Abstract
The activity of tigecycline, 9-(t-butylglycylamido)-minocycline, against Escherichia coli KAM3 (acrB) strains harboring plasmids encoding various tetracycline-specific efflux transporter genes, tet(B), tet(C), and tet(K), and multidrug transporter genes, acrAB, acrEF, and bcr, was examined. Tigecycline showed potent activity against all three Tet-expressing, tetracycline-resistant strains, with the MICs for the strains being equal to that for the host strain. In the Tet(B)-containing vesicle study, tigecycline did not significantly inhibit tetracycline efflux-coupled proton translocation and at 10 μM did not cause proton translocation. This suggests that tigecycline is not recognized by the Tet efflux transporter at a low concentration; therefore, it exhibits significant antibacterial activity. These properties can explain its potent activity against bacteria with a Tet efflux resistance determinant. Tigecycline induced the Tet(B) protein approximately four times more efficiently than tetracycline, as determined by Western blotting, indicating that it is at least recognized by a TetR repressor. The MICs for multidrug efflux proteins AcrAB and AcrEF were increased fourfold. Tigecycline inhibited active ethidium bromide efflux from intact E. coli cells overproducing AcrAB. Therefore, tigecycline is a possible substrate of AcrAB and its close homolog, AcrEF, which are resistance-modulation-division-type multicomponent efflux transporters.
One approach to overcoming the clinical drug resistance problem in bacterial infections is to modify existing antibiotics to avoid the presence of a resistance determinant. This approach is being used for tetracyclines. Tigecycline is a broad-spectrum glycylcycline derivative (2) and is efficacious against highly resistant bacteria (1), including methicillin-resistant Staphylococcus aureus and penicillin-resistant Streptococcus pneumoniae. However, it is less active against clinically problematic opportunistic pathogens, such as Pseudomonas aeruginosa (6) and Proteus mirabilis (3, 28).
The most common mechanism for tetracycline resistance is an efflux pump-mediated one in both gram-positive and gram-negative organisms (5), i.e., a metal-tetracycline/proton antiporter encoded on plasmids (4, 14, 32). Besides tetracycline-specific transporters, multidrug transporters are also becoming a problem in clinical situations (27).
We recently constructed a library of all 37 possible genes for efflux pumps in Escherichia coli and found that 20 of them conferred resistance to one or more antibiotics, detergents, or dyes (17). Although many of them are not expressed under the usual culture conditions, they may cause clinical resistance in the future, and it is thus meaningful to explore their characters and prepare for the possibility of the development of resistance before it becomes a real problem (17).
We were interested in determining whether in the future tigecycline will pose a resistance problem due to these efflux mechanisms. In this study, we studied the effects of efflux pumps on susceptibility to tigecycline using isogenic E. coli strains expressing plasmids encoding Tet proteins or endogenous chromosomally derived drug transporters. We chose the well-characterized organism E. coli as a model species and a drug-supersensitive strain of E. coli, KAM3 (acrB), as the host. KAM3 lacks the major intrinsic resistance determinant of the AcrAB transport system.
MATERIALS AND METHODS
Plasmids and strains.
