Abstract
Advances in research and technology has increased our quality of life, allowed us to combat diseases, and achieve increased longevity. Unfortunately, increased longevity is accompanied by a rise in the incidences of age-related diseases such as Alzheimer’s disease (AD). AD is the sixth leading cause of death, and one of the leading causes of dementia amongst the aged population in the USA. It is a progressive neurodegenerative disorder, characterized by the prevalence of extracellular Aβ plaques and intracellular neurofibrillary tangles, derived from the proteolysis of the amyloid precursor protein (APP) and the hyperphosphorylation of microtubule-associated protein tau, respectively. Despite years of extensive research, the molecular mechanisms that underlie the pathology of AD remain unclear. Model organisms, such as the nematode, Caenorhabditis elegans, present a complementary approach to addressing these questions. C. elegans has many advantages as a model system to study AD and other neurodegenerative diseases. Like their mammalian counterparts, they have complex biochemical pathways, most of which are conserved. Genes in which mutations are correlated with AD have counterparts in C. elegans, including an APP-related gene, apl-1, a tau homolog, ptl-1, and presenilin homologs, such as sel-12 and hop-1. Since the neuronal connectivity in C. elegans has already been established, C. elegans is also advantageous in modeling learning and memory impairments seen during AD. This article addresses the insights C. elegans provide in studying AD and other neurodegenerative diseases. Additionally, we explore the advantages and drawbacks associated with using this model.
Keywords: apl-1, C. elegans, Alzheimer’s disease, ALS, Parkinson disease, model systems
INTRODUCTION TO ALZHEIMER’S DISEASE
Alzheimer’s disease (AD) is the 6th leading cause of death in the US and affects more than 35 million people worldwide (Alzheimer’s Disease International, 2014). AD is a neurodegenerative disease characterized by a progressive loss of memory. Most cases of AD occur sporadically in aged people (>60 years, late-onset AD) without a clear inheritance pattern. However, in 5% of the cases (familial or early onset AD) AD symptoms appear earlier and are linked with gene mutations. Both forms of AD have two main neuropathologic features: the presence of extra-neuronal amyloid plaques, often referred to as senile plaques, and intraneuronal neurofibrillary tangles (Kidd, 1964; Luse and Smith, 1964; Terry et al., 1964; Krigman et al., 1965). Amyloid plaques are aggregates of the beta-amyloid peptide (Aβ), a cleavage product of the amyloid precursor protein (APP; Glenner and Wong, 1984; Masters et al., 1985; Kang et al., 1987). Hyperphosphorylation of the microtubule associated protein tau causes its polymerization into paired helical filaments (PHFs) and, presumably, its formation into neurofibrillary tangles (Goedert et al., 1989a).
Mutations in the APP gene and/or the enzymes involved in APP processing (γ-secretase components presenilins, PSEN1 and PSEN2; Chartier-Harlin et al., 1991; Goate et al., 1991; Murrell et al., 1991; Levy-Lahad et al., 1995; Rogaev et al., 1995; Sherrington et al., 1995) are correlated with early onset AD. These mutations increase the levels of toxic Aβ species and promote neurodegeneration. By contrast, a recently identified mutation in APP affects cleavage of APP, causing less Aβ production and conferring neuroprotective benefits (Jonsson et al., 2012). Despite the significant advances made using APP transgenic and knockout models in mammals, unraveling the cellular role of APP has been difficult. Alternative animal models provide complementary approaches to dissecting the function of APP and tau. In this review, we discuss the latest uses of the nematode Caenorhabditis elegans as a model system for the study of AD. We also include a brief review of a few representative examples of how C. elegans is being utilized to model other neurodegenerative diseases.
C. elegans AS A MODEL FOR ALZHEIMER’S DISEASE
Caenorhabditis elegans is a free-living, non-parasitic nematode that was first introduced as a model organism by Sydney Brenner in 1963 (Brenner, 1974). It is a small (1 mm in length), transparent roundworm, which makes it easy for manipulation, and has a short life cycle of 3 days from egg to adult at 25°C (Brenner, 1974). Under suitable growing conditions, hatched animals develop through four larval stages (L1–L4), each punctuated by a molt, to arise as an adult hermaphrodite with 959 somatic cells (Sulston and Horvitz, 1977). Its life span is between 2 and 3 weeks, which facilitates the study of its biology. Completion of the C. elegans genome sequence in 1998 (C. elegans Sequencing Consortium, 1998) demonstrated that roughly 38% of worm genes have a human ortholog, such as APP and tau (Shaye and Greenwald, 2011). Hence, C. elegans has many excellent advantages as an in vivo model for the study of AD and other neurodegenerative diseases.
MOLECULAR PATHWAYS OF APP. SIMILARITIES AND DIFFERENCES BETWEEN MAMMALS AND C. elegans
FUNCTION AND PROCESSING OF APP: NON-AMYLOIDOGENIC AND AMYLOIDOGENIC Aβ PATHWAY
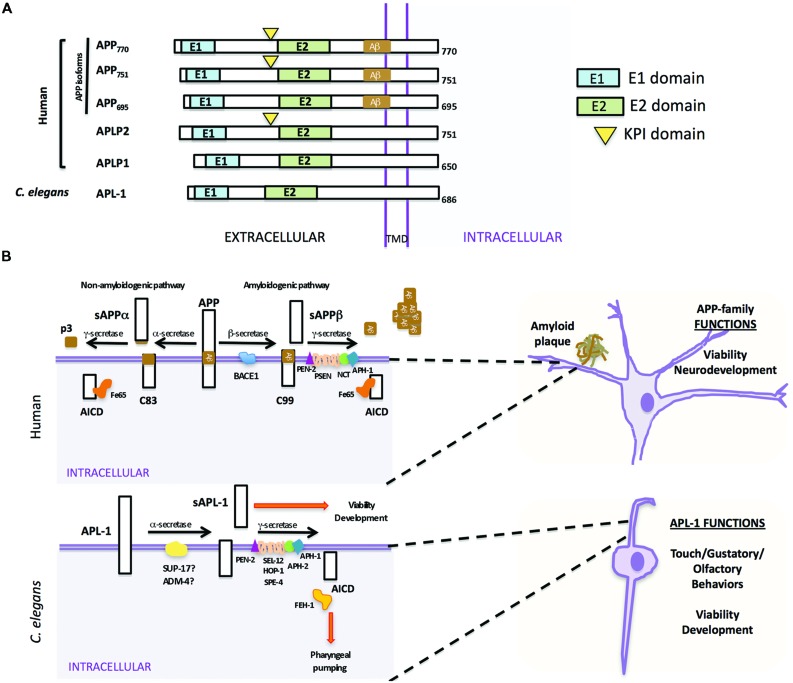
The APP family of proteins contains three members, APP, APLP1, and APLP2 (Wasco et al., 1992, 1993; Sprecher et al., 1993; Sandbrink et al., 1994; Slunt et al., 1994), which are characterized by a large extracellular region containing conserved E1 and E2 domains, a single transmembrane domain, and a small cytosolic domain (Kang et al., 1987). APLP1 and APLP2 do not contain the Aβ sequence and, hence, do not produce Aβ (Figure 1A; Wasco et al., 1992, 1993). The APP gene family is required for viability and brain development. APP mouse knockouts were viable, but had behavioral and cognitive defects (Ring et al., 2007). While knockout of APLP1 resulted in postnatal growth defects (Heber et al., 2000), mice in which APLP2 was inactivated appeared wild type (von Koch et al., 1997). Double knockouts of APLP2 and either APP or APLP1, however, resulted in postnatal lethality (von Koch et al., 1997; Heber et al., 2000); the lethality of APP/APLP2 double knockouts could be rescued by knock-in of an APP extracellular fragment, sAPPα (Weyer et al., 2011), suggesting that sAPPα is sufficient for viability. The triple knockout caused lethality and a type II lissencephaly and cortical disorganization (Herms et al., 2004). Collectively, these results suggest that APP family members have essential and redundant functions during development, including proper brain development, and these functions do not require Aβ.
FIGURE 1.
Similarities and differences between human APP and Caenorhabditis elegans APL-1. (A) Schematic representation of human APP isoforms, other members of the APP family (APLP1 and APLP2), and C. elegans APL-1. (B) Comparison between human APP proteolytic pathways (top) and C. elegans APL-1 proteolytic pathway (bottom). Top. APP can be cleaved by two different pathways. In the anti-amyloidogenic pathway, APP is first cleaved by the α-secretase to release an extracellular fragment sAPPα. The remaining APP fragment (APP-CTFα or C83) is then cleaved by the γ-secretase complex to release p3 extracellularly and the APP intracellular domain (AICD) to the cytosol. In the amyloidogenic pathway, β-secretase first cleaves APP, releasing the sAPPβ fragment. The APP-CTFβ (C99) fragment is subsequently cleaved by the γ-secretase complex, liberating the AICD to the cytosol and Aβ to the lumen. Aβ will aggregate to form amyloid plaques. Bottom. In C. elegans, APL-1 is first cleaved by the α-secretase homologs SUP-17/ADM-4, liberating the extracellular sAPL-1 that is known to regulate worm viability and development. The γ-secretase complex will then cleave the remaining APL-1-CTF to release AICD into the cytosol. General functions of the APP family and APL-1 are indicated.
In mammals, APP is processed through two proteolytic pathways, only one of which produces Aβ (Figure 1B; Haass et al., 1992, 1994a,b). In the non-amyloidogenic pathway, APP is first cleaved by an α-secretase within the Aβ sequence to release an extracellular fragment, sAPPα (Figure 1B). The remaining APP fragment (known as APP-CTFα or C83) is then cleaved by the γ-secretase complex to release the APP intracellular domain (AICD) to the cytosol. By contrast, in the amyloidogenic pathway, after cleavage by the β-secretase (BACE) to release sAPPβ, the remaining APP fragment (known as APP-CTFβ or C99) is then cleaved by the γ-secretase complex, liberating Aβ to the lumen and AICD to the cytosol (Gu et al., 2001; Sastre et al., 2001; Weidemann et al., 2002). This latter pathway is likely favored in AD patients.
Mammalian γ-secretase is a protease complex consisting of several components: presenilins 1 and 2 (PSEN1 and PSEN2), nicastrin (NCT), anterior pharynx defective (APH-1), and the presenilin enhancer (PEN-2; Yu et al., 2000; Francis et al., 2002). PSEN1 and PSEN2 are the catalytic components of the γ-secretase complex. NCT works as a stabilizing cofactor required for γ-secretase complex assembly and trafficking (Li et al., 2003; Zhang et al., 2005) and PEN-2 and APH-1 have a role in the maturation process of PSEN1 and PSEN2 (Luo et al., 2003). Besides APP, the γ-secretase complex is also involved in the proteolysis of Notch receptors, and the first identification of any PEN-2 or APH-1 ortholog was in C. elegans as the result of a genetic screen for modifiers of the Notch pathway (Francis et al., 2002; Goutte et al., 2002). Within the γ-secretase complex, only mutations in PSEN1 and PSEN2 have been associated with early onset AD (Levy-Lahad et al., 1995; Rogaev et al., 1995; Sherrington et al., 1995).
PROCESSING OF C. elegans APL-1
In C. elegans there is only one APP-related gene, apl-1. Like human APP (Kang et al., 1987), APL-1 contains a large extracellular region whose conserved E1 and E2 domains share 46 and 49% sequence similarity to human APP, respectively, a transmembrane domain, and a relatively small cytosolic domain, which shares 71% sequence similarity to human APP (Figure 1A; Daigle and Li, 1993). Notably, unlike APP but similar to APLP1 and APLP2, APL-1 does not contain the Aβ sequence (Daigle and Li, 1993).
Two α-secretase proteins are present in C. elegans, SUP-17 and ADM-4 (Jarriault and Greenwald, 2005). They work redundantly in the cleavage of the C. elegans Notch homologs, LIN-12 and GLP-1 (Jarriault and Greenwald, 2005). However, no experiments thus far have tested whether SUP-17 or ADM-4 cleaves APL-1. No BACE ortholog has been identified by bioinformatic searches and no β-secretase activity that cleaves human APP has been detected in C. elegans, suggesting that APL-1 is only processed by the α/γ-secretase processing pathway (Link, 2006). α-secretase cleavage of APL-1 releases the extracellular fragment, sAPL-1; subsequent cleavage of APL-1-CTFα by the γ-secretase complex liberates the intracellular domain (AICD; Figure 1B).
The initial characterizations of human PSEN1 (then called S182) and PSEN2 (first named E5-1) described them as novel proteins with multiple transmembrane domains (Rogaev et al., 1995; Sherrington et al., 1995). The cellular functions of the presenilins were determined by their homology to the C. elegans protein, SEL-12 (Levitan and Greenwald, 1995). The two C. elegans Notch genes, lin-12 and glp-1, are involved in many cell fate decisions during development, including vulval cell specification and germline development (Greenwald et al., 1983; Lambie and Kimble, 1991; Newman et al., 1995; Levitan et al., 1996) sel-12/PSEN was identified in a genetic screen to isolate suppressors of a dominant Lin-12/Notch multivulva phenotype (Levitan and Greenwald, 1995). Loss of sel-12/PSEN suppressed the Lin-12/Notch multivulva phenotype and produced a defect in egg laying that was rescued by introducing human PSEN1 or PSEN2, suggesting a conserved function between human and C. elegans presenilins (Levitan and Greenwald, 1995; Levitan et al., 1996). Like human PSENs (Thinakaran et al., 1996), SEL-12/PSEN is cleaved to attain its final topology (Li and Greenwald, 1996). A C. elegans PSEN gene family was identified and includes sel-12, hop-1, and spe-4 (L’Hernault and Arduengo, 1992; Levitan and Greenwald, 1995; Li and Greenwald, 1997; Westlund et al., 1999); spe-4 is exclusively expressed in male gonadal cells and will not be further discussed (Arduengo et al., 1998). Knockdown of hop-1/PSEN in sel-12/PSEN mutants showed maternal effect lethality, germline defects, and missing anterior pharynx, defects associated with loss of glp-1/Notch function, suggesting that sel-12 and hop-1 function redundantly in the LIN-12 and GLP-1/Notch pathways (Westlund et al., 1999). Similarly, mice carrying a null mutation in PSEN1 showed embryonic lethality, skeletal defects, and disrupted somite boundaries (Shen et al., 1997), similar to the phenotypes seen in Notch1 knockouts (Krebs et al., 2000, 2003, 2004; Duarte et al., 2004; Gale et al., 2004).
In screening for novel mutants showing the glp-1/Notch phenotype of defective anterior pharynx, Goutte et al. (2000, 2002) identified two genes, aph-1 and aph-2, which encodes the C. elegans NCT ortholog. Independently, Francis et al. (2002) screened for enhancers of sel-12/PSEN activity and identified pen-2. aph-2/NCT, pen-2, and aph-1 are all required for proper Notch signaling. Human PSEN, NCT, Aph1α2, and PEN-2 were subsequently shown to physically associate and cooperatively regulate the maturation of individual components to form a proteolytically active γ-secretase complex (Kimberly et al., 2003).
FUNCTION AND REGULATION OF APL-1
Like the mammalian APP family (Slunt et al., 1994; Lorent et al., 1995; Thinakaran et al., 1995), apl-1 is expressed in multiple tissue types. apl-1 expression is observed in neurons, supporting cells, and head muscles throughout development, while expression in vulval muscles, vulval cells, and hypodermal seam cells is not detected until the L4 stage to adult (Hornsten et al., 2007; Niwa et al., 2008).
Inactivation of apl-1, such as with the yn10 null allele, results in a completely penetrant lethality during the first to second larval (L1–L2) transition due to a molting defect (Hornsten et al., 2007). apl-1 activity is also necessary for later larval transitions, as RNAi knockdown of apl-1 in an RNAi-sensitized background showed animals with molting defects during the L3–L4 and L4 to adult transitions (Wiese et al., 2010). This lethality was rescued by microinjection of an apl-1 genomic fragment or cDNA. Hence, similar to the mammalian APP family, apl-1 has an essential function. High levels of apl-1 expression caused an incompletely penetrant L1 lethality (70% lethality), shortened body length, and morphogenetic, reproductive, and locomotory defects (Hornsten et al., 2007; Ewald et al., 2012b). These results indicate that levels of APL-1 must be tightly regulated as loss of APL-1 as well as high levels of APL-1 result in lethality. When sel-12/PSEN activity was reduced in transgenic animals with APL-1 overexpression, the 70% lethality was partially rescued, suggesting that SEL-12/PSEN regulates APL-1 cleavage and/or trafficking (Hornsten et al., 2007). The underlying basis of the loss- and gain-of-function apl-1 lethality is still unclear, but is not dependent on activation of CED-3/ caspase or necrotic cell death pathways (Hornsten et al., 2007). Characterization of apl-1 function may provide insights into the general function and pathways of APP, of which much is still unknown.
