Background: The pneumococcal endo-β-N-acetylglucosaminidase LytB is required for cell division and colonization.
Results: Structural analysis revealed that the catalytic domain of LytB consists of three structurally distinct modules.
Conclusion: All three modules of LytB are necessary for its optimal activity toward peptidoglycan hydrolysis and for pneumococcal adhesion to respiratory epithelial cells.
Significance: Provided is the structural insight into LytB-mediated pneumococcal cell wall remodeling and pathogenesis.
Keywords: Bacterial Pathogenesis, Enzyme Structure, Peptidoglycan, Streptococcus, Structural Biology, Cell Wall Remodeling
Abstract
Streptococcus pneumoniae causes a series of devastating infections in humans. Previous studies have shown that the endo-β-N-acetylglucosaminidase LytB is critical for pneumococcal cell division and nasal colonization, but the biochemical mechanism of LytB action remains unknown. Here we report the 1.65 Å crystal structure of the catalytic domain (residues Lys-375–Asp-658) of LytB (termed LytBCAT), excluding the choline binding domain. LytBCAT consists of three structurally independent modules: SH3b, WW, and GH73. These modules form a “T-shaped” pocket that accommodates a putative tetrasaccharide-pentapeptide substrate of peptidoglycan. Structural comparison and simulation revealed that the GH73 module of LytB harbors the active site, including the catalytic residue Glu-564. In vitro assays of hydrolytic activity indicated that LytB prefers the peptidoglycan from the lytB-deficient pneumococci, suggesting the existence of a specific substrate of LytB in the immature peptidoglycan. Combined with in vitro cell-dispersing and in vivo cell separation assays, we demonstrated that all three modules are necessary for the optimal activity of LytB. Further functional analysis showed that the full catalytic activity of LytB is required for pneumococcal adhesion to and invasion into human lung epithelial cells. Structure-based alignment indicated that the unique modular organization of LytB is highly conserved in its orthologs from Streptococcus mitis group and Gemella species. These findings provided structural insights into the pneumococcal cell wall remodeling and novel hints for the rational design of therapeutic agents against pneumococcal growth and thereby the related diseases.
Introduction
The cell wall, a multi-molecular coat, is essential for bacterial survival and growth. The cell walls of Gram-negative and Gram-positive bacteria have considerable structural and functional differences. Although the Gram-negative cell walls are relatively thin (7–8 nm) and covered by the outer membrane, those of Gram-positive bacteria are much thicker (20–80 nanometers) and exposed to the environment unless they are covered by other accessory structures (e.g. capsules or slime layers) (1). The bacterial cell wall mostly consists of repeating peptidoglycan (PGN,5 also known as murein) structures. PGN is composed of alternating residues of β(1,4)-linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), and cross-linking peptide chains of three to five amino acid residues (2). Delicate “destruction” or remodeling of the cell wall is crucial for bacterial cell growth and division (3). There are multiple hydrolases capable of cleaving the covalent bonds in PGN. According to the target chemical bonds, these PGN hydrolases are grouped into five classes: amidases, glucosaminidases, muramidases (lysozymes), endopeptidases and transglycosylases (4).
The Gram-positive bacterium Streptococcus pneumoniae (pneumococcus) causes multiple forms of infections in humans such as community pneumonia, otitis media, and meningitis (5). More than 90 pneumococcal serotypes have been identified based on the diversity of capsular polysaccharides, which are used as vaccines to limit the impact of pneumococcal disease. However, S. pneumoniae remains a major bacterial pathogen worldwide due to the high cost and incomplete serotype coverage of the current vaccines. Thus, there is a need for developing protein-based vaccines that can cover all capsular serotypes of pneumococci (6). Choline-binding proteins are among the most extensively studied surface proteins in terms of their contributions to pneumococcal pathogenesis and immuno-protection potentials for improving the existing pneumococcal vaccines. Among these are the choline-binding proteins that possess PGN hydrolase activities, including LytA (7), LytB (8), LytC (9), and CbpD (10). All the four proteins have a choline binding domain (CBD) beyond the catalytic domain. The CBDs of LytA and CbpD locate at the C termini, whereas those of LytB and LytC are at the N termini (11). LytA, an N-acetylmuramoyl-l-alanine amidase, is the first characterized autolysin in S. pneumoniae (7). It is required for pneumococcal autolysis in both stationary phase and the penicillin-induced conditions (12) or daughter cell separation (13). However, deletion of lytA does not alter the bacterial growth rate but results in pneumococci with short chains (6–8 cells) (13). A structural study of the LytA CBD suggests that active LytA adopts a dimer (14). Intranasal immunization with a recombinant LytA conferred significant protection against experimental infection of five different pneumococcal serotypes (15). LytC is a lysozyme toward the β(1,4)-glycosidic bond between NAM and NAG (9). The crystal structure of LytC shows that its hydrolase domain is oriented toward the CBD to form an unusual hook-shaped conformation that restricts LytC to hydrolyze only the non-cross-linked PGN (16). CbpD is a murein hydrolase that is required for killing of noncompetent pneumococci by the competent counterparts, a phenomenon called fratricide (10). Both LytA and LytC are also involved in fratricide due to their role of lysis of non-competent sister pneumococcal cells (17).
LytB was originally identified as a PGN hydrolase as the ΔlytB pneumococci were deficient in cell separation and formed long chains (∼100 cells) (8); however, the ΔlytB mutants displayed normal autolysis and genetic transformation (18). A subsequent study demonstrated that the purified recombinant LytB is able to disperse the long chains of ΔlytB pneumococci, suggesting an endo-β-N-acetylglucosaminidase activity that cleaves the β(1,4)-glycosidic bond between NAG and NAM (18). Both LytB and LytC are necessary for pneumococcal colonization at the nasopharynx of mice (19, 20). Recently, Atilano et al. (21) reported that the glucosaminidase domain of staphylococcal major autolysin Atl, which corresponds to the pneumococcal LytB, can conceal the bacteria from being detected by innate immune system of Drosophila. Altogether, it suggested that LytB takes an important role in cell separation and pathogenicity. However, the biochemical mechanism of LytB as a PGN hydrolase remains undefined. Here we report the 1.65 Å crystal structure of the catalytic domain (residue Lys-375–Asp-658) of LytB. Structural analysis revealed that the catalytic domain of LytB consists of three structurally independent modules: SH3b, WW domain-like, and the glycoside hydrolase family 73 (GH73). All of the three modules were proved to be necessary for the optimal activity of LytB by mutagenesis and enzymatic tests. Glu-564 in the GH73 module was further identified as the key residue for catalytic activity of LytB in the PGN hydrolysis, in vitro cell dispersing, and in vivo cell separation. Finally, we demonstrated that the full-length LytB is required for pneumococcal adhesion to and invasion into human lung epithelial cells.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Chemical Reagents
All S. pneumoniae strains used in this study are listed in Table 1. The strain TIGR4 (serotype 4) (22) was used as the parental strain. Pneumococcal strains were cultured in Todd-Hewitt broth (Oxoid Ltd., London, UK) containing 0.5% yeast extract (termed THY) or on tryptic soy agar (Difco) plates containing 5% (v/v) sheep blood. When necessary, kanamycin (200 μg/ml) or streptomycin (150 μg/ml) was included in the broth and agar media for selection purposes. Pneumococci were cultured at 37 °C with 5% CO2. Unless indicated otherwise, all bacterial culture media and chemicals used in this work were purchased from Sigma.
TABLE 1.