The strains and plasmids used in this study are summarized in Table 1. E. coli KAM3 (15), a drug-hypersensitive, acrB-deficient derivative of TG1 (25), was used as the host strain for drug susceptibility testing and the ethidium bromide efflux assay. E. coli W3104 (34) was used for Tet(B) induction and the quinacrine fluorescence study involving membrane fractions or vesicles. We used plasmids pLGT2 (31), pBR322 (24), and pTZ1252 (18), which encode tet(B), tet(C), and tet(K), respectively. pUCacrAB, pTrcHacrEF, and pTrcHbcr carry the acrRAB, acrEF, and bcr genes, respectively, and were cloned from the chromosome of E. coli W3104 as described by Nishino and Yamaguchi (17). Plasmid pAc8 (acrAB) (7) was used to measure the ethidium bromide efflux activities of the cells.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristica | Reference |
|---|---|---|
| E. coli strains | ||
| TG1 | supE hsdΔ5 thi Δ(lac proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 25 |
| KAM3 | acrB-deficient mutant of TG1 | 15 |
| W3104 (W3104rif) | galT12 rpoB λ− F−, rifampin-resistant derivative of W3104 | 34 |
| Plasmids | ||
| pLGT2 | Tn10-tetA(B) and tetR genes cloned into pLG339 | 31 |
| pBR322 | Cloning vector containing the tetA(C) gene as a selection marker for Tetr | 24 |
| pTZ1252 | 2.3-kb HindIII fragment of pNS1, which carries the tet(K) gene of S. aureus, in pUC119, in which the initiation codon and the ribosome binding sequence were changed from TTG to ATG and from GAGG to GGAGG, respectively, and the distance between the ribosome binding sequence and the initiation codon was altered from 4 to 11 bases | 18 |
| pUCacrAB | pUC119 into which the acrR (regulator), acrA (MFP), and acrB (multidrug transporter) genes were cloned | 17 |
| pTrcHacrEF | pTrc6His into which the acrE and acrF genes were cloned; derived from pTrc99A with C-terminal Six-His tag | 17 |
| pTrcHbcr | pTrc6His into which the bcr gene was cloned | 17 |
| pAc8 | pUC118 into which the acrR (regulator), acrA (MFP), and acrB (multidrug transporter) genes were cloned | 7 |
Tet, tetracycline; MFP, membrane fusion protein.
Chemicals.
Tigecycline (GAR-936; 9-[t-butylglycylamido]-minocycline), 9-N,N-dimethylglycylamido-6-demethyl-6-deoxytetracycline (DMG-DMDOT), minocycline, doxycycline, and tetracycline were obtained from Wyeth-Lederle Japan Ltd. (Tokyo, Japan). The structures of these tetracyclines and glycylcyclines are shown in Table 2. Other chemicals were of reagent grade and were from commercial sources.
TABLE 2.
Structures of tigecycline and tetracycline derivatives
In vitro susceptibility testing.
The MIC was determined by the serial dilution method with Mueller-Hinton agar. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at 0.1 mM to induce AcrEF and Bcr cloned under the trc promoter. Approximately 5 × 104 cells were spotted onto agar plates containing various concentrations of drugs, and the plates were incubated for 18 h. The MIC was taken as the lowest drug concentration with which no visible growth was observed.
Western blot analysis of Tet(B) induction by tigecycline and tetracyclines.
Tet(B) induction was determined as described previously (23). E. coli W3104/pLGT2 [tet(B)] cells were grown in 10 ml of medium A (14) supplemented with 0.2% glucose and 0.1% Casamino Acids (Difco) at 37°C with shaking. When the growth in the culture medium reached an optical density at 530 nm (OD530) of 0.4, tigecycline or a control tetracycline was added at various concentrations, followed by incubation for an additional 2 h. The cells were harvested, washed once with 50 mM morpholinepropanesulfonic acid (MOPS)-KOH (pH 7.0) containing 0.1 M KCl, and then disrupted by sonication with a Branson Sonifier 200 (Microson) for 1.5 min. After the undisrupted cells were removed by low-speed centrifugation (10,000 × g, 10 min), the membrane fraction was collected by ultracentrifugation (200,000 × g, 30 min). The membrane pellet was then resuspended in 50 mM MOPS-KOH (pH 7.0) containing 0.1 M KCl. The protein concentration was determined by the method of Lowry et al. (11) with bovine serum albumin as the standard. Cell membrane fractions were standardized to 10 μg of protein and were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12.3% polyacrylamide gel, followed by electroblotting onto a polyvinylidene difluoride membrane. The Tet(B) protein was detected with rabbit antiserum raised against the Tet(B) C-terminal 14 peptides (30) and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (heavy and light chains; Zymed Laboratories Inc., South San Francisco, Calif.), as described previously (23).