The apl-1(yn5) mutant, which contains a deletion of the region encoding the APL-1 transmembrane and cytosolic domains, produces only the extracellular domain of APL-1 (APL-1EXT) and is viable. Because APL-1EXT is not further cleaved by α-secretase, APL-1EXT is slightly larger than sAPL-1 and is expressed at high levels in apl-1(yn5) mutants (Hornsten et al., 2007). Hence, the APL-1 extracellular domain is sufficient for viability, similar to the rescue of APP/APLP2 double mutants by the knock-in of sAPPα (Weyer et al., 2011). However, although apl-1(yn5) mutants are viable, they display several phenotypes, including a slower developmental progression, decreased body length, reproductive defects, and temperature-sensitive lethality (Hornsten et al., 2007; Ewald et al., 2012b). Because these defects can be phenocopied by microinjection of APL-1EXT transgenes into wild-type animals, the phenotypes are due to overexpression of APL-1EXT and not due to loss of APL-1 signaling through its cytoplasmic domain (Ewald et al., 2012b). Interestingly, pan-neuronal expression of APL-1EXT, but not expression from muscle or hypodermal cells, is sufficient to rescue the lethality observed in apl-1 null mutants (Hornsten et al., 2007), suggesting that the cells (i.e., neurons) from which sAPL-1 is released as well as the extracellular milieu in which sAPL-1 travels is functionally relevant. We suggest that high levels of sAPP may also contribute to the pathology seen in AD patients. Down’s syndrome patients, whose chromosome 21 trisomy includes trisomy of APP, display a high incidence of AD and intellectual disability (Zigman, 2013), perhaps contributed in small part by the high levels of APP expression.
Decreasing apl-1 activity by RNAi resulted in hypersensitivity to aldicarb, an acetylcholinesterase inhibitor (Wiese et al., 2010). Using apl-1 knockouts to test different apl-1 deletion constructs, Wiese et al. (2010) determined that lack of sAPL-1 is responsible for the aldicarb hypersensitivity. These findings are consistent with mammalian studies, which show that a lack of APP and APLP2 impairs synaptic function at cholinergic neuromuscular junctions (Wang et al., 2005).
Heterochronic genes, whose differential spatiotemporal expression ensures proper progression through larval stages and transition into adulthood (Chalfie et al., 1981; Ambros and Horvitz, 1984), regulate expression of apl-1 in hypodermal seam cells (Niwa et al., 2008). Loss of let-7 microRNA (miRNA) function caused precocious seam cell development and vulval bursting at the adult stage, leading to death (Reinhart et al., 2000). These let-7 phenotypes can be rescued by knockdown of apl-1 (Niwa et al., 2008). apl-1, however, is not a direct target of let-7 miRNA. NHR-25/Ftz-F1, which is a nuclear hormone receptor (NHR) that is required for completion of larval molts (Asahina et al., 2000; Gissendanner and Sluder, 2000), binds an enhancer element in the promoter of apl-1 to regulate apl-1 expression in seam cells (Niwa and Hada, 2010). nhr-25/Ftz-F1 transcripts are possible targets of the let-7 family of miRNAs for downregulation (Hayes et al., 2006). Regulation of continued apl-1 expression in adult seam cells and other cell types is unknown.
Pathways through which apl-1 functions
The apl-1(yn5) phenotypes require activity of the DAF-16/FOXO transcription factor, which is negatively regulated by the insulin pathway. C. elegans has only one insulin/IGF-1 receptor, DAF-2 (Kimura et al., 1997). Under favorable environmental conditions, such as when adequate food is present, signaling through the insulin pathway activates a conserved PI 3-kinase/AKT cascade (Morris et al., 1996; Kimura et al., 1997; Paradis and Ruvkun, 1998; Paradis et al., 1999), which causes phosphorylation of DAF-16/FOXO, thereby allowing reproductive development (Larsen et al., 1995; Gems et al., 1998; Henderson and Johnson, 2001). Phosphorylation of DAF-16/FOXO causes its sequestration in the cytoplasm (Lin et al., 2001), thereby preventing it from entering the nucleus to activate its target genes (Henderson and Johnson, 2001; Lee et al., 2001), which regulate longevity (Lin et al., 1997; Ogg et al., 1997), stress resistance (McElwee et al., 2003, 2004; Murphy et al., 2003), and dauer formation (Riddle et al., 1981; Vowels and Thomas, 1992). Environmental conditions also affect other metabolic functions, such as reproductive behavior, which is inhibited under starvation conditions (Seidel and Kimble, 2011), and body size. Starvation survival behavior is regulated by DAF-16/FOXO activity (Lee and Ashrafi, 2008) and the insulin (So et al., 2011) and DAF-7/TGFβ (Savage-Dunn et al., 2003) pathways work in parallel to regulate body length via daf-16/FOXO activity.
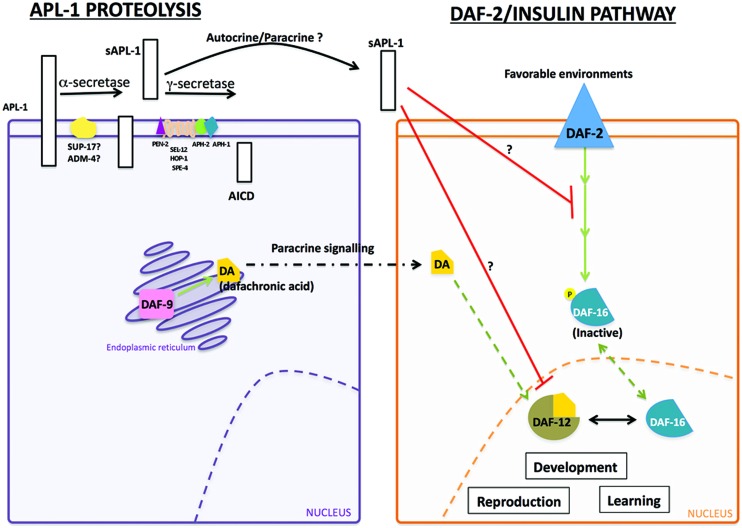
The slowed development, decreased body size, and decreased reproductive rates of apl-1(yn5) mutants are dependent on daf-16/FOXO activity. At 20°C, mutants with decreased insulin signaling or apl-1(yn5) mutants showed a delayed developmental progression and shorter body length, which were enhanced when insulin signaling was decreased in apl-1(yn5) mutants [i.e., daf-2(e1370); apl-1(yn5) double mutants]; at 25°C, the apl-1(yn5) mutants with decreased insulin signaling went into L1 arrest (Ewald et al., 2012b). By contrast, when daf-16/FOXO activity was removed from apl-1(yn5) mutants, the delayed developmental progression, decreased reproductive rate, and smaller body length of apl-1(yn5) single mutants were suppressed. Furthermore, loss of daf-16/FOXO activity in apl-1(yn5) mutants with decreased insulin signaling rescued the short body length and L1 arrest phenotypes (Ewald et al., 2012b). These results suggest that sAPL-1 signals in a parallel pathway to the insulin pathway or modulates the DAF-2/insulin/IGF-1 pathway to activate daf-16/FOXO activity to affect developmental progression, reproductive rates, and body length (Figure 2). Mammalian sAPP may have similar roles in development.
FIGURE 2.
Interaction between sAPL-1 and DAF-2 insulin/IGF-1 receptor and DAF-12/NHR pathways. Schematic representation of APL-1 proteolytic pathway and how sAPL-1 may modulate DAF-2 insulin/IGF-1 receptor and DAF-12/NHR signaling pathways. APL-1 is cleaved by the α/γ-secretase pathway in C. elegans. Released sAPL-1 could act as a signaling molecule in the same cell (autocrine regulation) or in neighboring cells (paracrine regulation) to inhibit daf-2 insulin/IGF-1 receptor and daf-12/NHR pathways to affect worm viability and development. The exact mechanism by which sAPL-1inhibits daf-2 insulin/IGF-1 receptor and daf-12/NHR is still unknown and labeled as a question mark (see Ewald et al., 2012b).
Activity of the daf-12/NHR signals in multiple pathways to integrate environmental stimuli with metabolic needs and can modulate the insulin pathway as well as function in an independent pathway (Gerisch et al., 2001; Dumas et al., 2010). Decreasing daf-12/NHR activity in apl-1(yn5) mutants rescued the slow development, low reproductive rate, and decreased body length phenotypes (Ewald et al., 2012b). Hence, decreased insulin signaling and signaling through a parallel daf-12/NHR pathway converge to activate daf-16/FOXO for the phenotypes seen in apl-1(yn5) mutants. Noteworthy, levels of insulin/IGF-1 receptors are decreased in AD brains (Steen et al., 2005), and APP processing and Aβ production in vitro was modulated by insulin signaling (Gasparini et al., 2001). Analogous to C. elegans, sAPP may also act to modulate the insulin pathway.
Pan-neuronal APL-1 expression affects learning
In transgenic mice expressing human or mouse APP, animals showed an increased lethality and learning defects that were not correlated with Aβ deposition (Hsiao et al., 1995); similarly, doubly transgenic mice carrying transgenes with APP and PSEN1 mutations showed learning defects that were not correlated with the number of Aβ plaques (Holcomb et al., 1999). The mechanisms underlying these defects are unclear. Use of the C. elegans model could be informative. C. elegans has many sensory modalities, including smell and taste. They respond to volatile and water soluble chemicals by moving toward or away from chemoattractive or chemorepulsive stimuli, respectively. Many chemoattractants and chemorepellants have been identified and the neural circuits mediating the chemosensory response identified (Bargmann and Horvitz, 1991). For instance, when C. elegans is given the choice between a neutral compound and a chemoattractant, such as benzaldehyde, animals will move toward benzaldehyde; this response is mediated by the AWC neurons (Bargmann et al., 1993); similarly, ASEL, a gustatory neuron, mediates chemoattraction to sodium acetate (Bargmann and Horvitz, 1991; Pierce-Shimomura et al., 2001).
Although apl-1 is not expressed in AWC neurons and the morphology of sensory neurons appears wild type with GFP markers, the overall chemoattractive response to benzaldyhyde and sodium acetate was decreased in apl-1(yn10/+) heterozygotes and transgenic animals that overexpress APL-1 [e.g., ynIs79 (Papl-1::apl-1::GFP)] (Ewald et al., 2012a). The chemotaxis response was restored in APL-1 overexpression lines [e.g., ynIs79 (Papl-1::apl-1::GFP)] when insulin signaling was decreased, but not when daf-16/FOXO activity was decreased, suggesting that daf-16/FOXO activity is needed for normal chemotaxis in transgenic lines overexpressing APL-1 (Figure 2). Pan-neuronal expression of APL-1 or targeted overexpression of APL-1 in the AWC or ASEL neurons resulted in wild-type chemotaxis responses (Ewald et al., 2012a). By contrast, ectopic expression of apl-1 with the snb-1 promoter, which drives pan-neuronal and multi-cell type expression, resulted in no chemotaxis response to benzaldehyde or sodium acetate (Ewald et al., 2012a). When signaling through the DAF-2/insulin/IGF-1 receptor, DAF-12/NHR, or DAF-7/TGFβ was decreased, the chemotaxis response toward benzaldehyde and sodium acetate in these transgenic lines was restored, indicating that the loss of the chemotaxis response due to ectopic apl-1 signaling in cells outside the nervous system is dependent on insulin and TGFβ signaling.
In addition to chemosensory responses, C. elegans is also capable of associative chemosensory plasticity (Wen et al., 1997). For example, when benzaldehyde was paired with starvation for as short as 30 min, C. elegans showed a significant reduction in preference for benzaldehyde; persistence of this plasticity was positively correlated with the length of pairing time (Colbert and Bargmann, 1995; Tomioka et al., 2006; Lin et al., 2010), suggestive of stable memory formation. Both chemotaxis and associative plasticity are dependent on insulin signaling (Tomioka et al., 2006; Lin et al., 2010). Little associative plasticity, however, was observed after pairing benzaldehyde with starvation for 60 min in animals with pan-neuronal APL-1 expression (Ewald et al., 2012a). The plasticity could be restored when daf-16/FOXO, daf-12/NHR, or daf-7/TGFβ activity was decreased, indicating that the impaired associative plasticity with pan-neuronal APL-1 expression requires daf-16/FOXO, daf-12/NHR, and daf-7/TGFβ activity (Ewald et al., 2012a).
Touch habituation is another sensory characteristic affected by pan-neuronal APL-1 expression. When a gentle touch is applied to the head of the animal, C. elegans responds by moving backward; conversely, when touched on the tail, the animal moves forward (Chalfie and Sulston, 1981). This response to gentle body touch is mediated by six mechanosensory touch neurons (Chalfie et al., 1985). After six cycles of alternating head/tail touches, wild-type animals habituated and became unresponsive (Ewald et al., 2012a). apl-1(yn10/+) heterozygotes or transgenic animals that overexpress APL-1 showed touch habituation. By contrast, animals with pan-neuronal APL-1 expression were slow to habituate and required more alternating head/tail touch cycles before becoming habituated (Ewald et al., 2012a). Collectively, these results indicate that pan-neuronal overexpression of APL-1 causes learning defects. These results parallel those seen in mammalian models in which overexpression of APP leads to cognitive defects, independent of Aβ aggregates (Hsiao et al., 1995; Simón et al., 2009), thereby suggesting that sAPP activity, in addition to Aβ aggregates, contributes to cognitive defects. Whether these cognitive defects depend on the DAF-16/FOXO transcription factor and/or TGFβ signaling remains to be tested.
APL-1 TRAFFICKING IS IMPORTANT FOR SYNAPTIC TRANSMISSION
APL-1, like APP (Koo et al., 1990), is transported from the cell body to synapses (Wiese et al., 2010). UNC-108, which is a neuronally expressed GTPase and localizes to the Golgi complex and early endosomes (Mangahas et al., 2008; Edwards et al., 2009), is involved in the maturation of dense core vesicles (Borgonovo et al., 2006; Mangahas et al., 2008; Edwards et al., 2009; Sumakovic et al., 2009) and the packaging of APL-1 into mature dense core vesicles (Wiese et al., 2010). Both UNC-116/kinesin-1 and UNC-104/KIF1A/kinesin-3 are involved in the anterograde transport of APL-1 (Wiese et al., 2010; Arimoto et al., 2011), but only UNC-116/kinesin-1 and dynein motors are responsible for the retrograde transport of APL-1 back to the cell body (Arimoto et al., 2011). The rates of anterograde and retrograde transport of APL-1 vesicles are 1.1 μm/s and 1.6 μm/s, respectively (Arimoto et al., 2011). Hence, APL-1 is transported similarly as in mammalian models where kinesin-1 is responsible for APP axonal transport (Koo et al., 1990). Surprisingly, mutations in unc-116/kinesin-1 and unc-104/KIF1A/kinesin-3 both caused decreased levels of APL-1 expression without affecting transcript levels, suggesting that without transport motors, APL-1 does not accumulate in cell bodies because of protein degradation (Wiese et al., 2010; Arimoto et al., 2011). APL-1 is also internalized from the cell surface of neurons via a RAB-5-dependent endocytosis (Wiese et al., 2010).
AICD INTRACELLULAR TRAFFICKING
The Fe65 family of proteins binds the cytoplasmic YENPTY sequence of APP, APLP1, and APLP2, via their PTB2 domain (Guénette et al., 1996; Zambrano et al., 1997; Duilio et al., 1998; Russo et al., 1998). Likewise, the sole family member ortholog in C. elegans, FEH-1, has a WW domain and PTB1 and PTB2 domains, which closely resemble those of the Fe65 family, and the PTB2 domain of FEH-1 interacts with APL-1 (Zambrano et al., 2002).
FEH-1 is expressed in pharyngeal muscle and neuronal processes and is necessary for survival. Inactivation of feh-1 caused an incompletely penetrant embryonic lethality. Survivors showed little pharyngeal pumping and were unable to feed, thereby resulting in L1 arrest (Zambrano et al., 2002). Decreasing feh-1 activity or decreasing feh-1 dosage caused pharyngeal pumping rates to increase, suggesting that the rate of pharyngeal pumping is feh-1 dosage dependent. However, the functional significance of FEH-1 and APL-1 interactions is unclear as apl-1(yn5) mutants, which do not have an AICD domain, and apl-1(yn10/+) heterozygotes do not have defective pumping rates (Ewald et al., 2012b).
INVESTIGATING THE AMYLOID HYPOTHESIS OF AD IN C. elegans
Aβ peptide, the cleavage product of APP believed to underlie the pathology of AD (Glenner and Wong, 1984; Masters et al., 1985; Gorevic et al., 1986; Selkoe et al., 1986), is not present in APL-1 (Daigle and Li, 1993) nor does C. elegans possess β-secretase activity to produce Aβ (Link, 2006). Nevertheless, C. elegans provides a powerful in vivo genetic system to study the effects of neurotoxic Aβ through transgene analysis (Shankar et al., 2008). Many transgenic strains have been generated in which a signal sequence followed by the human Aβ sequence is expressed in all cells, in all neurons, in specific subsets of neurons, or in muscle cells (Figure 4). These strains produce either Aβ1-42 or Aβ3-42.
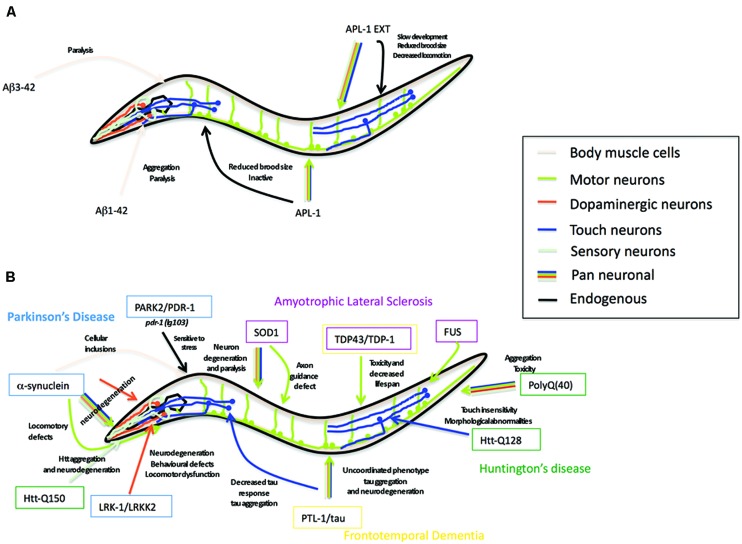
FIGURE 4.