Bacterial strains and primers used in this study
Kanr, kanamycin resistant; Kans, kanamycin sensitive; Strepr, streptomycin resistant; Streps, streptomycin-sensitive.
| Strains or primers | Description | Source or reference |
|---|---|---|
| Strains | ||
| TIGR4 | S. pneumoniae clinical isolate, serotype 4, encapsulated; rpsL+, lytB+; Streps, Kans | (22) |
| CP1200 | Rx derivative; hex mal, rpsL1; Hex+, Kans, RecA+ | (38) |
| CP1296 | CP1200 derivative; bgl-1, cbp3::kan-rpsL+; Streps, Kanr | (38) |
| ST588 | ST594 derivative; rpsL1, cbpA null, ΔcbpA::kan-rpsL+, the entire coding sequence of the cbpA gene was replaced by transformation with the Janus (kan-rpsL+) amplified from CP1296 chromosomal DNA using primers Pr311/Pr312; ΔCbpA1–701, Streps, Kanr | (22) |
| ST001 | TIGR4 derivative; ΔlytB::Janus cassette; Streps, Kanr | This study |
| ST002 | TIGR4 derivative; rpsL1 (by transformation with the rpsL1 allele, which was amplified by PCR from CP1200 chromosomal DNA using primers Pr387/Pr388 (Ref. 22) and gel purified), lytB+; Strepr, Kans | This study |
| ST003 | ST002 derivative; rpsL1, lytB null, ΔlytB::Janus cassette; the entire coding sequence of the lytB gene was replaced by transformation with the Janus (kan-rpsL+) amplified from CP1296 chromosomal DNA using primers Pr1097/Pr1098 (22); Streps, Kanr | This study |
| ST004 | ST003 derivative; rpsL1, lytB; LytBΔSH3b-WW, the coding region correspond to Lys-375--Asp-493 of LytB (the SH3b and WW modules) were deleted by allelic exchange in strain ST003; Strepr, Kans | This study |
| ST005 | ST003 derivative; rpsL1, lytB; LytBΔSH3b, the coding region correspond to Lys375-Lys451 of LytB (the SH3b module) were deleted by allelic exchange in strain ST003; Strepr, Kans | This study |
| ST007 | ST003 derivative; rpsL1, lytB; the entire coding region of lytB gene was amplified from the wild-type TIGR4 for complementation by allelic exchange in strain ST003; Strepr, Kans | This study |
| ST008 | ST003 derivative; rpsL1, lytBE564Q; the entire coding sequence of the lytB gene with E564Q mutant was amplified from the mutant plasmid for transformation by allelic exchange in strain ST003; Strepr, Kans | This study |
| Primers | ||
| PR0013 | 5′-AGGTTCTATTATGATCAAGGCGAC-3′ | This study |
| PR0014 | 5′-GAGATCTAGATGCACCCTCTGGACTAGCTAAG-3′ | This study |
| PR0015 | 5′-GCTCTCGAGACAAGGCTTCTGGTATGAATGTG-3′ | This study |
| PR0016 | 5′-GACACCTACTATAAGGATAAGGCTGAG-3′ | This study |
| PR0018 | 5′-TCAAAATGCAGGCCATCTTTACCATCGCTTCCAA-3′ | This study |
| PR0019 | 5′-TTGGAAGCGATGGTAAAGATGGCCTGCATTTTGA-3′ | This study |
| PR0049 | 5′-TAATAAGGGATAAAGTCACCATCGCTTCCAAGCT-3′ | This study |
| PR0050 | 5′-AGCTTGGAAGCGATGGTGACTTTATCCCTTATTA-3′ | This study |
| PR0054 | 5′-TCTTCTACCCATTCTGATTTGGCCA-3′ | This study |
| PR0055 | 5′-TGGCCAAATCAGAATGGGTAGAAGA-3′ | This study |
| PR0056 | 5′-CTAATCTTTGCCACCTAGCTTCTCA-3′ | This study |
| PR0057 | 5′-TGAGAAGCTAGGTGGCAAAGATTAG-3′ | This study |
Protein Expression and Purification
The coding region of the full-length LytB and its truncated versions were amplified from the genomic DNA of wild-type S. pneumoniae TIGR4 or the deletion strain ST004 (Table 1). All of them were cloned into a pET28a-derived expression vector with an N-terminal His6 tag. The recombination plasmids were transformed into Escherichia coli strain BL21 (DE3) (Novagen) growing at 37 °C in LB culture medium (10 g of NaCl, 10 g of Bacto-Tryptone, and 5 g of yeast extract/liter) containing 30 μg/ml kanamycin until the A600 nm reached 0.6. Expression of the recombinant proteins was then induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for another 20 h at 16 °C before harvesting. Cells were collected and resuspended in 40 ml of lysis buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl). After 30 min of sonication and centrifugation at 12,000 × g for 30 min, the supernatant containing the soluble target protein was collected and loaded onto a nickel-nitrilotriacetic (GE Healthcare) equilibrated with the binding buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl). The target protein was eluted with 300 mm imidazole and further loaded onto a Superdex 75 column (GE Healthcare) pre-equilibrated with 20 mm Tris-Cl, pH 8.0, 100 mm NaCl. Fractions containing the target protein were combined and concentrated to 10 mg/ml for crystallization.
The selenium-methionine (Se-Met)-labeled LytBCAT protein was expressed in E. coli strain B834 (DE3) (Novagen). Transformed cells were grown at 37 °C in Se-Met medium (M9 medium with 25 μg/ml Se-Met and the other essential amino acids at 50 μg/ml) containing 30 μg/ml kanamycin until the A600 nm reached 0.6 and were then induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for 4 h at 37 °C. Se-Met substituted His6-LytBCAT was purified in the same manner as native His6-LytBCAT.
The LytB site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the plasmid encoding the full-length LytB without the signal peptide as the template. The mutant protein was expressed, purified, and stored in the same manner as the wild-type protein.
Crystallization, Data Collection, and Processing
Both native and Se-Met-substituted LytBCAT were concentrated to 10 mg/ml by ultrafiltration (Millipore Amicon) for crystallization. Crystals were grown at 289 K using the sitting drop vapor diffusion method, with the initial condition of mixing 1 μl of protein solution with an equal volume of the reservoir solution (15% polyethylene glycol 6000, 0.1 m sodium cacodylate, pH 6.0). The crystals were transferred to cryoprotectant (reservoir solution supplemented with 30% glycerol) and flash-cooled with liquid nitrogen. Both native and Se-Met derivative data for a single crystal were collected at 100 K in a liquid nitrogen stream using beamline 17U with a Quantum 315r CCD detector (Area Detector Systems Corp., Poway, CA) at the Shanghai Synchrotron Radiation Facility. All diffraction data were integrated and scaled with the program HKL2000 (23).
Structure Determination and Refinement
The crystal structure of LytBCAT was determined using the single-wavelength anomalous dispersion phasing (SAD) (24) method from a single Se-Met-substituted protein crystal. The Autosol program from PHENIX (25) was used to locate the heavy atoms, and the phase was calculated and further improved with the program SOLVE/RESOLVE (26, 27). Automatic model building was carried out using Autobuild in PHENIX. The initial model was refined using the maximum likelihood method implemented in REFMAC5 (28) as part of the CCP4i (29) program suite and rebuilt interactively using the program COOT (30). The final model was evaluated with the programs MOLPROBITY (31) and PROCHECK (32). Crystallographic parameters were listed in Table 2. All structure figures were prepared with PyMOL.
TABLE 2.
Crystal parameters, data collection, and structure refinement
| Se-Met substituted LytBCAT | |
|---|---|
| Data collection | |
| Space group | C2 |
| Unit cell | |
| a, b, c (Å) | 123.83, 44.59, 52.22 |
| α, β, γ (°) | 90.00, 94.52, 90.00 |
| Resolution range (Å) | 50.00-1.65 |
| Unique reflections | 33,952 (3,070)a |
| Completeness (%) | 98.4 (89.7) |
| 〈I/σ(I)〉 | 14.9 (2.2) |
| Rmergeb (%) | 9.4 (65.6) |
| Average redundancy | 5.3 (4.2) |
| Structure refinement | |
| Resolution range (Å) | 33.10-1.65 |
| R-factorc/R-freed (%) | 18.0/21.7 |
| Number of protein atoms | 2,095 |
| Number of water atoms | 173 |
| r.m.s.d.e bond lengths (Å) | 0.008 |
| r.m.s.d. bond angles (°) | 1.196 |
| Mean B factors (Å2) | 37.7 |
| Ramachandran plotf (residues, %) | |
| Most favored (%) | 95.8 |
| Additional allowed (%) | 4.2 |
| Outliers (%) | 0 |
| PDB entry | 4q2w |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation, and 〈I(hkl)〉 is the mean value for its unique reflection; summations are over all reflections.
c R-factor = Σh|Fo(h) − Fc(h)|/ΣhFo(h), where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
d R-free was calculated with 5% of the data excluded from the refinement.
e r.m.s.d. from ideal values.
f Categories were defined by Molprobity.
