Bordetella Bacteriophage (BPP-1) infects both Bordetella bronchiseptica, and B. pertussis, the bacterium that causes whooping cough. BPP-1 is a short-tailed dsDNA virus and a member in the Podoviridae family. It has a T=7 icosahedral head with a diameter of ~700 Å and a tail on one of the twelve icosahedral vertices [1]. The capsid head is composed of two proteins: a major capsid protein (MCP) and a cement protein, and neither has recognizable sequence similarity with proteins in other well studied dsDNA bacteriophages like HK97 and PRD1 [2, 3].
We have determined the structure of the icosahedral head of BPP-1 to 3.4Å resolution using single particle cryo-electron microscopy (cryoEM) (Fig. 1a) and derived an atomic model of BPP-1. Briefly, cryoEM images were recorded on Kodak SO-163 micrographs at 59K magnification in an FEI Titan Krios microscope at 300kV with a dosage of ~25 e−2/Å on the specimen. ~43,000 particles from 340 micrographs were selected for image processing and three dimensional (3D) reconstruction (Fig. 1b, c). Local averaging of the 7 copies of the cement or MCP protein in the asymmetric unit was performed to further improve resolution, model building was performed using Coot, and atomic models were refined using CCP4 (Fig. 1c–e) [4].
Fig. 1. Structure of BPP-1 at 3.4Å resolution determined by cryoEM.
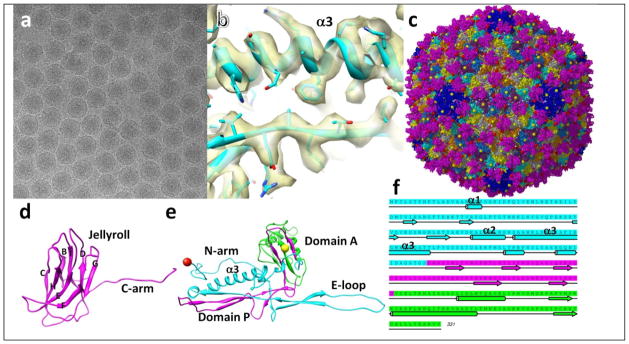
(a) cryoEM image of BPP. (b) Density of a portion of MCP protein. (c) atomic model of BPP capsid. Balls indicate Thr6 (red) and Val330 (yellow) exposed on the external surface of the BPP capsid. (d) ribbon model of the cement protein. (e) ribbon model of MCP. The three building blocks were indicated by cyan (N-terminal), purple (two strands in domain P, and two strands in domain A), and green (major part of domain A). (f) amino acid sequence and secondary structure of MCP, with segments colored according to structures of the building blocks in (e).
The BPP-1 head reconstruction has an angular shape with 420 MCP subunits making up the capsid shell and 210 dimers of the cement protein joining neighboring MCPs, together forming chainmail (Fig. 1c). Side chains of amino acids are clearly resolved in both proteins and were used for atomic model building. All but the last amino acid of the 140 amino acid residues of the cement protein were visible in the cryoEM density. The atomic model of the cement protein shows a jellyroll fold and a ~40Å C-terminal arm (Fig. 1d). The interactions between the two monomers in the cement protein dimer are limited to the F-strand of the jellyroll (Fig. 1d), primarily through a pair of Pro residues, one on each monomer. In contrast, each cement protein monomer interacts with five underlying MCP molecules, mediated through loops of the jellyroll and through a β augmentation of the C-terminal arm (Fig. 1c). This use of the jellyroll fold for an auxiliary stabilizing role is quite remarkable because such fold has only been observed as a building block for viral capsids.
Of the 331 residues of the BPP-1 MCP, we traced Thr6 to Val330 in our density map. The N and C termini are both located on the surface, but the first 5 and the last one amino acids are not visible, suggesting their flexibility (Fig. 1e). The BPP-1 MCP has the fold/architecture of the capsid protein of HK97 bacteriophage. We hereby divide it into three building blocks from N to C termini: chainmail-forming (cyan), β sheet-forming (purple) and capsomer-forming (green) (Fig. 1e) [2]. Surprisingly, the topologies of folding these three building blocks into this architecture are different between BPP-1 and HK97 (Fig. 1e, f): The last two of the three building blocks in gp5 of HK97 are swapped as compared to the sequence in BPP-1, in a rare, non-circular permutation, as found in DNA methyltransferases [5]. The difference in the topologies in folding suggests convergent evolution in selecting the characteristic HK97-like fold to build chainmail in the two viruses.
It has been found that the internal pressure inside bacteriophages is very high due to the high packing density of the viral genome inside the viral capsid [6], and that different bacteriophages adopt distinctive strategies to stabilize their capsids to withstand the high pressure. HK97 uses covalent isopeptide bonds to link adjacent gp5 proteins to stabilize its capsid [2]. Having the same building blocks but lacking isopeptide bonds to join the chainmail-forming building blocks, BPP-1 uses an extra cement protein with the jellyroll fold to complete the chainmail (Fig. 1c). Our structure highlights the diverse strategies to build a highly stable dsDNA virus and suggests different maturation processes between the two bacteriophages.
Acknowledgments
Supported in part by grants from the National Institutes of Health (GM071940 and AI069015 to ZHZ).
References
- 1.Dai W, et al. Proc Natl Acad Sci U S A. 2010;107:4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wikoff WR, et al. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 3.Abrescia NG, et al. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki JM. BMC Evol Biol. 2002;2:3. doi: 10.1186/1471-2148-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith DE, et al. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]