Caenorhabditis elegans as a transgenic model for AD and other neurodegenerative diseases. (A) Summary of the Alzheimer’s disease models in C. elegans expressing human Aβ peptide or C. elegans full-length APL-1 or APL-1 extracellular domain (APL-1 EXT). Arrow color represents the tissues where transgenes were expressed. Phenotypes observed are next to the arrow. (B) Summary of the C. elegans models for Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and Huntington’s disease (HD). Genes modeling PD are shown in blue boxes, ALS in pink boxes, FTD in yellow boxes, and HD in green boxes. Arrow color represents tissues where transgenes were expressed. Phenotypes observed are written close to the arrow.
AD is a late onset neurodegenerative disease. C. elegans expressing human Aβ3-42 in muscle tissue (Link, 1995; Link et al., 2001) showed an age-dependent paralysis at 20°C (Cohen et al., 2006; McColl et al., 2009); paralysis occurred more rapidly and more severely when Aβ1-42 was produced at 25°C (McColl et al., 2009). The level of muscle paralysis was significantly decreased when insulin signaling was decreased (Cohen et al., 2006). Furthermore, inhibiting daf-16/FOXO and hsf-1, which encodes a heat shock protein transcription factor (Hsu et al., 2003; Morley and Morimoto, 2004), reversed the effects of decreased insulin signaling (Cohen et al., 2006). Hence, the paralysis effects of Aβ correlates with age and is dependent on insulin signaling.
Since aggregated Aβ is toxic to neurons (Glenner and Wong, 1984; Masters et al., 1985; Gorevic et al., 1986; Selkoe et al., 1986) and causes muscle paralysis in C. elegans, molecules and pathways that can prevent the formation or promote the disassembly of Aβ aggregates can be screened for in C. elegans. For instance, when C. elegans extracts are incubated with aggregated human Aβ3-42 in the presence or absence of protease inhibitors, disaggregation occurred, but disaggregation did not occur when extracts were either heated to denature proteins or incubated with proteinases (Bieschke et al., 2009). Hence, an unidentified protein or protein complex in C. elegans extracts can disaggregate Aβ3-42 aggregates.
Several orthologs to human heat shock (HSP) chaperone proteins were found to interact directly with Aβ3-42 in C. elegans. C. elegans HSP-16 proteins, HSP-16-1, HSP-16-2, and HSP-16–48, orthologs of αB-crystallin, bound intracellular Aβ3-42 monomers and soluble Aβ3-42 oligomers, but not fibrillar Aβ3-42 (Fonte et al., 2002). Moreover, hsp-16 transcript levels were upregulated in Aβ3-42 transgenic lines, but whether these chaperone proteins protect against or promote Aβ paralysis is unclear (Fonte et al., 2002). By contrast, increased expression of the HSP70 chaperones had a protective role by suppressing paralysis (Fonte et al., 2008). These results are consistent with human studies showing that HSP70 and αB-crystallin were upregulated in AD brains (Hamos et al., 1991; Perez et al., 1991; Shinohara et al., 1993; Renkawek et al., 1994; Yoo et al., 2001) and binds Aβ (Liang, 2000).
Transgenic lines in which Aβ is expressed in glutamatergic neurons showed age-dependent neurodegeneration, whereby 7-day adults showed 75% glutamatergic neurodegeneration (Treusch et al., 2011). This degeneration was suppressed when genes involved in clathrin-mediated endocytosis, such as unc-11, unc-26, Y44E3A.4, C. elegans RTS1 ortholog, C. elegans ADE12 ortholog, and human CRMI, were co-expressed with Aβ, and enhanced when a PBS2/MAP2K4 mitogen-activated protein kinase transgene was co-expressed with Aβ in glutamatergic neurons (Treusch et al., 2011). Interestingly, mutations in the C. elegans human REST ortholog spr-4, which suppressed the sel-12/PSEN egg-laying defect (Lakowski et al., 2003), also enhanced the degeneration seen in the transgenic animals expressing Aβ in glutamatergic neurons (Lu et al., 2014). Modifying clathrin-mediated endocytosis in rat cortical neurons was similarly neuroprotective against Aβ aggregates (Treusch et al., 2011). In addition, early stage AD brains showed higher expression of REST target genes, while late stage AD and frontotemporal dementia (FTD) brains showed lower expression (Lu et al., 2014). Hence, REST may confer neuroprotective benefits in C. elegans and in humans (Lu et al., 2014).
The C. elegans Aβ model also proves useful in screens to identify drugs that disaggregate Aβ. The drug PBT2, an 8-hydroxy quinoline analog, reversed AD phenotypes in mice within days (Adlard et al., 2008). Similarly, C. elegans expressing inducible Aβ1-42, which become paralyzed within 48 h after induction, were protected against paralysis when exposed to PBT2 (McColl et al., 2012).
C. elegans lrp-1 FUNCTIONS SIMILARLY TO LRP2/MEGALIN, AN LDL RECEPTOR FAMILY MEMBER
In mammals, the LDL receptor family is responsible for many functions, including binding ligands for internalization and degradation and cholesterol metabolism (Brown and Goldstein, 1986; Mahley, 1988; Herz et al., 1992; Willnow et al., 1994). Binding of LRP1 to sAPP770 or full-length APP770, one of the APP isoforms (Figure 1), causes its internalization and degradation (Kounnas et al., 1995; Knauer et al., 1996); disrupting cell surface APP internalization with an LRP-antagonist increases sAPPα processing and full-length APP at the cell surface and decreases Aβ formation, suggesting that LRP1-APP interactions favor APP processing through the amyloidogenic pathway (Ulery et al., 2000). Apolipoprotein E and LRP2/megalin have also been implicated in Aβ clearance (Zlokovic et al., 1996; Deane et al., 2004; Carro et al., 2005).
C. elegans LRP-1 most closely resembles mammalian LRP2/megalin (Yochem and Greenwald, 1993; Yochem et al., 1999). C. elegans does not have the ability to synthesize cholesterol and, therefore, must rely on dietary sources (Hieb and Rothstein, 1968). Inactivation of lrp-1 resulted in late larval lethality due to molting defects during the L3–L4 transition (Yochem et al., 1999). When wild-type C. elegans were grown in the absence of cholesterol, the molting defects of the lrp-1 knockouts were phenocopied (Yochem et al., 1998; Wiese et al., 2012), suggesting that LRP-1 is involved in cholesterol uptake from the environment. LRP-1 is expressed in the epithelial hypodermal cells, hyp6 and hyp7, where it localizes to their apical surface (Yochem et al., 1999) and where apl-1 is also expressed in adults (Niwa et al., 2008). Similarly, its mammalian counterpart, LRP2/megalin, is mainly expressed at the apical surface of epithelial cells (Cui et al., 1996; Morales et al., 1996; Willnow et al., 1996; Nielsen et al., 1998; Zheng et al., 1998; Hermo et al., 1999; Mizuta et al., 1999).
LRP2/megalin interacts with different domains of APP (Zlokovic et al., 1996; Pietrzik et al., 2004; Carro et al., 2005; Yoon et al., 2005; Cam and Bu, 2006). A physical interaction between APL-1 and LRP-1 has not been determined. Expression of LRP-1 with an N-terminal domain truncation did not rescue the lethality of apl-1 null mutants, suggesting that sAPL-1 is not activating an lrp-1 pathway (Hornsten et al., 2007). When lrp-1 expression is decreased or when wild-type animals are deprived of dietary cholesterol, neurotransmission is affected (Wiese et al., 2012). However, apl-1 null mutants die at an earlier stage in development than lrp-1 null mutants, suggesting that apl-1 functions in earlier developmental pathways that are necessary for survival.
C. elegans AS A MODEL FOR OTHER NEURODEGENERATIVE DISEASES
PTL-1 AS A TAU MODEL
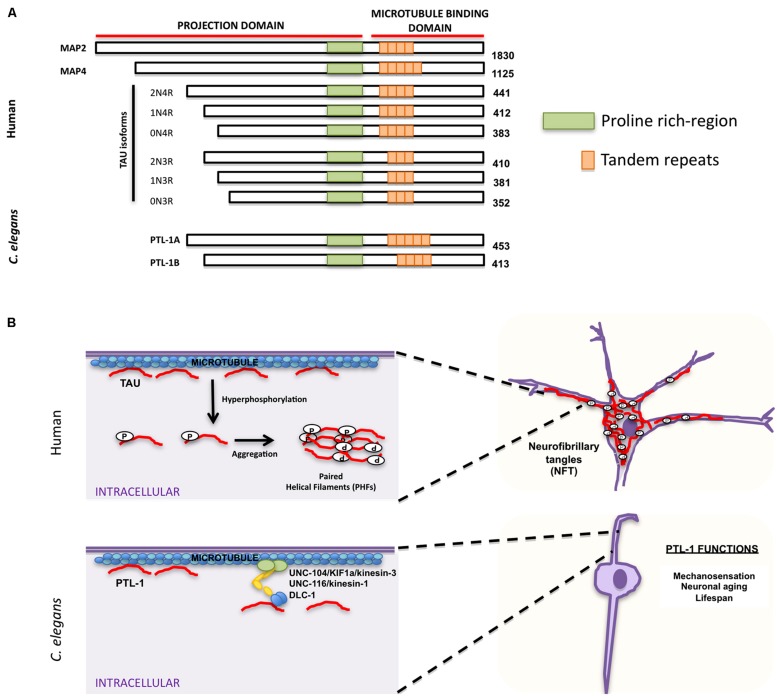
Accumulation of neurofibrillary tangles in cell bodies is another hallmark characteristic of AD and other neurodegenerative disorders. The major component of these tangles is tau, which belongs to the family of microtubule-associated proteins (MAPs) that includes MAP2 and MAP4 (Lee et al., 1988; Lewis et al., 1988; Goedert et al., 1989b; Chapin and Bulinski, 1991). MAPs share characteristic homology domains, including a proline-rich domain and a region of a variable number of tandem amino acid repeats (Figure 3A; Goedert et al., 1988, 1989a,b, 1992a; Lee et al., 1988; Lewis et al., 1988; Aizawa et al., 1990). Tau is the predominant MAP expressed in axons, while MAP2 is expressed in dendrites (Matus et al., 1981; Binder et al., 1985) and MAP4 is expressed in dividing cells (Bulinski and Borisy, 1980). MAPs bind microtubules and are responsible for promoting microtubule assembly and stability (reviewed in Amos and Schlieper, 2005). MAP family members appear to have redundant functions; mice in which tau was knocked out were viable, but showed increased levels of MAP1A (Harada et al., 1994), suggesting that upregulation of MAP1A can compensate for the lack of tau in vivo. Tau phosphorylation affects its ability to bind microtubules and can cause a conformational change that favors tubulin assembly (Figure 3B; Feijoo et al., 2005). Aberrant hyperphosphorylation of tau, however, impairs its ability to bind microtubules, thus resulting in their disassembly (Lindwall and Cole, 1984; Biernat et al., 1993; Bramblett et al., 1993). In addition, phosphorylated tau self-aggregates into PHFs and presumably generates the intracellular neurofibrillary tangles characteristic of AD patients (Figure 3B; Goedert et al., 1992b; Alonso et al., 1996, 1997, 2001; Billingsley and Kincaid, 1997).
FIGURE 3.
Similarities and differences between human tau and Caenorhabditis elegans PTL-1. (A) Schematic representation of human microtubule binding proteins (MAPs) family, including tau isoforms and C. elegans PTL-1 isoforms. (B) Comparison between tau functions in humans (top) and C. elegans PTL-1/tau functions (bottom). Top. Tau has a physiological role in promoting and maintaining microtubule stability. In pathological conditions tau is hyperphosphorylated and self-aggregates into paired helical filaments (PHFs) that can form intracellular neurofibrillary tangles (NFT). Bottom. C. elegans PTL-1/tau binds microtubules and induces microtubule assembly. It also affects synaptic transport through motor proteins UNC-104/KIF1a/kinesin-3, UNC-116/kinesin-1, and DLC-1/dynein. PTL-1/tau is also important for C. elegans mechanosensation and aging.
Because of the functional redundancy of MAPs, their specific functions have been difficult to determine. C. elegans has only one tau homolog protein with tau-like repeats 1 (PTL-1; Goedert et al., 1996; McDermott et al., 1996). PTL-1 exists as two isoforms, PTL-1A and PTL-1B, with five or four tandem repeats, respectively (Goedert et al., 1996; Figure 3A). They have a high level of sequence homology with mammalian tau, especially in the C-terminal microtubule binding region (Goedert et al., 1996; McDermott et al., 1996). Both PTL-1A and PTL-1B bound microtubules in vitro and induced tubulin polymerization (Goedert et al., 1996; McDermott et al., 1996). PTL-1 is initially expressed embryonically in the epidermis of elongating embryos and in head neurons; in larval and adult animals PTL-1 is expressed mainly in the mechanosensory neurons mediating gentle body touch (Figure 3B; Goedert et al., 1996; Gordon et al., 2008), although transcriptional fusions show a wider expression pattern in neurons and stomatointestinal cells (Gordon et al., 2008).
Loss of ptl-1/tau results in an incompletely penetrant lethality at the same stage of embryogenesis (Gordon et al., 2008) at which PTL-1/tau expression is first observed (Goedert et al., 1996). ptl-1/tau mutants that escaped lethality showed normal development, but had a shortened lifespan, and although the overall integrity of microtubule structure appeared unaffected at the light microscopic level, there was a significant reduction in gentle touch sensitivity as compared to wild-type (Gordon et al., 2008; Chew et al., 2013). These touch defects were enhanced in mutants with defects in β- and α-tubulin, indicating that the absence of full-length PTL-1/tau disrupts mechanosensation, but it does so independently of tubulin (Gordon et al., 2008). In mutants in which only the C-terminal microtubule binding repeats of PTL-1/tau are deleted, touch sensitivity was identical to that of wild-type (Chew et al., 2013), suggesting that the N-terminal domain of PTL-1/tau is sufficient for gentle touch responses.
As C. elegans ages, mechanosensory touch neurons exhibit age-related morphological changes: cell bodies initially elaborate branches and axons subsequently display blebbing and branching (Pan et al., 2011; Tank et al., 2011; Toth et al., 2012). Strikingly, mechanosensory touch neurons in ptl-1/tau mutants displayed these aging characteristics at higher incidences and at an earlier stage than wild-type animals; GABAergic neurons also showed age-related phenotypes, such as ectopic branching (Chew et al., 2013). Expression of a human tau isoform (htau40) in ptl-1/tau mutants rescued the touch insensitivity, but not the morphological aging defects, indicating that htau40 shares some functional conservation with PTL-1/tau (Chew et al., 2013). Interestingly, when ptl-1/tau was expressed in Sf9 cells, cells projected neurite-like processes that were positive for PTL-1/tau immunoreactivity (Goedert et al., 1996) and that were indistinguishable from those visualized when htau40 was expressed in Sf9 cells (Knops et al., 1991; Chen et al., 1992). Although wild-type PTL-1/tau has not been reported to aggregate into fibrils, PTL-1/tau, like human tau, clearly has an essential role in maintaining neuronal integrity, controlling neuronal aging, and affecting lifespan (Goedert et al., 1996; Gordon et al., 2008). While there are no known mutations in tau that are associated with AD, tau mutations are associated with FTD with parkinsonism (FTPD-17), another form of dementia (Hutton et al., 1998; Poorkaj et al., 1998; Spillantini et al., 1998; see Section “Frontotemporal Dementia”).
FRONTOTEMPORAL DEMENTIA
Frontotemporal dementia (FTD) is a group of neurodegenerative disorders characterized by severe brain frontotemporal lobar degeneration (reviewed in Rabinovici and Miller, 2010). In some cases it may be hard to distinguish between FTD and AD; however, FTD usually develops earlier in life and is more likely to have a genetic component (Lindau et al., 2000; Pasquier, 2005). Many mutations can cause FTD with or without motor neuron disease (Cruts et al., 2012). Two mutations have been well characterized and are associated with specific types of FTD: tau-positive FTD linked to chromosome 17 (FTD-17) and FTD caused by TDP43 proteinopathy (FTD-TDP43). Patients with FTD-17 suffer behavioral changes and often Parkinson-like motor problems. While mutations in the tau gene MAPT are not described in familial or sporadic AD, MAPT tau mutations are linked with FTD-17 (Hutton et al., 1998; Poorkaj et al., 1998; Spillantini et al., 1998).