Preparation of inverted membrane vesicles.
The preparation of membrane vesicles from W3104/pLGT2 [tet(B)] was carried out as described previously (8, 33). Cells were grown in 1 liter of medium A (22) supplemented with 0.2% glucose and 0.1% Casamino Acids until the OD530 reached 0.4. Tet(B) was induced by incubation for 2 h with 0.25 μg of heat-inactivated chlortetracycline per ml. Inverted (inside-out) vesicles were prepared by disruption of the cells with a French pressure cell (Amicon) at 5,000 lb/in2 in 50 mM MOPS-KOH buffer (pH 6.6) containing 0.1 M KCl and 10 mM EDTA. Then, the vesicles were collected by ultracentrifugation (200,000 × g, 60 min), washed once with 50 mM MOPS-KOH (pH 7.0) containing 0.1 M KCl, resuspended in the same buffer, and stored at −80°C until use.
Quinacrine fluorescence analysis of proton translocation in Tet(B)-containing membrane vesicles.
Proton translocation across inverted membrane vesicles was measured as the change in quinacrine fluorescence (8). Vesicles were diluted to 0.05 mg of protein per ml with 2 ml of 50 mM MOPS-KOH (pH 7.0) containing 0.1 M KCl and 10 mM MgSO4. Ten microliters of 160 μM quinacrine and then 5 μl of 250 mM β-NADH were added. Fluorescence was measured with a Perkin-Elmer LS-55 luminescence spectrophotometer at an excitation wavelength of 440 nm and an emission wavelength of 500 nm at a constant temperature of 30°C with stirring. Tetracycline or tigecycline was added after the fluorescence approached equilibrium or at the indicated time point. Finally, the change in pH (ΔpH) was disrupted by adding 20 μl of 1 M NH4Cl.
Fluorescence measurement of ethidium bromide efflux mediated by AcrAB in E. coli KAM3/pAc8 (acrAB) whole cells.
Ethidium bromide transport in intact E. coli cells was measured by a modification of the method of Lewinson et al. (10). E. coli KAM3 or E. coli KAM3/pAc8 (acrAB) (7) was grown in 10 ml of 2× yeast extract-tryptone medium (22) at 37°C. When the OD600 reached 0.6, the cells were chilled on ice. Two milliliters of each culture was removed, and the cells were pelleted by centrifugation at 6,700 × g for 1 min at room temperature. The supernatant was removed, and the cells were suspended in 2 ml of 50 mM potassium phosphate buffer (pH 7.5) containing 5 μM ethidium bromide and the indicated concentration of tigecycline in the presence of 100 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP). The cells were incubated at 37°C for 5 min to load the ethidium bromide and deplete the energy and were pelleted again by centrifugation; then the supernatant was removed. The cell pellet was stored on ice until use. Prior to use, the cell pellet was suspended in 2 ml of the same buffer containing 5 μM ethidium bromide and various concentrations of tigecycline without CCCP just before fluorescence measurement. Fluorescence was measured with a Perkin-Elmer LS-55 luminescence spectrophotometer at an excitation wavelength of 500 nm and an emission wavelength of 588 nm and at a constant temperature of 37°C with stirring. As the fluorescence had reached equilibrium, the efflux was initiated by adding 0.2% (wt/vol; final concentration) glucose. Finally, CCCP was added at a final concentration of 50 μM to disrupt the proton motive force.
RESULTS AND DISCUSSION
Antibacterial activities of tigecycline against strains harboring plasmids carrying genes encoding tetracycline-specific efflux pumps.
