Abstract
A colorimetric yield reduction assay, ELVIRA (enzyme-linked virus inhibitor reporter assay) HSV, was developed to determine the antiviral drug susceptibilities of herpes simplex virus (HSV). It uses an HSV-inducible reporter cell line. This simple and rapid assay has an objective readout, low inoculum size, and good reproducibility. The results correlate well with those of the plaque reduction assay.
The prevalence of herpes simplex virus (HSV) infections caused by a drug-resistant virus in immunocompromised patients has been demonstrated to be significant (3.5 to 7.1%) (1-4). This underlines the clinical importance of HSV drug susceptibility determinations for this patient group. The “gold standard,” the plaque reduction assay (PRA), is laborious and time-consuming and has a subjective endpoint, and the results are often obtained too late to play a role in therapeutic decision making (5). There has been a considerable effort to develop less laborious and more rapid assays (8). One of the strategies was a modified PRA, which used a transgenic cell line expressing β-galactosidase upon infection with HSV and microscopic counting of blue plaques as a readout (9, 10).
We describe a rapid, quantitative colorimetric antiviral drug susceptibility assay, ELVIRA (enzyme-linked virus inhibitor reporter assay) HSV (Diagnostic Hybrids, Inc.). The assay is based on the HSV-inducible reporter cell line BHKICP6LacZ-5 (ELVIRA cells) stably transformed with the Escherichia coli lacZ gene under the control of the HSV type 1 (HSV-1) early promoter ICP6, which expresses β-galactosidase upon HSV infection (6). A yield reduction assay was set up in which virus is inoculated on human fibroblasts in the presence of antiviral drug. Subsequently, reporter ELVIRA cells, which represent an overlay readout cell line, are added. The β-galactosidase activity in the cell lysates reflects the number of infected reporter cells and, thereby, the yield of infectious virus after drug action.
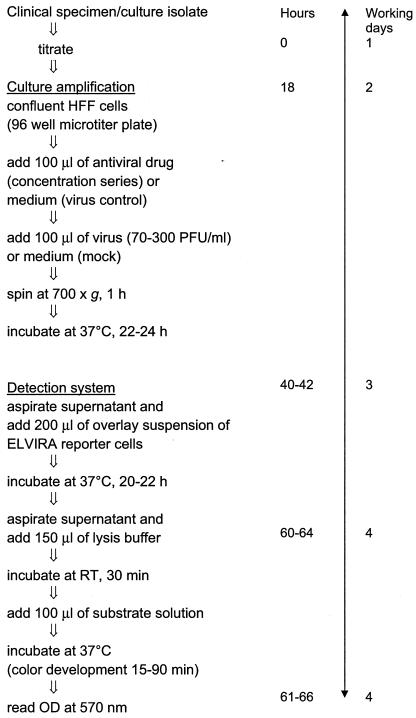
Confluent HFF cells were inoculated in triplicate with 0.1 ml of virus suspension and 0.1 ml of culture medium containing antiviral drugs (acyclovir [ACV] and foscarnet [PFA]) at different concentrations (7). After centrifugation (700 × g)-enhanced virus adsorption for 1 h and incubation overnight at 37°C, a suspension of reporter ELVIRA cells (Diagnostic Hybrids, Inc., Athens, Ohio) was prepared from frozen stocks (final concentration, 29,000 cells/ml). The culture supernatant was aspirated, and 0.2 ml of the ELVIRA cell suspension was added and was allowed to settle. After overnight incubation, the culture supernatant was aspirated, 0.15 ml of 0.03% sodium desoxycholate solution was added, and cell cultures were lysed for 30 min. The β-galactosidase activity in the lysates was determined spectrophotometrically (optical density at 570 nm) after incubation for 15 to 90 min at 37°C with 0.1 ml of substrate solution (chlorophenol red-β-d-galactopyranoside monosodium salt [3 mg/ml; Roche Diagnostics, Almere, The Netherlands] and 4.35 mM magnesium chloride in phosphate-buffered saline) (Fig. 1).
FIG. 1.
Schematic overview of ELVIRA HSV procedure. RT, room temperature; OD, optical density.
