Abstract
All six penicillin-binding protein (PBP) genes, namely, pbp1a, pbp1b, pbp2a, pbp2b, pbp2x, and pbp3, of 40 Streptococcus pneumoniae clinical isolates, including penicillin-resistant S. pneumoniae isolates collected in Japan, were completely sequenced. The MICs of penicillin for these strains varied between 0.015 and 8 μg/ml. In PBP 2X, the Thr550Ala mutation close to the KSG motif was observed in only 1 of 40 strains, whereas the Met339Phe mutation in the STMK motif was observed in six strains. These six strains were highly resistant (MICs ≧ 2 μg/ml) to cefotaxime. The MICs of cefotaxime for 27 strains bearing the Thr338Ala mutation tended to increase, but the His394Leu mutation next to the SSN motif did not exist in these strains. In PBP 2B, the Thr451Ala/Phe/Ser and Glu481Gly mutations close to the SSN motif were observed in 24 strains, which showed penicillin resistance and intermediate resistance, and the Thr624Gly mutation close to the KTG motif was observed in 2 strains for which the imipenem MIC (0.5 μg/ml) was the highest imipenem MIC detected. In PBP 1A, the Thr371Ser/Ala mutation in the STMK motif was observed in all 13 strains for which the penicillin MICs were ≧1 μg/ml. In PBP 2A, the Thr411Ala mutation in the STIK motif was observed in one strain for which with the cefotaxime MIC (8 μg/ml) was the highest cefotaxime MIC detected. On the other hand, in PBPs 1B and 3, no mutations associated with resistance were observed. The results obtained here support the concept that alterations in PBPs 2B, 2X, and 1A are mainly involved in S. pneumoniae resistance to β-lactam antibiotics. Our findings also suggest that the Thr411Ala mutation in PBP 2A may be associated with β-lactam resistance.
Streptococcus pneumoniae is a crucial pathogen that causes community-acquired infections such as pneumonia, otitis media, and meningitis. Since an S. pneumoniae strain showing intermediate resistance to penicillin was first isolated in 1967 in Australia (13), the emergence of penicillin-resistant S. pneumoniae (PRSP) strains has been reported throughout the world.
One of the common mechanisms of resistance to β-lactam antibiotics is the alterations of penicillin-binding proteins (PBPs). PBPs are the enzymes that catalyze polymerization and cross-linking of peptidoglycan precursors in the bacterial cell wall biosynthesis step (20). The penicillin-binding module contains three conserved motifs that form the active cavity. They are the Ser-X-X-Lys (SXXK), the Ser-X-Asn (SXN), and the Lys-Thr/Ser-Gly (KT/SG) motifs (11). Alterations of the PBPs reduce the affinities of β-lactam antibiotics, resulting in resistance. In S. pneumoniae, it has been documented that among the six PBPs, PBPs 1A, 2B, and 2X are the most often associated with penicillin resistance (4, 6, 11).
Although several studies have described the mutations of PBPs and some amino acids residues that must be responsible for resistance to β-lactam antibiotics, only one or two PBP genes or their partial sequences were investigated and compared to the sequences of the genes of penicillin-sensitive S. pneumoniae strains in those studies (2, 3, 10, 16, 21, 22, 23, 24, 28). The pbp1a, pbp2b, and pbp2x genes of 2 and 18 clinical isolates collected mainly in the United States (5, 17), 8 clinical isolates collected in France (9), and 15 clinical isolates collected in Canada (21) were sequenced. In the present study, we completely sequenced the six PBP genes of 40 clinical isolates of S. pneumoniae, including PRSP isolates, collected in Japan and describe the mutations in these genes.
MATERIALS AND METHODS
Bacterial strains.
Forty clinical isolates of S. pneumoniae were selected from among more than 6,600 isolates collected between 1998 and 2000 by the Community-Acquired Bacterial Infections Working Group in Japan (27). The isolates were selected on the basis of of penicillin, cefotaxime, and imipenem MICs (see Results). To find out the common features of the penicillin-susceptible S. pneumoniae, penicillin-intermediate S. pneumoniae, and PRSP strains spread throughout Japan, we chose a variety of strains in terms of the diseases that they caused and the districts where they were isolated. Strain R6, obtained from American Type Culture Collection, was used as a reference strain.
Identification of S. pneumoniae.