The antibacterial activities of tigecycline against isogenic strains expressing known tetracycline-specific transporters, Tet(B) and Tet(C) from gram-negative species and Tet(K) from the gram-positive species S. aureus, were examined. Plasmid pTZ1252, in which the original staphylococcal start codon and Shine-Dalgarno sequence had been modified by Noguchi et al. (18), was used to express Tet(K) effectively in E. coli. Three pumps were expressed in acrB-deficient drug-sensitive host, E. coli KAM3, a derivative of the TG1 strain (15). The expression was confirmed by the 32- to 256-fold increases in the MICs of tetracycline and the tetracycline derivatives compared to those for the host strain, KAM3, as shown in Table 3, although the resistance levels were different for each transporter. The tigecycline MIC was 0.125 μg/ml for both the host strain and the Tet-expressing E. coli strain, demonstrating these three types of tetracycline efflux transporters failed to confer resistance to tigecycline. This property is similar to that of another glycylcycline, DMG-DMDOT (4, 23), but tigecycline is more potent than DMG-DMDOT and is also active against Tet(C)-producing bacteria, while the DMG-DMDOT MIC for this strain was increased significantly (Table 3). In accord with the results for isogenic E. coli strains mentioned here, Petersen et al. (20) found that the introduction of tet-type efflux determinants did not confer any tigecycline resistance in a study with prototype strains and clinical isolates. An investigation of how tigecycline escapes this Tet-mediated efflux is described below.
TABLE 3.
MICs of tigecycline and tetracyclines for isogenic E. coli strains harboring plasmids carrying efflux-pump related genes
| Compound | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| TG1 (acrAB+) | KAM3 (acrB) | KAM3 harboringa:
|
||||||
| pLGT2 Tet(B) | pBR322 Tet(C) | pTZ1252 Tet(K) | pUCacrAB | pTrcHacrEF | pTrcHbcr | |||
| Tigecycline | 0.5 | 0.125 | 0.125 | 0.125 | 0.125 | 0.5 | 0.5 | 0.125 |
| DMG-DMDOT | 1 | 0.5 | 0.5 | 2 | 0.25 | 0.5 | 1 | 0.5 |
| Minocycline | 2 | 0.25 | 2 | 0.5 | 0.25 | 1 | 2 | 0.25 |
| Doxycycline | 2 | 0.25 | 8 | 2 | 1 | 1 | 2 | 0.25 |
| Tetracycline | 2 | 0.5 | 128 | 32 | 16 | 1 | 1 | 2 |
Values in boldface differ significantly (more than fourfold) from the value for the KAM3 control strain.
Induction of Tet(B) by tigecycline in E. coli W3104/pLGT2 [tet(B)].
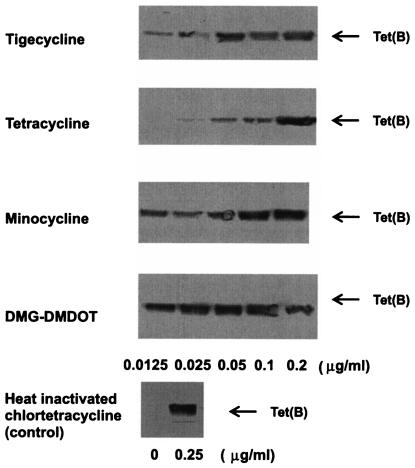
It is known that some Tet efflux proteins are induced by their substrate tetracyclines through binding to the TetR repressor. Therefore, it is possible that the potent activity of tigecycline against Tet(B)-expressing E. coli is due to the lack of Tet(B) induction with this compound. Induction of Tet(B) transporter expression was examined in the membrane fraction by Western blotting analysis after the cells had been incubated with different concentrations of drugs for 2 h. As shown in Fig. 1, tigecycline at a concentration of 0.05 μg/ml significantly induced Tet(B), which makes it more potent than tetracycline and which suggests that tigecycline can bind to the TetR repressor and induce the Tet(B) efflux pump. The potencies of induction were DMG-DMDOT > minocycline ≈ tigecycline > tetracycline, with estimated 50% effective doses of 0.01, 0.04, 0.04, and 0.14 μg/ml, respectively, under the conditions used in this study. These results indicate that tigecycline has properties similar to those of other tetracycline derivatives from the viewpoint of Tet induction, and the mechanism by which tigecycline escapes from tetracycline efflux protein-mediated resistance must be by one other than Tet induction.
