Abstract
We have examined the antipneumococcal activities of novel quinolone dimers in which ciprofloxacin was tethered to itself or to pipemidic acid by linkage of C-7 piperazinyl rings. Symmetric 2,6-lutidinyl- and trans-butenyl-linked ciprofloxacin dimers (dimers 1 and 2, respectively) and a pipemidic acid-ciprofloxacin dimer (dimer 3) had activities against Streptococcus pneumoniae strain 7785 that were comparable to that of ciprofloxacin, i.e., MICs of 2, 1, and 4 to 8 μg/ml versus an MIC of 1 to 2 μg/ml, respectively. Surprisingly, unlike ciprofloxacin (which targets topoisomerase IV), several lines of evidence revealed that the dimers act through gyrase in S. pneumoniae. First, ciprofloxacin-resistant parC mutants of strain 7785 remained susceptible to dimers 1 to 3, whereas a gyrA mutation conferred a four- to eightfold increase in the dimer MIC but had little effect on ciprofloxacin activity. Second, dimer 1 selected first-step gyrA (S81Y or S81F) mutants (MICs, 8 to 16 μg/ml) that carried wild-type topoisomerase IV parE-parC genes. Third, dimers 1 and 2 promoted comparable DNA cleavage by S. pneumoniae gyrase and topoisomerase IV, whereas ciprofloxacin-mediated cleavage was 10-fold more efficient with topoisomerase IV than with gyrase. Fourth, the GyrA S81F and ParC S79F enzymes were resistant to dimers, confirming that the resistance phenotype is largely silent in parC mutants. Although a dimer molecule could bind very tightly by bridging quinolone binding sites in the enzyme-DNA complex, the greater potency of ciprofloxacin against gyrase and topoisomerase IV suggests that dimers 1 to 3 bind in a monomeric fashion. The bulky C-7 side chain may explain dimer targeting of gyrase and activity against efflux mutants. Tethered quinolones have potential as mechanistic tools and as novel antimicrobial agents.
Fluoroquinolones such as levofloxacin, gatifloxacin, and moxifloxacin are now widely used as first-line therapy for community-acquired pneumonia caused by Streptococcus pneumoniae (11). These agents act through the enzymes DNA gyrase and topoisomerase IV and are effective against both penicillin-susceptible and -resistant isolates. The success of the “respiratory” quinolones has prompted efforts to understand their mechanisms of action in S. pneumoniae and to uncover more potent analogues, especially those effective against quinolone-resistant strains associated with treatment failures.
A key to understanding quinolone action in S. pneumoniae is the observation that the drugs can act preferentially through topoisomerase IV, through gyrase, or through both targets in a manner dependent on quinolone structure (21). Gyrase, a GyrA2GyrB2 tetramer encoded by gyrA and gyrB genes, catalyzes the introduction of negative supercoils into DNA (9, 28), whereas topoisomerase IV, a ParC2ParE2 complex (specified by parC and parE), is an efficient DNA decatenase (12). Both these ATP-dependent enzyme reactions occur by a double-stranded DNA break (15) and are coordinated to allow DNA unlinking during DNA replication and chromosome segregation (1, 32). Quinolones form a ternary drug-topoisomerase-DNA complex and interfere with DNA breakage-reunion mediated by the two GyrA (ParC) subunits. Addition of a protein denaturant in vitro or collision with a DNA replication fork in vivo converts the ternary complex into a lethal double-stranded DNA break (4, 8, 10). Mutations conferring quinolone resistance usually map to a limited domain of the GyrA (ParC) and GyrB (ParE) proteins termed the “quinolone resistance-determining region” (QRDR) (15, 30, 31). Many quinolones, including ciprofloxacin, select first-step parC or parE mutants of S. pneumoniae, indicating that topoisomerase IV is the preferred intracellular target (17, 19, 20). By contrast, sparfloxacin selects first-step gyrA mutants, implicating gyrase as the primary target (21). Other quinolones, such as clinafloxacin, gatifloxacin, gemifloxacin, and moxifloxacin, select gyrA mutants but act substantially through both gyrase and topoisomerase IV (7, 22). Although the rules governing selectivity are poorly understood, it has been shown (2) that the intracellular targeting of ciprofloxacin can be switched from topoisomerase IV to gyrase by the addition of a benzenesulfonylamido moiety to the quinolone C-7 position.
Much of the improved potency of modern fluoroquinolones has been achieved by tinkering with the N-1, C-7, and C-8 substituents on the quinolone ring system. These alterations promote more efficient target binding and, in the case of the C-7 substitutions, can overcome efflux-mediated resistance, at least in Staphylococcus aureus (26). We have sought a radically different approach by synthesizing covalently linked dimeric quinolones (13, 14) that can potentially bridge drug binding sites in the topoisomerase-DNA complex. Models based on biochemical considerations envisage either two enzyme-bound drug molecules positioned to interfere with enzymatic DNA breakage-reunion on strands (see, e.g., reference 3) or four quinolones bound as two pairs of noncovalently associated drug dimers in a single-stranded DNA bubble opened up by topoisomerase action (25). Although the crystal structure of a dimeric GyrA fragment containing the QRDR is available (16), it lacks DNA and quinolones and therefore does not discriminate the models. However, based on either model, two quinolones tethered through a linker would be expected to achieve tight binding by bridging adjacent quinolone binding pockets. In the study described in this paper, we have examined the modes of action of novel piperazinyl-linked ciprofloxacin dimers (Fig. 1) by testing their activities against S. pneumoniae and its quinolone-resistant parC and gyrA mutants, by analyzing first-step mutants selected by challenge with the dimers, and by studying the inhibition of wild-type and quinolone-resistant S. pneumoniae gyrase and topoisomerase IV. We show that three different dimers display activities comparable to that of ciprofloxacin against wild-type S. pneumoniae and retain activity against parC mutants, although not against parC gyrA mutants. Surprisingly, whereas ciprofloxacin acts through topoisomerase IV, the dimers act through gyrase as the primary target in S. pneumoniae.
