M. Divizia, R. Gabrieli, A. Pana, and M. L. Stein. 1989. Effect of isoflavans and isoflavenes on the infection of Frp/3 cells by hepatitis A virus. Antivir. Res. 11:247-254. [DOI] [PubMed] [Google Scholar]
- 52.Tisdale, M., and W. T. Selway. 1984. Effect of dichloroflavan (BW683C) on the stability and uncoating of rhinovirus type 1B. J. Antimicrob. Chemother. 14(Suppl. A):97-105. [DOI] [PubMed] [Google Scholar]
- 53.Toyoda, H., M. Kohara, Y. Kataoka, T. Suganuma, T. Omata, N. Imura, and A. Nomoto. 1984. Complete nucleotide sequence of all three serotype genomes. Implications for genetic relationship, gene function and antigenic determinants. J. Mol. Biol. 174:561-585. [DOI] [PubMed] [Google Scholar]
- 54.Vance, L., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webster, D. 2000. The potential use of anti-enteroviral drugs in the immunocompromised. Curr. Opin. Infect. Dis. 13:625-629. [DOI] [PubMed] [Google Scholar]
- 56.Zeichardt, H., M. J. Otto, M. A. McKinley, P. Willingmann, and K. O. Habermehl. 1987. Inhibition of poliovirus uncoating by disoxaril (WIN51711). Virology 160:281-285. [DOI] [PubMed] [Google Scholar]