Several transgenic lines expressing human tau harboring FTD-17 mutations (htau-FTD-17) have been generated in C. elegans (Figure 4B) (see also Section “PTL-1 as a Tau Model”; Kraemer et al., 2003; Miyasaka et al., 2005; Brandt et al., 2009; Fatouros et al., 2012). Pan-neuronal transgene expression of wild type or htau-FTD-17 caused an uncoordinated phenotype that progressively worsened with age, an accumulation of insoluble tau, and neurodegeneration (Kraemer et al., 2003). Similarly, expression of htau-FTD-17 in touch neurons resulted in a decrease in the touch response due to neuritic abnormalities and tau accumulation (Miyasaka et al., 2005). Using these C. elegans models of tauopathy in forward genetic screens, Kraemer and co-workers identified two new factors, SUT-1 and SUT-2, that may participate in the pathological pathway activated by tau (Kraemer et al., 2003; Kraemer and Schellenberg, 2007; Guthrie et al., 2009). Moreover, down-regulation of the human SUT-2 homolog (MSUT-2) in mammalian cell lines caused a marked decrease in tau aggregation, suggesting that MSUT-2 may be a good candidate target for FTD therapies (Wheeler et al., 2010; Guthrie et al., 2011). More recently, Fatouros et al. (2012) generated two htau-FTD-17 transgenic models: one with a pro-aggregant mutated form of human tau (deletion of K280) and a second with mutated forms of human tau (I277P and I308P) that prevented tau aggregation. The tau (ΔK280) transgenic line had high levels of tau aggregation, which caused uncoordinated movement in adults, axonal defects, and alterations in presynaptic structures (Fatouros et al., 2012); the locomotory defects could be partially suppressed by a compound of the aminothioenopyridazine (ATPZ) class cmp16, suggesting that this compound may be neuroprotective (Fatouros et al., 2012). The tau (I277P and I308P) transgenic lines had low levels of tau aggregates and displayed only mild phenotypes with significantly less morphological abnormalities.
Accumulation of TDP-43 [transactive response (TAR) DNA-binding protein] is found in ∼50% of the cases of FTD (The Association for Frontotemporal Degeneration, 2014) and has numerous genetic causes. However, only one case has been reported with mutations in the TDP-43 gene (Borroni et al., 2009). C. elegans models overexpressing human TDP-43 or its C. elegans ortholog TDP-1 recapitulates some of the FTD phenotypes, including neurotoxicity and protein aggregation (see also Section “Amyotrophic Lateral Sclerosis”).
C9orf72 encodes a protein that regulates endosomal trafficking and autophagy in primary neurons and neuronal cells (Farg et al., 2014). It is expressed in multiple tissue types, including cerebellar cortex and spinal cord (DeJesus-Hernandez et al., 2011). Hexanucleotide (GGGGCC) repeat expansions in a non-coding region of C9orf72 are found in patients with amyotrophic lateral sclerosis (ALS) and FTD (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Majounie et al., 2012), providing the first genetic link between the two diseases, although it remains unclear how C9orf72 hexanucleotide expansion triggers ALS and FTD pathology. Mutations in the C. elegans C9ORF72 ortholog alfa-1 caused age-dependent motility defects, leading to paralysis and degeneration of GABAergic motoneurons (Therrien et al., 2013), suggesting that a loss-of-function mechanism is involved in the C9ORF72-dependent pathogenesis.
PARKINSON’S DISEASE
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that affects the control of body movements. The impaired motor control in PD is the result of the death of dopaminergic (DA) neurons (Hughes et al., 1992; Fahn and Sulzer, 2004). The disease is characterized by the accumulation of α-synuclein into neuronal inclusions called Lewy bodies (Lewy, 1912; Tretiakoff, 1919). Most PD cases are of unknown cause. However, ∼5–10% of PD cases are familial (Wood-Kaczmar et al., 2006) and include mutations in the following genes: α-synuclein, parkin (PRKN), leucine-rich repeat kinase 2 (LRRK2), PTEN-induced putative kinase 1 (PINK1), and ATP13A2 (Hardy, 2010).
α-synuclein is a presynaptic neuronal protein whose cellular function is not well understood, but may include controlling the supply of synaptic vesicles in neuronal terminals and regulating dopamine release. It is a small acidic protein (14 kDa) whose sequence can be divided into three domains: the N-terminal α-helical domain (amino acids 1–65), the central hydrophobic domain (residues 66–95), and the acidic carboxyl-terminal domain (residues 96–140; Recchia et al., 2004). Three mutations in the α-helical domain (A53T, A30P, E46K) are linked with autosomal dominant early onset PD, suggesting that these mutations can predispose to oligomer and fibril formation (Polymeropoulos et al., 1997; Krüger et al., 1998; Conway et al., 2000a,b,c; Zarranz et al., 2004). Because C. elegans has no α-synuclein ortholog, C. elegans models are based on transgenic worms overexpressing wild-type or mutant forms of human α-synuclein (Figure 4B). Although different transgenic lines showed some differences, most lines with pan-neuronal or DA neuronal expression of wild-type or mutated α-synuclein (A53T and/or A30P mutations) displayed locomotory defects and degeneration of dopamine neurons (Lakso et al., 2003; Kuwahara et al., 2006; Cao et al., 2010). Furthermore, downregulating the activity of the nuclease EndoG decreased α-synuclein toxicity in DA neurons (Büttner et al., 2013). EndoG is a mitochondria-specific endonuclease that mediates cellular death by apoptosis (Li et al., 2001). When cell death is induced, EndoG translocates from the mitochondria to the nucleus to fragment DNA (Li et al., 2001). Approximately 50% of the dopamine neurons expressing α-synuclein degenerate, whereas a mutation in cps-6, which encodes the C. elegans EndoG ortholog, rescues this degeneration (Büttner et al., 2013). Similar results were found in yeast, flies, and human cells suggesting that EndoG is a conserved requirement for α-synuclein toxicity (Büttner et al., 2013).
C. elegans models of α-synuclein overexpression-induced toxicity have also been examined by whole genome RNAi knockdown and microarray screenings (Vartiainen et al., 2006; Hamamichi et al., 2008; Kuwahara et al., 2008; van Ham et al., 2008). These screens highlighted the importance of endocytosis for ameliorating α-synuclein-dependent neurotoxicity (Kuwahara et al., 2008). Transgenic lines expressing α-synuclein specifically in body wall muscle cells produced inclusions as animals aged, resembling a feature of neurons in patients with PD; the number of inclusions decreased when genes affecting different biological processes, such as vesicle and lysosomal trafficking (W08D2.5/ATP13A2), lipid metabolism, and lifespan control (sir-2.1,lagr-1), were knocked down (van Ham et al., 2008). In a genome-wide microarray analysis to identify genes that were modulated in C. elegans overexpressing wild-type or A53T human α-synuclein, seven genes encoding components of the ubiquitin-proteasome machinery and 35 mitochondrial function genes were found to be upregulated, while nine genes encoding histones H1, H2B, and H4 were down regulated (Vartiainen et al., 2006). These data provide support for the role of the proteasome complex and mitochondrial proteins in mediating neurotoxicity.
Parkin (human PARK2) is a component of an ubiquitin E3 ligase that is part of the proteasome complex (Shimura et al., 2000). Mutations in PARK2 have been associated with early onset recessive forms of PD (Kitada et al., 1998; Poorkaj et al., 2004). C. elegans has a parkin ortholog, PDR-1. A truncated form of PDR-1(Δaa24–247) encoded by the in-frame deletion null allele pdr-1(lg103) had altered solubility and propensity to aggregate when expressed in cell lines, resembling parkin mutant proteins in PD (Springer et al., 2005). Furthermore, pdr-1 mutants were hypersensitive to different proteotoxic stress conditions, suggesting that PDR-1/PARK2 mutations act to block the proteostasis machinery, thereby making it easier for proteins to abnormally fold and aggregate (Springer et al., 2005).
Mutations in LRRK2/leucine-rich repeat kinase 2 are the most common known cause of late-onset PD. LRRK2 belongs to the LRRK family; gain-of-function LRRK2 mutations interfere with chaperone-mediated autophagic functions and presumably decrease levels of α-synuclein degradation (Orenstein et al., 2013). Transgenic worms overexpressing pathogenic mutant forms of LRRK2 in DA neurons caused DA neurodegeneration (Liu et al., 2011; Yao et al., 2013). Interestingly, treatment of the transgenic worms with kinase inhibitors resulted in arrested neurodegeneration, suggesting that LRRK2 kinase activity is important for its pathogenesis (Liu et al., 2011; Yao et al., 2013).
Many studies have reported a link between toxin exposure and increased risk of PD. C. elegans has been used to test different toxins and help elucidate the mechanism by which they produce neurotoxicity. Administration of the 6-hydroxydopamine (6-OHDA) neurotoxin to C. elegans produced specific degeneration of dopamine neurons (Nass et al., 2002). By performing forward genetic and high-throughput chemical screens, mutations within the dopamine transporter dat-1 were found to suppress 6-OHDA sensitivity (Nass et al., 2005) and bromocriptine, quinpirole, and acetaminophen, and plant extracts from Bacopa monnieri and Uncaria tomentosa were found to be neuroprotective (Marvanova and Nichols, 2007; Locke et al., 2008; Ruan et al., 2010; Jadiya et al., 2011; Shi et al., 2013). These data demonstrate that pathological characteristics of PD can be recapitulated in C. elegans models and used to investigate the mechanism by which α-synuclein and other PD proteins produce neurotoxicity and cause motor defects.
AMYOTROPHIC LATERAL SCLEROSIS
Amyotrophic lateral sclerosis is a neurodegenerative disease characterized by the death of motor neurons in brain and spinal cord and progressive paralysis of the body (Hardiman et al., 2011). Approximately 10% of ALS cases are familial and associated with mutations in several genes. The most common mutation in familial ALS is found in the superoxide dismutase enzyme (SOD1) with more than 160 different mutations identified (Wroe et al., 2008). SOD1 is a ubiquitously expressed protein that converts the toxic radical superoxide anion to hydrogen peroxide. Although it is not clear yet how SOD1 mutations causes motor neuron degeneration, toxicity is likely generated by a gain-of-function mechanism (Valentine et al., 2005) and associated with misfolding and aggregation of the enzyme (Pasinelli and Brown, 2006).
Transgenic lines expressing mutant human SOD1 proteins have been successfully generated in C. elegans and recapitulate the motor neuron degeneration and paralysis characteristic of ALS patients (Figure 4B) (Witan et al., 2008; Gidalevitz et al., 2009; Wang et al., 2009; Li et al., 2014). The locomotion defect caused by pan-neuronal expression of the SOD1(G85R) mutant isoform was reduced when insulin signaling was decreased (Boccitto et al., 2012), suggesting that decreased insulin signaling increases the capacity of cells to prevent the accumulation of toxic non-soluble proteins and opening the possibility of finding new therapeutic targets. Similarly, when wild-type or mutant SOD1(G93A) was expressed exclusively in GABAergic motor neurons, animals showed an age-dependent paralysis and accumulation of wild-type and mutant SOD1(G93A), although defects were more severe in the mutant lines; interestingly, the SOD1 aggregates were soluble in the wild-type SOD1 lines and insoluble in the SOD1(G93A) lines (Li et al., 2014). In addition, motor neurons showed axonal guidance defects during development and caspase-independent cell death in adulthood in the wild-type and SOD1(G93A) lines (Li et al., 2014).
Other genes associated with ALS have also been modeled using C. elegans. TDP-43 [TAR DNA-binding protein] is a 43 kDa RNA binding protein identified as the main component of ubiquitinated protein aggregates (Tsuda et al., 2008; Murakami et al., 2012; Vaccaro et al., 2012a,b; Han et al., 2013; Therrien et al., 2013) found in patients with sporadic ALS (Neumann et al., 2006) and also in some cases of FTD (see Section “Frontotemporal Dementia”). TDP-43 is normally located in the nucleus of neurons, but dominant mutations in TDP-43 cause aberrant localization of TDP-43 in the cytoplasm, thereby preventing it from functioning in the nucleus (Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008; Van Deerlin et al., 2008; Yokoseki et al., 2008; Ash et al., 2010). C. elegans has one TDP-43 ortholog, TDP-1. TDP-1 controls longevity and oxidative stress in the worm by regulating the insulin pathway (Vaccaro et al., 2012b). Overexpression of tdp-1/TDP-43 resulted in toxicity and decreased lifespan, analogous to the phenotypes found in ALS patients (Vaccaro et al., 2012b). In transgenic worms expressing TDP-43 harboring ALS-associated mutations, proteotoxicity affecting neuronal functions was induced. Similar results were found when the RNA binding protein FUS with ALS-related mutations was expressed in the nematode (Murakami et al., 2012; Vaccaro et al., 2012b).
Excess exposure to some pesticides and chemicals, such as the metalloid selenium, have been implicated in the etiology of ALS (Vinceti et al., 2009; Kamel et al., 2012; Malek et al., 2012). Exposure to high levels of sodium selenite in the worm induced neurodegeneration and resulted in paralysis (Estevez et al., 2012, 2014). When insulin pathway activity was reduced, the adverse effects of environmental selenium exposure was altered (Estevez et al., 2014). Overall, the C. elegans models have highlighted the possible importance of the insulin and autophagy pathways in the generation of ALS.
HUNTINGTON’S DISEASE
Huntington’s disease (HD) is a progressive neurodegenerative disorder inherited through autosomal dominant mutations of the IT15 gene. ITI5 encodes the huntingtin protein, whose functions remain unknown (The Huntington’s Disease Collaborative Research Group, 1993). The mutations result in an N-terminal polyglutamine (polyQ) expansion (Goldberg et al., 1996; Mangiarini et al., 1996). In normal individuals, up to 34 repeats have been reported, whereas in HD afflicted individuals, up to 100 polyQ repeats have been recorded (The Huntington’s Disease Collaborative Research Group, 1993). The huntingtin-polyQ (HdhQ) proteins form aggregates, whose toxicity is determined by the length of the polyQ expansion and which cause swollen, disorganized, and ribosome-deficient endoplasmic reticulum and chromatin irregularities (Martindale et al., 1998). Eventually, cellular defects caused by the aggregates culminate in HD symptoms, which include involuntary movement, cognitive impairment, and loss of neurons in the striatum and deep layers of the frontal cortex (Martin and Gusella, 1986).
Although C. elegans does not have a huntingtin homolog, transgenic C. elegans models that express an N-terminal human huntingtin (htt) fragment with different numbers of CAG repeats have been used to model HD and identify genes that prevent polyQ aggregates (Figure 4B). The models generally express the repeats in specific neurons, such as the ASH sensory neurons, which are multi-modal sensory neurons that mediate avoidance to chemo- and mechanosensory stimuli. In transgenic animals expressing htt171 with 150 CAG repeats (htt171-Q150), 13% of the ASH neurons began to lose function after 8 days, suggesting an age-dependent degeneration (Faber et al., 1999). This loss of ASH function was reversed in a ced-3/caspase (Faber et al., 1999) or hda-3/HDAC (Bates et al., 2006) mutant background, suggesting that processes characteristic of apoptotic cell death and histone deacetylases play a role in HD (Dragunow et al., 1995). By contrast, the number of htt171-Q150 aggregates and neurodegeneration were enhanced when genes mediating autophagy, CREB, CREB binding proteins, and pqe-1 were disrupted, suggesting that autophagy and activation of CREB target genes decreases htt171-Q150 aggregation and are neuroprotective (Faber et al., 2002; Bates et al., 2006; Jia et al., 2007). Transgenic animals expressing fewer CAG repeats (2, 23, and 95 polyQ) showed normal ASH function (Faber et al., 1999). The onset of behavioral defects are consistent with most cases of HD, in which symptoms usually appear during midlife (Vonsattel et al., 1985; Martin and Gusella, 1986; Strong et al., 1993; The Huntington’s Disease Collaborative Research Group, 1993; Gusella and MacDonald, 1995) and fewer than 10% of reported cases occur before the age of 21 (Farrer and Conneally, 1985; van Dijk et al., 1986; Nance, 1997; Siesling et al., 1997).
In a different HD model, htt57-Q128 was expressed in the touch mechanosensory neurons (Parker et al., 2001). These transgenic animals did not show neurodegeneration, but had a significantly reduced response to posterior touch and a milder defect in anterior touch response (Parker et al., 2001). The touch neurons contained polyQ aggregates and morphological abnormalities primarily along axonal processes (Parker et al., 2001). The touch insensitivity could be rescued by activating Sir2 sirtuins (Parker et al., 2005), which act through the DAF-16/FOXO transcription factor to promote longevity (Tissenbaum and Guarente, 2001). Similarly, in neuronal cell lines derived from knockin HdhQ111 mice, activation of sirtuins reduced the level of cell death (Parker et al., 2005). Additionally, in an RNAi based screen for genes that suppressed htt57-Q128 defects, identified C. elegans genes were also upregulated in the striatum of mouse HD models (Lejeune et al., 2012). Thus, C. elegans is a useful model to identify additional genes that may protect against or contribute to defects caused by polyQ expansions.
RNAi knockdown of dnj-27/ERdj5, an ER luminal protein upregulated in response to ER stress, exacerbated the impaired mobility observed when a Q40 transgene is expressed in body wall muscles, suggesting that dnj-27 interacts with polyQ and protects against polyQ induced paralysis (Muñoz-Lobato et al., 2014).
C. elegans has the advantage that it is transparent, allowing visualization of the formation of that aggregates, including aggregates made by shorter polyQ tracts, whereas only longer tracts are visible in mammals (Brignull et al., 2006). To determine the threshold number of polyQ repeats needed to elicit a morphological and behavioral response, varying lengths of polyQ repeats were tested in C. elegans. Pan-neuronal expression of more than 40 polyQ led to variable protein aggregation and paralysis (Brignull et al., 2006). These data suggest that 40 polyQ may be the critical number of repeats to elicit HD symptoms and are consistent with unaffected humans who have up to 34 polyQ repeats and HD patients who have as few as 42 repeats (The Huntington’s Disease Collaborative Research Group, 1993).
Overall, C. elegans HD models illustrate that human huntingtin polyQs disrupt the morphology and function of sensory neurons. The genetic and RNAi screens highlight candidate genes that may be involved in HD pathogenesis in mammalian models and provide insights into genes that may serve a protective role against polyQ toxicity. In addition to HD, other diseases caused by polyQ repeats include spinocerebellar ataxias and spinal and bulbar muscular atrophy (Orr and Zoghbi, 2007). Hence, using C. elegans provides another approach toward determining how polyQ pathogenicity contributes to neurodegeneration.