To identify S. pneumoniae, the lytA gene encoding pneumococcal autolysin was detected by PCR (18). The primers used to detect the lytA gene are shown in Table 1. They were designed on the basis of the nucleotide sequence reported by Kawamura et al. (14). Furthermore, the partial sequence of the sodA gene, which encodes superoxide dismutase, was determined to confirm the identification (14). The primers were designed as shown in Table 1, and the DNA sequences were compared with those of other streptococci obtained from the GenBank database.
TABLE 1.
Oligonucleotide primers for PCR amplification
| Gene | Sense primer sequence | Antisense primer sequence |
|---|---|---|
| pbp1a | 5′-CCAGCAACAGGTGAGAGTC-3′ | 5′-GTAAACACAAGCCAAGACAC-3′ |
| pbp1aa | 5′-GAACTTCAAGACAAGGCAGT-3′ | 5′-GTAAACACAAGCCAAGACAC-3′ |
| pbp1b | 5′-TTCATACCCTTATTGTAACACG-3′ | 5′-TAGCTCATCAAAAATGTGCATG-3′ |
| pbp2a | 5′-CCGCTGATCTTGATTGAATAG-3′ | 5′-ATGCGTTTTCATCCCCTCTG-3′ |
| pbp2b | 5′-CCGTCTTAATCCCGATACC-3′ | 5′-ATTTTTGGGTGACTTGTTGAG-3′ |
| pbp2x | 5′-GGAATTGGTGTCCCGTAAGC-3′ | 5′-CATCTGCTGGCCTGTAATTTG-3′ |
| pbp3 | 5′-ATACGCAGTTGAGCTGAACC-3′ | 5′-GCACGCTAACAAAAGGGTTAA-3′ |
| lytA | 5′-CTGTTTCAATCGTCAAGCCG-3′ | 5′-TAAGAACAGATTTGCCTCAAGT-3′ |
| sodA | 5′-ACAATGCATTTGCACCATGAC-3′ | 5′-GGTTTGCTGTTGAAGTCACTT-3′ |
Used for strains SP00081, SP00087, and SP00090.
Antimicrobial susceptibility testing.
MICs were determined by the microdilution method recommended by the National Committee for Clinical Laboratory Standards (19). Antibiotics were obtained from Eiken Chemical Co., Ltd. (Tokyo, Japan).
PCR and nucleotide sequencing.
The six sets of primers used for PCR were designed on the basis of the DNA sequences of strain R6 (DDBJ/EMBL/GenBank accession number NC003098). The oligonucleotide sequences used for DNA amplification are shown in Table 1. The pbp1a gene was not successfully amplified from strains SP00081, SP00087, and SP00090 with the set of primers described above. Therefore, an additional set of primers was designed to amplify the pbp1a gene; the primers were based on the DNA sequence obtained by direct sequencing with a GenomicTip G20 kit (Qiagen K. K., Tokyo, Japan). The oligonucleotide sequences used to amplify the DNA of these three strains are shown in Table 1. PCR amplification was performed with a PCR System 9700 (Applied Biosystems, Foster City, Calif.). The PCR products were sequenced with an ABI PRISM 3700 DNA analyzer (Applied Biosystems). Both strands were amplified by PCR, and a given position was sequenced at least three times with different primers.
Nucleotide sequence accession numbers.
The complete sequences of the six PBP genes from 40 clinical isolates appear in the DDBJ/EMBL/GenBank nucleotide sequence database. The serial numbers for the 40 clinical isolates are SP00051 to SP00092, with SP00066 and SP00071 missing. The corresponding accession numbers for the sequences of the PBP genes from the 40 clinical isolates in the DDBJ/EMBL/GenBank database are as follows: AB119753 to AB119792 for pbp1a, AB119793 to AB119832 for pbp1b, AB119833 to AB119872 for pbp2a, AB119873 to AB119912 for pbp2b, AB119913 to AB119952 for pbp2x, and AB119953 to AB119992 for pbp3.
RESULTS AND DISCUSSION
Amino acid substitutions in PBPs and β-lactam antibiotic susceptibilities.
