Abstract
Understanding the mechanisms driving the extraordinary diversification of parasites is a major challenge in evolutionary biology. Co-speciation, one proposed mechanism that could contribute to this diversity is hypothesized to result from allopatric co-divergence of host–parasite populations. We found that island populations of the Galápagos hawk (Buteo galapagoensis) and a parasitic feather louse species (Degeeriella regalis) exhibit patterns of co-divergence across variable temporal and spatial scales. Hawks and lice showed nearly identical population genetic structure across the Galápagos Islands. Hawk population genetic structure is explained by isolation by distance among islands. Louse population structure is best explained by hawk population structure, rather than isolation by distance per se, suggesting that lice tightly track the recent population histories of their hosts. Among hawk individuals, louse populations were also highly structured, suggesting that hosts serve as islands for parasites from an evolutionary perspective. Altogether, we found that host and parasite populations may have responded in the same manner to geographical isolation across spatial scales. Allopatric co-divergence is likely one important mechanism driving the diversification of parasites.
Keywords: population genetic structure, co-speciation, coevolution, co-divergence
1. Introduction
Parasitism is among the most successful life-history strategies [1], but the evolutionary processes responsible for parasite diversification remain the subject of debate [2]. Co-speciation (a process in which hosts and parasites undergo synchronous speciation) is thought to be an important mechanism driving parasite diversification [3,4]. This process is inferred when independent phylogenetic trees of host and parasite species exhibit mirror-image branching patterns (Farhenholz's rule) [4–6] and is hypothesized to result from allopatric co-divergence of pairs of host–parasite populations across the landscape [7].
Evolutionarily congruent histories of hosts and parasites can occur under a variety of conditions, but are most likely when parasites show a high degree of host specificity, vertical transmission, short generation times and small effective population sizes [6,8,9]. Given the high prevalence of these traits, congruent evolutionary histories are common between birds and their chewing lice [10,11]. Thus, bird–louse interactions are attractive systems to test for patterns of allopatric co-divergence. Because of their isolation and simplified faunas, oceanic archipelagoes are useful natural evolutionary laboratories in which to examine allopatric co-divergence of host–parasite pairs.
Here, we studied patterns of allopatric co-divergence among geographical islands and among individual hosts within islands in the Galápagos. The Galápagos hawk (Buteo galapagoensis) lineage diverged from within the Swainson's hawk (B. swainsoni) lineage ca 125 000 years before present and island populations exhibit strong isolation by distance across the archipelago [12,13]. The feather louse Degeeriella regalis (Ischnocera: Philopteridae), an obligate, host-specialist ectoparasite, is found on the Galápagos and Swainson's hawks and likely accompanied the ancestor of the Galápagos hawk during the initial founding event [14,15]. A preliminary analysis suggests that the colonization patterns of hawks and D. regalis among islands are tightly linked [16,17], providing a likely scenario under which co-divergence could occur. Here, we used hawk and louse microsatellite markers to investigate patterns of allopatric co-divergence across multiple scales; first, at the level of geographical island and then at the level of individual hosts.
2. Material and methods
Blood was sampled from Galápagos hawks trapped on the eight major islands within the archipelago on which they nest. To determine population genetic structure of hawks at the level of geographical island, 193 individuals were sampled from all eight island-populations and genotyped at 20 variable microsatellite loci [18,19]. Lice were sampled from a subset of these hawks by dust-ruffling. For comparisons at the level of geographical island, a single louse from each hawk on eight islands (158 lice in total) was genotyped at six polymorphic microsatellite markers [20]. Within the louse dataset, multiple loci were found to have an excess of homozygotes (Wahlund effect) across island populations. To avoid pseudo-replication from using genotypes of multiple lice per hawk, a single louse genotype was haphazardly chosen from each hawk on an island for use in the island-level analysis. Because geographical structure among louse populations found on individual birds was so strong, a single louse representative is likely to serve as a good proxy for population structure comparisons across geographical islands. For comparisons at the level of individual hosts, louse infrapopulations (population of lice on a single host individual) were collected from 19 hawks on two islands (Fernandina and Santiago), which served as source material for replicate tests of the hypothesis that louse infrapopulation structure among hosts is also high. The small size of individual lice (less than 3 mm in length) restricted the amount of extractable DNA and subsequently limited genotyping to six loci. Variation across all microsatellite loci for each species was tested for departures from Hardy–Weinberg equilibrium and linkage disequilibrium using Arlequin v. 3.5 [21]. Mantel tests, partial-Mantel tests [22] and analysis of molecular variances (AMOVAs) to estimate pairwise F-statistics were also performed using Arlequin v. 3.5. The most likely number of genetically distinct population clusters of hawks and lice was estimated using Structure v. 2.3.4 [23]. See the electronic supplementary material for additional information.