ADVANTAGES AND LIMITATIONS OF THE C. elegans MODEL
The use of C. elegans to study AD and other neurodegenerative diseases has, as many other models, many advantages as well as some drawbacks. Major advantages of C. elegans include the ability to perform forward genetic, RNAi, and high throughput chemical screens and the ease of generating transgenic lines. These benefits have been effective in informing the role of APP and the presenilins and identifying components of the γ-secretase complex. The function of APP and the pathways in which it acts are still unclear. C. elegans presents a complementary system to understand the function and pathways of an APP-related protein, APL-1. Furthermore, overexpression of APL-1 by mutation or by transgene induces phenotypes that converge on the insulin/DAF-16/FOXO pathways, similar to what has been found in mammals. Although APL-1 does not contain the Aβ sequence and C. elegans does not have β-secretase activity, transgenic lines that produce Aβ expression pan-neuronally or in muscle are being used to identify pathways that detoxify the Aβ aggregates, some of which also involve the insulin/DAF-16/FOXO pathways. Whether these models are relevant to human pathology or whether the pathways will be conserved in humans are unknown; however, they present alternative approaches to understanding neurodegenerative diseases for which there are currently few effective therapies. Human tau, as well as mutant tau isoforms, have also been expressed in the worm to recapitulate AD and FTD phenotypes. Recent findings have shown that PTL-1 regulates neuronal aging in the worm. These findings may be important to link aging and tau pathology in AD and FTD patients. Although C. elegans transgene models have many advantages, they also have several disadvantages. In C. elegans, transgenes are present as extrachromosomal arrays and are not integrated into the genome as they are in other systems; a few copies to several hundred copies of the transgene are present in the arrays, so the level of overexpression can be much higher than what is found in vivo. Fortunately, methods for single copy insertions have now been developed (Frøkjaer-Jensen et al., 2008).
AD is considered a multifactorial disease in which other risk factors, such as neuroinflammation, head trauma, and diabetes, may be important in the development of the disease. The C. elegans nervous system is simple compared to the human nervous system. This simplicity allows researchers to study neuronal function and neural circuits in a tractable system. However, the complex network of connections and cell interactions found in humans is not mimicked in C. elegans and this complexity may underlie some of the pathology of neurodegenerative diseases. Nevertheless, most of the pathways and signaling molecules in C. elegans are conserved between worms and mammals. The goal is to translate some of the C. elegans insights into understanding the pathology of AD and other neurodegenerative diseases and designing effective strategies to treat the diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Collin Ewald for reviewing our manuscript. This work was supported by grants from the NIH and NSF.
REFERENCES
- Adlard P. A., Cherny R. A., Finkelstein D. I., Gautier E., Robb E., Cortes M., et al. (2008). Rapid restoration of cognition in Alzheimer’s transgenic mice with 8-hydroxy quinoline analogs is associated with decreased interstitial Abeta. Neuron 59 43–55 10.1016/j.neuron.2008.06.018 [DOI] [PubMed] [Google Scholar]
- Aizawa H., Emori Y., Murofushi H., Kawasaki H., Sakai H., Suzuki K. (1990). Molecular cloning of a ubiquitously distributed microtubule-associated protein with Mr 190,000. J. Biol. Chem. 265 13849–13855 [PubMed] [Google Scholar]
- Alonso A., Zaidi T., Novak M., Grundke-Iqbal I., Iqbal K. (2001). Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc. Natl. Acad. Sci. U.S.A. 98 6923–6928 10.1073/pnas.121119298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A. C., Grundke-Iqbal I., Iqbal K. (1996). Alzheimer’s disease hyperphosphorylated tau sequesters normal tau into tangles of filaments and disassembles microtubules. Nat. Med. 2 783–787 10.1038/nm0796-783 [DOI] [PubMed] [Google Scholar]
- Alonso A. D., Grundke-Iqbal I., Barra H. S., Iqbal K. (1997). Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc. Natl. Acad. Sci. U.S.A. 94 298–303 10.1073/pnas.94.1.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Disease International. (2014). World Alzheimer report 2013 [Online]. Available at: http://www.alz.co.uk/ (accessed March 05, 2014) [Google Scholar]
- Ambros V., Horvitz H. R. (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226 409–416 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Amos L. A., Schlieper D. (2005). Microtubules and maps. Adv. Protein Chem. 71 257–298 10.1016/S0065-3233(04)71007-4 [DOI] [PubMed] [Google Scholar]
- Arduengo P. M., Appleberry O. K., Chuang P., L’hernault S. W. (1998). The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J. Cell Sci. 111 3645–3654 [DOI] [PubMed] [Google Scholar]
- Arimoto M., Koushika S. P., Choudhary B. C., Li C., Matsumoto K., Hisamoto N. (2011). The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31 2216–2224 10.1523/JNEUROSCI.2653-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M., Ishihara T., Jindra M., Kohara Y., Katsura I., Hirose S. (2000). The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells 5 711–723 10.1046/j.1365-2443.2000.00361.x [DOI] [PubMed] [Google Scholar]
- Ash P. E., Zhang Y. J., Roberts C. M., Saldi T., Hutter H., Buratti E., et al. (2010). Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum. Mol. Genet. 19 3206–3218 10.1093/hmg/ddq230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., Hartwieg E., Horvitz H. R. (1993). Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74 515–527 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R. (1991). Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7 729–742 10.1016/0896-6273(91)90276-6 [DOI] [PubMed] [Google Scholar]
- Bates E. A., Victor M., Jones A. K., Shi Y., Hart A. C. (2006). Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J. Neurosci. 26 2830–2838 10.1523/JNEUROSCI.3344-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat J., Gustke N., Drewes G., Mandelkow E. M., Mandelkow E. (1993). Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron 11 153–163 10.1016/0896-6273(93)90279-Z [DOI] [PubMed] [Google Scholar]
- Bieschke J., Cohen E., Murray A., Dillin A., Kelly J. W. (2009). A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis. Protein Sci. 18 2231–2241 10.1002/pro.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley M. L., Kincaid R. L. (1997). Regulated phosphorylation and dephosphorylation of tau protein: effects on microtubule interaction, intracellular trafficking and neurodegeneration. Biochem. J. 323 577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder L. I., Frankfurter A., Rebhun L. I. (1985). The distribution of tau in the mammalian central nervous system. J. Cell Biol. 101 1371–1378 10.1083/jcb.101.4.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccitto M., Lamitina T., Kalb R. G. (2012). Daf-2 signaling modifies mutant SOD1 toxicity in C. elegans. PLoS ONE 7:e33494 10.1371/journal.pone.0033494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgonovo B., Ouwendijk J., Solimena M. (2006). Biogenesis of secretory granules. Curr. Opin. Cell Biol. 18 365–370 10.1016/j.ceb.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Borroni B., Bonvicini C., Alberici A., Buratti E., Agosti C., Archetti S., et al. (2009). Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 30 E974–E983 10.1002/humu.21100 [DOI] [PubMed] [Google Scholar]
- Bramblett G. T., Goedert M., Jakes R., Merrick S. E., Trojanowski J. Q., Lee V. M. (1993). Abnormal tau phosphorylation at Ser396 in Alzheimer’s disease recapitulates development and contributes to reduced microtubule binding. Neuron 10 1089–1099 10.1016/0896-6273(93)90057-X [DOI] [PubMed] [Google Scholar]
- Brandt R., Gergou A., Wacker I., Fath T., Hutter H. (2009). A Caenorhabditis elegans model of tau hyperphosphorylation: induction of developmental defects by transgenic overexpression of Alzheimer’s disease-like modified tau. Neurobiol. Aging 30 22–33 10.1016/j.neurobiolaging.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull H. R., Moore F. E., Tang S. J., Morimoto R. I. (2006). Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J. Neurosci. 26 7597–7606 10.1523/JNEUROSCI.0990-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. (1986). A receptor-mediated pathway for cholesterol homeostasis. Science 232 34–47 10.1126/science.3513311 [DOI] [PubMed] [Google Scholar]
- Bulinski J. C., Borisy G. G. (1980). Microtubule-associated proteins from cultured HeLa cells. Analysis of molecular properties and effects on microtubule polymerization. J. Biol. Chem. 255 11570–11576 [PubMed] [Google Scholar]
- Büttner S., Habernig L., Broeskamp F., Ruli D., Vögtle F. N., Vlachos M., et al. (2013). Endonuclease G mediates α-synuclein cytotoxicity during Parkinson’s disease. EMBO J. 32 3041–3054 10.1038/emboj.2013.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282 2012–2018 [DOI] [PubMed] [Google Scholar]
- Cam J. A., Bu G. (2006). Modulation of beta-amyloid precursor protein trafficking and processing by the low density lipoprotein receptor family. Mol. Neurodegener. 1:8 10.1186/1750-1326-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P., Yuan Y., Pehek E. A., Moise A. R., Huang Y., Palczewski K., et al. (2010). Alpha-synuclein disrupted dopamine homeostasis leads to dopaminergic neuron degeneration in Caenorhabditis elegans. PLoS ONE 5:e9312 10.1371/journal.pone.0009312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro E., Spuch C., Trejo J. L., Antequera D., Torres-Aleman I. (2005). Choroid plexus megalin is involved in neuroprotection by serum insulin-like growth factor I. J. Neurosci. 25 10884–10893 10.1523/JNEUROSCI.2909-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R., Sulston J. E. (1981). Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24 59–69 10.1016/0092-8674(81)90501-8 [DOI] [PubMed] [Google Scholar]
- Chalfie M., Sulston J. (1981). Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82 358–370 10.1016/0012-1606(81)90459-0 [DOI] [PubMed] [Google Scholar]
- Chalfie M., Sulston J. E., White J. G., Southgate E., Thomson J. N., Brenner S. (1985). The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 5 956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin S. J., Bulinski J. C. (1991). Non-neuronal 210 × 103 Mr microtubule-associated protein (MAP4) contains a domain homologous to the microtubule-binding domains of neuronal MAP2 and tau. J. Cell Sci. 98 27–36 [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin M. C., Crawford F., Houlden H., Warren A., Hughes D., Fidani L., et al. (1991). Early-onset Alzheimer’s disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene. Nature 353 844–846 10.1038/353844a0 [DOI] [PubMed] [Google Scholar]
- Chen J., Kanai Y., Cowan N. J., Hirokawa N. (1992). Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 360 674–677 10.1038/360674a0 [DOI] [PubMed] [Google Scholar]
- Chew Y. L., Fan X., Götz J., Nicholas H. R. (2013). PTL-1 regulates neuronal integrity and lifespan in C. elegans. J. Cell Sci. 126 2079–2091 10.1242/jcs.jcs124404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E., Bieschke J., Perciavalle R. M., Kelly J. W., Dillin A. (2006). Opposing activities protect against age-onset proteotoxicity. Science 313 1604–1610 10.1126/science.1124646 [DOI] [PubMed] [Google Scholar]
- Colbert H. A., Bargmann C. I. (1995). Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron 14 803–812 10.1016/0896-6273(95)90224-4 [DOI] [PubMed] [Google Scholar]
- Conway K. A., Harper J. D., Lansbury P. T. (2000a). Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry 39 2552–2563 10.1021/bi991447r [DOI] [PubMed] [Google Scholar]
- Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Harper J. D., Williamson R. E., et al. (2000b). Accelerated oligomerization by Parkinson’s disease linked alpha-synuclein mutants. Ann. N. Y. Acad. Sci. 920 42–45 10.1111/j.1749-6632.2000.tb06903.x [DOI] [PubMed] [Google Scholar]
- Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Williamson R. E., Lansbury P. T. (2000c). Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U.S.A. 97 571–576 10.1073/pnas.97.2.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M., Theuns J., Van Broeckhoven C. (2012). Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 33 1340–1344 10.1002/humu.22117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Verroust P. J., Moestrup S. K., Christensen E. I. (1996). Megalin/gp330 mediates uptake of albumin in renal proximal tubule. Am. J. Physiol. 271 F900–F907 [DOI] [PubMed] [Google Scholar]
- Daigle I., Li C. (1993). apl-1, a Caenorhabditis elegans gene encoding a protein related to the human beta-amyloid protein precursor. Proc. Natl. Acad. Sci. U.S.A. 90 12045–12049 10.1073/pnas.90.24.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R., Wu Z., Sagare A., Davis J., Du Yan S., Hamm K., et al. (2004). LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron 43 333–344 10.1016/j.neuron.2004.07.017 [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., Rutherford N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72 245–256 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M., Faull R. L., Lawlor P., Beilharz E. J., Singleton K., Walker E. B., et al. (1995). In situ evidence for DNA fragmentation in Huntington’s disease striatum and Alzheimer’s disease temporal lobes. Neuroreport 6 1053–1057 10.1097/00001756-199505090-00026 [DOI] [PubMed] [Google Scholar]
- Duarte A., Hirashima M., Benedito R., Trindade A., Diniz P., Bekman E., et al. (2004). Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 18 2474–2478 10.1101/gad.1239004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duilio A., Faraonio R., Minopoli G., Zambrano N., Russo T. (1998). Fe65L2: a new member of the Fe65 protein family interacting with the intracellular domain of the Alzheimer’s beta-amyloid precursor protein. Biochem. J. 330 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas K. J., Guo C., Wang X., Burkhart K. B., Adams E. J., Alam H., et al. (2010). Functional divergence of dafachronic acid pathways in the control of C. elegans development and lifespan. Dev. Biol. 340 605–612 10.1016/j.ydbio.2010.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Charlie N. K., Richmond J. E., Hegermann J., Eimer S., Miller K. G. (2009). Impaired dense core vesicle maturation in Caenorhabditis elegans mutants lacking Rab2. J. Cell Biol. 186 881–895 10.1083/jcb.200902095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez A. O., Morgan K. L., Szewczyk N. J., Gems D., Estevez M. (2014). The neurodegenerative effects of selenium are inhibited by FOXO and PINK1/PTEN regulation of insulin/insulin-like growth factor signaling in Caenorhabditis elegans. Neurotoxicology 41C, 28–43 10.1016/j.neuro.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez A. O., Mueller C. L., Morgan K. L., Szewczyk N. J., Teece L., Miranda-Vizuete A., et al. (2012). Selenium induces cholinergic motor neuron degeneration in Caenorhabditis elegans. Neurotoxicology 33 1021–1032 10.1016/j.neuro.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald C. Y., Cheng R., Tolen L., Shah V., Gillani A., Nasrin A., et al. (2012a). Pan-neuronal expression of APL-1, an APP-related protein, disrupts olfactory, gustatory, and touch plasticity in Caenorhabditis elegans. J. Neurosci. 32 10156–10169 10.1523/JNEUROSCI.0495-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald C. Y., Raps D. A., Li C. (2012b). APL-1, the Alzheimer’s Amyloid precursor protein in Caenorhabditis elegans, modulates multiple metabolic pathways throughout development. Genetics 191 493–507 10.1534/genetics.112.138768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P. W., Alter J. R., Macdonald M. E., Hart A. C. (1999). Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc. Natl. Acad. Sci. U.S.A. 96 179–184 10.1073/pnas.96.1.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P. W., Voisine C., King D. C., Bates E. A., Hart A. C. (2002). Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc. Natl. Acad. Sci. U.S.A. 99 17131–17136 10.1073/pnas.262544899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn S., Sulzer D. (2004). Neurodegeneration and neuroprotection in Parkinson disease. NeuroRx 1 139–154 10.1602/neurorx.1.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg M. A., Sundaramoorthy V., Sultana J. M., Yang S., Atkinson R. A., Levina V., et al. (2014). C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 23 3579–3595 10.1093/hmg/ddu068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer L. A., Conneally P. M. (1985). A genetic model for age at onset in Huntington disease. Am. J. Hum. Genet. 37 350–357 [PMC free article] [PubMed] [Google Scholar]
- Fatouros C., Pir G. J., Biernat J., Koushika S. P., Mandelkow E., Mandelkow E. M., et al. (2012). Inhibition of tau aggregation in a novel Caenorhabditis elegans model of tauopathy mitigates proteotoxicity. Hum. Mol. Genet. 21 3587–3603 10.1093/hmg/dds190 [DOI] [PubMed] [Google Scholar]
- Feijoo C., Campbell D. G., Jakes R., Goedert M., Cuenda A. (2005). Evidence that phosphorylation of the microtubule-associated protein Tau by SAPK4/p38delta at Thr50 promotes microtubule assembly. J. Cell Sci. 118 397–408 10.1242/jcs.01655 [DOI] [PubMed] [Google Scholar]
- Fonte V., Kapulkin W. J., Kapulkin V., Taft A., Fluet A., Friedman D., et al. (2002). Interaction of intracellular beta amyloid peptide with chaperone proteins. Proc. Natl. Acad. Sci. U.S.A. 99 9439–9444 10.1073/pnas.152313999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonte V., Kipp D. R., Yerg J., Merin D., Forrestal M., Wagner E., et al. (2008). Suppression of in vivo beta-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J. Biol. Chem. 283 784–791 10.1074/jbc.M703339200 [DOI] [PubMed] [Google Scholar]