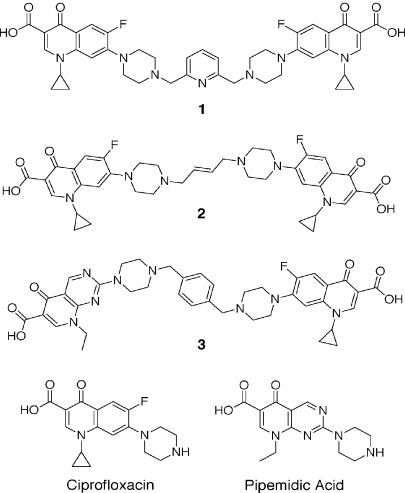
FIG. 1.
Structures of quinolone dimers used in this study.
MATERIALS AND METHODS
Bacterial strains.
Quinolone-susceptible S. pneumoniae strains 7785 and R6 and isogenic 7785 mutants 1C1, 2C6, 2C7, 1S1, 1S4, 2S1, and 2S4 have been described previously (21, 22).
Drugs and drug susceptibilities.
Ciprofloxacin was kindly supplied by Bayer- UK, Newbury, United Kingdom. Pipemidic acid and the three quinolone dimers used in this work were synthesized as described previously (13, 14). MICs were determined by twofold agar dilution on brain heart infusion (BHI) agar plates supplemented with 10% horse blood (21). Approximately 104 bacteria were spotted onto the plates, which were examined following overnight incubation at 37°C.
Selection of S. pneumoniae mutants resistant to dimer 1.
Mutants were obtained by plating approximately 2 × 108 CFU of S. pneumoniae strain 7785 on BHI agar plates containing 10% horse blood and various concentrations of dimer 1. The plates were incubated aerobically at 37°C for 48 h, and colonies were restreaked on plates containing the same level of drug. Mutants were then maintained under drug-free conditions.
PCR and RFLP analysis.
Genomic DNA was isolated from S. pneumoniae strains and used as a template for PCR amplification of the DNA fragments encompassing the QRDRs of the gyrA, gyrB, parC, and parE genes (22). Restriction fragment length polymorphism (RFLP) analysis of the gyrA and parC PCR products was carried out as described previously (22). The PCR products were sequenced by Lark Technologies, Ltd., Saffron Walden, United Kingdom
Analysis of topoisomerase IV genes.
The parE-parC locus of each of the strains 1D1 and 1D2, which were selected for resistance to dimer 1, was amplified and sequenced in its entirety as described previously (24).
Enzymes and DNA substrates.
Recombinant S. pneumoniae GyrA, GyrB, ParC, ParE, S81F GyrA, and S79F ParC proteins were overexpressed and purified to >95% homogeneity as described previously (23, 29). Supercoiled pBR322 was obtained from Helena Biosciences, Tyne and Wear, United Kingdom. Relaxed pBR322 DNA was prepared as described previously by incubating the plasmid with calf thymus topoisomerase I (Invitrogen, Paisley, Scotland) (23). Kinetoplast DNA (kDNA) from Crithidia fasciculata was purchased from Topogen Inc., Columbus, Ohio.
Topoisomerase and DNA cleavage assays.
Enzyme assays were carried out as described previously (23). DNA products were resolved by electrophoresis in 1% agarose, stained with ethidium bromide, photographed, and quantified with an Alpha Innotech digital camera and software. All enzyme assays were done at least twice, with reproducible results.
Dimer-DNA interactions.
The abilities of the quinolone dimers to interact with DNA were tested by a DNA unwinding assay. Relaxed pBR322 (0.8 μg) was incubated with calf thymus topoisomerase I (12 U) in 50 mM Tris-HCl (pH 7.5)-50 mM KCl-10 mM MgCl2-0.5 mM dithiothreitol-30 μg of bovine plasma albumin per ml-0.1 mM EDTA at room temperature for 10 min, prior to the addition of ciprofloxacin or dimer and further incubation at 37°C for 1 h (total reaction volume, 50 μl). The reactions were stopped by addition of sodium dodecyl sulfate (SDS; 0.2% [wt/vol]), and the samples were extracted twice with an equal volume of phenol followed by an equal volume of chloroform. DNA was precipitated with ethanol, dried, and redissolved in TE buffer (20 μl; 10 mM Tris-HCl [pH 8.0], 1 mM EDTA). DNA samples (10 μl) were analyzed by electrophoresis in 1% agarose gels run in Tris-phosphate-EDTA buffer in the absence or presence of chloroquine at 3 μg/ml (27). DNA was visualized by staining with ethidium bromide.
RESULTS
Antipneumococcal activities of quinolone dimers.