The deduced amino acid substitutions in PBPs 2X, 2B, 1A, and 2A are shown in Table 2, which summarizes the mutations for groups of strains with identical or similar mutations. The activities of the β-lactam antibiotics against the S. pneumoniae isolates used in this study are shown in Table 3, which provides the range of MICs for each group in Table 2. The activities of carbapenems and faropenem were more potent than those of the penicillins and cephalosporins against PRSP strains. In particular, panipenem showed the most potent activity, and its geometric mean MIC was 0.091 μg/ml. Among the cephalosporins, cefditoren, the active form of cefditoren pivoxil, showed the most potent activity against PRSP strains.
TABLE 2.
Deduced amino acid substitutions in PBPs
| Strain | Amino acid substitutiona
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBP 2X
|
PBP 2B
|
PBP 1A
|
PBP 2A
|
||||||||||||||||
| 338T | 339M | 394H | 550T | 431T | Insertion | 432Q | 451T | 481E | 624A | 371T | 432P | 574T | 575S | 576Q | 577F | 411T | 461S | 586V | |
| R6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| SP00051, SP00053, SP00054 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A/. | I/. | |
| SP00052 | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | A | I | |
| SP00058, SP00059, SP00060, SP00061 | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | I/. | |
| SP00069, SP00074 | A | . | . | . | . | . | A | G | . | . | . | . | . | . | . | . | A | . | |
| SP00055, SP00067, SP00073 | . | . | L | . | . | . | . | . | . | . | . | . | . | . | . | . | A | I/. | |
| SP00072 | . | . | L | . | . | . | A | G | . | . | . | N | T | G | Y | . | A | . | |
| SP00057, SP00070 | . | . | L | . | . | P/− | ./A | S/P | G | . | . | . | N | T | G | Y | . | A | I/. |
| SP00081 | . | . | L | . | K | L | A | G | . | S | T | N | T | G | Y | . | A | I | |
| SP00056, SP00063, SP00064, SP00068 | A | . | . | . | . | . | . | . | . | S/. | T/. | N | T | G | Y | . | A | I/. | |
| SP00085, SP00088 | A | . | . | . | . | . | A | G | . | S/. | T | N | T | G | Y | . | A | I | |
| SP00075, SP00076, SP00078, SP00079, SP00082, SP00083, SP00086, SP00087, SP00089 | A | . | . | . | K | L | A | G | . | S/A | T | N | T | G | Y | . | A | I/. | |
| SP00062 | P | . | . | . | . | WYT | . | S | G | . | . | . | N | T | G | Y | . | A | I |
| SP00065 | A | F | . | . | . | . | . | . | . | S | . | N | T | G | Y | . | A | I | |
| SP00077 | A | F | . | . | . | . | A | G | . | A | T | N | T | G | Y | . | A | . | |
| SP00080, SP00091 | A | F | . | . | K | L | A | G | . | S | T | N | T | G | Y | . | A | I | |
| SP00090 | A | F | . | . | . | WYT | . | S | G | . | S | T | N | T | G | Y | . | A | I |
| SP00092 | A | . | . | . | K | L | A | G | G | A | T | N | T | G | Y | . | A | . | |
| SP00084 | A | F | . | . | K | L | A | G | G | S | T | N | T | G | Y | A | A | . | |
Identity with the amino acid from strain R6 is indicated by a dot. The strains were typed by the mutations indicated in boldface, which are mainly close to the conserved motifs: 337STMK, 395SSN, and 547KSG for PBP 2X; 391SVVK, 448SSN, and 619KTG for PBP 2B; 370STMK, 428SRN, and 557KTG for PBP 1A; and 410STIK, 465SLN, and 590KTG for PBP 2A.
TABLE 3.