Computational Docking
The tetrasaccharide-pentapeptide docking with LytBCAT was performed with AutoDock Vina software (Version 1.0) (33), which uses a unique algorithm that implements a machine learning approach to its scoring function. The docking allowed us to obtain a population of possible conformations and orientations for the ligand at the binding site. Using AutoDock Tools (ADT) 1.5.4 (34), polar hydrogen atoms were added to LytBCAT structure, and its non-polar hydrogen atoms were merged. The protein LytBCAT, and the ligand were converted from a PDB format to a PDBQT format. All single bonds within the ligand were set to allow rotation. A grid box with dimensions of 40 × 45 × 50 points was used around the active site to cover the entire active site and allow ligand to move freely. The results were sorted by binding affinity and visually analyzed using PyMOL.
Purification of Pneumococcal PGN and Hydrolytic Activity Assay
Mature and immature PGN were purified from the wild-type S. pneumoniae TIGR4 strain and the lytB knock-out strain, respectively, according a previous protocol (35). The purified PGN was labeled with Remazol Brilliant Blue (Sigma) (termed RBB-labeled) as previously described (36).
Recombinant LytB truncations were incubated with RBB-labeled mature or immature PGN substrate and assayed for the activity by measuring RBB-labeled PGN release as previously reported (36). Each assay was performed at 37 °C in a system of 150 μl containing the buffer of 50 mm Na2HPO4/NaH2PO4, pH 7.0, 10 μm purified protein, and 1 mg/ml RBB-labeled PGN. For LytB and mutant LytBE564Q, 10 mm choline chloride was added. After incubating at 37 °C for 10 h, the insoluble substrate was removed by centrifugation at 130,000 × g for 20 min, and the amount of soluble RBB-labeled PGN fragments that was released to the supernatant by hydrolysis was determined by measuring the optical density at 595 nm with a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA). The buffer without the protein was used as the negative control.
Construction of lytB Knock-out S. pneumoniae and the Chain-dispersing Assay
The chromosomal lytB deletion mutant of S. pneumoniae was generated from the strain TIGR4 by allelic replacement as described previously (37). The upstream (978 bp) and downstream (1046 bp) lytB flanking regions in strain TIGR4 were separately amplified from the prepared genomic DNA by PCR with primer pairs PR0013/PR0014 and PR0015/PR0016 (Table 1), respectively. The Janus cassette was amplified from the prepared genomic DNA of S. pneumoniae strain ST588 (22) using primers Pr1097 and Pr1098 (Table 1). The PCR products of the Janus cassette, upstream and downstream of lytB sequences were purified from agarose gels using the DNA gel purification kit (Qiagen, Valencia, CA), digested by XbaI and XhoI, and ligated by T4 ligase (New England Biolabs, Beverly, MA). The ligation mixtures were then transformed into the strain TIGR4 to select kanamycin-resistant colonies on blood agar plates as described previously (37). The loss of the whole lytB coding region and the presence of the Janus cassette in the lytB locus in kanamycin-resistant colonies were detected by PCR amplification, restriction digestion, and DNA sequencing.
The chain-dispersing activity assay was performed as described previously (18). The S. pneumoniae TIGR4 ΔlytB (strain ST001) was grown in THY to an A620 nm 0.3, incubated in the presence of 1 μm full-length LytB or its mutant forms at 37 °C for 30 min, and diluted with PBS before Gram staining. Bacterial cells were photographed under a Zeiss light microscope (Carl Zeiss, Thornwood, CA) with an Axion Vision camera (Axion Technologies, Houston, TX).
Construction of S. pneumoniae Mutants, Complementation and Cell Separation Activity Assay
The chromosomal lytB complement and point mutant lytBE564Q S. pneumoniae strains were generated in the streptomycin-resistant derivative strain ST003 by allelic replacement using the counter selectable Janus cassette as described previously (38). In brief, the upstream and downstream regions of lytB locus in strain TIGR4 were separately amplified from the preparation genomic DNA by PCR using primer pairs PR0013/PR0054 and PR0057/PR0016 (Table 1), respectively. The mutant coding region of LytBE564Q was amplified from the LytBE564Q recombination plasmid pET28a using primer pairs PR0055/PR0056 (Table 1). All these PCR products were purified from agarose gels with the DNA gel purification kit (Qiagen). The PCR fragments of the upstream region, the mutant lytB coding region, and downstream were fused by overlap extension PCR with primers PR0013 and PR0016 (39). The fusion PCR products purified from agarose gels were used to transform the lytB knock-out streptomycin-resistant derivative strain ST003 to select streptomycin-resistant colonies on blood agar plates.
In-frame deletion in the lytB gene of strain TIGR4 was generated by allelic exchange at the streptomycin-resistant lytB-null mutant strain ST003 as described previously (38). DNA fragments flanking the deleted sequences of the lytB gene were initially amplified by PCR using primers listed in Table 1 according a previously reported procedure (22). The purified initial PCR products were subsequently joined together by overlap extension PCR (39), then used to transform strain ST003, respectively. Transformants, which are resistant to streptomycin and sensitive to kanamycin due to the loss of the Janus cassette (38), were selected and confirmed by DNA sequencing.
The in-frame deletions lytB strains were grown in THY medium at 37 °C with 5% CO2 to an A620 nm of 0.3, and bacterial samples were taken and dyed by Gram's stain, respectively. Pictures were taken with a Zeiss microscope as above.
Cell Adhesion and Invasion Assays
Pneumococcal adhesion was assessed as described previously (40). A549 human lung epithelial cells were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum, penicillin (5 μg/ml), and streptomycin (100 μg/ml). Briefly, ∼5 × 104 cells were seeded into 24-well tissue culture plates (Nunc, Naperville, IL) and grown in 5% CO2 at 37 °C for 48 h. The cell monolayers with ∼2 × 105 cells were washed twice with PBS and then inoculated in a standardized assay with 2 × 107 pneumococci, which were grown in THY medium to an optical density at 620 nm of 0.3∼0.4 and resuspended in the DMEM medium without serum. After 3 h of incubation at 37 °C in 5% CO2, the infected monolayers were washed 5 times with sterile PBS to remove unbound bacteria and treated with 200 μl of 0.25% trypsin, 0.02% EDTA for 3 min at 37 °C, and then lysed by the addition of Triton X-100 (0.025% in PBS). The number of adhered bacteria was determined by plating serial dilutions of the recovered bacterial suspensions onto tryptic soy agar plates containing 5% (v/v) sheep blood.
Pneumococcal adhesion was also assessed by fluorescence microscopy. A549 cells were grown on glass coverslips in 6-well plates (Nunc) and infected with fluorescein isothiocyanate-labeled pneumococci as described previously (19). The infected monolayers were fixed with paraformaldehyde (4.0% in PBS for 10 min at 4 °C) and permeabilized with Triton X-100 (0.1% for 5 min). After washing with PBS for three times, propidium iodide (Sigma) was used for nuclear staining according to the manufacturer's instructions. Slides were examined with a confocal laser scanning biological microscope FV1000-IX81 (Olympus, Japan) with an UPlanFLN 100×/1.3NA objective. Confocal parameters set for immunofluorescence detection were taken as standard settings. Images were processed with the ImageJ software (imagej.nih.gov).