We have investigated the antibacterial activities of quinolone dimers (Fig. 1), in which ciprofloxacin monomers were joined through their C-7 piperazinyl groups by 2,6-lutidinyl (dimer 1) or trans-butenyl linkers (dimer 2) or a ciprofloxacin monomer was similarly joined to pipemidic acid by a 1,4-xylenyl linkage (dimer 3) (13, 14). Except for the three unique linkers that allow different geometries, the monomer constituents of the dimers were unaltered. The activities of the dimers were compared with those of ciprofloxacin and pipemidic acid against quinolone-susceptible S. pneumoniae clinical isolate 7785 and its mutants bearing defined parC, gyrA, and parC gyrA quinolone resistance mutations (Table 1). The ciprofloxacin MIC for parental strain 7785 (1 to 2 μg/ml) was in accordance with that found earlier (2). Symmetric dimers 1 and 2 had potencies equivalent to that of ciprofloxacin against strain 7785 (MICs, 2 and 1 μg/ml, respectively). Asymmetric dimer 3 was less active against 7785 (MIC, 4 to 8 μg/ml) than ciprofloxacin but was more potent than pipemidic acid, whose MIC was >32 μg/ml. Similar results were seen for S. pneumoniae R6, the well-known laboratory strain (Table 1). The presence of an efflux mutation in strain 1C1 had little effect on susceptibility to any of the dimers. Single gyrA mutations in strains 1S1 and 1S4 did not significantly affect the ciprofloxacin MICs, whereas a parC mutation in 2C6 and 2C7 elevated the MIC some fourfold, consistent with drug action through topoisomerase IV (19). By contrast, for all three dimers, a parC mutation increased the MICs twofold or less, whereas a gyrA change in 1S1 and 1S4 elevated the MICs by four- to eightfold. Surprisingly, these results suggest that the dimers act through gyrase as the primary target rather than through topoisomerase IV. parC gyrA mutants 2S1 and 2S4 were highly resistant to all three dimers and to ciprofloxacin (MICs, 32 to 64 μg/ml or greater). Thus, high dimer or ciprofloxacin concentrations engage both gyrase and topoisomerase IV in cell growth inhibition.
TABLE 1.
Fluoroquinolone susceptibilities of S. pneumoniae mutants
| Strain | Mutation in QRDR of:
|
MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| GyrA | ParC | CIP | Dimer 1 | Dimer 2 | Dimer 3 | PIP | |
| 7785 | 1-2 | 2 | 1 | 4-8 | >32 | ||
| R6 | 1 | 2 | 1 | 4-8 | >32 | ||
| 1C1 | 4 | 2-4 | 1-2 | 4-8 | >32 | ||
| 2C6 | S79Y | 16 | 4 | 2 | 8-16 | >32 | |
| 2C7 | S79F | 8-16 | 4 | ND | ND | ND | |
| 1S1 | S81F | 2 | 16 | 4 | 16-32 | >32 | |
| 1S4 | S81Y | 2-4 | 8-16 | 4 | 16-32 | >32 | |
| 2S1 | S81F | S79Y | 32-64 | >32 | >32 | >32 | >32 |
| 2S4 | S81F | D83N | 32 | >32 | >32 | >32 | >32 |
CIP, ciprofloxacin; dimers 1, 2, and 3 are shown in Fig. 1; PIP, pipemidic acid. ND, not determined. MICs were determined in two or three independent experiments, with reproducible results. When two values are given, the growth of all but a few colonies was inhibited at the lower drug concentration.
Symmetric dimer 1 selects S. pneumoniae gyrA S81F or S81Y mutants that retain wild-type parE-parC genes.
To ascertain the primary intracellular target, we isolated and characterized mutants of S. pneumoniae 7785 selected by exposure to dimer 1, the compound to which a gyrA mutation conferred the largest (eightfold) degree of resistance. After 48 h of aerobic growth at 37°C, 11, 6, and 0 mutants were recovered when 2 × 108 CFU of strain 7785 was plated on BHI agar containing 10% horse blood and dimer 1 at 2 μg/ml (the MIC), 4 μg/ml (two times the MIC), or 6 μg/ml (three times the MIC), respectively. Colonies were restreaked on fresh plates containing the selecting concentration of dimer 1 before growth on drug-free plates and subsequent analysis. The gyrA, gyrB, parC, and parE QRDRs were amplified from each mutant strain by PCR with genomic DNA as the template (22). RFLP analysis by HinfI digestion was performed to detect mutations affecting codon 81 in gyrA and codon 79 in parC (22). All 17 mutants selected with dimer 1 yielded a 382-bp gyrA QRDR PCR product that did not undergo cleavage with HinfI at an internal site overlapping codon 81. Thus, gyrA codon 81 was mutated in every mutant recovered. By contrast, all the parC QRDR products yielded the wild-type HinfI digestion pattern, consistent with the absence of changes at the Ser-79 ParC hot spot. DNA sequence analysis of the PCR products from three mutants, mutant 1D1 (isolated with 2 μg/ml) and mutants 1D2 and 1D3 (recovered with 4 μg/ml), confirmed the presence of mutations in gyrA leading to alterations of Ser81 to Phe or Tyr (Table 2). These changes were associated with four- to eightfold increases in the MICs of dimer 1. No alterations were found in the parC, gyrB, or parE QRDR sequences.
TABLE 2.
Properties of S. pneumoniae mutants selected for resistance to dimer 1.
| Strain | MIC (μg/ml)
|
Mutation in QRDR ofa:
|
||||
|---|---|---|---|---|---|---|
| CIPb | Dimer 1 | GyrA | ParC | GyrB | ParE | |
| 7785 | 1-2 | 2 | ||||
| 1D1c | 2 | 8-16 | S81F | None | None | None |
| 1D2d | 2 | 8-16 | S81Y | None | None | None |
| 1D3d | 2 | 8-16 | S81Y | None | None | None |
The GyrA mutation resulted from the following nucleotide changes: S81F, TCC to TTC; S81Y, TCC to TAC.
CIP, ciprofloxacin.
Derived from S. pneumoniae strain 7785 by selection with dimer 1 at 2 μg/ml.
Derived from S. pneumoniae strain 7785 by selection with dimer 1 at 4 μg/ml.