Antibacterial activities of β-lactam antibiotics
| Strain | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | AMX | CTX | CRO | CPD | CDR | CDN | IPM | MEM | PAM | FRM | |
| R6 | 0.016 | 0.016 | 0.016 | 0.031 | 0.031 | 0.008 | 0.063 | 0.008 | 0.016 | 0.004 | 0.002 |
| SP00051, SP00053, SP00054 | 0.016-0.063 | 0.016-0.031 | 0.031-0.125 | 0.031-0.125 | 0.063-0.125 | 0.125-0.25 | 0.016-0.063 | 0.008 | 0.016 | 0.004 | 0.008-0.016 |
| SP00052 | 0.008 | 0.008 | 0.25 | 0.125 | 0.5 | 0.5 | 0.063 | 0.016 | 0.016 | 0.002 | 0.008 |
| SP00058, SP00059, SP00060, SP00061 | 0.031-0.063 | 0.031-0.063 | 0.25-0.5 | 0.25-0.5 | 0.5-1 | 0.25-0.5 | 0.125-0.25 | 0.008-0.016 | 0.016-0.031 | 0.002-0.008 | 0.008-0.016 |
| SP00069, SP00074 | 0.125-0.25 | 0.125 | 0.5 | 0.25-0.5 | 1-2 | 0.5-1 | 0.25 | 0.016 | 0.063 | 0.008 | 0.016-0.063 |
| SP00055, SP00067, SP00073 | 0.031-0.125 | 0.031-0.063 | 0.125-0.25 | 0.125-0.25 | 0.25 | 0.25-0.5 | 0.063-0.125 | 0.008 | 0.016 | 0.002-0.004 | 0.008-0.016 |
| SP00072 | 0.031 | 0.25 | 0.25 | 0.25 | 0.5 | 0.5 | 0.25 | 0.031 | 0.063 | 0.008 | 0.031 |
| SP00057, SP00070 | 0.25 | 0.25 | 0.125-0.25 | 0.25 | 0.25-0.5 | 0.5 | 0.125 | 0.031-0.063 | 0.063-0.125 | 0.008 | 0.063 |
| SP00081 | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 | 0.125 | 0.25 | 0.031 | 0.125 |
| SP00056, SP00063, SP00064, SP00068 | 0.063-0.5 | 0.031-0.25 | 0.5-1 | 0.5-1 | 1-2 | 2-8 | 0.25-1 | 0.008-0.063 | 0.016-0.125 | 0.002-0.016 | 0.008-0.063 |
| SP00085, SP00088 | 0.5-0.1 | 0.25 | 1-2 | 1-2 | 2 | 2-8 | 0.5-1 | 0.063 | 0.125 | 0.016-0.031 | 0.125 |
| SP00075, SP00076, SP00078, SP00079, SP00082, SP00083, SP00086, SP00087, SP00089 | 0.5-2 | 0.5-1 | 0.5-1 | 0.5-2 | 1-4 | 1-8 | 0.25-1 | 0.125-0.25 | 0.25-0.5 | 0.031-0.125 | 0.125-0.5 |
| SP00062 | 0.25 | 0.125 | 0.063 | 0.063 | 0.125 | 0.125 | 0.031 | 0.063 | 0.063 | 0.008 | 0.063 |
| SP00065 | 0.063 | 0.063 | 2 | 2 | 16 | 2 | 0.5 | 0.016 | 0.031 | 0.004 | 0.016 |
| SP00077 | 1 | 0.5 | 2 | 2 | 16 | 8 | 1 | 0.125 | 0.25 | 0.031 | 0.125 |
| SP00080, SP00091 | 2-4 | 1-2 | 4 | 2-4 | 4-16 | 16 | 1-2 | 0.25-0.5 | 0.5 | 0.063-0.25 | 0.25-0.5 |
| SP00090 | 4 | 1 | 4 | 4 | 16 | 32 | 4 | 0.25 | 0.5 | 0.125 | 0.25 |
| SP00092 | 2 | 8 | 4 | 2 | 4 | 16 | 1 | 0.5 | 1 | 0.25 | 1 |
| SP00084 | 4 | 2 | 8 | 4 | 16 | 16 | 2 | 0.5 | 2 | 0.125 | 1 |
Abbreviations: PEN, penicillin; AMX, amoxicillin; CTX, cefotaxime; CRO, ceftriaxone; CPD, cefpodoxime; CDR, cefdinir; CDN, cefditoren; IPM, imipenem; MEM, meropenem; PAM, panipenem; and FRM, faropenem.
Substitutions in PBP 2X.