FIG. 1.
Induction of the Tet(B) protein by tigecycline and tetracyclines. E. coli W3104/pLGT2 [tet(B)] was grown to an OD530 of 0.4, and then the indicated concentrations of drugs were added for induction. After 2 h, the cells were harvested and membrane fractions were prepared. Ten micrograms of membrane proteins was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Tet(B) was visualized by Western blotting, as described in Materials and Methods.
Effect of tigecycline on proton translocation coupled to tetracycline efflux in E. coli W3104/pLGT2 [tet(B)] membrane vesicles.
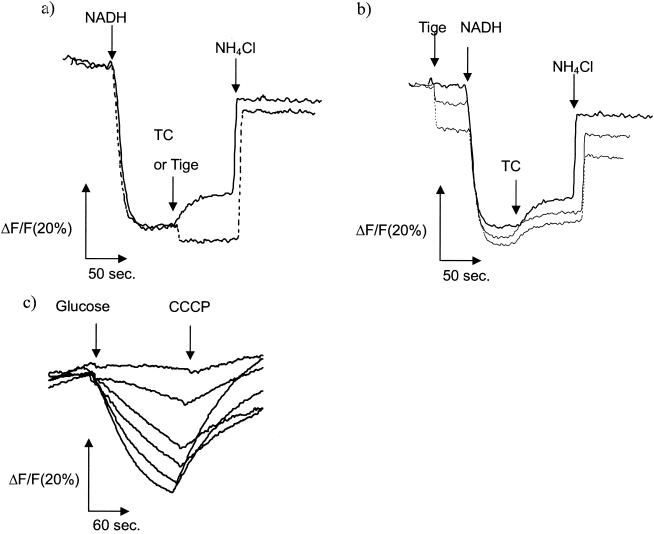
We then studied tigecycline inhibition of proton translocation using Tet(B)-containing inverted (inside-out) membrane vesicles prepared from membranes of E. coli W3104/pLGT2 [tet(B)] by using quinacrine fluorescence as a probe for proton movement. As shown in Fig. 2a, the fluorescence decreased after energization with NADH, reflecting proton movement inward. After equilibrium had been reached, 10 μM tetracycline was added. The increase in fluorescence can be explained by the outward movement of protons due to tetracycline/proton antiport mediated by Tet(B) (9), which was not observed for control vesicles without Tet(B) (29) (data not shown). In contrast, fluorescence did not increase after the addition of an identical concentration of tigecycline, indicating no tigecycline/proton antiport at this concentration. The sharp and small decrease in fluorescence after tigecycline addition represents the quenching effect of tigecycline on the fluorescence of quinacrine. Although the strain used possesses the acrAB+ genetic background, the effect of AcrB is negligible because, in a separate experiment, no proton translocation activity was observed by the addition of 10 μM tigecycline to vesicles in which AcrB is overexpressed and which were prepared in a similar way, probably due to the difficulty in the reconstitution of AcrB in the absence of AcrA and TolC under the conditions used (data not shown). A competition study was carried out, as shown in Fig. 2b, in which tigecycline was added before membrane energization and tetracycline was added later. There was no significant inhibition of the decrease in fluorescence after NADH addition, indicating that tigecycline does not have a strong inhibitory effect on the formation of ΔpH. The proton antiport activity coupled to tetracycline efflux mediated by Tet(B) was not significantly inhibited by tigecycline, being slightly inhibited by 25 μM tigecycline (Fig. 2b). Because of the intrinsic quenching, a higher concentration could not be examined with this fluorescence system. By use of a radiolabeled substrate, [3H]tetracycline uptake into Tet(B)-containing membrane vesicles was found to be inhibited by tigecycline at high concentrations (more than 100 μM) (data not shown). The Ki value was determined to be 238 ± 36 μM by nonregression curve analysis, revealing the considerably low affinity of Tet(B) for tigecycline (data not shown). These results indicate that tigecycline is not practically recognized by Tet(B) at low concentrations; therefore, tigecycline exhibits potent antibacterial activity, although at high concentrations it would be recognized with a low affinity. Tuckman et al. (26) reported that a tet(A) mutation, i.e., a double frameshift, in the interdomain loop region resulted in resistance to glycylcyclines, although tigecycline was the least affected. It might be possible that such a mutant with a higher affinity would emerge from a clinical situation in the future.