To examine the possibility of non-QRDR mutations in topoisomerase IV, the parE-parC gene locus of each of strains 1D1 and 1D2 was amplified as three overlapping PCR products spanning a 5.5-kb region (from 300 bp upstream of the parE start codon to 300 bp downstream of the parC termination signal) and sequenced in full (24). No mutations were found in these genes or their promoter region. It appears that the four- to eightfold increase in the dimer 1 MICs for independently selected strains 1D1 and 1D2 arises from the gyrA mutations encoding S81F or S81Y changes, two well-established quinolone resistance mutations in S. pneumoniae (Table 2). These findings concur with the identical susceptibility data for strains 1S1 and 1S4 (Table 1), which carry the same gyrA changes as strains 1D1 and 1D2 and which are known to express wild-type topoisomerase IV (24). The results show unequivocally that ciprofloxacin dimer 1 preferentially targets gyrase in S. pneumoniae, in contrast to ciprofloxacin itself, which acts through topoisomerase IV as the primary target.
Ciprofloxacin dimers inhibit the catalytic activities of S. pneumoniae gyrase and topoisomerase IV.
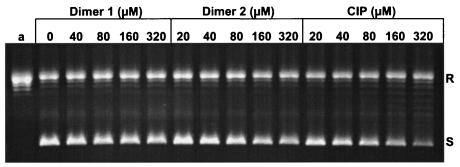
To gain information on how the dimers interact with their topoisomerase targets, we studied the inhibition of ATP-dependent DNA supercoiling by recombinant S. pneumoniae gyrase and of ATP-dependent decatenation of kDNA by topoisomerase IV. Drug potencies were expressed as the drug concentration that resulted in 50% inhibition of enzyme activity (IC50). Figure 2 shows the results of a representative experiment involving inhibition of S. pneumoniae gyrase. Relaxed pBR322 DNA (lane a) was incubated with sufficient gyrase (1 U) to cause 50% conversion of substrate to the supercoiled form in the absence of drug. Inclusion of ciprofloxacin caused a dose-dependent inhibition of supercoiling, with an IC50 of 40 to 80 μM (14.7 to 29.4 μg/ml), in line with that found in previous studies (24). Dimers 1 and 2 also inhibited gyrase activity, although less potently than ciprofloxacin, with IC50s of >320 μM (>245 μg/ml) and 80 to 160 μM (57 to 114 μg/ml), respectively.
FIG. 2.
Ciprofloxacin dimers inhibit DNA supercoiling by S. pneumoniae DNA gyrase. Relaxed pBR322 DNA (0.4 μg) was incubated with S. pneumoniae GyrA (1 U), GyrB (1 U), and 1.4 mM ATP in the absence or presence of dimer 1, dimer 2, or ciprofloxacin (CIP) at the indicated concentrations. The DNA reaction products were analyzed by electrophoresis in a 1% agarose gel. The concentrations of dimers and ciprofloxacin present in the reaction mixtures are indicated above the wells. Lane a, relaxed pBR322. All reaction mixtures contained 5% dimethyl sulfoxide. R and S, relaxed and supercoiled pBR322 DNA, respectively.
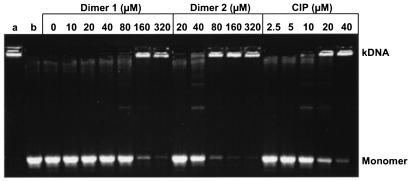
Figure 3 compares the inhibitory activities of the symmetric dimers with ciprofloxacin against topoisomerase IV. Incubation with 1 U of enzyme was just sufficient to resolve 0.4 μg of kDNA (lane a) into its monomeric circles (lane b) under the reaction conditions. Dimers 1 and 2 and ciprofloxacin all caused dose-dependent inhibition of decatenation, with IC50s of ∼120, ∼60, and 10 to 20 μM, respectively (92, 43, and 3.7 to 7.4 μg/ml, respectively). Thus, similar to the results obtained with gyrase, the ciprofloxacin dimers were less efficient inhibitors of topoisomerase IV than ciprofloxacin. Moreover, despite their in vivo action through gyrase as the primary target, the dimers were more potent against purified topoisomerase IV than against purified gyrase.
FIG. 3.
Ciprofloxacin is more potent than its dimers as an inhibitor of DNA decatenation by S. pneumoniae topoisomerase IV. kDNA (0.4 μg) was incubated with recombinant ParC and ParE (1 U each) in the absence or presence of dimers and ciprofloxacin (CIP) at the indicated concentrations. DNA products were separated by agarose gel electrophoresis. Lane a, kDNA; lane b, kDNA plus ParC, ParE, and ATP in the absence of drug and dimethyl sulfoxide. All other reaction mixtures contained 5% dimethyl sulfoxide. Monomer, released minicircles.
Dimers induce DNA cleavage by S. pneumoniae gyrase and topoisomerase IV.
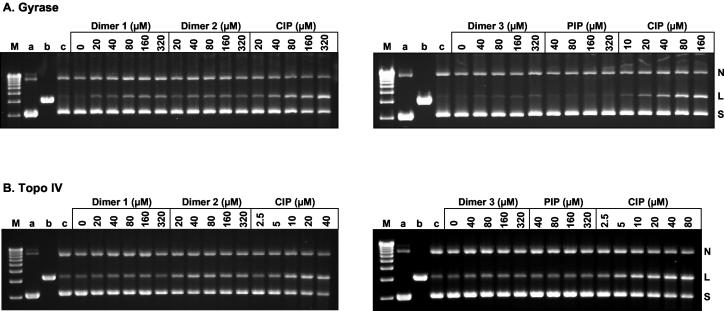
It is thought that the antibacterial activities of quinolones arise from their stabilization of cleavage complexes on DNA (29). To examine the ability of quinolone dimers to mediate cleavage complex formation, supercoiled pBR322 was incubated with either recombinant S. pneumoniae gyrase or topoisomerase IV in the presence of various concentrations of dimer 1, 2, or 3. Parallel assays were also set up in which either ciprofloxacin or pipemidic acid was included for comparison. After SDS treatment to induce DNA breakage and incubation with proteinase K to remove DNA-linked GyrA (or ParC) proteins, the DNA products were separated by agarose gel electrophoresis and quantified. The results of representative cleavage experiments are shown in Fig. 4, in which drug concentrations were chosen to generate comparable extents of DNA breakage.