The deduced amino acid substitutions in PBP 2X are shown in Table 2. Mutations at positions close to each of three conserved motifs, SXXK, SXN, and KT/SG, or those which seemed to be related to susceptibility are indicated in Table 2. The Met339Phe mutation in the STMK motif was observed in 6 of the 40 strains, and all of them were highly resistant to cefotaxime (MICs ≧ 2 μg/ml). On the other hand, the Thr338Ala mutation was observed in 28 strains, but not all of them were necessarily highly resistant to cefotaxime. Asahi et al. (2) reported that both the Met339Phe and the Thr338Ala mutations in PBP 2X are important substitutions leading to high levels of resistance to cefotaxime. However, our findings do not completely support their results in the case of strains bearing the Thr338Ala mutation. Interestingly, the His394Leu mutation just before the SSN motif did not exist in the strains bearing the Thr338Ala mutation. Although there might be an association between Thr338Ala and His394Leu mutations, the influences of these substitutions on β-lactam resistance are not clear. The Thr550Ala mutation just after the KSG motif was observed in only 1 of the 40 strains, and no strain bore the Gln552Glu mutation. It has been clearly shown by site-directed mutagenesis that these two mutations in PBP 2X are responsible for β-lactam resistance (5, 9). However, these mutations are not found in clinical isolates too frequently (9).
Substitutions in PBP 2B.
The deduced amino acids substitutions in PBP 2B are shown in Table 2. The insertion of Trp-Tyr-Thr between residues 431and 432, described by Yamane et al. (28), was observed in two strains. The insertion of Phe between residues 431 and 432 was observed in one strain. The short mosaic sequence Ala-Phe-Ser-Arg/Val-Pro-Asn from residues 432 to 437 described by Dowson et al. (7) was observed as Ala-Phe-Ser-Gly-Ala-Met in one strain in this study.
The Thr446Ala mutation adjacent to the SSN motif was not observed in this study. This mutation was confirmed by site-directed mutagenesis to be associated with β-lactam resistance (10). However, no clinical isolates bearing this mutation were found in Japan or France (9), similar to the case of the Thr550Ala and the Gln552Glu mutations in PBP 2X. The Thr451Ala/Phe/Ser and the Glu481Gly mutations, close to the SSN motif, were observed in 18 of the 19 strains for which penicillin MICs were ≧0.5 μg/ml. These findings are consistent with the observation that these mutations are common in resistant strains isolated in both South Africa and France (14, 23). The Thr624Gly mutation, close to the KTG motif, was observed in two strains; and the imipenem MICs for these two strains were the highest (0.5 μg/ml) among those for the strains tested. This mutation was reported in a β-lactam-resistant recombinant of S. pneumoniae obtained by transformation with chromosomal DNA prepared from a Streptococcus mitis isolate (12). Although strains bearing the Thr624Gly mutation are rare among clinical isolates, this mutation seems to be important for the acquisition of high-level resistance to carbapenems.
Substitutions in PBP 1A.
The deduced amino acids substitutions in PBP 1A are shown in Table 2. The Thr371Ser/Ala mutation in the STMK motif was observed in 18 of the 19 strains for which the penicillin MICs were ≧0.5 μg/ml. Four consecutive substitutions, Thr574Asn, Ser575Thr, Gln576Gly, and Phe577Tyr, were found in the 19 strains for which the penicillin MICs were ≧0.5 μg/ml and in 3 of 6 strains for which they were 0.12 to 0.25 μg/ml. These two types of mutations were confirmed to be essential for penicillin resistance by using reverse amino acid substitution by site-directed mutagenesis (24, 25). Interestingly, one isolate had a 16-amino-acid deletion, from residues 670 to 685, which did not appear to be necessary for resistance to β-lactams. In strains SP00081, SP00087, and SP00090, the DNA from the pbp1a genes was not amplified when the set of primers designed from the sequence data for strain R6 were used. We determined the DNA sequences of downstream regions by direct sequencing and detected a rearrangement of the mosaic elements, which would be the partial pena gene of S. mitis. The PBP encoded by the pena gene has been known as a low-affinity PBP (1). Therefore, these three strains probably acquired the mosaic element on the pbp1a gene by interspecies gene transfer and, as a result, showed high-level resistance to β-lactams.
An insertion sequence (IS) of about 1.4 kb was detected in three strains (strains SP00055, SP00065, and SP00073) downstream (from 10 to 20 bases) from the pbp1a gene. The DNA sequences of these IS elements corresponded to IS1167, which encodes the transposase of S. pneumoniae (30). Compared with the DNA sequence of IS1167 (DDBJ/EMBL/GenBank accession number AE07425), the percent identities for SP00055 and SP00073 (98.0% for both) were higher than that for SP00065 (88.5%). Furthermore, the DNA sequences at the 3′ region of the pbp1a gene were also identical between strains SP00055 and SP00073, as were those of the IS elements. This observation suggests that the IS element contributes to horizontal gene transfer and the genetic rearrangement of pbp genes.