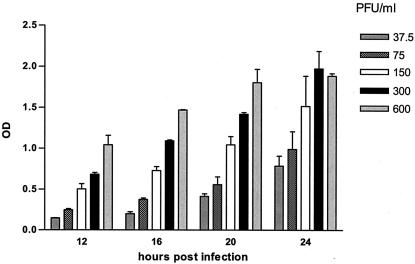
The optimal incubation period of the culture amplification step was determined with virus inocula ranging from 37.5 to 600 PFU/ml (multiplicities of infection [MOI], 0.00013 to 0.00200 PFU/cell) and by addition of ELVIRA cells after 12, 16, 20, and 24 h. An increased incubation period postinfection resulted in an increased sensitivity of the assay (Fig. 2). Similar data were obtained for HSV-2 (data not shown). An incubation period of 22 to 24 h was chosen for further use. The incubation period of the detection system was based on previously determined β-galactosidase expression kinetics (6, 9). The ELVIRA HSV had a linear range from approximately 70 to 300 PFU/ml (∼MOI, 0.0002 to 0.0010 PFU/cell).
FIG. 2.
The effect of duration of infection of HFF cells on sensitivity of ELVIRA HSV. HFF cell monolayers were infected with a range of HSV-1 inocula; and ELVIRA reporter cells were added after 12, 16, 20, and 24 h. Results are representative of four independent experiments performed in triplicate with HSV-1; error bars indicate SDs. OD, optical density.
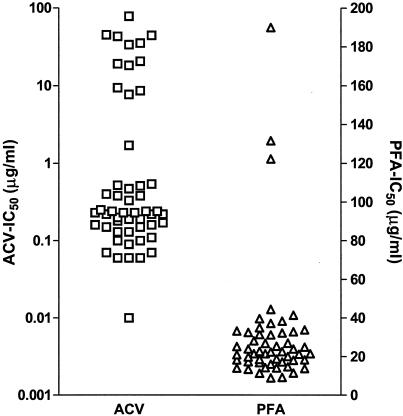
The utility of the assay for susceptibility testing was evaluated (in parallel with PRA) with well-characterized ACV- and PFA-sensitive and -resistant HSV strains (n = 6) and clinical isolates (n = 16) with various susceptibilities to ACV and PFA (Table 1; Fig. 3). The antiviral effects of ACV and PFA measured by ELVIRA HSV corresponded to the results of PRA. The data in Table 1 clearly demonstrate the ability of ELVIRA HSV to discriminate between HSV strains sensitive and resistant to both drugs examined. The ACV 50% inhibitory concentrations (IC50s) obtained by ELVIRA HSV showed a good correlation with the results of PRA (rACV = 0.94). The IC50s for ACV-sensitive strains obtained by ELVIRA HSV were lower than those obtained by PRA (P = 0.002). For ACV-resistant strains, ELVIRA HSV showed higher mean IC50s than PRA (P = 0.0005). The PFA IC50s obtained by ELVIRA HSV did not differ from those obtained by PRA for sensitive (P = 0.21) and resistant strains (P = 0.75); however, the number of strains tested was small.
TABLE 1.
Results of ACV and PFA susceptibility testing by ELVIRA HSV and PRA with well-characterized reference strains and clinical isolates
| Virus straina | HSV type | IC50 (μg/ml)b
|
|||
|---|---|---|---|---|---|
| ACV
|
PFA
|
||||
| ELVIRA HSV | PRA | ELVIRA HSV | PRA | ||
| Reference strains | |||||
| ACVs | |||||
| KOS | 1 | 0.52 ± 0.17 | 1.26 ± 0.37 | 33.2 ± 8.9 | 50.3 ± 1.8 |
| McIntyre | 1 | 0.33 ± 0.09 | 1.91 ± 0.56 | 32.9 ± 6.8 | NDc |
| R39 | 1 | 0.38 ± 0.12 | 0.46 ± 0.04 | 41.5 ± 5.3 | ND |
| MS | 2 | 0.51 ± 0.16 | 1.63 ± 0.63 | 21.5 ± 8.3 | 50.6 ± 7.2 |
| ACVr, PFAr | |||||
| AraAr8 | 1 | 1.70 ± 0.51 | 4.50 ± 1.65 | 122.3 ± 15.5 | 133.0 ± 49.8 |
| 97.1218 | 2 | 78.10 ± 12.28 | 70.50 ± 15.3 | 190.0 ± 10.1 | 189.4 ± 20.3 |
| Clinical isolates | |||||
| ACVs, PFAs | |||||
| 97.11896 | 1 | 0.19 ± 0.05 | 0.24 ± 0.19 | 22.6 ± 5.6 | 14.5 ± 2.1 |
| 97.09227 | 1 | 0.24 ± 0.01 | 0.87 ± 0.19 | 26.9 ± 5.6 | 14.9 ± 3.6 |