FIG. 4.
Quinolone-mediated DNA cleavage by S. pneumoniae gyrase (A) and topoisomerase IV (Topo IV) (B). Supercoiled pBR322 (0.4 μg) was incubated with S. pneumoniae GyrA (0.45 μg) and GyrB (1.7 μg) in the absence and presence of dimers 1, 2, and 3, ciprofloxacin (CIP), and pipemidic acid (PIP) at the indicated concentrations. After treatment with SDS and proteinase K, the DNA products were examined by electrophoresis in a 1% agarose gel. Lanes M, linear markers; lanes a and b, supercoiled and linear pBR322 DNA, respectively; lanes c, supercoiled pBR322 plus enzyme in the absence of drug but with 5 mM NaOH added. All the dimer reaction mixtures contained 5% dimethyl sulfoxide, and all ciprofloxacin reaction mixtures contained 5 mM NaOH. N, L, and S, nicked, linear, and supercoiled plasmid pBR322 DNA, respectively.
For gyrase, little or no DNA breakage was seen when drugs were omitted (Fig. 4A, lanes c). Inclusion of ciprofloxacin resulted in a dose-dependent stimulation of DNA cleavage, with the drug concentration that induces 25% linearization of input DNA (CC25) being ∼80 μM (Fig. 4A). This value compares favorably with the CC25 of 40 μM determined previously (29). Symmetric ciprofloxacin dimers 1 and 2 also induced DNA breakage that reached a plateau of 13 to 14% cleavage of input DNA at 80 and 40 μM, respectively. This amount of DNA breakage was induced by ciprofloxacin at 20 to 40 μM (Fig. 4A). Therefore, dimers 1 and 2 were some two- to fourfold less effective than ciprofloxacin in stabilizing a cleavage complex with gyrase. Asymmetric dimer 3 induced very weak DNA breakage by gyrase, amounting to only 8% of the input DNA at 320 μM (Fig. 4A). The poor activity of dimer 3 resembled that of pipemidic acid (Fig. 4A).
In the case of topoisomerase IV (Fig. 4B) and as observed previously (29), the enzyme induced some DNA cleavage even in the absence of added drug. Inclusion of ciprofloxacin gave a dose-dependent stimulation of cleavage, with a CC25 of 5 μM. Dimer 1 caused a weak stimulation of DNA breakage that was maximal (1.5-fold over the background level) at 40 to 160 μM and that was reduced at higher concentrations (Fig. 4B). Dimer 2 showed the same nonlinear response but was more effective than dimer 1, with maximal cleavage (2.5-fold over background) at 40 μM and a CC25 of 20 μM. Thus, dimer 2 was some fourfold less effective than ciprofloxacin. Dimer 3, like pipemidic acid, produced only a marginal stimulation of DNA cleavage (Fig. 4B). It appears that dimers 1 and 2 stimulate more efficient DNA cleavage with both gyrase and topoisomerase IV than dimer 3, in line with their somewhat lower MICs for strain 7785.
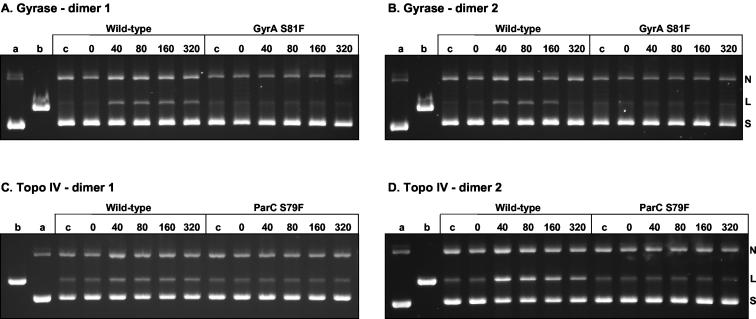
GyrA S81F and ParC S79F mutations block cleavage complex formation by dimers.
The genetic studies described above show that mutant strains expressing GyrA S81F or GyrA S81Y are resistant to all three dimers, dimers 1 to 3 (Tables 1 and 2). By contrast, a single parC (S79F) mutation did not offer much resistance to these compounds (Table 1). To examine the effects of mutations at the enzyme level, S. pneumoniae gyrase and topoisomerase IV were reconstituted with wild-type GyrB and GyrA S81F subunits and wild-type ParE and ParC S79F subunits, respectively, and were tested for their abilities to form cleavage complexes in the presence of the dimers. Figure 5 shows the results of a representative experiment for dimers 1 and 2. As shown above, concentrations of dimer 1 between 40 and 160 μM stimulated DNA breakage by wild-type gyrase and topoisomerase IV (Fig. 5A and C). However, for the mutant enzymes there was no detectable stimulation of DNA breakage by dimer 1 over background levels, even at 320 μM (Fig. 5A and C). Similar results were observed for dimer 2 (Fig. 5B and D). It is clear that both the GyrA S81F and ParC S79F mutations render the respective gyrase and topoisomerase IV complexes resistant to trapping as a cleavage complex. That the dimer resistance phenotype of the parC (S79F) gene is largely silent in the presence of the wild-type gyrA gene (Table 1) confirms that the dimers act preferentially through gyrase in vivo.