3. Results
A hierarchical AMOVA and pairwise permutation tests showed that Galápagos hawk populations are highly genetically differentiated among islands (overall FST = 0.353, p < 0.0001; figure 1). Even after correcting for false-discovery rate (FDR) [24], all pairwise FST estimates among islands differed significantly from 0. We found strong support for isolation by distance among hawk populations (Mantel test, r = 0.59, p = 0.003; figure 2), even after correcting for island size. The optimal estimated number of genetic clusters was K = 8, which was determined by the point at which the posterior probabilities plateau for independent runs of K from 6 to 10 (figure 1 also see the electronic supplementary material). Further analysis of the genetic clusters revealed a clear correspondence with the geographical islands on which populations were sampled.
Figure 1.
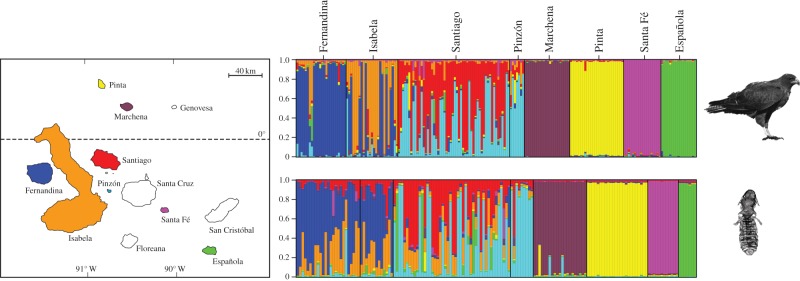
Map and population genetic structure plots for Galápagos hawks (Buteo galapagoensis) and their feather lice (Degeeriella regalis). Map of the Galápagos Islands shows sampled islands in colour. Breeding populations of hawks have been extirpated from Santa Cruz, San Cristobal and Floreana. There is no evidence that hawks have ever inhabited Genovesa. To the right of the map are results of Structure analyses estimating the most probable number of genetic clusters of hawks (top) and lice (bottom; K = 8). Each individual is represented by a thin vertical line, and island populations are separated by thicker black lines. Analysis of genetic clusters for both hawks and lice revealed groupings that correspond to the eight geographical islands sampled. Thus, colours of the genetic clusters correspond to colours of the islands on the map.
Figure 2.
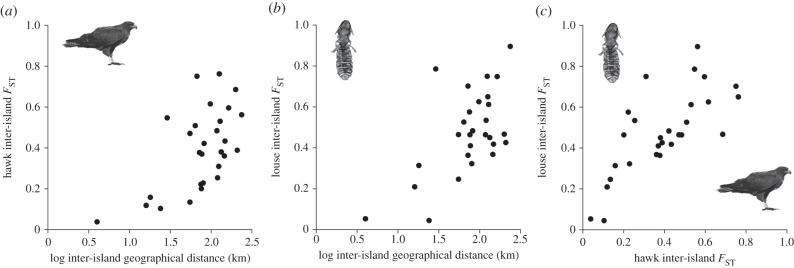
Tests of isolation by distance for (a) hawks and (b) lice. (c) Relationship between hawk and lice inter-island FST. Results of a partial-Mantel test indicate that hawk interisland FST predicted louse interisland FST better than geographical distance between islands.
Similarly, D. regalis showed an extraordinarily high degree of population genetic structure among geographical islands. AMOVA and pairwise permutation tests showed that populations of lice on each island were highly differentiated (overall FST = 0.429, p < 0.0001). Even after correcting for FDR, all of the pairwise population FST comparisons differed from 0 except for Santiago and Pinzón (p = 0.06). The optimal estimated number of genetic clusters was nearly identical for K = 7 and K = 8 (figure 1), approximately the same number of islands sampled and the same number of genetic clusters found for the hawks. We also found evidence of geographical isolation by distance for louse populations among islands (Mantel, r = 0.60, p = 0.004; figure 2). However, hawk population genetic structure (pairwise FST estimates) explained louse population genetic structure (partial-Mantel, r = 0.59, p = 0.03; figure 2) better than geographical distance (partial-Mantel, r = 0.31, p = 0.09).
Louse infrapopulations were genetically differentiated in 93% of pairwise FST comparisons among host individuals on Fernandina after correcting for FDR (FST = 0.183, p < 0.0001; see the electronic supplementary material). On Santiago, 67% of pairwise infrapopulation FST comparisons among host individuals differed after correcting for FDR (FST = 0.145, p < 0.0001). The difference in the total number of significant pairwise infrapopulation FST comparisons between Fernandina and Santiago did not correlate with the number of lice sampled within an infrapopulation or with differences in the number of male hawks in a social group. Observed genetic differences between louse infrapopulations did not reflect isolation caused by distance between hosts on Fernandina (r = 0.14, p = 0.33) or Santiago (r = 0.24, p = 0.06).