FIG. 2.
Effect of tigecycine on Tet(B)-mediated proton translocation and AcrAB-mediated ethidium bromide transport. (a) Effect of tigecycline on proton translocation measured as quinacrine fluorescence in membrane vesicles of E. coli W3104/pLGT2 [tet(B)]. Solid line, tetracycline (TC) at 10 μM; dotted line, tigecycline (Tige) at 10 μM. Note that tigecycline quenched quinacrine fluorescence. (b) Effect of tigecycline on tetracycline efflux-coupled proton translocation in membrane vesicles of E. coli W3104/pLGT2 [tet(B)]. Tetracycline was used at 10 μM; and tigecycline was used at 0, 10, and 25 μM (from top to bottom). (c) Inhibition of ethidium bromide transport by tigecycline in E. coli KAM3/pAc8 (acrAB) whole cells. The tigecycline concentrations (top to bottom) were 200, 100, 50, 25, 12.5, and 0 μM. Glucose (0.2%) was added to start the efflux for ethidium bromide-preloaded and energy-depleted cells.
Antibacterial activities of tigecycline against strains harboring plasmids carrying genes encoding multidrug efflux pumps.
We also examined the antibacterial activities of tigecycline against strains expressing three transporters, AcrAB and AcrEF, which are of the resistance-nodulation-division (RND) type, and Bcr, which is a major facilitator superfamily-type transporter (17, 19). These three transporters were chosen from our E. coli transporter library because they conferred tetracycline resistance in a previous study (17). The native promoter was used for AcrAB expression. The trc promoter, which requires induction by IPTG, was used for AcrEF and Bcr because AcrEF is not normally expressed by the native promoter under laboratory culture conditions (12, 21). This also eliminates the effect of the putative regulator (yeiD) contained in our clone of pUCbcr containing the Bcr native promoter (17). As shown in Table 3, the three pumps conferred decreased (two- to eightfold) susceptibility to tetracycline and the other tetracycline derivatives compared to that of the host strain lacking acrB, confirming the proper expression of each transporter. Note that Bcr conferred decreased susceptibility only to tetracycline, i.e., not the tetracycline derivatives (17). Both AcrAB and AcrEF expression resulted in an increase in the tigecycline MIC from 0.125 to 0.5 μg/ml (fourfold), indicating that both pumps could confer decreased susceptibility to this compound. Likewise, other tetracycline derivatives were similarly affected by AcrAB and/or AcrEF, with two- to fourfold increases in the MICs (Table 3). AcrB is the highly potent MDR transporter in E. coli, and AcrF is its close homologue; the amino acid identity between the two is 77% (13). Bcr expression did not result in any change in the tigecycline MIC. Recently, it was reported that the AcrB homologue is involved in the tigecycline resistance of P. mirabilis (28), which is in good agreement with the result obtained here. The MICs for the strains expressing AcrAB and AcrEF were restored to the MIC for strain TG1, which has an intact AcrB pump, but did not reach the level of clinical resistance. This also suggests that AcrB, which is usually expressed in wild-type cells, is responsible for the decreased activity against tigecycline in E. coli. This antibacterial property of tigecycline in vitro was further characterized biochemically by an ethidium bromide efflux assay, as described below.
Inhibition of ethidium bromide transport by tigecycline in AcrAB-overproducing E. coli whole cells.