Invasion assay was used to quantify the number of viable intracellular pneumococci as described (40). The cell monolayers were infected as described above. After 3 h of infection, the cells were washed 5 times with PBS to remove unbound bacteria, and 0.7 ml of fresh DMEM containing 100 μg/ml gentamicin and 10 μg/ml penicillin G was added per well. The plates were incubated for 1 h at 37 °C to kill the extracellular bacteria. Cell monolayers were washed 3 times with sterile PBS and treated with 200 μl of 0.25% trypsin, 0.02% EDTA for 3 min at 37 °C and then lysed by the addition of Triton X-100 (0.025% in PBS). Invasive bacteria were determined by plating serial dilutions of the recovered bacterial suspensions onto tryptic soy agar plates containing 5% (v/v) sheep blood. Experiments were repeated three times. Data corresponding to adhesion and invasion were compared using analysis of variance. Statistical analyses were performed with GraphPad InStat Version 5.0 (GraphPad Software, San Diego, CA).
All adhesion and invasion experiments were repeated at least twice. The data are presented as the means ± S.E. from the results of three or more replicate wells or cover slips.
RESULTS
Overall Structure
The full-length LytB is composed of an N-terminal signal peptide and a CBD of 13 putative choline binding repeats followed by a C-terminal catalytic domain (Fig. 1A). The recombinant LytB without the signal peptide exists as a monomer in solution, which was applied to crystallization in the presence of choline. After exhaustive screening, we did not find diffraction-quality crystals, which might be due to flexibility of the CBD. Using limited proteolysis and liquid chromatography-mass spectrometry analyses, we identified a stable 32-kDa segment that corresponds to the C-terminal catalytic domain (residues Lys-375–Asp-658, termed LytBCAT hereafter). We thus overexpressed this segment and subjected it to crystallization. We obtained the high quality crystals and solved the structure at 1.65 Å resolution (PDB code 4q2w). Each asymmetric unit contains one protein molecule, most residues of which could be well defined in the final model, except for 10 residues from Lys-375 to Glu-384 at the very N terminus and an 11-residue region (residue Ala-584–Lys-594) between helices α4 and α5, due to the missing of electronic density.
FIGURE 1.
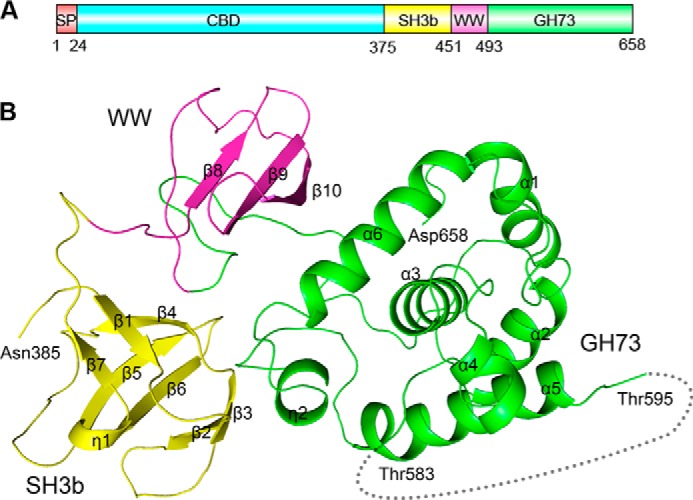
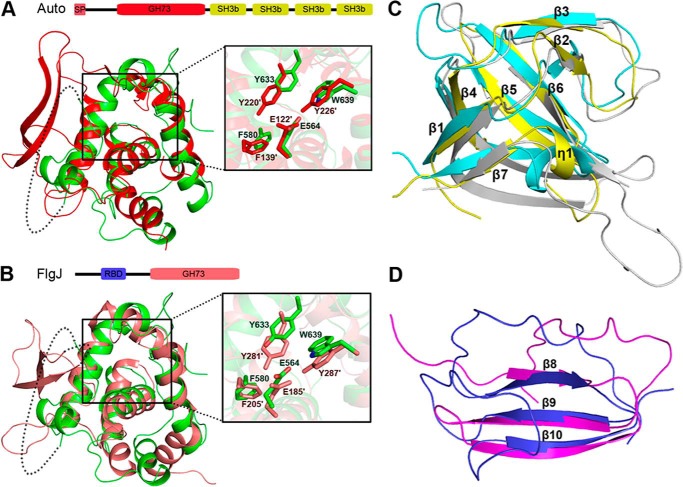
Domain organization and overall structure of LytB. A, domain organization of the full-length LytB. SP, signal peptide; SH3b, bacterial SH3b module; WW, WW domain-like module; GH73, the catalytic module of GH73. B, schematic representation of LytBCAT overall structure. The SH3b, WW, and GH73 modules are colored in yellow, magenta, and green, respectively.
The overall structure is composed of three distinct modules packing against each other: two all-β modules (residues Asn-385–Ser-450 and Lys-451–Asp-493, respectively) followed by the C-terminal catalytic module (residues Gly-494–Asp-658). The three modules are linked by two loops, forming a triangle structure (Fig. 1B). The first all-β module (residues Asn-385–Ser-450) adopts a β-barrel fold consisting of one twisted antiparallel β-sheet (β1-β4-β7) and two β-hairpins (β2-β3 and β5-β6) packing against each other. A 310 helix η1 is located between strands β6 and β7. The second all-β module contains a three-stranded antiparallel β-sheet (β8-β10). The C-terminal catalytic core module contains six α-helices (α1-α6) and a 310 helix η2. Notably, the helix α3 is located at the center and adopts an orientation almost perpendicular to the surrounding helices.
Structural Comparison
Based on the primary sequence analysis, the catalytic module of LytB has been previously annotated to the GH73 in the Carbohydrate Active EnZymes database (41). To date, two members in this family have known structures. They are the surface-associated autolysin Auto from Listeria monocytogenes (42) and the flagellar protein FlgJ from Sphingomonas sp. (43). Auto consists of a GH73 domain followed by four bacterial SH3 domains (termed SH3b), whereas FlgJ contains a GH73 following a rod binding domain (Fig. 2, A and B). Despite a sequence identity of less than 12%, the catalytic core module of LytB shares a very similar overall structure to the GH73 domains of Auto and FlgJ; thus we termed it LytBGH73. Superposition of LytBGH73 against the GH73 domains of Auto and FlgJ yielded an r.m.s.d. of 2.12 and 1.96 Å over 94 and 86 Cα atoms, respectively. Beyond the core all-α domain, both GH73 domains of Auto and FlgJ have an extra thumb-like β-hairpin (Fig. 2, A and B), which was proposed to contribute to the extended substrate binding groove (42, 43). In contrast, the corresponding segment of LytBGH73 is missing in the crystal structure due to its flexibility (Fig. 2, A and B). In addition, LytBGH73 possesses an active site similar to that of Auto, with Glu-564 of LytBGH73 exactly superimposed onto the catalytic residue Glu-122′ of Auto (Fig. 2A, inset). Moreover, the aromatic residues Phe-580, Tyr-633, and Trp-639 of LytBGH73 also adopt a conformation similar to that of three corresponding residues of Auto (Phe-139′, Tyr-220′, and Tyr-226′), which were proposed to create a hydrophobic environment to increase the pKa of the catalytic residue Glu-122′ for ensuring the protonation of its carboxyl group (42). In addition, sequence alignment indicated that the GH73 domains of the major autolysin Atl from Staphylococcus aureus and Staphylococcus epidermidis (44) share a sequence identity of ∼30% to the GH73 of LytB.
FIGURE 2.
Structural comparison of the GH73, SH3b, and WW modules. Superposition of LytBGH73 to the GH73 domain of L. monocytogenes (A) autolysin Auto and Sphingomonas sp. flagellar protein FlgJ (B). The GH73 domains of LytB, Auto, and FlgJ are colored in green, red, and pink, respectively. RBD, rod binding domain. The domain organizations of Auto and FlgJ are shown above the corresponding superimposed structures. Structural superposition of the LytBSH3b module (yellow) against the SH3b domain of A. variabilis AvPCP (cyan) and S. capitis ALE-1 (gray) (C) and the WW module of LytB (magenta) against the chitin binding domain (blue) of ChiB from S. marcescens (D).