4. Discussion
Given the intimate ecological association between Galápagos hawks and D. regalis, we expected to find similar degrees of inter-island population genetic structure for both species. Our results support the hypothesis that water acts as a major dispersal barrier for Galápagos hawks, severely restricting gene flow among islands [12,25]. Host dispersal is limited by geographical distance among islands [25] and lice rarely move independently of their hosts [26]. Thus, we expected to find that D. regalis would show similar patterns of population divergence between islands as observed in hawks (co-divergence). Our data may indicate that lice track hawk movements and histories across the Galápagos Islands. One reasonable biogeographic scenario is that as hawk populations became isolated, so did the louse populations living on those hawks, a process consistent with a model of allopatric co-divergence. However, other explanations cannot be ruled out by our analysis. For example, these same patterns of apparent co-divergence could arise if gene flow among islands was rare for both hawks and lice. Thus, the possibility exists that colonization of the islands by the lice was uncoupled from hawk colonization—perhaps via other bird species. To date, though, this louse species has been documented exclusively from Galapagos hawks within the archipelago.
Similar to the isolation and diversification of populations following establishment on an island, louse infrapopulations may undergo additional isolation events when establishing on new host individuals. Epidemiological evidence indicates that D. regalis is transmitted primarily from parent to offspring host [26]. The number of lice transmitted among hosts is expected to be small. Thus, founder events resulting in strong genetic drift within louse infrapopulations on single birds may contribute to the rapid divergence of louse infrapopulations among hosts. As such, we expected that louse infrapopulations would be further structured at the level of individual hosts within a geographical island. Our results suggest that from the perspective of louse infrapopulations, individual birds serve as islands, limiting louse gene flow between bird individuals in a similar manner as that observed among geographical island populations. Such processes of rapid divergence may explain, at least in part, the patterns consistent with allopatric co-divergence we observed for hawks and lice at the level of geographical islands.
Oceanic archipelagoes continue to serve as irreplaceable natural evolutionary laboratories. Remarkably, an ecological interaction at the very top of the relatively simple terrestrial trophic cascade in the Galápagos Islands has provided a context in which to illuminate the potential for co-divergence as one mechanism that contributes to the extraordinary diversification of parasites.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Jennifer Bollmer for leading field collections of samples, and Jessica Rabenold and Cindee Rettke for help in the laboratory.
All procedures were approved by the University of Missouri-St Louis Institutional Animal Care and Use Committee and by the Galápagos National Park.
Funding statement
The study was facilitated by grants from the US National Science Foundation (INT-030759 to N.K.W. and PGP; DEB 1256758 to N.K.W.), the Saint Louis Zoo's Field Research for Conservation (to N.K.W. and P.G.P.), the University of Missouri Research Board (to N.K.W. and P.G.P.), The John Templeton Foundation (grant ID no. 41855 to N.K.W.), The University of Arizona (Faculty Seed Grant to N.K.W.) and National Institutes of Health-PERT postdoctoral fellowship (K12-GM000708 J.A.H.K.).
References
- 1.Price P. 1980. Evolutionary biology of parasites. Princeton, NJ: Princeton University Press. [Google Scholar]
- 2.Poulin R, Morand S. 2000. The diversity of parasites. Q. Rev. Biol. 75, 277–293. ( 10.1086/393500) [DOI] [PubMed] [Google Scholar]
- 3.Hafner MS, Nadler SA. 1988. Phylogenetic trees support the coevolution of parasites and their hosts. Nature 332, 258–259. ( 10.1038/332258a0) [DOI] [PubMed] [Google Scholar]
- 4.Eichler W. 1948. Some rules in ectoparasitism. Annu. Mag. Nat. Hist. 12, 588–598. ( 10.1080/00222934808653932) [DOI] [Google Scholar]
- 5.Page RDM. 2003. Tangled trees: phyogeny, cospeciation, and coevolution. Chicago, IL: University of Chicago Press. [Google Scholar]