FIG. 5.
Comparison of dimer-promoted DNA breakage by wild-type and quinolone-resistant S. pneumoniae gyrase (A and B) and topoisomerase IV (Topo IV) (C and D). Supercoiled plasmid pBR322 (0.4 μg) was incubated with recombinant S. pneumoniae GyrB (1.7 μg) and either wild-type GyrA or GyrA S81F (in each case 0.4 μg) (A and B) or ParE (1.7 μg) and either wild-type ParC or ParC S79F (0.45 μg) (C and D). Samples were processed and analyzed as described in the legend to Fig. 4; and lanes a, b, and c are as described in the legend to Fig. 4.
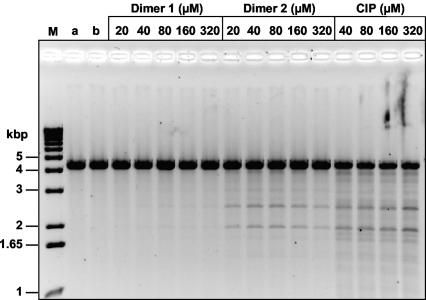
Ciprofloxacin dimers mediate DNA breakage at the same sites as ciprofloxacin.
To determine whether the dimers induced cleavage at the same spectrum of sites as ciprofloxacin, pBR322 DNA was linearized by digestion with EcoRI and used as a substrate in cleavage assays with gyrase and topoisomerase IV. In contrast to the results obtained with the supercoiled substrate (Fig. 4), dimers 1 and 2 induced very little cleavage of linear DNA by gyrase (data not shown). In the case of topoisomerase IV, cleavage of linear DNA mediated by dimers 1 and 2 occurred at the same spectrum of sites as induced by ciprofloxacin (Fig. 6). Interestingly, the cleavage of linear DNA promoted by dimer 1 was relatively weak and was much weaker than that mediated by dimer 2.
FIG. 6.
Site-specific DNA cleavage by S. pneumoniae topoisomerase IV mediated by ciprofloxacin and its dimers. Plasmid pBR322 DNA linearized at its EcoRI site (0.4 μg) was incubated with ParC (0.45 μg) and GyrB (1.7 μg) in the absence or presence of dimer 1 or 2 or ciprofloxacin (CIP) at the indicated concentrations. After addition of SDS and proteinase K treatment, the DNA samples were analyzed by electrophoresis in a 1% agarose gel. Lanes M, a, and b, DNA size markers, linear pBR322, and linear pBR322 incubated with ParC and ParE in the absence of drug, respectively.
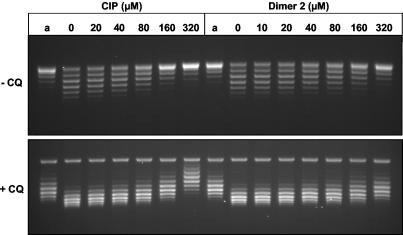
Dimer 2 interacts with DNA to cause helix unwinding.
We noted that dimer 2 showed a nonlinear response in DNA cleavage assays: DNA breakage increased to a maximum and then progressively declined at higher drug levels (Fig. 4 to 6). This behavior could arise from progressive conformational effects on the DNA substrate caused by DNA intercalation at high drug levels. Quinolones are known to unwind duplex DNA in a magnesium-dependent manner (18, 27). To test whether dimer 2 could be acting as a powerful intercalator, we incubated relaxed pBR322 DNA with calf thymus topoisomerase I (to remove the positive turns arising from DNA unwinding) in the presence of various concentrations of ciprofloxacin or dimer 2. The reactions were stopped, the drugs were removed by phenol extraction, and the DNA was analyzed by agarose gel electrophoresis (Fig. 7) in the absence or presence of chloroquine. Under the conditions used for the gel shown in Fig. 7, all reaction products ran as positively supercoiled species, as confirmed by their greater migration in the presence of chloroquine, which introduces additional positive supercoils into DNA (Fig. 7, +CQ [in the presence of chloroquine]). It can be seen that DNA relaxed in the presence of ciprofloxacin or dimer 2 is underwound compared to the DNA of the drug-free control (see Fig. 7 and the legend to Fig. 7). The linking difference caused by 320 μM ciprofloxacin was modest at about −5, with 320 μM dimer 2 inducing about half that degree of unwinding. It appears that dimer 2 interacts with DNA but is less effective than ciprofloxacin as an unwinding agent. This is consistent with the results of spectroscopic investigations with structurally similar C-7-linked ciprofloxacin dimers, in which no significant hyperbathochromic or hypobathochromic shifts were observed with various concentrations of calf thymus DNA, indicating no significant intercalation of the dimers tested compared with that for ciprofloxacin (data not shown). Reduced cleavage complex formation at high dimer levels could arise from additional DNA or enzyme binding modes.
FIG. 7.
Ciprofloxacin and its symmetric dimer (dimer 2) induce unwinding of closed circular DNA. Relaxed plasmid pBR322 (0.8 μg) was incubated with calf thymus DNA topoisomerase I (12 U) in the presence of 10 mM MgCl2 at room temperature for 10 min before addition of ciprofloxacin (CIP) and dimer 2 at the indicated concentrations. After incubation for 1 h at 37°C, the topoisomerase I was inactivated and the DNA was extracted with phenol to remove any bound ligand prior to agarose gel electrophoresis in Tris-phosphate-EDTA buffer. Half of each DNA sample was run in the absence of chloroquine (−CQ); the other half was electrophoresed in the presence of chloroquine (+CQ). Under the conditions of the gel without chloroquine, all the DNA samples are positively supercoiled and are made more positively coiled by the inclusion of chloroquine. Lanes a, pBR322 DNA (previously relaxed in the absence of Mg2+ ions), used as the substrate.