Substitutions in PBP 2A.
The deduced amino acid substitutions in PBP 2A are shown in Table 2. The Thr411Ala mutation in the STIK motif was observed in only one strain, which showed a high level of resistance to cefotaxime (MIC = 8 μg/ml). This mutation was also seen in β-lactam-resistant recombinants of S. pneumoniae obtained through transformation of chromosomal DNA from S. mitis strain B6, resulting in low-affinity variants (12). Although Zhao et al. (29) and Du Plessis et al. (8) reported that the substitutions in PBP 2A would not be associated with the acquisition of β-lactam resistance, our findings suggest that the Thr411Ala mutation in PBP 2A would be important for additional resistance.
As was the case for pbp1a, the IS element was observed downstream from the pbp2a gene in three strains (strains SP00065, SP00069, and SP00084). However, the DNA sequences of the IS element and its upstream region were slightly different among these strains. The significance of these insertions near the pbp2a gene is not clear at present.
Substitutions in PBP 1B and PBP 3.
The deduced amino acids substitutions in PBP 1B and PBP 3 were also investigated. No mutations at positions close to their conserved motifs that would be associated with β-lactam resistance were noted in this study. However, it has not been clarified whether PBP 1B plays an important role in the development of resistance (8, 12). As for PBP 3, Krauss and Hakenbeck (15) detected the Thr242Ile mutation in laboratory mutants resistant to cefotaxime. We believe that the Thr242Ile mutation is not necessarily common in clinical isolates, since this mutation was not observed in this study.
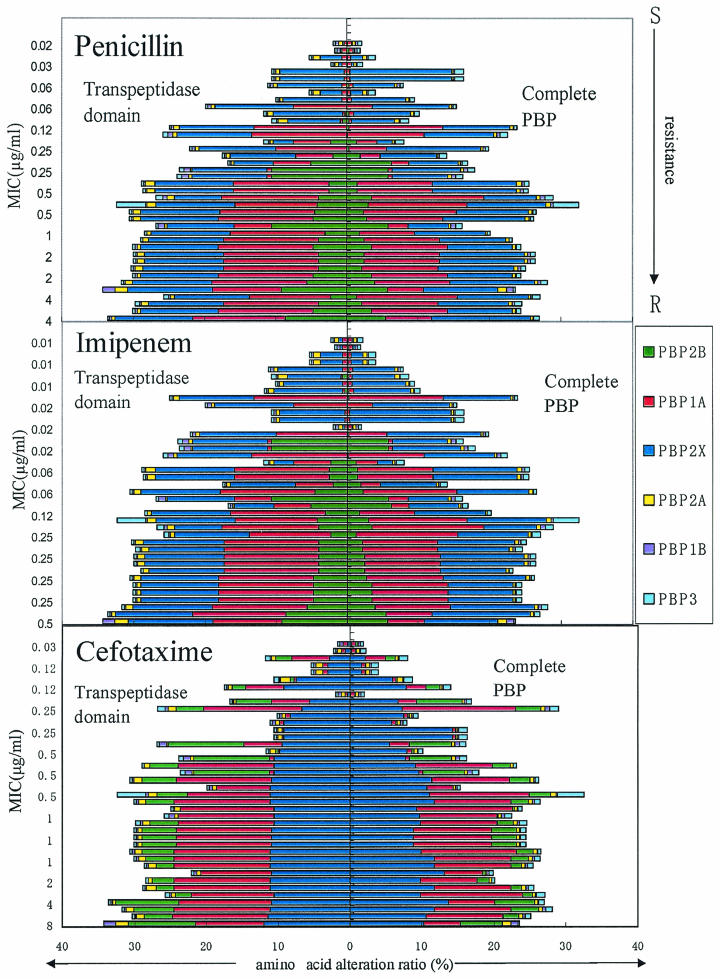
Alteration ratio of the TP domain and complete PBPs.