- Francis R., Mcgrath G., Zhang J., Ruddy D. A., Sym M., Apfeld J., et al. (2002). aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev. Cell 3 85–97 10.1016/S1534-5807(02)00189-2 [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S. P., et al. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40 1375–1383 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale N. W., Dominguez M. G., Noguera I., Pan L., Hughes V., Valenzuela D. M., et al. (2004). Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. U.S.A. 101 15949–15954 10.1073/pnas.0407290101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini L., Gouras G. K., Wang R., Gross R. S., Beal M. F., Greengard P., et al. (2001). Stimulation of beta-amyloid precursor protein trafficking by insulin reduces intraneuronal beta-amyloid and requires mitogen-activated protein kinase signaling. J. Neurosci. 21 2561–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D., Sutton A. J., Sundermeyer M. L., Albert P. S., King K. V., Edgley M. L., et al. (1998). Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150 129–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V., Antebi A. (2001). A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1 841–851 10.1016/S1534-5807(01)00085-5 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Krupinski T., Garcia S., Morimoto R. I. (2009). Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 5:e1000399 10.1371/journal.pgen.1000399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner C. R., Sluder A. E. (2000). nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev. Biol. 221 259–272 10.1006/dbio.2000.9679 [DOI] [PubMed] [Google Scholar]
- Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., et al. (2008). TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 63 535–538 10.1002/ana.21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner G. G., Wong C. W. (1984). Alzheimer’s disease and Down’s syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem. Biophys. Res. Commun. 122 1131–1135 10.1016/0006-291X(84)91209-9 [DOI] [PubMed] [Google Scholar]
- Goate A., Chartier-Harlin M. C., Mullan M., Brown J., Crawford F., Fidani L., et al. (1991). Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349 704–706 10.1038/349704a0 [DOI] [PubMed] [Google Scholar]
- Goedert M., Baur C. P., Ahringer J., Jakes R., Hasegawa M., Spillantini M. G., et al. (1996). PTL-1, a microtubule-associated protein with tau-like repeats from the nematode Caenorhabditis elegans. J. Cell Sci. 109 2661–2672 [DOI] [PubMed] [Google Scholar]
- Goedert M., Cohen E. S., Jakes R., Cohen P. (1992a). p42 MAP kinase phosphorylation sites in microtubule-associated protein tau are dephosphorylated by protein phosphatase 2A1. Implications for Alzheimer’s disease [corrected]. FEBS Lett. 312 95–99 10.1016/0014-5793(92)81418-L [DOI] [PubMed] [Google Scholar]
- Goedert M., Spillantini M. G., Cairns N. J., Crowther R. A. (1992b). Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8 159–168 10.1016/0896-6273(92)90117-V [DOI] [PubMed] [Google Scholar]
- Goedert M., Spillantini M. G., Jakes R., Rutherford D., Crowther R. A. (1989a). Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3 519–526 10.1016/0896-6273(89)90210-9 [DOI] [PubMed] [Google Scholar]
- Goedert M., Spillantini M. G., Potier M. C., Ulrich J., Crowther R. A. (1989b). Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 8 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M., Wischik C. M., Crowther R. A., Walker J. E., Klug A. (1988). Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc. Natl. Acad. Sci. U.S.A. 85 4051–4055 10.1073/pnas.85.11.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Y. P., Kalchman M. A., Metzler M., Nasir J., Zeisler J., Graham R., et al. (1996). Absence of disease phenotype and intergenerational stability of the CAG repeat in transgenic mice expressing the human Huntington disease transcript. Hum. Mol. Genet. 5 177–185 10.1093/hmg/5.2.177 [DOI] [PubMed] [Google Scholar]
- Gordon P., Hingula L., Krasny M. L., Swienckowski J. L., Pokrywka N. J., Raley-Susman K. M. (2008). The invertebrate microtubule-associated protein PTL-1 functions in mechanosensation and development in Caenorhabditis elegans. Dev. Genes Evol. 218 541–551 10.1007/s00427-008-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorevic P. D., Goñi F., Pons-Estel B., Alvarez F., Peress N. S., Frangione B. (1986). Isolation and partial characterization of neurofibrillary tangles and amyloid plaque core in Alzheimer’s disease: immunohistological studies. J. Neuropathol. Exp. Neurol. 45 647–664 10.1097/00005072-198611000-00004 [DOI] [PubMed] [Google Scholar]
- Goutte C., Hepler W., Mickey K. M., Priess J. R. (2000). aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development 127 2481–2492 [DOI] [PubMed] [Google Scholar]
- Goutte C., Tsunozaki M., Hale V. A., Priess J. R. (2002). APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. U.S.A. 99 775–779 10.1073/pnas.022523499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R. (1983). The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34 435–444 10.1016/0092-8674(83)90377-X [DOI] [PubMed] [Google Scholar]
- Gu Y., Misonou H., Sato T., Dohmae N., Takio K., Ihara Y. (2001). Distinct intramembrane cleavage of the beta-amyloid precursor protein family resembling gamma-secretase-like cleavage of Notch. J. Biol. Chem. 276 35235–35238 10.1074/jbc.C100357200 [DOI] [PubMed] [Google Scholar]
- Gusella J. F., MacDonald M. E. (1995). Huntington’s disease: CAG genetics expands neurobiology. Curr. Opin. Neurobiol. 5 656–662 10.1016/0959-4388(95)80072-7 [DOI] [PubMed] [Google Scholar]
- Guthrie C. R., Greenup L., Leverenz J. B., Kraemer B. C. (2011). MSUT2 is a determinant of susceptibility to tau neurotoxicity. Hum. Mol. Genet. 20 1989–1999 10.1093/hmg/ddr079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. R., Schellenberg G. D., Kraemer B. C. (2009). SUT-2 potentiates tau-induced neurotoxicity in Caenorhabditis elegans. Hum. Mol. Genet. 18 1825–1838 10.1093/hmg/ddp099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénette S. Y., Chen J., Jondro P. D., Tanzi R. E. (1996). Association of a novel human FE65-like protein with the cytoplasmic domain of the beta-amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 93 10832–10837 10.1073/pnas.93.20.10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C., Hung A. Y., Selkoe D. J., Teplow D. B. (1994a). Mutations associated with a locus for familial Alzheimer’s disease result in alternative processing of amyloid beta-protein precursor. J. Biol. Chem. 269 17741–17748 [PubMed] [Google Scholar]
- Haass C., Koo E. H., Teplow D. B., Selkoe D. J. (1994b). Polarized secretion of beta-amyloid precursor protein and amyloid beta-peptide in MDCK cells. Proc. Natl. Acad. Sci. U.S.A. 91 1564–1568 10.1073/pnas.91.4.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C., Koo E. H., Mellon A., Hung A. Y., Selkoe D. J. (1992). Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature 357 500–503 10.1038/357500a0 [DOI] [PubMed] [Google Scholar]
- Hamamichi S., Rivas R. N., Knight A. L., Cao S., Caldwell K. A., Caldwell G. A. (2008). Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc. Natl. Acad. Sci. U.S.A. 105 728–733 10.1073/pnas.0711018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamos J. E., Oblas B., Pulaski-Salo D., Welch W. J., Bole D. G., Drachman D. A. (1991). Expression of heat shock proteins in Alzheimer’s disease. Neurology 41 345–350 10.1212/WNL.41.3.345 [DOI] [PubMed] [Google Scholar]
- Han S. M., El Oussini H., Scekic-Zahirovic J., Vibbert J., Cottee P., Prasain J. K., et al. (2013). VAPB/ALS8 MSP ligands regulate striated muscle energy metabolism critical for adult survival in Caenorhabditis elegans. PLoS Genet. 9:e1003738 10.1371/journal.pgen.1003738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A., Oguchi K., Okabe S., Kuno J., Terada S., Ohshima T., et al. (1994). Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 369 488–491 10.1038/369488a0 [DOI] [PubMed] [Google Scholar]
- Hardiman O., Van Den Berg L. H., Kiernan M. C. (2011). Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 7 639–649 10.1038/nrneurol.2011.153 [DOI] [PubMed] [Google Scholar]
- Hardy J. (2010). Genetic analysis of pathways to Parkinson disease. Neuron 68 201–206 10.1016/j.neuron.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G. D., Frand A. R., Ruvkun G. (2006). The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133 4631–4641 10.1242/dev.02655 [DOI] [PubMed] [Google Scholar]
- Heber S., Herms J., Gajic V., Hainfellner J., Aguzzi A., Rülicke T., et al. (2000). Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J. Neurosci. 20 7951–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., Johnson T. E. (2001). daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11 1975–1980 10.1016/S0960-9822(01)00594-2 [DOI] [PubMed] [Google Scholar]
- Hermo L., Lustig M., Lefrancois S., Argraves W. S., Morales C. R. (1999). Expression and regulation of LRP-2/megalin in epithelial cells lining the efferent ducts and epididymis during postnatal development. Mol. Reprod. Dev. 53 282–293 [DOI] [PubMed] [Google Scholar]
- Herms J., Anliker B., Heber S., Ring S., Fuhrmann M., Kretzschmar H., et al. (2004). Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J. 23 4106–4115 10.1038/sj.emboj.7600390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J., Clouthier D. E., Hammer R. E. (1992). LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 71 411–421 10.1016/0092-8674(92)90511-A [DOI] [PubMed] [Google Scholar]
- Hieb W. F., Rothstein M. (1968). Sterol requirement for reproduction of a free-living nematode. Science 160 778–780 10.1126/science.160.3829.778 [DOI] [PubMed] [Google Scholar]
- Holcomb L. A., Gordon M. N., Jantzen P., Hsiao K., Duff K., Morgan D. (1999). Behavioral changes in transgenic mice expressing both amyloid precursor protein and presenilin-1 mutations: lack of association with amyloid deposits. Behav. Genet. 29 177–185 10.1023/A:1021691918517 [DOI] [PubMed] [Google Scholar]
- Hornsten A., Lieberthal J., Fadia S., Malins R., Ha L., Xu X., et al. (2007). APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc. Natl. Acad. Sci. U.S.A. 104 1971–1976 10.1073/pnas.0603997104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K. K., Borchelt D. R., Olson K., Johannsdottir R., Kitt C., Yunis W., et al. (1995). Age-related CNS disorder and early death in transgenic FVB/N mice overexpressing Alzheimer amyloid precursor proteins. Neuron 15 1203–1218 10.1016/0896-6273(95)90107-8 [DOI] [PubMed] [Google Scholar]
- Hsu A. L., Murphy C. T., Kenyon C. (2003). Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 1142–1145 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- Hughes A. J., Ben-Shlomo Y., Daniel S. E., Lees A. J. (1992). What features improve the accuracy of clinical diagnosis in Parkinson’s disease: a clinicopathologic study. Neurology 42 1142–1146 10.1212/WNL.42.6.1142 [DOI] [PubMed] [Google Scholar]
- Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., et al. (1998). Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393 702–705 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- Jadiya P., Khan A., Sammi S. R., Kaur S., Mir S. S., Nazir A. (2011). Anti-Parkinsonian effects of Bacopa monnieri: insights from transgenic and pharmacological Caenorhabditis elegans models of Parkinson’s disease. Biochem. Biophys. Res. Commun. 413 605–610 10.1016/j.bbrc.2011.09.010 [DOI] [PubMed] [Google Scholar]
- Jarriault S., Greenwald I. (2005). Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev. Biol. 287 1–10 10.1016/j.ydbio.2005.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K., Hart A. C., Levine B. (2007). Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy 3 21–25 10.4161/auto.3528 [DOI] [PubMed] [Google Scholar]
- Jonsson T., Atwal J. K., Steinberg S., Snaedal J., Jonsson P. V., Bjornsson S., et al. (2012). A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488 96–99 10.1038/nature11283 [DOI] [PubMed] [Google Scholar]
- Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., Mcconkey B. J., Vande Velde C., et al. (2008). TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40 572–574 10.1038/ng.132 [DOI] [PubMed] [Google Scholar]
- Kamel F., Umbach D. M., Bedlack R. S., Richards M., Watson M., Alavanja M. C., et al. (2012). Pesticide exposure and amyotrophic lateral sclerosis. Neurotoxicology 33 457–462 10.1016/j.neuro.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., et al. (1987). The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325 733–736 10.1038/325733a0 [DOI] [PubMed] [Google Scholar]
- Kidd M. (1964). Alzheimer’s disease–An electron microscopical study. Brain 87 307–320 10.1093/brain/87.2.307 [DOI] [PubMed] [Google Scholar]
- Kimberly W. T., Lavoie M. J., Ostaszewski B. L., Ye W., Wolfe M. S., Selkoe D. J. (2003). Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc. Natl. Acad. Sci. U.S.A. 100 6382–6387 10.1073/pnas.1037392100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. (1997). daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 942–946 10.1126/science.277.5328.942 [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., et al. (1998). Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392 605–608 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Knauer M. F., Orlando R. A., Glabe C. G. (1996). Cell surface APP751 forms complexes with protease nexin 2 ligands and is internalized via the low density lipoprotein receptor-related protein (LRP). Brain Res. 740 6–14 10.1016/S0006-8993(96)00711-1 [DOI] [PubMed] [Google Scholar]
- Knops J., Kosik K. S., Lee G., Pardee J. D., Cohen-Gould L., Mcconlogue L. (1991). Overexpression of tau in a nonneuronal cell induces long cellular processes. J. Cell Biol. 114 725–733 10.1083/jcb.114.4.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo E. H., Sisodia S. S., Archer D. R., Martin L. J., Weidemann A., Beyreuther K., et al. (1990). Precursor of amyloid protein in Alzheimer disease undergoes fast anterograde axonal transport. Proc. Natl. Acad. Sci. U.S.A. 87 1561–1565 10.1073/pnas.87.4.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounnas M. Z., Moir R. D., Rebeck G. W., Bush A. I., Argraves W. S., Tanzi R. E., et al. (1995). LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell 82 331–340 10.1016/0092-8674(95)90320-8 [DOI] [PubMed] [Google Scholar]
- Kraemer B. C., Schellenberg G. D. (2007). SUT-1 enables tau-induced neurotoxicity in C. elegans. Hum. Mol. Genet. 16 1959–1971 10.1093/hmg/ddm143 [DOI] [PubMed] [Google Scholar]
- Kraemer B. C., Zhang B., Leverenz J. B., Thomas J. H., Trojanowski J. Q., Schellenberg G. D. (2003). Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc. Natl. Acad. Sci. U.S.A. 100 9980–9985 10.1073/pnas.1533448100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs L. T., Shutter J. R., Tanigaki K., Honjo T., Stark K. L., Gridley T. (2004). Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 18 2469–2473 10.1101/gad.1239204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs L. T., Xue Y., Norton C. R., Shutter J. R., Maguire M., Sundberg J. P., et al. (2000). Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14 1343–1352 [PMC free article] [PubMed] [Google Scholar]
- Krebs L. T., Xue Y., Norton C. R., Sundberg J. P., Beatus P., Lendahl U., et al. (2003). Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 37 139–143 10.1002/gene.10241 [DOI] [PubMed] [Google Scholar]
- Krigman M. R., Feldman R. G., Bensch K. (1965). Alzheimer’s presenile dementia. A histochemical and electron microscopic study. Lab. Invest. 14 381–396 [PubMed] [Google Scholar]
- Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., et al. (1998). Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 18 106–108 10.1038/ng0298-106 [DOI] [PubMed] [Google Scholar]
- Kuwahara T., Koyama A., Gengyo-Ando K., Masuda M., Kowa H., Tsunoda M., et al. (2006). Familial Parkinson mutant alpha-synuclein causes dopamine neuron dysfunction in transgenic Caenorhabditis elegans. J. Biol. Chem. 281 334–340 10.1074/jbc.M504860200 [DOI] [PubMed] [Google Scholar]
- Kuwahara T., Koyama A., Koyama S., Yoshina S., Ren C. H., Kato T., et al. (2008). A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in alpha-synuclein transgenic C. elegans. Hum. Mol. Genet. 17 2997–3009 10.1093/hmg/ddn198 [DOI] [PubMed] [Google Scholar]
- L’Hernault S. W., Arduengo P. M. (1992). Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J. Cell Biol. 119 55–68 10.1083/jcb.119.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B., Eimer S., Göbel C., Böttcher A., Wagler B., Baumeister R. (2003). Two suppressors of sel-12 encode C2H2 zinc-finger proteins that regulate presenilin transcription in Caenorhabditis elegans. Development 130 2117–2128 10.1242/dev.00429 [DOI] [PubMed] [Google Scholar]
- Lakso M., Vartiainen S., Moilanen A. M., Sirviö J., Thomas J. H., Nass R., et al. (2003). Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alpha-synuclein. J. Neurochem. 86 165–172 10.1046/j.1471-4159.2003.01809.x [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J. (1991). Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112 231–240 [DOI] [PubMed] [Google Scholar]
- Larsen P. L., Albert P. S., Riddle D. L. (1995). Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139 1567–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Ashrafi K. (2008). A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 4:e1000213 10.1371/journal.pgen.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y., Hench J., Ruvkun G. (2001). Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 11 1950–1957 10.1016/S0960-9822(01)00595-4 [DOI] [PubMed] [Google Scholar]