Besides the GH73 module, LytBCAT consists of two all-β modules. Structural comparison using the Dali server (45) revealed that the first all-β module closely resembles SH3b domains, with a Z-score of ≥5.1. Thus we termed this module LytBSH3b. All SH3b domains in the output were listed as predicted or hypothetical bacterial cell wall hydrolases in the form of single or multiple modules. The only two well characterized hits were the SH3b of the γ-d-glutamyl-l-diamino acid endopeptidase AvPCP from Anabaena variabilis (46) and that of Staphylococcus capitis peptidoglycan hydrolase ALE-1 (47), both of which appear to contribute to substrate binding. SH3b modules have also been found in fusion with NlpC/P60 domain of cell wall peptidases (46) or CHAP domain of cell wall amidases (48). Superposition of LytBSH3b against the SH3b of AvPCP (PDB code 2hbw) and ALE-1 (PDB code 1r77) yielded an r.m.s.d. of 2.5 and 2.1 Å over 60 and 58 Cα atoms with a Z-score 7.6 and 5.1, respectively. The SH3b modules of LytB, AvPCP, and ALE-1 share a quite similar fold, with most secondary-structure elements well superimposed (Fig. 2C). In addition, the SH3b of S. capitis PGN hydrolase ALE-1 was reported to specifically recognize the pentaglycine interpeptide of S. aureus PGN (47). This suggests that LytBSH3b might also contribute to the peptide recognition despite only sharing a sequence identity of <15% to the currently structure-known SH3b domains.
A homology search of the second all-β module with the Dali server produced an output of several functionally unrelated proteins with a Z-score of ≤2.5. Structural superposition yielded an r.m.s.d. of about 2.7 Å over 43 Cα atoms against a segment (Arg-68 to Pro-125) of the first hit, an elongation factor P from Thermus thermophilus (49). Alternatively, a search against the SCOP indicated that it mostly resembles the three-stranded β-sheet of the chitin binding domain of Serratia marcescens chitinase ChiB (PDB code 1e15) (50). The chitin binding domain belongs to the carbohydrate binding domain superfamily in WW-like fold (50); thus we termed this module LytBWW. Structural superposition of LytBWW and the chitin binding domain yields an r.m.s.d. of 1.6 Å over 26 Cα atoms. Except that the loops vary a lot, the β-sheet could be well superimposed onto each other (Fig. 2D).
A Simulated Model of LytB Binding to the Putative Substrate
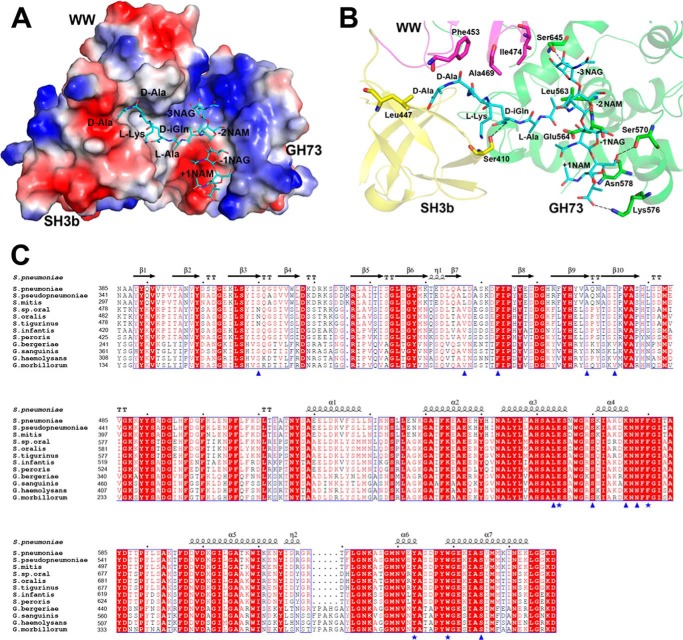
LytB has been proved to be a glycosidase toward the NAG-(β-1,4)-NAM glycosidic bond, functioning at the septum to separate the cell walls of two daughter cells (18). However, its bona fide physiological substrate remains undefined. Electrostatistic potential of LytBCAT surface revealed a T-shaped pocket that is composed of two parts: a groove through the catalytic module LytBGH73 and a cleft between LytBSH3b and LytBWW (Fig. 3A). This putative substrate binding pocket is reminiscent of the structure of an extended repetitive unit of PGN, the tetrasaccharide-pentapeptide NAM-NAG-NAM(-l-Ala-d-iGln-l-Lys-d-Ala-d-Ala)-NAG (termed TSPP). Due to the commercial unavailability of TSPP, we docked the structure of TSPP generated by the program of PRODRG server (51) to our LytBCAT structure by AutoDock (33). The simulated model showed that the carbohydrate moiety is located in the groove of LytBGH73, whereas the pentapeptide stretches into the cleft between LytBSH3b and LytBWW (Fig. 3A). The tetrasaccharide moiety is stabilized by hydrogen bonds with four polar residues of LytBGH73. In detail, residues Ser-570, Lys-576, and Asn-578 form three hydrogen bonds with +1 subsite NAM, whereas Ser-645 forms a hydrogen bond with O6 of −3 subsite NAG. In addition, several hydrophobic residues including Ile-474 in LytBWW and Leu-563 in LytBGH73 form hydrophobic interactions with the carbohydrate moiety of −3 subsite NAG (Fig. 3B). Notably, the residue Glu-564 is ∼4.5 Å to the C1 atom of −1 subsite NAG; moreover, it is well superimposed to the catalytic Glu-122′ of Auto (Fig. 2A). The pentapeptide moiety is mainly stabilized by hydrophobic interactions with three residues: Leu-447 in LytBSH3b and Phe-453 and Ala-469 in LytBWW. In addition, Ser-410 of LytBSH3b forms a hydrogen bond with d-iGln. Multiple sequence alignment indicated all these putative substrate binding residues are highly conserved among the Streptococcus mitis group and Gemella species (Fig. 3C). Beyond the previous reports (46, 47), this model provided another example that the SH3b domain participates in the binding to PGN peptide.
FIGURE 3.
A simulated model of LytBCAT binding to the putative substrate TSPP. A, surface representation of LytBCAT with TSPP in the substrate binding pocket. B, interactions between LytBCAT and TSPP. TSPP is shown in cyan sticks. Residues involved in hydrogen bonds and van der Waals interactions with TSPP are assigned using the PISA server (64) and are shown as sticks and colored in yellow, magenta, and green. Hydrogen bonds are indicated with dashed lines. C, multiple-sequence alignment of LytBCAT and homologs. Sequences of proteins are downloaded from the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov) with accession numbers: Streptococcus pseudopneumoniae, ZP_09990767.1; S. mitis, ZP_13525443.1; Streptococcus sp. oral, ZP_07458768.1; Streptococcus oralis, ZP_12441552.1; Streptococcus sanguinis, ZP_07887886.1; Streptococcus tigurinus, ZP_23320820.1; Streptococcus infantis, ZP_08523398.1; Streptococcus peroris, ZP_08065421.1; Gemella bergeriae, WP_021753068.1; Gemella haemolysans, WP_003144609.1; Gemella morbillorum, WP_004633969.1; Gemella sanguinis, WP_016359661.1. The alignment was performed with the programs ESPript. The residues participating in TSPP binding are labeled with blue triangles, and residues involved in catalysis are labeled with blue star.