We measured the preloaded ethidium bromide efflux activity of AcrB from intact AcrAB-overproducing E. coli cells by the modified method of Lewinson et al. (10). pAc8, an AcrAB expression plasmid (7), was introduced into E. coli KAM3. Log-phase cells were prepared and preloaded with 5 μM ethidium bromide in the presence of CCCP to deplete their energy. CCCP was then removed by washing. An identical concentration of ethidium bromide was added to cells without CCCP; and after the fluorescence had reached equilibrium, glucose was added to start the efflux reaction; i.e., the efflux of ethidium bromide started, as indicated by the decrease in fluorescence due to the dissociation of ethidium bromide from DNA or RNA (Fig. 2c, bottom trace line, no drug) (10). The fluorescence returned to a plateau when CCCP was added to dissipate the proton motive force. The decrease in fluorescence was not observed in the acrB-deficient E. coli host strain (data not shown), indicating that this change in fluorescence is due to the efflux activity of AcrB expressed from the plasmid that was introduced, pAc8 (7). When tigecycline was added simultaneously with ethidium bromide both at preloading and at the efflux reaction, it inhibited the efflux activity of AcrB in a dose-dependent manner, with a 50% inhibitory concentration of about 25 μM (Fig. 2c). This revealed that tigecycline is recognized by AcrB and could possibly be its substrate. Since AcrF is a close homologue of AcrB, it is highly possible that tigecycline is a substrate for both pumps. It is unlikely that this inhibition is due to disruption of ΔpH by tigecycline because tigecycline did not significantly affect the formation of the proton motive force in the membrane vesicle study, as already described for the quinacrine fluorescence assay. These results are in good agreement with those in a recent paper by Dean et al. (6), who reported the tigecycline resistance conferred by MexXY and possibly MexAB, which are Pseudomonas homologues of AcrAB.
In conclusion, although tigecycline is a promising antibiotic for the treatment of infections caused by problematic pathogens as well as those caused by the existing tetracycline-resistant bacteria, it has been revealed to be recognized by RND-type transporters; and there is still room to escape this resistance mechanism by further modification. For this purpose, crystallographic analysis with information on the structure of AcrB (16) would facilitate drug improvement. We should also carefully watch for the emergence of efflux-mediated resistance to tigecycline with clinical use to prolong the life span of this useful antibiotic resource.
Acknowledgments
We thank T. Tsuchiya for strain KAM3 and N. Noguchi for plasmid pTZ1252.
K. Nishino and N. Tamura were supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists. This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by a grant-in-aid for the Zoonosis Control Project of the Ministry of Agriculture, Forestry and Fisheries of Japan, by a grant for the COE Program in the 21st Century of the Japan Society for the Promotion of Science, and by a grant for the Core Research Evolutional Science and Technology program of the Japan Science and Technology Corporation.
REFERENCES
- 1.Abbanat, D., M. Macielag, and K. Bush. 2003. Novel antibacterial agents for the treatment of serious gram-positive infections. Expert Opin. Investig. Drugs 12:379-399. [DOI] [PubMed] [Google Scholar]
- 2.Bryskier, A. 1999. Novelties in the field of anti-infectives in 1998. Clin. Infect. Dis. 29:632-658. [DOI] [PubMed] [Google Scholar]
- 3.Chopra, I. 2001. Glycylcyclines: third-generation tetracycline antibiotics. Curr. Opin. Pharmacol. 1:464-469. [DOI] [PubMed] [Google Scholar]
- 4.Chopra, I. 2002. New developments in tetracycline antibiotics: glycylcyclines and tetracycline efflux pump inhibitors. Drug Resist. Update 5:119-125. [DOI] [PubMed] [Google Scholar]
- 5.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, C. R., M. A. Visalli, S. J. Projan, P. E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujihira, E., N. Tamura, and A. Yamaguchi. 2002. Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia coli. J. Biochem. (Tokyo) 131:145-151. [DOI] [PubMed] [Google Scholar]