DISCUSSION
To explore the potential of tethered quinolones as antipneumococcal agents, we have determined the activities of novel C-7 piperazinyl-linked quinolone dimers against S. pneumoniae and against its topoisomerase targets in vitro. Symmetric ciprofloxacin dimers 1 and 2 (which use 2,6-lutidinyl and trans-butenyl linkers, respectively) were about as active as ciprofloxacin (weight for weight) against wild-type S. pneumoniae, whereas an asymmetric dimer of ciprofloxacin and pipemidic acid (dimer 3) was four- to eightfold less potent (Table 1). Unexpectedly, we found that the dimers act through gyrase in S. pneumoniae, unlike the ciprofloxacin monomer, which preferentially targets topoisomerase IV. This conclusion is based on the responses of defined gyrA or parC mutants to the dimers and the selection by dimer 1 of gyrA mutants bearing demonstrably wild-type parE-parC genes. In combination with the enzymatic characterization of dimer interactions with S. pneumoniae gyrase and topoisomerase IV, these results open new aspects of structure-function involving monomeric and tethered quinolones.
It is recognized that drug dimers have the potential to bind to and bridge two quinolone binding sites on gyrase or topoisomerase IV, so that a single dimer molecule would overcome the unfavorable entropy associated with the binding of two separate quinolone monomers. Synergistic binding by each half of the dimer could be extremely tight, facilitating the stabilization of a cleavable complex and thereby producing a potent antipneumococcal drug. Despite these aims, the genetic and enzyme inhibition data for S. pneumoniae suggest that it is unlikely that any of dimers 1 to 3 act by bridging quinolone sites on the target. First, none of the dimers was more effective than ciprofloxacin as a growth inhibitor of S. pneumoniae: the effectiveness of dimers 1 and 2 was at best only comparable to that of ciprofloxacin (Table 1). Second, on a molar basis, ciprofloxacin was some two- to sixfold better as a catalytic inhibitor of gyrase and topoisomerase IV than any of the dimers (Fig. 2 and 3) and was also severalfold more effective in stabilizing the cleavable complex with either enzyme (Fig. 4). Bridging of quinolone pockets is expected to yield much tighter enzyme binding. In short, the marked improvement in the activities of the dimers over that of ciprofloxacin that would be anticipated from bridging of quinolone binding pockets was not observed. Third, we note that the activities of dimers 1 and 2 were not greatly influenced by the different geometries imposed by different linkers. These various considerations argue against bridging of quinolone binding pockets. Instead, the data are consistent with a monomeric binding mode in which one half of the dimer contacts DNA and the other monomeric component is accommodated as a bulky C-7 group, perhaps interacting with the enzyme. Interestingly, in a previous study of quinolone dimers (aimed at testing aspects of the Shen model) (25), dimers of norfloxacin were made by linkage of monomers through their N-1 groups by using methylene spacers of various chain lengths (—CH2)n—, where n = 3 to 5). These dimers were equally potent (n = 4) or less potent (n = 3 or 5) than norfloxacin in inhibiting the DNA supercoiling activity of Escherichia coli DNA gyrase. It appears that the optimal linkage of monomers that permits bridging of quinolone sites in gyrase and topoisomerase IV remains to be discovered.
The most surprising outcome of our study was that dimers 1 to 3 act through gyrase as the primary target in S. pneumoniae, whereas monomeric ciprofloxacin targets topoisomerase IV. This situation is reminiscent of that in earlier work showing that addition of various benzenesulfonylamido groups to the C-7 piperazinyl group of ciprofloxacin also switches its intracellular target to gyrase (2). It may be significant that all the dimers tested here involve linkage of monomers through their C-7 piperazinyl groups. In the context of monomeric binding, dimers could be viewed as being ciprofloxacin but carrying a very bulky C-7 side chain, which may favor the targeting of gyrase. Some support for this idea is suggested by a comparison of cleavage complex formation by ciprofloxacin and dimer 1 (Fig. 4). In the case of ciprofloxacin, topoisomerase IV is trapped as a cleavage complex about 10 times more efficiently than gyrase. By contrast, dimer 1 traps gyrase more efficiently than topoisomerase IV (Fig. 4A, B). For dimer 2, the enzymes are trapped comparably. If this situation is reproduced in S. pneumoniae, the relatively more potent trapping of gyrase by dimers 1 and 2 might account for the switching of the intracellular target. Whatever the explanation, dimer 1 can be added to the growing list of quinolones, including sparfloxacin and NSFQ-105, for which genetic and DNA sequence analysis show unequivocally that gyrase is the target in vivo (2, 21, 24). Observations with the dimers again highlight the key role of the C-7 group in determining target preference.
Despite their different intracellular targets, ciprofloxacin dimers were about as active as ciprofloxacin against S. pneumoniae (Table 1). Presumably, some other cellular factor such as impaired efflux compensates for the much weaker target interactions measured for the dimers in vitro (Fig. 2 to 6). The dimer MICs for the putative efflux mutant 1C1 could be consistent with this idea (Table 1). Interestingly, unlike their relatively modest antipneumococcal activities, dimers 1 to 3 have been shown to exhibit much greater potencies against S. aureus. In fact, the dimers were each at least 4-fold more active than ciprofloxacin against wild-type S. aureus and, moreover, remained fully active against isogenic norA and norA/parC mutants that were 8- and 64-fold more resistant to ciprofloxacin than the wild type, respectively (13, 14). Consistent with other work, a bulky C-7 side chain may account for the lack of effective dimer efflux through the norA pump (26). Similarly, the susceptibility of the S. aureus parC mutant could be consistent with dimer targeting of gyrase rather than topoisomerase IV (5, 6), as we show here for S. pneumoniae. Further work will be needed on these aspects.