The SH3b and WW Modules Are Indispensable for the Activity of LytB
It was previously reported that LytB could hydrolyze the cell wall at a much lower level compared with the autolysin LytA (18). Moreover, LytB is enriched at the septum of dividing pneumococci (18), indicating LytB prefers the immature PGN. We first compared the hydrolytic activity of LytB toward PGN prepared from the wild-type TIGR4 strain (termed mature PGN) and that from the lytB knock-out strain (termed immature PGN), according to a previously reported procedure (36). In agreement with the previous data (18), LytB hydrolyzed the mature PGN at a very low velocity. By using the PGN from the lytB knock-out bacteria as the substrate, which was proposed to consist of more immature branches of PGN, the hydrolytic velocity of LytB is increased by about 50% (Fig. 4). Moreover, compared with the ∼30% retained activity toward the mature PGN, LytBCAT retained about 60% of the activity of the full-length LytB toward the immature PGN. In consequence, we systematically compared the hydrolytic activity of wild-type LytB and its mutants toward the immature PGN. In contrast, deletion of the SH3b module resulted in a hydrolytic activity of about one-fifth that of LytBCAT, whereas deletion of both SH3b and WW modules almost completely abolished the activity (Fig. 4). In addition, the hydrolytic activity of the LytBE564Q mutant protein was almost undetectable (Fig. 4), further proving the crucial role of Glu-564 for the catalysis.
FIGURE 4.
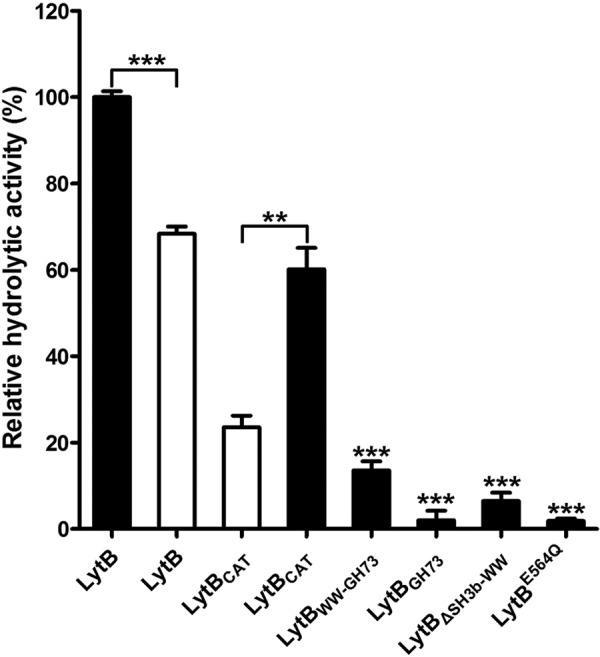
Hydrolytic activity of LytB and mutants toward purified PGN. The hydrolytic activity toward the immature and mature PGN are shown as solid and open bars, respectively. The enzymes applied to the assays include 1) the full-length LytB (LytB), 2) the catalytic domain (LytBCAT), 3) the WW and GH73 modules (LytBWW-GH73), 4) the GH73 module (LytBGH73), 5) the CBD fused with GH73 (LytBΔSH3b-WW), and 6) the single E564Q mutant of the full-length (LytBE564Q). The hydrolytic activities of all mutants are shown as a percentage to that of the full-length LytB. Data are presented as the means ± S.D. from three independent assays. Two-tailed Student's t test is used for the comparison of statistical significance. The p values of <0.05, 0.01, and 0.001 are indicated with *, **, and ***, respectively.
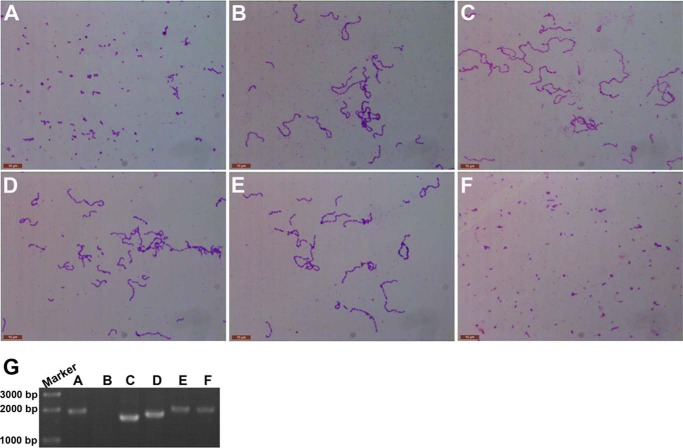
Deletion of LytB resulted in the formation of long-chain pneumococci, which could be dispersed by incubation with recombinant LytB (18). To check if SH3b and WW modules contribute to the chain-dispersing activity of LytB, we compared the morphology of pneumococci incubated with LytB and various truncated versions. The wild-type pneumococci usually appear in pairs of cocci or diplococci (Fig. 5A), whereas the lytB knock-out strain ST001 form long chains (Fig. 5B), as reported previously (8, 18). Compared with the negative control with phosphate buffer (Fig. 5C), incubation with the wild-type LytB or LytBCAT could separate most long-chain cells (strain ST001) into diplococci (Fig. 5, D and E). In contrast, very few of long-chain pneumococci could be hydrolyzed by LytBWW-GH73 or LytBGH73 protein (Fig. 5, F and G). Moreover, mutation of E564Q at the full-length LytB would almost completely abolish the chain-dispersing activity of LytB (Fig. 5H).
FIGURE 5.
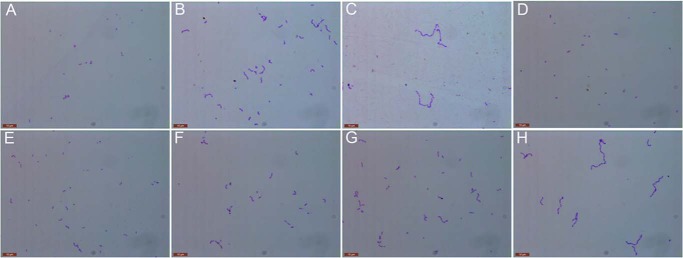
The chain-dispersing activity of LytB and mutants. The cell morphology of the wild-type S. pneumoniae TIGR4 strain (A) and lytB knock-out strain (B) is shown. Incubation of the lytB knock-out strain with the phosphate buffer (C), the full-length LytB (D), LytBCAT, the catalytic domain of LytB (E), LytBWW-GH73 (F), LytBGH73 (G), and LytBE564Q (H) mutant. Images of the bacteria dyed by Gram's stain are taken after incubation at 37 °C for 30 min.
To further demonstrate the in vivo contribution of each LytB module to cell separation, we compared the morphology of a series of strains with the coding region of full-length LytB or individual modules deleted (Table 1). Similar to the wild-type TIGR4 strain (Fig. 5A), the streptomycin-resistant derivative S. pneumoniae strain also formed diplococci (Fig. 6A), whereas the corresponding lytB knock-out strain exhibited long chains (Fig. 6B). Deletion of either SH3b and WW modules or only the SH3b module led to the phenotype of forming long-chain pneumococci (Fig. 6, C and D), similar to the lytB knock-out strain (Fig. 6B). All these results clearly suggested that the SH3b and WW modules are indispensable for LytB in cell separation. Remarkably, a single nucleotide mutation of G → C (resulting in an E564Q mutation in LytB) on the chromosome of S. pneumoniae TIGR4 resulted in the long chain morphology (Fig. 6E). Moreover, the strain with the chromosomal complementation of lytB gene could restore the phenotype of diplococci (Fig. 6F). These results indicated that an active LytB is necessary for the cell separation of pneumococci.
FIGURE 6.
Morphology of pneumococci with deletion of the coding region for LytB or module(s). A, the streptomycin-resistant derivative S. pneumoniae strain ST002 (Table 1). B, the lytB knock-out streptomycin-resistant derivative S. pneumoniae strain ST003; complement of coding region for lytBΔSH3b-WW (C), lytBΔSH3b (D), lytBE564Q (E), and wild-type lytB (F). Images of bacteria dyed with Gram's stain are taken when the A620 nm reached 0.3; G, PCR validation of the deletion and complement of various strains.