| 97.06348 | 1 | 0.22 ± 0.01 | 1.05 ± 0.28 | 21.5 ± 1.9 | 36.8 ± 4.1 |
| 97.07631 | 1 | 0.07 ± 0.02 | 0.76 ± 0.49 | 14.3 ± 2.4 | 27.3 ± 2.3 |
| 01.14606 | 1 | 0.23 ± 0.01 | 0.37 ± 0.14 | 24.0 ± 5.5 | 24.4 ± 3.1 |
| ACVr, PFAs | |||||
| 96.05940 | 1 | 44.18 ± 6.96 | 10.74 ± 6.89 | 24.9 ± 8.9 | 30.5 ± 3.3 |
| 96.07922 | 1 | 43.14 ± 6.76 | 9.93 ± 4.17 | 31.3 ± 3.3 | 28.3 ± 4.5 |
| 97.10788 | 1 | 35.18 ± 3.56 | 14.15 ± 2.26 | 33.9 ± 6.2 | 32.7 ± 5.3 |
| 97.07632 | 1 | 8.60 ± 2.35 | 4.21 ± 1.88 | 18.4 ± 0.2 | 27.6 ± 1.6 |
| 01.22388 | 1 | 9.38 ± 2.96 | 6.23 ± 2.30 | 14.7 ± 3.6 | 31.5 ± 7.0 |
| 01.22733 | 1 | 18.21 ± 3.10 | 2.31 ± 1.10 | 38.4 ± 10.6 | 20.4 ± 4.8 |
| 98.14742-PE/1 | 1 | 19.12 ± 0.96 | 8.65 ± 1.37 | ND | ND |
| 99.16237 | 1 | 45.15 ± 13.0 | 11.52 ± 0.86 | ND | ND |
| 99.17213 | 1 | 20.61 ± 6.73 | 5.73 ± 1.76 | 9.2 ± 3.1 | 10.3 ± 2.8 |
| 01.28565 | 2 | 33.56 ± 3.00 | 14.97 ± 3.10 | 11.5 ± 1.4 | 16.4 ± 2.3 |
| ACVr, PFAr 01.24080 | 1 | 7.68 ± 2.31 | 4.31 ± 1.60 | 131.7 ± 4.1 | 111.6 ± 7.8 |
s and r, sensitive and resistant, respectively.
The IC50s are expressed as means ± SDs from at least two independent experiments.
ND, not determined.
FIG. 3.
Susceptibilities to ACV and PFA of well-characterized HSV strains and clinical isolates and clinical isolates from untreated patients determined by ELVIRA HSV (the results represent data combined from Table 1 and the Results).
The interassay variability of the IC50s, expressed as a coefficient of variation (CV), ranged from 4 to 33% (Table 1), with similar mean CVs for sensitive and resistant strains (21.2 and 20.6%, respectively).
The natural variation in the susceptibilities of HSV strains to ACV and PFA was tested with HSV-1 clinical isolates from untreated patients. Isolates from 18 patients were tested for their susceptibilities to both drugs, and additional isolates from 12 and 15 patients were tested for their susceptibilities to ACV and PFA, respectively. The median ACV IC50 was 0.17 μg/ml (range, 0.01 to 0.57 μg/ml; standard deviation [SD], 0.12 μg/ml), and the median PFA IC50 was 20.98 μg/ml (range, 9 to 47.1 μg/ml; SD, 8.5 μg/ml). On the basis of the susceptibility data for the clinical isolates from untreated patients, the well-characterized strains, and clinical isolates (Table 1) and on the basis of the reproducibility of ELVIRA HSV, preliminary ACV and PFA IC50 cutoffs were set at 0.8 and 60 μg/ml, respectively.
ELVIRA HSV demonstrated a 100% correlation with PRA for the discrimination between ACV- and PFA-sensitive and -resistant HSV of both serotypes. The reproducibility of the assay was acceptable, with interassay CVs ranging from 4 to 43%. The early expression of β-galactosidase in reporter ELVIRA cells results in rapid (pre-cytopathic effect [CPE]) virus detection (6). The whole assay is thus more rapid than CPE- or DNA-based assays, which require more replication cycles. In a diagnostic laboratory setting, the ELVIRA HSV susceptibility result can be available within 5 days from the time of specimen arrival, which also includes the time needed for determination of the virus titer in the specimen.