Finally, it is interesting to reflect on the similarities and differences between ciprofloxacin and its dimers occasioned by linkage through the C-7 piperazinyl group. In common with ciprofloxacin, the dimers stimulate cleavage complex formation (Fig. 4) and do so at the same DNA sites (Fig. 6), and their effects on cleavage are resisted by GyrA S81F and ParC S79F mutations (Fig. 5). As might be expected, these features indicate that monomers and dimers share binding features in common. However, linkage through C-7 switches intracellular specificity, weakens target binding in vitro, and alters Mg2+-dependent dimer interactions with DNA, as judged by DNA unwinding (Fig. 7), a characteristic that could bear on drug mechanism. To understand these effects, more information will be needed on quinolone binding, especially the role of the C-7 group. Clearly, rational design of dimeric quinolones would be aided by detailed knowledge of the number and disposition of drug binding sites in the topoisomerase target. Nonetheless, our initial results for S. pneumoniae and those for S. aureus establish that tethered quinolones can display altered mechanisms of action and improved activities against drug-resistant strains.
Acknowledgments
K.A.G. was supported in part by Pfizer, Ltd. X.-S.P. was funded by project grant 117/C16747 from the Biotechnology and Biological Sciences Research Council of the United Kingdom
REFERENCES
- 1.Adams, D. E., E. M. Shekman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning DNA replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Alovero, F. L., X.-S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen, M. E., A. W. Wyke, R. Kuroda, and L. M. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrero, L., B. Cameron, B. Manse, D. Lagneux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 6.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher, L. M., K. A. Gould, X.-S. Pan, S. Patel, and V. J. Heaton. 2003. Analysis of dual active fluoroquinolones in Streptococcus pneumoniae. J. Antimicrob. Chemother. 52:312-313. [DOI] [PubMed] [Google Scholar]
- 8.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J.-I. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiasa, H., D. O. Yousef, and K. J. Marians. 1996. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 271:26424-26429. [DOI] [PubMed] [Google Scholar]
- 11.Jones, R. N., and L. A. Mandell. 2002. Fluoroquinolones for the treatment of outpatient community-acquired pneumonia. Diagn. Microbiol. Infect. Dis. 44:69-76. [DOI] [PubMed] [Google Scholar]
- 12.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Higara, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 13.Kerns, R. J., M. R. Rybak, G. W. Kaatz, F. Vaka, R. Cha, R. G. Gruz, and V. U. Diwadka. 2003. Structural features of piperazinyl-linked ciprofloxacin dimers required for activity against drug-resistant strains of Staphylococcus aureus. Bioorg. Med. Chem. Lett. 13:2109-2112. [DOI] [PubMed] [Google Scholar]
- 14.Kerns, R. J., M. R. Rybak, G. W. Kaatz, F. Vaka, R. Cha, R. G. Gruz, V. U. Diwadkar, and T. D. Ward. 2003. Piperazinyl-linked fluoroquinolone dimers possessing potent antibacterial activity against drug-resistant strains of Staphylococcus aureus. Bioorg. Med. Chem. Lett. 13:1745-1749. [DOI] [PubMed] [Google Scholar]
- 15.Mizuuchi, K., L. M. Fisher, M. H. O'Dea, and M. Gellert. 1980. DNA gyrase action involves the introduction of transient double strand breaks into DNA. Proc. Natl. Acad. Sci. USA 77:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morais-Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 17.Munoz, R., and A. G. de la Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of quinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palu, G., S. Valisena, G. Ciarrochi, B. Gatto, and M. Palumbo. 1992. Quinolone binding to DNA is mediated by magnesium ions. Proc. Natl. Acad. Sci. USA 89:9671-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan, X.-S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan, X.-S., and L. M. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan, X.-S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan, X.-S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, X.-S., and L. M. Fisher. 1999. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob. Agents Chemother. 43:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan, X.-S., G. Yague, and L. M. Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, L. L., L. A. Mitscher, P. N. Sharma, T. J. O'Donnell, W. T. Chu, C. S. Cooper, T. Rosen, and A. G. Pernet. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a co-operative drug-DNA binding model. Biochemistry 28:3886-3894. [DOI] [PubMed] [Google Scholar]
- 26.Takenouchi, T., F. Tabata, Y. Iwata, H. Hanzawa, M. Sugawara, and S. Ohya. 1996. Hydrophilicity of quinolones is not an exclusive factor for decreased activity in efflux-mediated resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tornaletti, S., and A. M. Pedrini. 1988. Studies on the interaction of 4-quinolones with DNA by DNA unwinding experiments. Biochim. Biophys. Acta 949:279-287. [DOI] [PubMed] [Google Scholar]
- 28.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 29.Yague, G., J. E. Morris, X.-S. Pan, K. A. Gould, and L. M. Fisher. 2002. Cleavable complex formation by wild-type and quinolone-resistant Streptococcus pneumoniae type II topoisomerases mediated by gemifloxacin and other fluoroquinolones. Antimicrob. Agents Chemother. 46:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zechiedrich, E. L., and N. R. Cozzarelli. 1995. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication of Escherichia coli. Genes Dev. 9:2859-2869. [DOI] [PubMed] [Google Scholar]