The Catalytic Activity of LytB Is Crucial for S. pneumoniae Adhesion to and Invasion into Human Lung Epithelial Cells
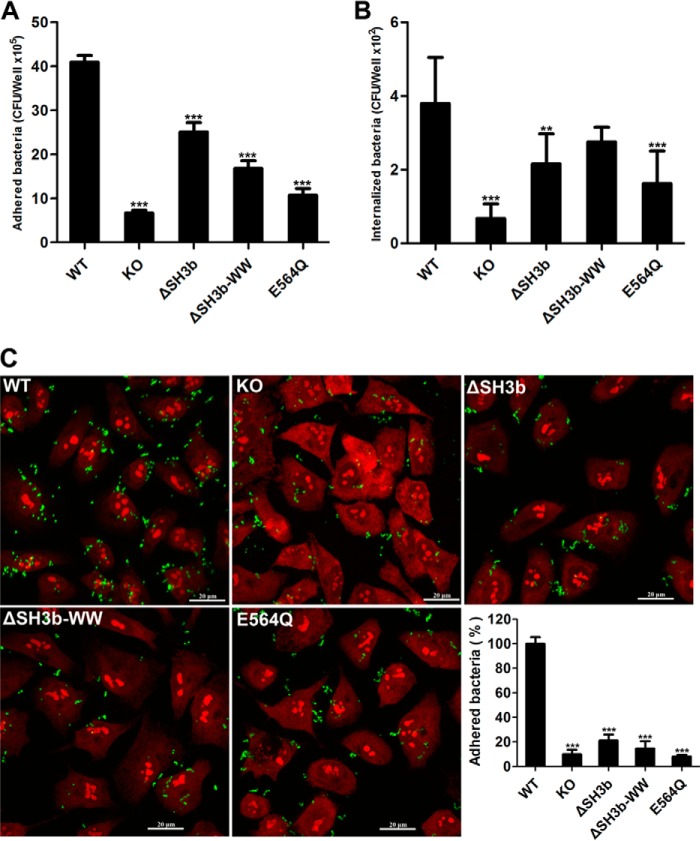
As a common colonizer of the human nasopharynx, S. pneumoniae can radiate from the nasopharynx to the lung for further pathogenesis (52). It has been reported that LytB is required for pneumococcal colonization in nasopharynx (20), but little is known for its infection to the lung. To address this question, we detected the ability of various pneumococci to adhere to and invade into human lung epithelial A549 cells. Compared with the wild-type TIGR4 strain, the lytB knock-out strain lost ∼90% of adhesion and invasion capacity (Fig. 7, A and B). The amount of mutant bacteria for both strains of lytBΔSH3b and lytBΔSH3b-WW adhered to and invaded into A549 cells were reduced to ∼60% (Fig. 7, A and B). These data revealed that the SH3b and WW modules of LytB are important for pneumococcal adhesion. In addition, the lytBE564Q mutant strain also showed a significant decrease of adhesion and invasion (Fig. 7, A and B), indicating the putative catalytic residue Glu-564 is critical for LytB. The reduced adhesion rates of various lytB mutants were further validated by fluorescence microscopy. The experiments showed that bacterial adhesion was significantly compromised in the strains lacking the entire lytB or various modules of its catalytic domain (Fig. 7C). Together, these data demonstrated that the full activity and integrity of LytB play an important role in pneumococcal adhesion to and invasion into respiratory epithelial cells.
FIGURE 7.
Efficiency of adhesion to and invasion into human lung epithelial cells by S. pneumoniae TIGR4 and its isogenic lytB mutants. Quantification of bacteria adhesion to (A) and invasion into (B) human lung epithelial A549 cells. C, comparison of the wild-type and various lytB-mutant strains of pneumococci adhering to A549 cells by fluorescence microscopy. Cells grown on coverslips were incubated with wild-type S. pneumoniae and its isogenic lytB mutants. The strains used are listed in Table 1. WT, wild-type S. pneumoniae ST002 was used as the positive control; KO, the lytB knock-out strain ST003; ΔSH3b, the lytBΔSH3b strain ST005; ΔSH3b-WW, the lytBΔSH3b-WW strain ST004; E564Q, the lytBE564Q mutant strain ST008. Bacteria were label with fluorescein isothiocyanate. A549 cells were labeled with propidium iodide. The adhesion rates are shown as the percentage of wild-type pneumococci. Data are presented as the means ± S.D. for three independent experiments. One-way analysis of variance with a post hoc The Dunnett test was used for the comparison of statistical significance. The p values of <0.05, 0.01, and 0.001 are indicated with *, **, and ***, respectively.
DISCUSSION
The GH73 module represents a common catalytic domain for many cell wall PGN hydrolases, which are ubiquitously found in both Gram-positive and -negative bacteria according to the Carbohydrate Active EnZymes database. It is usually fused with the varying cell wall binding modules, such as CBD, LysM, and SH3b (see the Pfam protein families database) (53). However, GH73 can adopt completely opposite physical organizations and functional relationships with the cell wall binding modules in various bacterial cell wall hydrolases. For instance, GH73 is fused to the N terminus of four SH3b modules in L. monocytogenes autolysin Auto (42, 54), whereas the Clostridium perfringens autolysin Acp adopts an opposite modular organization in which GH73 is linked to the C terminus of 10 SH3b modules (55). In both the Auto and Acp autolysins, the GH73 module is catalytically active in the absence of the SH3b modules (42, 54, 55), suggesting that the SH3b modules are involved in another functional aspect(s) of these enzymes, such as cell wall positioning and regulation of autolytic activity as described for the SH3b modules of the invasion protein InlB of L. monocytogenes (42, 56). In contrast, the major cell wall hydrolase AtlA of Enterococcus faecalis requires both the GH73 and the cell wall binding LysM modules for optimal activity (57). These observations have indicated that the precise physical organizations of the catalytic and cell wall binding domains dictate the functional properties of bacterial cell wall hydrolases. However, to date, only the GH73 structures of L. monocytogenes Auto (42) and flagellar protein FlgJ of Sphingomonas sp. (43) have been characterized. This study represents the first report of the crystal structure of GH73 together with SH3b and WW modules from the endo-β-N-acetylglucosaminidase LytB of S. pneumoniae.
Previous studies have shown that LytB is important for pneumococcal cell division, immune evasion, and infectivity (8, 20). De Las Rivas et al. (18) have shown that a recombinant LytB hydrolyzed pneumococcal PGN and dispersed the long chains of pneumococcal ΔlytB mutant, suggesting an endo-β-N-acetylglucosaminidase activity that cleaves the β(1,4)-glycosidic bond between NAG and NAM. The same study also described that LytB tends to accumulate in the cell poles or septal boundary of S. pneumoniae, whereas LytA, another cell wall hydrolase and the major autolysin of S. pneumoniae, preferentially accumulates to the equatorial regions of the cells (18). These studies have unequivocally indicated important and unique features of S. pneumoniae LytB, but the structural and biochemical mechanism of LytB as a PGN hydrolyze remains to be defined. Our structure-guided activity assays clearly elucidated the relationship between the modular organization and molecular functions of LytB.