In conclusion, we describe the development and evaluation of a novel colorimetric, transgenic cell-based assay, ELVIRA HSV, that can be used for rapid determination of HSV drug susceptibility and also for evaluation of new antiviral compounds. The assay can be used to determine the susceptibilities of HSV-1 and HSV-2 to antiviral drugs even in specimens with low virus titers. The short turn-around time, easy-to-use format, objective endpoint, and good reproducibility make ELVIRA HSV amenable for use in the routine diagnostic virology laboratory. The testing of larger number of sensitive and resistant clinical isolates is warranted for further validation of the assay.
Acknowledgments
We thank M. Aymard (Universite Claude Bernard, Lyon, France), D. M. Coen (Harvard Medical School, Boston, Mass.), and A. Linde (Swedish Institute for Infectious Disease Control, Solna, Sweden) for providing HSV control strains and clinical isolates.
REFERENCES
- 1.Chen, Y., C. Scieux, V. Garrait, G. Socie, V. Rocha, J. M. Molina, D. Thouvenot, F. Morfin, L. Hocqueloux, L. Garderet, H. Esperou, F. Selimi, A. Devergie, G. Leleu, M. Aymard, F. Morinet, E. Gluckman, and P. Ribaud. 2001. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clin. Infect. Dis. 31:927-935. [DOI] [PubMed] [Google Scholar]
- 2.Christophers, J., J. Clayton, J. Craske, R. Ward, P. Collins, M. Trowbridge, and G. Darby. 1998. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob. Agents Chemother. 42:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danve-Szatanek, C., M. Aymard, D. Thouvenot, F. Morfin, G. Agius, I. Bertin, S. Billaudel, B. Chanzy, M. Coste-Burel, L. Finkielsztejn, H. Fleury, T. Hadou, C. Henquell, H. Lafeuille, M. E. Lafon, A. Le Faou, M. C. Legrand, L. Maille, C. Mengelle, P. Morand, F. Morinet, E. Nicand, S. Omar, B. Picard, B. Pozzetto, J. Puel, D. Raoult, C. Scieux, M. Segondy, J. M. Seigneurin, R. Teyssou, and C. Zandotti. 2004. Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J. Clin. Microbiol. 42:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morfin, F., and D. Thouvenot. 2003. Herpes simplex virus resistance to antiviral drugs. J. Clin. Virol. 26:29-37. [DOI] [PubMed] [Google Scholar]
- 5.Safrin, S., T. Elbeik, L. Phan, D. Robinson, J. Rush, A. Elbaggari, and J. Mills. 1994. Correlation between response to acyclovir and foscarnet therapy and in vitro susceptibility result for isolates of herpes simplex virus from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 38:1246-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stabell, E. C., and P. D. Olivo. 1992. Isolation of a cell line for rapid and sensitive histochemical assay for the detection of herpes simplex virus. J. Virol. Methods 38:195-204. [DOI] [PubMed] [Google Scholar]
- 7.Stranska, R., A. M. van Loon, R. G. Bredius, M. Polman, E. Nienhus, M. F. C. Beersma, A. C. Lankester, and R. Schuurman. 2004. Sequential switching of DNA polymerase and thymidine kinase mediated HSV-1 drug resistance in an immunocompromised child. Antivir. Ther. 9:99-104. [PubMed] [Google Scholar]
- 8.Swierkosz, E. M. 2000. Antiviral susceptibility testing, p. 154-168. In S. Specter, R. L. Hodinka, and S. A. Young (ed.), Antiviral drug susceptibility testing. ASM Press, Washington, D.C.
- 9.Tebas, P., D. Scholl, J. Jollick, K. McHarg, M. Arens, and P. D. Olivo. 1998. A rapid assay to screen for drug-resistant herpes simplex virus. J. Infect. Dis. 177:217-220. [DOI] [PubMed] [Google Scholar]
- 10.Tebas, P., E. C. Stabell, and P. D. Olivo. 1995. Antiviral susceptibility testing with a cell line which expresses β-galactosidase after infection with herpes simplex virus. Antimicrob. Agents Chemother. 39:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]