- Lee V. M., Otvos L., Carden M. J., Hollosi M., Dietzschold B., Lazzarini R. A. (1988). Identification of the major multiphosphorylation site in mammalian neurofilaments. Proc. Natl. Acad. Sci. U.S.A. 85 1998–2002 10.1073/pnas.85.6.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F. X., Mesrob L., Parmentier F., Bicep C., Vazquez-Manrique R. P., Parker J. A., et al. (2012). Large-scale functional RNAi screen in C. elegans identifies genes that regulate the dysfunction of mutant polyglutamine neurons. BMC Genomics 13:91 10.1186/1471-2164-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D., Doyle T. G., Brousseau D., Lee M. K., Thinakaran G., Slunt H. H., et al. (1996). Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 93 14940–14944 10.1073/pnas.93.25.14940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D., Greenwald I. (1995). Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature 377 351–354 10.1038/377351a0 [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E., Wasco W., Poorkaj P., Romano D. M., Oshima J., Pettingell W. H., et al. (1995). Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269 973–977 10.1126/science.7638622 [DOI] [PubMed] [Google Scholar]
- Lewis S. A., Wang D. H., Cowan N. J. (1988). Microtubule-associated protein MAP2 shares a microtubule binding motif with tau protein. Science 242 936–939 10.1126/science.3142041 [DOI] [PubMed] [Google Scholar]
- Lewy F. H. (1912). Handbuch der Neurologie. (Berlin: Springer; ) 920–933 [Google Scholar]
- Li J., Li T., Zhang X., Tang Y., Yang J., Le W. (2014). Human superoxide dismutase 1 overexpression in motor neurons of Caenorhabditis elegans causes axon guidance defect and neurodegeneration. Neurobiol. Aging 35 837–846 10.1016/j.neurobiolaging.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Li L. Y., Luo X., Wang X. (2001). Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412 95–99 10.1038/35083620 [DOI] [PubMed] [Google Scholar]
- Li T., Ma G., Cai H., Price D. L., Wong P. C. (2003). Nicastrin is required for assembly of presenilin/gamma-secretase complexes to mediate Notch signaling and for processing and trafficking of beta-amyloid precursor protein in mammals. J. Neurosci. 23 3272–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Greenwald I. (1996). Membrane topology of the C. elegans SEL-12 presenilin. Neuron 17 1015–1021 10.1016/S0896-6273(00)80231-7 [DOI] [PubMed] [Google Scholar]
- Li X., Greenwald I. (1997). HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc. Natl. Acad. Sci. U.S.A. 94 12204–12209 10.1073/pnas.94.22.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. J. (2000). Interaction between beta-amyloid and lens alphaB-crystallin. FEBS Lett. 484 98–101 10.1016/S0014-5793(00)02136-0 [DOI] [PubMed] [Google Scholar]
- Lin C. H., Tomioka M., Pereira S., Sellings L., Iino Y., Van Der Kooy D. (2010). Insulin signaling plays a dual role in Caenorhabditis elegans memory acquisition and memory retrieval. J. Neurosci. 30 8001–8011 10.1523/JNEUROSCI.4636-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Dorman J. B., Rodan A., Kenyon C. (1997). daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 1319–1322 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- Lin K., Hsin H., Libina N., Kenyon C. (2001). Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28 139–145 10.1038/88850 [DOI] [PubMed] [Google Scholar]
- Lindau M., Almkvist O., Kushi J., Boone K., Johansson S. E., Wahlund L. O., et al. (2000). First symptoms–frontotemporal dementia versus Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 11 286–293 10.1159/000017251 [DOI] [PubMed] [Google Scholar]
- Lindwall G., Cole R. D. (1984). Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 259 5301–5305 [PubMed] [Google Scholar]
- Link C. D. (1995). Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 92 9368–9372 10.1073/pnas.92.20.9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. D. (2006). C. elegans models of age-associated neurodegenerative diseases: lessons from transgenic worm models of Alzheimer’s disease. Exp. Gerontol. 41 1007–1013 10.1016/j.exger.2006.06.059 [DOI] [PubMed] [Google Scholar]
- Link C. D., Johnson C. J., Fonte V., Paupard M., Hall D. H., Styren S., et al. (2001). Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol. Aging 22 217–226 10.1016/S0197-4580(00)00237-2 [DOI] [PubMed] [Google Scholar]
- Liu Z., Hamamichi S., Lee B. D., Yang D., Ray A., Caldwell G. A., et al. (2011). Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Hum. Mol. Genet. 20 3933–3942 10.1093/hmg/ddr312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke C. J., Fox S. A., Caldwell G. A., Caldwell K. A. (2008). Acetaminophen attenuates dopamine neuron degeneration in animal models of Parkinson’s disease. Neurosci. Lett. 439 129–133 10.1016/j.neulet.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Lorent K., Overbergh L., Moechars D., De Strooper B., Van Leuven F., Van Den Berghe H. (1995). Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the alpha-2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, alpha-2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience 65 1009–1025 10.1016/0306-4522(94)00555-J [DOI] [PubMed] [Google Scholar]
- Lu T., Aron L., Zullo J., Pan Y., Kim H., Chen Y., et al. (2014). REST and stress resistance in ageing and Alzheimer’s disease. Nature 507 448–454 10.1038/nature13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W. J., Wang H., Li H., Kim B. S., Shah S., Lee H. J., et al. (2003). PEN-2 and APH-1 coordinately regulate proteolytic processing of presenilin 1. J. Biol. Chem. 278 7850–7854 10.1074/jbc.C200648200 [DOI] [PubMed] [Google Scholar]
- Luse S. A., Smith K. R. (1964). The ultrastructure of senile plaques. Am. J. Pathol. 44 553–563 [PMC free article] [PubMed] [Google Scholar]
- Mahley R. W. (1988). Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240 622–630 10.1126/science.3283935 [DOI] [PubMed] [Google Scholar]
- Majounie E., Abramzon Y., Renton A. E., Keller M. F., Traynor B. J., Singleton A. B. (2012). Large C9orf72 repeat expansions are not a common cause of Parkinson’s disease. Neurobiol. Aging 33 e2521–e2522 10.1016/j.neurobiolaging.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek A. M., Barchowsky A., Bowser R., Youk A., Talbott E. O. (2012). Pesticide exposure as a risk factor for amyotrophic lateral sclerosis: a meta-analysis of epidemiological studies: pesticide exposure as a risk factor for ALS. Environ. Res. 117 112–119 10.1016/j.envres.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Mangahas P. M., Yu X., Miller K. G., Zhou Z. (2008). The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J. Cell Biol. 180 357–373 10.1083/jcb.200708130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., et al. (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87 493–506 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- Martin J. B., Gusella J. F. (1986). Huntington’s disease. Pathogenesis and management. N. Engl. J. Med. 315 1267–1276 10.1056/NEJM198611133152006 [DOI] [PubMed] [Google Scholar]
- Martindale D., Hackam A., Wieczorek A., Ellerby L., Wellington C., Mccutcheon K., et al. (1998). Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat. Genet. 18 150–154 10.1038/ng0298-150 [DOI] [PubMed] [Google Scholar]
- Marvanova M., Nichols C. D. (2007). Identification of neuroprotective compounds of Caenorhabditis elegans dopaminergic neurons against 6-OHDA. J. Mol. Neurosci. 31 127–137 [DOI] [PubMed] [Google Scholar]
- Masters C. L., Simms G., Weinman N. A., Multhaup G., Mcdonald B. L., Beyreuther K. (1985). Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. U.S.A. 82 4245–4249 10.1073/pnas.82.12.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A., Bernhardt R., Hugh-Jones T. (1981). High molecular weight microtubule-associated proteins are preferentially associated with dendritic microtubules in brain. Proc. Natl. Acad. Sci. U.S.A. 78 3010–3014 10.1073/pnas.78.5.3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G., Roberts B. R., Gunn A. P., Perez K. A., Tew D. J., Masters C. L., et al. (2009). The Caenorhabditis elegans A beta 1-42 model of Alzheimer disease predominantly expresses A beta 3-42. J. Biol. Chem. 284 22697–22702 10.1074/jbc.C109.028514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G., Roberts B. R., Pukala T. L., Kenche V. B., Roberts C. M., Link C. D., et al. (2012). Utility of an improved model of amyloid-beta (Aβ1-42) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol. Neurodegener. 7:57 10.1186/1750-1326-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J. B., Aamodt S., Aamodt E. (1996). ptl-1, a Caenorhabditis elegans gene whose products are homologous to the tau microtubule-associated proteins. Biochemistry 35 9415–9423 10.1021/bi952646n [DOI] [PubMed] [Google Scholar]
- McElwee J., Bubb K., Thomas J. H. (2003). Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2 111–121 10.1046/j.1474-9728.2003.00043.x [DOI] [PubMed] [Google Scholar]
- McElwee J. J., Schuster E., Blanc E., Thomas J. H., Gems D. (2004). Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279 44533–44543 10.1074/jbc.M406207200 [DOI] [PubMed] [Google Scholar]
- Miyasaka T., Ding Z., Gengyo-Ando K., Oue M., Yamaguchi H., Mitani S., et al. (2005). Progressive neurodegeneration in C. elegans model of tauopathy. Neurobiol. Dis. 20 372–383 10.1016/j.nbd.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Mizuta K., Saito A., Watanabe T., Nagura M., Arakawa M., Shimizu F., et al. (1999). Ultrastructural localization of megalin in the rat cochlear duct. Hear. Res. 129 83–91 10.1016/S0378-5955(98)00221-4 [DOI] [PubMed] [Google Scholar]
- Morales C. R., Igdoura S. A., Wosu U. A., Boman J., Argraves W. S. (1996). Low density lipoprotein receptor-related protein-2 expression in efferent duct and epididymal epithelia: evidence in rats for its in vivo role in endocytosis of apolipoprotein J/clusterin. Biol. Reprod. 55 676–683 10.1095/biolreprod55.3.676 [DOI] [PubMed] [Google Scholar]
- Morley J. F., Morimoto R. I. (2004). Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 15 657–664 10.1091/mbc.E03-07-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. Z., Tissenbaum H. A., Ruvkun G. (1996). A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382 536–539 10.1038/382536a0 [DOI] [PubMed] [Google Scholar]
- Murakami T., Yang S. P., Xie L., Kawano T., Fu D., Mukai A., et al. (2012). ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism. Hum. Mol. Genet. 21 1–9 10.1093/hmg/ddr417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., Mccarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., et al. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 277–283 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Murrell J., Farlow M., Ghetti B., Benson M. D. (1991). A mutation in the amyloid precursor protein associated with hereditary Alzheimer’s disease. Science 254 97–99 10.1126/science.1925564 [DOI] [PubMed] [Google Scholar]
- Muñoz-Lobato F., Rodríguez-Palero M. J., Naranjo-Galindo F. J., Shephard F., Gaffney C. J., Szewczyk N. J., et al. (2014). Protective role of DNJ-27/ERdj5 in Caenorhabditis elegans models of human neurodegenerative diseases. Antioxid. Redox. Signal. 20 217–235 10.1089/ars.2012.5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance M. A. (1997). Genetic testing of children at risk for Huntington’s disease. US Huntington Disease Genetic Testing Group. Neurology 49 1048–1053 10.1212/WNL.49.4.1048 [DOI] [PubMed] [Google Scholar]
- Nass R., Hahn M. K., Jessen T., Mcdonald P. W., Carvelli L., Blakely R. D. (2005). A genetic screen in Caenorhabditis elegans for dopamine neuron insensitivity to 6-hydroxydopamine identifies dopamine transporter mutants impacting transporter biosynthesis and trafficking. J. Neurochem. 94 774–785 10.1111/j.1471-4159.2005.03205.x [DOI] [PubMed] [Google Scholar]
- Nass R., Hall D. H., Miller D. M., Blakely R. D. (2002). Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 99 3264–3269 10.1073/pnas.042497999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314 130–133 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- Newman A. P., White J. G., Sternberg P. W. (1995). The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development 121 263–271 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Birn H., Moestrup S. K., Nielsen M., Verroust P., Christensen E. I. (1998). Characterization of a kidney proximal tubule cell line, LLC-PK1, expressing endocytotic active megalin. J. Am. Soc. Nephrol. 9 1767–1776 [DOI] [PubMed] [Google Scholar]
- Niwa R., Hada K. (2010). Identification of a spatio-temporal enhancer element for the Alzheimer’s amyloid precursor protein-like-1 gene in the nematode Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 74 2497–2500 10.1271/bbb.100450 [DOI] [PubMed] [Google Scholar]
- Niwa R., Zhou F., Li C., Slack F. J. (2008). The expression of the Alzheimer’s amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev. Biol. 315 418–425 10.1016/j.ydbio.2007.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., et al. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 994–999 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- Orenstein S. J., Kuo S. H., Tasset I., Arias E., Koga H., Fernandez-Carasa I., et al. (2013). Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 16 394–406 10.1038/nn.3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. T., Zoghbi H. Y. (2007). Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30 575–621 10.1146/annurev.neuro.29.051605.113042 [DOI] [PubMed] [Google Scholar]
- Pan C. L., Peng C. Y., Chen C. H., Mcintire S. (2011). Genetic analysis of age-dependent defects of the Caenorhabditis elegans touch receptor neurons. Proc. Natl. Acad. Sci. U.S.A. 108 9274–9279 10.1073/pnas.1011711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S., Ailion M., Toker A., Thomas J. H., Ruvkun G. (1999). A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13 1438–1452 10.1101/gad.13.11.1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S., Ruvkun G. (1998). Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12 2488–2498 10.1101/gad.12.16.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. A., Arango M., Abderrahmane S., Lambert E., Tourette C., Catoire H., et al. (2005). Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet. 37 349–350 10.1038/ng1534 [DOI] [PubMed] [Google Scholar]
- Parker J. A., Connolly J. B., Wellington C., Hayden M., Dausset J., Neri C. (2001). Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc. Natl. Acad. Sci. U.S.A. 98 13318–13323 10.1073/pnas.231476398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P., Brown R. H. (2006). Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 7 710–723 10.1038/nrn1971 [DOI] [PubMed] [Google Scholar]
- Pasquier F. (2005). Telling the difference between frontotemporal dementia and Alzheimer’s disease. Curr. Opin. Psychiatry 18 628–632 10.1097/01.yco.0000185988.05741.2a [DOI] [PubMed] [Google Scholar]
- Perez N., Sugar J., Charya S., Johnson G., Merril C., Bierer L., et al. (1991). Increased synthesis and accumulation of heat shock 70 proteins in Alzheimer’s disease. Mol. Brain Res. 11 249–254 10.1016/0169-328X(91)90033-T [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J. T., Faumont S., Gaston M. R., Pearson B. J., Lockery S. R. (2001). The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410 694–698 10.1038/35070575 [DOI] [PubMed] [Google Scholar]
- Pietrzik C. U., Yoon I. S., Jaeger S., Busse T., Weggen S., Koo E. H. (2004). FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J. Neurosci. 24 4259–4265 10.1523/JNEUROSCI.5451-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., et al. (1997). Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276 2045–2047 10.1126/science.276.5321.2045 [DOI] [PubMed] [Google Scholar]
- Poorkaj P., Bird T. D., Wijsman E., Nemens E., Garruto R. M., Anderson L., et al. (1998). Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol. 43 815–825 10.1002/ana.410430617 [DOI] [PubMed] [Google Scholar]
- Poorkaj P., Nutt J. G., James D., Gancher S., Bird T. D., Steinbart E., et al. (2004). parkin mutation analysis in clinic patients with early-onset Parkinson [corrected] disease. Am. J. Med. Genet. A 129A, 44–50 10.1002/ajmg.a.30157 [DOI] [PubMed] [Google Scholar]
- Rabinovici G. D., Miller B. L. (2010). Frontotemporal lobar degeneration: epidemiology, pathophysiology, diagnosis and management. CNS Drugs 24 375–398 10.2165/11533100-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A., Debetto P., Negro A., Guidolin D., Skaper S. D., Giusti P. (2004). Alpha-synuclein and Parkinson’s disease. FASEB J. 18 617–626 10.1096/fj.03-0338rev [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 901–906 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Renkawek K., Voorter C. E., Bosman G. J., Van Workum F. P., De Jong W. W. (1994). Expression of alpha B-crystallin in Alzheimer’s disease. Acta Neuropathol. 87 155–160 10.1007/BF00296185 [DOI] [PubMed] [Google Scholar]
- Renton A. E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J. R., et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72 257–268 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Swanson M. M., Albert P. S. (1981). Interacting genes in nematode dauer larva formation. Nature 290 668–671 10.1038/290668a0 [DOI] [PubMed] [Google Scholar]
- Ring S., Weyer S. W., Kilian S. B., Waldron E., Pietrzik C. U., Filippov M. A., et al. (2007). The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J. Neurosci. 27 7817–7826 10.1523/JNEUROSCI.1026-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaev E. I., Sherrington R., Rogaeva E. A., Levesque G., Ikeda M., Liang Y., et al. (1995). Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 376 775–778 10.1038/376775a0 [DOI] [PubMed] [Google Scholar]
- Ruan Q., Harrington A. J., Caldwell K. A., Caldwell G. A., Standaert D. G. (2010). VPS41, a protein involved in lysosomal trafficking, is protective in Caenorhabditis elegans and mammalian cellular models of Parkinson’s disease. Neurobiol. Dis. 37 330–338 10.1016/j.nbd.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T., Faraonio R., Minopoli G., De Candia P., De Renzis S., Zambrano N. (1998). Fe65 and the protein network centered around the cytosolic domain of the Alzheimer’s beta-amyloid precursor protein. FEBS Lett. 434 1–7 10.1016/S0014-5793(98)00941-7 [DOI] [PubMed] [Google Scholar]