- 6.Nadler SA, Hafner MS, Hafner JC, Hafner DJ. 1990. Genetic differentiation among chewing louse populations (Mallophaga, Trichodectidae) in a pocket gopher contact zone (Rodentia, Geomyidae). Evolution 44, 942–951. ( 10.2307/2409557) [DOI] [PubMed] [Google Scholar]
- 7.Rannala B, Michalakis Y. 2002. Population genetics and co-speciation. In Tangled trees: phyogeny, cospeciation, and coevolution (ed. Page RDM.), pp. 120–143. Chicago, IL: University of Chicago Press. [Google Scholar]
- 8.Huyse T, Poulin R, Theron A. 2005. Speciation in parasites: a population genetics approach. Trends Parasitol. 21, 469–475. ( 10.1016/j.pt.2005.08.009) [DOI] [PubMed] [Google Scholar]
- 9.Nieberding CM, Olivieri I. 2007. Parasites: proxies for host genealogy and ecology? Trends Ecol. Evol. 22, 156–165. ( 10.1016/j.tree.2006.11.012) [DOI] [PubMed] [Google Scholar]
- 10.Hughes J, Kennedy M, Johnson KP, Palma RL, Page RDM. 2007. Multiple cophylogenetic analyses reveal frequent cospeciation between pelecaniform birds and Pectinopygus lice. Syst. Biol. 56, 232–251. ( 10.1080/10635150701311370) [DOI] [PubMed] [Google Scholar]
- 11.Stefka J, Hoeck PEA, Keller LF, Smith VS. 2011. A hitchhikers guide to the Galápagos: co-phylogeography of Galápagos mockingbirds and their parasites. BMC Evol. Biol. 11, 284 ( 10.1186/1471-2148-11-284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollmer JL, Kimball RT, Whiteman NK, Sarasola JH, Parker PG. 2006. Phylogeography of the Galápagos hawk (Buteo galapagoensis): a recent arrival to the Galápagos islands. Mol. Phylogenet. Evol. 39, 237–247. ( 10.1016/j.ympev.2005.11.014) [DOI] [PubMed] [Google Scholar]
- 13.Hull JM, Savage WK, Bollmer JL, Kimball RT, Parker PG, Whiteman NK, Ernest HB. 2008. On the origin of the Galápagos hawk: an examination of phenotypic differentiation and mitochondrial paraphyly. Biol. J. Linn. Soc. 95, 779–789. ( 10.1111/j.1095-8312.2008.01082.x) [DOI] [Google Scholar]
- 14.Clay T. 1958. Revisions of Mallophaga genera. Degeeriellla from the Falconiformes. Bull. Brit. Mus. Nat. Hist. Entomol. 7, 121–207. [Google Scholar]
- 15.Price RD, Hellenthal RA, Palma RL. 2003. World checklist of chewing lice with host associations and keys to families and genera. Champaign, IL: Illinois Natural History Survey Special Publication. [Google Scholar]
- 16.Whiteman NK, Kimball RT, Parker PG. 2007. Co-phylogeography and comparative population genetics of the threatened Galápagos hawk and three ectoparasite species: ecology shapes population histories within parasite communities. Mol. Ecol. 16, 4759–4773. ( 10.1111/j.1365-294X.2007.03512.x) [DOI] [PubMed] [Google Scholar]
- 17.Parker PG, Whiteman NK. 2012. Evolution of pathogens and parasites on the Galapagos islands. In The role of science for conservation (eds Wolff M, Gardener M.), pp. 35–51. New York, NY: Routledge. [Google Scholar]
- 18.Bollmer JL, Hull JM, Ernest HB, Sarasola JH, Parker PG. 2011. Reduced MHC and neutral variation in the Galápagos hawk, an island endemic. BMC Evol. Biol. 11, 143 ( 10.1186/1471-2148-11-143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hull JM, Tufts D, Topinka JR, May B, Ernest HB. 2007. Development of 19 microsatellite loci for Swainson's hawks (Buteo swainsoni) and other buteos. Mol. Ecol. Notes 7, 346–349. ( 10.1111/j.1471-8286.2006.01604.x) [DOI] [Google Scholar]
- 20.Peters MB, Hagen C, Whiteman NK, Parker PG, Glenn TC. 2009. Characterization of 10 microsatellite loci in an avian louse, Degeeriella regalis (Phthiraptera: Ischnocera: Philopteridae). Mol. Ecol. Resour. 9, 882–884. ( 10.1111/j.1755-0998.2008.02363.x) [DOI] [PubMed] [Google Scholar]
- 21.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 22.Legendre P, Fortin MJ. 2010. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol. Ecol. Resour. 10, 831–844. ( 10.1111/j.1755-0998.2010.02866.x) [DOI] [PubMed] [Google Scholar]
- 23.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B (Stat. Methodol.) 57, 289–300. [Google Scholar]
- 25.Bollmer JL, Whiteman NK, Cannon MD, Bednarz JC, De Vries T, Parker PG. 2005. Population genetics of the Galápagos Hawk (Buteo galapagoensis): genetic monomorphism within isolated populations. Auk 122, 1210–1224. ( 10.1642/0004-8038(2005)122[1210:PGOTGH]2.0.CO;2) [DOI] [Google Scholar]
- 26.Whiteman NK, Parker PG. 2004. Effects of host sociality on ectoparasite population biology. J. Parasitol. 90, 939–947. ( 10.1645/GE-310R) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