We investigated the relationship between the ratio of mutations in the PBPs and susceptibility to penicillin, cefotaxime, and imipenem. The alteration ratio within the transpeptidase (TP) domain and complete PBPs was calculated on the basis of the sequence of reference strain R6, and the results are shown in Fig. 1. The alteration ratios increased in the following order: PBP 2B (alteration ratio for TP domain, 0 to 10.6%; alteration ratio for complete PBP, 0 to 6.3%), PBP 2X (alteration ratio for TP domain, 0.28 to 11.9%; alteration ratio for complete PBP, 0.13 to 14.3%), and PBP 1A (alteration ratio for TP domain, 0.31 to 17.3%; alteration ratio for complete PBP, 0.42 to 15.7%). Furthermore, the alteration ratios for the TP domain tended to be higher than those for the complete PBP. On the other hand, the alteration ratios for the other three PBPs, namely, PBPs 2A, 1B, and 3, were low, regardless of whether the strain was susceptible or resistant to β-lactams (for PBP 1B, the alteration ratio for the TP domain was 0 to 1.2% and that for the complete PBP was 0 to 1.0%; for PBP 2A, the alteration ratio for the TP domain was 0 to 1.8% and that for the complete PBP was 0 to 1.2%; and for PBP 3, the alteration ratio for the TP domain was 0 to 4.0% and that for the complete PBP was 0.24 to 3.6%). Interestingly, the alteration ratios for PBPs 1A, 2B, and 2X, which are responsible for β-lactam resistance, were higher than those for PBPs 1B, 2A, and 3. These findings suggest that antibiotic pressure is capable of promoting genetic rearrangement in S. pneumoniae. The alteration ratios for PBPs have been reported by a few investigators (9, 21), but these pieces of information were restricted to the TP domains of PBPs. Clinical practitioners in Japan prefer to use cephalosporins instead of penicillins for the treatment of community-acquired infections, and the prevalence of cephalosporin-resistant S. pneumoniae strains in Japan has been increasing year by year (26). The sensitivity of S. pneumoniae to cephalosporins is related to the specific amino acid changes and also to the alteration ratio for PBP 2X.
FIG. 1.
Relationship between the amino acid alteration ratios for six PBPs and susceptibility to penicillin, imipenem, and cefotaxime for 40 clinical isolates of S. pneumoniae. The alteration ratios for the TP domain and the complete PBP were calculated on the basis of the sequence of reference strain R6.
Concluding remarks.
In conclusion, the results obtained from the complete sequencing of the pbp genes support the concept that mutations in PBPs 1A, 2B, and 2X play an important role in the development of resistance to β-lactam antibiotics by S. pneumoniae. We also detected a mutation which would be important for additional resistance to cefotaxime in the STIK motif of PBP 2A but did not detect such a mutation in PBP 1B or PBP 3. The MICs of penicillins and carbapenems tended to be strongly influenced by mutations in PBP 2B, but those of cephalosporins tended to be strongly influenced by mutations in PBP 2X. Mutations in PBP 1A were associated with additional increases in the MICs for strains bearing mutations in PBP 2X or PBP 2B. Panipenem, a parenteral carbapenem, and cefditren, an active form of an oral cephalosporin, showed the most potent activities against S. pneumoniae strains, including PRSP.
Acknowledgments
We thank Kimiko Ubukata, Kitasato University, for kindly providing clinical isolates of S. pneumoniae.