- Sandbrink R., Masters C. L., Beyreuther K. (1994). Similar alternative splicing of a non-homologous domain in beta A4-amyloid protein precursor-like proteins. J. Biol. Chem. 269 14227–14234 [PubMed] [Google Scholar]
- Sastre M., Steiner H., Fuchs K., Capell A., Multhaup G., Condron M. M., et al. (2001). Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2 835–841 10.1093/embo-reports/kve180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage-Dunn C., Maduzia L. L., Zimmerman C. M., Roberts A. F., Cohen S., Tokarz R., et al. (2003). Genetic screen for small body size mutants in C. elegans reveals many TGFbeta pathway components. Genesis 35 239–247 10.1002/gene.10184 [DOI] [PubMed] [Google Scholar]
- Seidel H. S., Kimble J. (2011). The oogenic germline starvation response in C. elegans. PLoS ONE 6:e28074 10.1371/journal.pone.0028074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D. J., Abraham C. R., Podlisny M. B., Duffy L. K. (1986). Isolation of low-molecular-weight proteins from amyloid plaque fibers in Alzheimer’s disease. J. Neurochem. 46 1820–1834 10.1111/j.1471-4159.1986.tb08501.x [DOI] [PubMed] [Google Scholar]
- Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., et al. (2008). Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 14 837–842 10.1038/nm1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I. (2011). OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6:e20085 10.1371/journal.pone.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Bronson R. T., Chen D. F., Xia W., Selkoe D. J., Tonegawa S. (1997). Skeletal and CNS defects in Presenilin-1-deficient mice. Cell 89 629–639 10.1016/S0092-8674(00)80244-5 [DOI] [PubMed] [Google Scholar]
- Sherrington R., Rogaev E. I., Liang Y., Rogaeva E. A., Levesque G., Ikeda M., et al. (1995). Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375 754–760 10.1038/375754a0 [DOI] [PubMed] [Google Scholar]
- Shi Z., Lu Z., Zhao Y., Wang Y., Zhao-Wilson X., Guan P., et al. (2013). Neuroprotective effects of aqueous extracts of Uncaria tomentosa: insights from 6-OHDA induced cell damage and transgenic Caenorhabditis elegans model. Neurochem. Int. 62 940–947 10.1016/j.neuint.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., et al. (2000). Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25 302–305 10.1038/77060 [DOI] [PubMed] [Google Scholar]
- Shinohara H., Inaguma Y., Goto S., Inagaki T., Kato K. (1993). Alpha B crystallin and HSP28 are enhanced in the cerebral cortex of patients with Alzheimer’s disease. J. Neurol. Sci. 119 203–208 10.1016/0022-510X(93)90135-L [DOI] [PubMed] [Google Scholar]
- Siesling S., Vegter-Van Der Vlis M., Roos R. A. (1997). Juvenile Huntington disease in the Netherlands. Pediatr. Neurol. 17 37–43 10.1016/S0887-8994(97)00069-6 [DOI] [PubMed] [Google Scholar]
- Simón A. M., Schiapparelli L., Salazar-Colocho P., Cuadrado-Tejedor M., Escribano L., López De Maturana R., et al. (2009). Overexpression of wild-type human APP in mice causes cognitive deficits and pathological features unrelated to Abeta levels. Neurobiol. Dis. 33 369–378 10.1016/j.nbd.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Slunt H. H., Thinakaran G., Von Koch C., Lo A. C., Tanzi R. E., Sisodia S. S. (1994). Expression of a ubiquitous, cross-reactive homologue of the mouse beta-amyloid precursor protein (APP). J. Biol. Chem. 269 2637–2644 [PubMed] [Google Scholar]
- So S., Miyahara K., Ohshima Y. (2011). Control of body size in C. elegans dependent on food and insulin/IGF-1 signal. Genes Cells 16 639–651 10.1111/j.1365-2443.2011.01514.x [DOI] [PubMed] [Google Scholar]
- Spillantini M. G., Crowther R. A., Kamphorst W., Heutink P., Van Swieten J. C. (1998). Tau pathology in two Dutch families with mutations in the microtubule-binding region of tau. Am. J. Pathol. 153 1359–1363 10.1016/S0002-9440(10)65721-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher C. A., Grant F. J., Grimm G., O’hara P. J., Norris F., Norris K., et al. (1993). Molecular cloning of the cDNA for a human amyloid precursor protein homolog: evidence for a multigene family. Biochemistry 32 4481–4486 10.1021/bi00068a002 [DOI] [PubMed] [Google Scholar]
- Springer W., Hoppe T., Schmidt E., Baumeister R. (2005). A Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum. Mol. Genet. 14 3407–3423 10.1093/hmg/ddi371 [DOI] [PubMed] [Google Scholar]
- Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319 1668–1672 10.1126/science.1154584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen E., Terry B. M., Rivera E. J., Cannon J. L., Neely T. R., Tavares R., et al. (2005). Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease – is this type 3 diabetes? J. Alzheimers Dis. 7 63–80 [DOI] [PubMed] [Google Scholar]
- Strong T. V., Tagle D. A., Valdes J. M., Elmer L. W., Boehm K., Swaroop M., et al. (1993). Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nat. Genet. 5 259–265 10.1038/ng1193-259 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56 110–156 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sumakovic M., Hegermann J., Luo L., Husson S. J., Schwarze K., Olendrowitz C., et al. (2009). UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans. J. Cell Biol. 186 897–914 10.1083/jcb.200902096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank E. M., Rodgers K. E., Kenyon C. (2011). Spontaneous age-related neurite branching in Caenorhabditis elegans. J. Neurosci. 31 9279–9288 10.1523/JNEUROSCI.6606-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry R. D., Gonatas N. K., Weiss M. (1964). Ultrastructural studies in Alzheimer’s presenile dementia. Am. J. Pathol. 44 269–297 [PMC free article] [PubMed] [Google Scholar]
- The Association for Frontotemporal Degeneration. (2014). The Association for Frontotemporal Degeneration [Online]. Available at: http://www.theaftd.org (accessed March 18, 2014) [Google Scholar]
- The Huntington’s Disease Collaborative Research Group. (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72 971–983 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- Therrien M., Rouleau G. A., Dion P. A., Parker J. A. (2013). Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS ONE 8:e83450 10.1371/journal.pone.0083450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran G., Borchelt D. R., Lee M. K., Slunt H. H., Spitzer L., Kim G., et al. (1996). Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17 181–190 10.1016/S0896-6273(00)80291-3 [DOI] [PubMed] [Google Scholar]
- Thinakaran G., Kitt C. A., Roskams A. J., Slunt H. H., Masliah E., Von Koch C., et al. (1995). Distribution of an APP homolog, APLP2, in the mouse olfactory system: a potential role for APLP2 in axogenesis. J. Neurosci. 15 6314–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H. A., Guarente L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410 227–230 10.1038/35065638 [DOI] [PubMed] [Google Scholar]
- Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W. R., Iino Y. (2006). The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51 613–625 10.1016/j.neuron.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Toth M. L., Melentijevic I., Shah L., Bhatia A., Lu K., Talwar A., et al. (2012). Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J. Neurosci. 32 8778–8790 10.1523/JNEUROSCI.1494-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretiakoff K. (1919). Contribution a l’Etude de L’Anatomie pathologique du Locus Niger de Soemmering avec quelques déductions relatives à la pathogénie des troubles du tonus musculaire et De La Maladie de Parkinson. University of Paris: Paris [Google Scholar]
- Treusch S., Hamamichi S., Goodman J. L., Matlack K. E., Chung C. Y., Baru V., et al. (2011). Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science 334 1241–1245 10.1126/science.1213210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H., Han S. M., Yang Y., Tong C., Lin Y. Q., Mohan K., et al. (2008). The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell 133 963–977 10.1016/j.cell.2008.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulery P. G., Beers J., Mikhailenko I., Tanzi R. E., Rebeck G. W., Hyman B. T., et al. (2000). Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer’s disease. J. Biol. Chem. 275 7410–7415 10.1074/jbc.275.10.7410 [DOI] [PubMed] [Google Scholar]
- Vaccaro A., Patten S. A., Ciura S., Maios C., Therrien M., Drapeau P., et al. (2012a). Methylene blue protects against TDP-43 and FUS neuronal toxicity in C. elegans and D. rerio. PLoS ONE 7:e42117 10.1371/journal.pone.0042117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro A., Tauffenberger A., Ash P. E., Carlomagno Y., Petrucelli L., Parker J. A. (2012b). TDP-1/TDP-43 regulates stress signaling and age-dependent proteotoxicity in Caenorhabditis elegans. PLoS Genet. 8:e1002806 10.1371/journal.pgen.1002806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine J. S., Doucette P. A., Zittin Potter S. (2005). Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 74 563–593 10.1146/annurev.biochem.72.121801.161647 [DOI] [PubMed] [Google Scholar]
- Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., et al. (2008). TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7 409–416 10.1016/S1474-4422(08)70071-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J. G., Van Der Velde E. A., Roos R. A., Bruyn G. W. (1986). Juvenile Huntington disease. Hum. Genet. 73 235–239 10.1007/BF00401235 [DOI] [PubMed] [Google Scholar]
- van Ham T. J., Thijssen K. L., Breitling R., Hofstra R. M., Plasterk R. H., Nollen E. A. (2008). C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 4:e1000027 10.1371/journal.pgen.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen S., Pehkonen P., Lakso M., Nass R., Wong G. (2006). Identification of gene expression changes in transgenic C. elegans overexpressing human alpha-synuclein. Neurobiol. Dis. 22 477–486 10.1016/j.nbd.2005.12.021 [DOI] [PubMed] [Google Scholar]
- Vinceti M., Maraldi T., Bergomi M., Malagoli C. (2009). Risk of chronic low-dose selenium overexposure in humans: insights from epidemiology and biochemistry. Rev. Environ. Health 24 231–248 10.1515/REVEH.2009.24.3.231 [DOI] [PubMed] [Google Scholar]
- von Koch C. S., Zheng H., Chen H., Trumbauer M., Thinakaran G., Van Der Ploeg L. H., et al. (1997). Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiol. Aging 18 661–669 10.1016/S0197-4580(97)00151-6 [DOI] [PubMed] [Google Scholar]
- Vonsattel J. P., Myers R. H., Stevens T. J., Ferrante R. J., Bird E. D., Richardson E. P. (1985). Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 44 559–577 10.1097/00005072-198511000-00003 [DOI] [PubMed] [Google Scholar]
- Vowels J. J., Thomas J. H. (1992). Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130 105–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Farr G. W., Hall D. H., Li F., Furtak K., Dreier L., et al. (2009). An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 5:e1000350 10.1371/journal.pgen.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Yang G., Mosier D. R., Chang P., Zaidi T., Gong Y. D., et al. (2005). Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J. Neurosci. 25 1219–1225 10.1523/JNEUROSCI.4660-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasco W., Bupp K., Magendantz M., Gusella J. F., Tanzi R. E., Solomon F. (1992). Identification of a mouse brain cDNA that encodes a protein related to the Alzheimer disease-associated amyloid beta protein precursor. Proc. Natl. Acad. Sci. U.S.A. 89 10758–10762 10.1073/pnas.89.22.10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasco W., Gurubhagavatula S., Paradis M. D., Romano D. M., Sisodia S. S., Hyman B. T., et al. (1993). Isolation and characterization of APLP2 encoding a homologue of the Alzheimer’s associated amyloid beta protein precursor. Nat. Genet. 5 95–100 10.1038/ng0993-95 [DOI] [PubMed] [Google Scholar]
- Weidemann A., Eggert S., Reinhard F. B., Vogel M., Paliga K., Baier G., et al. (2002). A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry 41 2825–2835 10.1021/bi015794o [DOI] [PubMed] [Google Scholar]
- Wen J. Y., Kumar N., Morrison G., Rambaldini G., Runciman S., Rousseau J., et al. (1997). Mutations that prevent associative learning in C. elegans. Behav. Neurosci. 111 354–368 10.1037/0735-7044.111.2.354 [DOI] [PubMed] [Google Scholar]
- Westlund B., Parry D., Clover R., Basson M., Johnson C. D. (1999). Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signaling. Proc. Natl. Acad. Sci. U.S.A. 96 2497–2502 10.1073/pnas.96.5.2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer S. W., Klevanski M., Delekate A., Voikar V., Aydin D., Hick M., et al. (2011). APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J. 30 2266–2280 10.1038/emboj.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J. M., Guthrie C. R., Kraemer B. C. (2010). The role of MSUT-2 in tau neurotoxicity: a target for neuroprotection in tauopathy? Biochem. Soc. Trans. 38 973–976 10.1042/BST0380973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese M., Antebi A., Zheng H. (2010). Intracellular trafficking and synaptic function of APL-1 in Caenorhabditis elegans. PLoS ONE 5:e12790 10.1371/journal.pone.0012790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese M., Antebi A., Zheng H. (2012). Regulation of neuronal APL-1 expression by cholesterol starvation. PLoS ONE 7:e32038 10.1371/journal.pone.0032038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willnow T. E., Hilpert J., Armstrong S. A., Rohlmann A., Hammer R. E., Burns D. K., et al. (1996). Defective forebrain development in mice lacking gp330/megalin. Proc. Natl. Acad. Sci. U.S.A. 93 8460–8464 10.1073/pnas.93.16.8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willnow T. E., Sheng Z., Ishibashi S., Herz J. (1994). Inhibition of hepatic chylomicron remnant uptake by gene transfer of a receptor antagonist. Science 264 1471–1474 10.1126/science.7515194 [DOI] [PubMed] [Google Scholar]
- Witan H., Kern A., Koziollek-Drechsler I., Wade R., Behl C., Clement A. M. (2008). Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation. Hum. Mol. Genet. 17 1373–1385 10.1093/hmg/ddn025 [DOI] [PubMed] [Google Scholar]
- Wood-Kaczmar A., Gandhi S., Wood N. W. (2006). Understanding the molecular causes of Parkinson’s disease. Trends Mol. Med. 12 521–528 10.1016/j.molmed.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Wroe R., Wai-Ling Butler A., Andersen P. M., Powell J. F., Al-Chalabi A. (2008). ALSOD: the Amyotrophic Lateral Sclerosis Online Database. Amyotroph. Lateral Scler. 9 249–250 10.1080/17482960802146106 [DOI] [PubMed] [Google Scholar]
- Yao C., Johnson W. M., Gao Y., Wang W., Zhang J., Deak M., et al. (2013). Kinase inhibitors arrest neurodegeneration in cell and C. elegans models of LRRK2 toxicity. Hum. Mol. Genet. 22 328–344 10.1093/hmg/dds431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Greenwald I. (1993). A gene for a low density lipoprotein receptor-related protein in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 90 4572–4576 10.1073/pnas.90.10.4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M. (1998). A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Tuck S., Greenwald I., Han M. (1999). A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development 126 597–606 [DOI] [PubMed] [Google Scholar]
- Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., et al. (2008). TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol. 63 538–542 10.1002/ana.21392 [DOI] [PubMed] [Google Scholar]
- Yoo B. C., Kim S. H., Cairns N., Fountoulakis M., Lubec G. (2001). Deranged expression of molecular chaperones in brains of patients with Alzheimer’s disease. Biochem. Biophys. Res. Commun. 280 249–258 10.1006/bbrc.2000.4109 [DOI] [PubMed] [Google Scholar]
- Yoon I. S., Pietrzik C. U., Kang D. E., Koo E. H. (2005). Sequences from the low density lipoprotein receptor-related protein (LRP) cytoplasmic domain enhance amyloid beta protein production via the beta-secretase pathway without altering amyloid precursor protein/LRP nuclear signaling. J. Biol. Chem. 280 20140–20147 10.1074/jbc.M413729200 [DOI] [PubMed] [Google Scholar]
- Yu G., Nishimura M., Arawaka S., Levitan D., Zhang L., Tandon A., et al. (2000). Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature 407 48–54 10.1038/35024009 [DOI] [PubMed] [Google Scholar]
- Zambrano N., Bimonte M., Arbucci S., Gianni D., Russo T., Bazzicalupo P. (2002). feh-1 and apl-1, the Caenorhabditis elegans orthologues of mammalian Fe65 and beta-amyloid precursor protein genes, are involved in the same pathway that controls nematode pharyngeal pumping. J. Cell Sci. 115 1411–1422 [DOI] [PubMed] [Google Scholar]
- Zambrano N., Buxbaum J. D., Minopoli G., Fiore F., De Candia P., De Renzis S., et al. (1997). Interaction of the phosphotyrosine interaction/phosphotyrosine binding-related domains of Fe65 with wild-type and mutant Alzheimer’s beta-amyloid precursor proteins. J. Biol. Chem. 272 6399–6405 10.1074/jbc.272.10.6399 [DOI] [PubMed] [Google Scholar]
- Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., et al. (2004). The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55 164–173 10.1002/ana.10795 [DOI] [PubMed] [Google Scholar]
- Zhang Y. W., Luo W. J., Wang H., Lin P., Vetrivel K. S., Liao F., et al. (2005). Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J. Biol. Chem. 280 17020–17026 10.1074/jbc.M409467200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Marino’ M., Zhao J., Mccluskey R. T. (1998). Megalin (gp330): a putative endocytic receptor for thyroglobulin (Tg). Endocrinology 139 1462–1465 10.1210/endo.139.3.5978 [DOI] [PubMed] [Google Scholar]
- Zigman W. B. (2013). Atypical aging in Down syndrome. Dev. Disabil. Res. Rev. 18 51–67 10.1002/ddrr.1128 [DOI] [PubMed] [Google Scholar]
- Zlokovic B. V., Martel C. L., Matsubara E., Mccomb J. G., Zheng G., Mccluskey R. T., et al. (1996). Glycoprotein 330/megalin: probable role in receptor-mediated transport of apolipoprotein J alone and in a complex with Alzheimer disease amyloid beta at the blood–brain and blood–cerebrospinal fluid barriers. Proc. Natl. Acad. Sci. U.S.A. 93 4229–4234 10.1073/pnas.93.9.4229 [DOI] [PMC free article] [PubMed] [Google Scholar]