- 8.Hirata, T., R. Wakatabe, J. Nielsen, Y. Someya, E. Fujihira, T. Kimura, and A. Yamaguchi. 1997. A novel compound, 1,1-dimethyl-5(1-hydroxypropyl)-4,6,7-trimethylindan, is an effective inhibitor of the tet(K) gene-encoded metal-tetracycline/H+ antiporter of Staphylococcus aureus. FEBS Lett. 412:337-340. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko, M., A. Yamaguchi, and T. Sawai. 1985. Energetics of tetracycline efflux system encoded by Tn10 in Escherichia coli. FEBS Lett. 193:194-198. [DOI] [PubMed] [Google Scholar]
- 10.Lewinson, O., J. Adler, G. J. Poelarends, P. Mazurkiewicz, A. J. Driessen, and E. Bibi. 2003. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl. Acad. Sci. USA 100:1667-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 12.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 13.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMurry, L., R. E. Petrucci, Jr., and S. B. Levy. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:3974-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 17.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noguchi, N., A. Emura, M. Sasatsu, and M. Kono. 1994. Expression in Escherichia coli of a TetK determinant from Staphylococcus aureus. Biol. Pharm. Bull. 17:352-355. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Someya, Y., A. Yamaguchi, and T. Sawai. 1995. A novel glycylcycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline, is neither transported nor recognized by the transposon Tn10-encoded metal-tetracycline/H+ antiporter. Antimicrob. Agents Chemother. 39:247-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutcliffe, J. G. 1979. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harbor Symp. Quant. Biol. 43:77-90. [DOI] [PubMed] [Google Scholar]
- 25.Taylor, J. W., J. Ott, and F. Eckstein. 1985. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 13:8765-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuckman, M., P. J. Petersen, and S. J. Projan. 2000. Mutations in the interdomain loop region of the tetA(A) tetracycline resistance gene increase efflux of minocycline and glycylcyclines. Microb. Drug Resist. 6:277-282. [DOI] [PubMed] [Google Scholar]
- 27.Van Bambeke, F., Y. Glupczynski, P. Plesiat, J. C. Pechere, and P. M. Tulkens. 2003. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J. Antimicrob. Chemother. 51:1055-1065. [DOI] [PubMed] [Google Scholar]
- 28.Visalli, M. A., E. Murphy, S. J. Projan, and P. A. Bradford. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob. Agents Chemother. 47:665-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi, A., K. Adachi, T. Akasaka, N. Ono, and T. Sawai. 1991. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon Tn10. Histidine 257 plays an essential role in H+ translocation. J. Biol. Chem. 266:6045-6051. [PubMed] [Google Scholar]
- 30.Yamaguchi, A., K. Adachi, and T. Sawai. 1990. Orientation of the carboxyl terminus of the transposon Tn10-encoded tetracycline resistance protein in Escherichia coli. FEBS Lett. 265:17-19. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi, A., M. Nakatani, and T. Sawai. 1992. Aspartic acid-66 is the only essential negatively charged residue in the putative hydrophilic loop region of the metal-tetracycline/H+ antiporter encoded by transposon Tn10 of Escherichia coli. Biochemistry 31:8344-8348. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi, A., N. Ono, T. Akasaka, T. Noumi, and T. Sawai. 1990. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon, Tn10. The role of the conserved dipeptide, Ser65-Asp66, in tetracycline transport. J. Biol. Chem. 265:15525-15530. [PubMed] [Google Scholar]
- 33.Yamaguchi, A., Y. Shiina, E. Fujihira, T. Sawai, N. Noguchi, and M. Sasatsu. 1995. The tetracycline efflux protein encoded by the tet(K) gene from Staphylococcus aureus is a metal-tetracycline/H+ antiporter. FEBS Lett. 365:193-197. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto, T., M. Tanaka, C. Nohara, Y. Fukunaga, and S. Yamagishi. 1981. Transposition of the oxacillin-hydrolyzing penicillinase gene. J. Bacteriol. 145:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]