REFERENCES
- 1.Amoroso, A., D. Demares, M. Mollerach, G. Gutkind, and J. Coyette. 2001. All detectable high-molecular-mass penicillin-binding proteins are modified in a high-level beta-lactam-resistant clinical isolate of Streptococcus mitis. Antimicrob. Agents Chemother. 45:2075-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahi, Y., Y. Takeuchi, and K. Ubukata. 1999. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 43:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahi, Y., and K. Ubukata. 1998. Association of Thr-371 substitution in a conserved amino acid motif of penicillin-binding protein 1A with penicillin resistance of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2267-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barcus, V. A., K. Ghanekar, M. Yeo, T. J. Coffey, and C. G. Dowson. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol. Lett. 126:299-304. [DOI] [PubMed] [Google Scholar]
- 5.Coffey, T. J., M. Daniels, L. K. McDougal, C. G. Dowson, R. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowson, C. G., T. J. Coffey, and B. G. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 2:361-366. [DOI] [PubMed] [Google Scholar]
- 7.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Linares, A. Tomasz, J. Maynard, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Plessis, M., A. M. Smith, and K. P. Klugman. 2000. Analysis of penicillin-binding protein 1B and 2a genes from Streptococcus pneumoniae. Microb. Drug Resist. 6:127-131. [DOI] [PubMed] [Google Scholar]
- 9.Ferroni, A., and P. Berche. 2001. Alterations to penicillin-binding proteins 1A, 2B and 2X amongst penicillin-resistant clinical isolates of Streptococcus pneumoniae serotype 23F from the nasopharyngeal flora of children. J. Med. Microbiol. 50:828-832. [DOI] [PubMed] [Google Scholar]
- 10.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakenbeck, R. 1998. Mosaic genes and their role in penicillin-resistant Streptococcus pneumoniae. Electrophoresis 19:597-601. [DOI] [PubMed] [Google Scholar]
- 12.Hakenbeck, R., A. Koenig, I. Kern, M. V. D. Linden, W. Keck, D. Bioot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansman, D., and M. M. Bullen. 1997. A resistant pneumococcus. Lancet ii:264-265. [DOI] [PubMed] [Google Scholar]
- 14.Kawamura, Y., R. A. Whiley, S. Shu, T. Ezaki, and J. M. Hardie. 1999. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 145:2605-2613. [DOI] [PubMed] [Google Scholar]
- 15.Krauss, J., and R. Hakenbeck. 1997. A mutation in d,d-carboxypeptidase penicillin-binding protein 3 of Streptococcus pneumoniae contributes to cefotaxime resistance of the laboratory mutant C604. Antimicrob. Agents Chemother. 41:936-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouz, N., A. M. D. Guilmi, E. Gordon, R. Hakenbeck, O. Diderberg, and T. Vernet. 1999. Mutations in the active site of penicillin-binding proteins PBP2x from Streptococcus pneumoniae. J. Biol. Chem. 274:19175-19180. [DOI] [PubMed] [Google Scholar]
- 17.Nagai, K., T. A. Davies, M. R. Jacobs, and P. C. Appelbaum. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai, K., Y. Shibasaki, K. Hasegawa, T. A. Davies, M. R. Jacobs, K. Ubukata, and P. C. Appelbaum. 2001. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and β-lactam resistance, and to detect common macrolide resistance determinants. J. Antimicrob. Chemother. 48:915-918. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 5th ed., M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Navarre, W. W., and O. Scheneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichol, K. A., G. G. Zhanel, and D. J. Hoban. 2002. Penicillin-binding protein 1A, 2B, and 2X alterations in Canadian isolates of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:3261-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reichmann, P., A. Koenig, A. Marton, and R. Hakenbeck. 1996. Penicillin-binding proteins as resistance determinants in clinical isolates of Streptococcus pneumoniae. Microb. Drug Resist. 2:177-181. [DOI] [PubMed] [Google Scholar]
- 23.Smith, A. M., and K. Klugman. 1995. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, A. M., and K. Klugman. 1998. Alterations in PBP 1A essential for high-level penicillin-resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:1329-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, A. M., and K. Klugman. 2003. Site-specific mutagenesis analysis of PBP 1A from a penicillin-cephalosporin-resistant pneumococcal isolate. Antimicrob. Agents Chemother. 47:387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki, K., T. Nishimura, and S. Baba. 2003. Current status of bacterial resistance in the otolaryngology field: results from the Second Nationwide Survey in Japan. J. Infect. Chemother. 9:46-52. [DOI] [PubMed] [Google Scholar]
- 27.Ubukata, K. 2001. Epidemiological study of pneumococcal infection and Haemophilus influenzae infection. Jpn. J. Antibiot. 54(Suppl. B):4-10. [PubMed] [Google Scholar]
- 28.Yamane, A., H. Nakano, Y. Asahi, K. Ubukata, and M. Konno. 1996. Directly repeated insertion of 9-nucleotide sequence detected in penicillin-binding protein 2B gene of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:1257-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, G., T. I. Meiler, J. Hoskins, and K. A. McAllister. 2000. Identification and characterization of the penicillin-binding protein 2a of Streptococcus pneumoniae and its possible role in resistance to β-lactam antibiotics. Antimicrob. Agents Chemother. 44:1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou, L., F. M. Hui, and D. A. Morrison. 1995. Characterization of IS1167, a new insertion sequence in Streptococcus pneumoniae. Plasmid 33:127-138. [DOI] [PubMed] [Google Scholar]