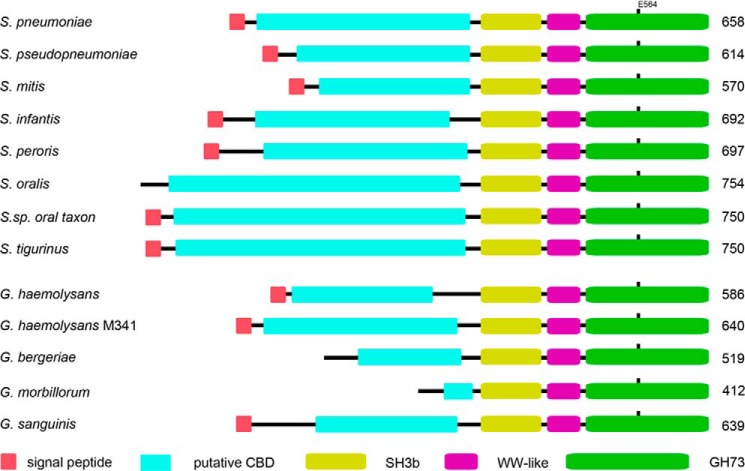
The modular structure and primary sequences of mature S. pneumoniae LytB are unusually similar among the LytB orthologs in many species of two distantly related Gram-positive bacterial groups: S. mitis group and Gemella species (Fig. 8). S. mitis group and Gemella species taxonomically belong to Lactobacillales and Bacillales (two different orders of the Firmicutes), respectively (58). Although the mitis group represents a fraction of streptococcal species that colonize the oral cavity and the upper respiratory tract of humans, Gemella species are common residents of mucosal surfaces in the oral cavity, upper respiratory tract, and digestive tract of humans and animals. Because the S. mitis group and Gemella species co-inhabit in the oral cavity and the upper respiratory tract, the sequence and structural similarities of the LytB orthologs may be a result of lateral gene exchange between the two bacterial groups.
FIGURE 8.
The modular organization of SH3b-WW-GH73 in S. mitis group and Gemella species. The term and boundaries of each choline binding repeat are defined according to the PROSITE database and/or sequence alignment.
The α-helical structure of the C-terminal GH73 module is the catalytic core of LytB. This module contains the catalytic glutamine residue (Glu-564) and three additional aromatic amino acids (residues Phe-580, Tyr-633, and Trp-639) that create a hydrophobic environment for catalysis. These residues and their arrangement are highly conserved in other GH73-containing cell wall hydrolases, such as L. monocytogenes autolysin Auto (42) and the flagellar protein FlgJ of Sphingomonas sp. (43). The importance of residue Glu-564 in LytB catalysis is validated by subsequent mutagenesis and biochemical analyses. The LytBE564Q mutant protein exhibited barely detectable activities in hydrolysis of purified pneumococcal PGN (Fig. 4) and chain dispersing of the ΔlytB mutant pneumococci (Fig. 5H). Consistently, the LytBE564Q mutant pneumococci displayed severe defects in cell division (Fig. 6E).
The two all-β modules of LytB are critical for its PGN hydrolase activity. Our structural results showed that the SH3b and WW segments (upstream of the GH73) form two all-β structural modules (Fig. 1). The LytB recombinant proteins lacking SH3b alone or both the SH3b and WW modules displayed a significant reduction in hydrolytic activity toward the immature PGN (Fig. 4) and chain dispersing of the ΔlytB mutant pneumococci (Fig. 5). In addition, the pneumococcal mutants lacking the SH3b and WW modules also displayed severe defects in cell division (Fig. 6). Comparative analysis revealed that the first all-β module formed by the SH3b segment resembles the structures of the SH3b domains in the γ-d-glutamyl-l-diamino acid endopeptidase AvPCP of A. variabilis (46) and peptidoglycan hydrolase ALE-1 of S. capitis (47). Because the SH3b domains of AvPCP and ALE-1 are necessary for peptide substrate recognition, it is reasonable to predict that the first all-β module formed by the SH3b segments contributes to the recognition of the PGN layer-bridging peptide in the context of LytB substrate binding. Additional analysis revealed that the second all-β module derived from the WW-like segments is structurally similar to the chitin binding domain of S. marcescens chitinase ChiB, implying a peptide binding function for this module. These predictions are consistent with the significant impact of both the SH3b and WW modules on the PGN hydrolase activity of LytB as discussed above. Taken together, we believed that the SH3b and WW modules of S. pneumoniae LytB and orthologs are involved in specifically recognizing the bridging peptide structures of the cell wall PGN.
The N-terminal CBD domain is not essential for the optimal catalytic activity of LytB, especially toward the PGN containing more nascent fragments. Although we were unable to obtain the structure of the full-length LytB with its N-terminal CBD, we attempted to understand the impact of the CBD on the PGN hydrolase activity of LytB by biochemical analysis of the full-length protein. The LytBCAT showed ∼60% of the hydrolase activity of the full-length LytB with the immature PGN preparations (Fig. 4), indicating a significant but not essential role of the CBD in the LytB function. The CBDs also contribute to the activities of other S. pneumoniae PGN hydrolases, such as LytA (59, 60), LytC (16), and CbpF (61). The CBD of LytA is essential for the formation of the homodimer and thereby its NAM amidase activity (59, 62, 63). In contrast, our data showed that the full-length LytB is a monomer. Thus, the CBD of LytB is likely to contribute to the LytB activity by anchoring the protein to the choline-containing cell wall.
Finally, it is intriguing that the LytB mutants exhibited significant impairment in epithelial adhesion and invasion. One potential reason behind this observation is that LytB may promote pneumococcal adhesion/invasion by direct physical interaction with its cognate receptor(s) on the host cells. Alternatively, LytB may enhance bacterial adhesion/invasion by contributing to appropriate cell division and cellular integrity through its cell wall remodeling activity. Additional studies will be needed to define the physiological mechanism by which LytB contributes to pneumococcal pathogenesis.
Acknowledgment
We thank the staff at the Shanghai Synchrotron Radiation Facility for technical assistance.
This work was supported by Ministry of Science and Technology of China Grants 2013CB835300, 2014CB910100, and 2012CB518702, National Natural Science Foundation of China Grants 31270781 and U1332114, Grand Challenges Exploration of the Bill and Melinda Gates Foundation Grant OPP1021992, Tsinghua University Collaborative Research Program Grant 2011Z23153, Center for Marine Medicine and Rescue of Tsinghua University Grant 20124812029, the 2011 Collaborative Innovation Center for Biotherapy, and the Fundamental Research Funds for the Central Universities.
The atomic coordinates and structure factors (code 4q2w) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PGN
- peptidoglycan
- NAG
- N-acetylglucosamine
- CBD
- choline binding domain
- GH73
- glycoside hydrolase family 73
- RBB
- Remazol Brilliant Blue
- NAM
- N-acetylmuramic acid
- TSPP
- tetrasaccharide-pentapeptide NAM-NAG-NAM(-l-Ala-d-iGln-l-Lys-d-Ala-d-Ala)-NAG
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Neidhardt F. C., Ingraham J. L., Schaechter M. (1990) Physiology of the Bacterial Cell: A Molecular Approach, pp. 507, American Society for Microbiology, Washington, D. C [Google Scholar]
- 2. Typas A., Banzhaf M., Gross C. A., Vollmer W. (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neuhaus F. C., Baddiley J. (2003) A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vollmer W., Joris B., Charlier P., Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 [DOI] [PubMed] [Google Scholar]
- 5. Musher D. M. (2010) Streptococcus pneumoniae. In Principles and Practice of Infectious Diseases. 7th Ed., pp. 2623–2641, Elsevier Churchill Livingstone, New York [Google Scholar]
- 6. Paton J. C., Boslego J. W. (2008) Protein vaccines. In Pneumococcal Vaccines: The Impact of Conjugate Vaccines. 6th Ed., pp. 421–435, American Society for Microbiology, Washington, D. C [Google Scholar]
- 7. Mosser J. L., Tomasz A. (1970) Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J. Biol. Chem. 245, 287–298 [PubMed] [Google Scholar]
- 8. García P., González M. P., García E., López R., García J. L. (1999) LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31, 1275–1281 [DOI] [PubMed] [Google Scholar]
- 9. García P., Paz González M., García E., García J. L., López R. (1999) The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33, 128–138 [DOI] [PubMed] [Google Scholar]
- 10. Eldholm V., Johnsborg O., Straume D., Ohnstad H. S., Berg K. H., Hermoso J. A., Håvarstein L. S. (2010) Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol. Microbiol. 76, 905–917 [DOI] [PubMed] [Google Scholar]
- 11. López R., González M. P., García E., García J. L., García P. (2000) Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol. 151, 437–443 [DOI] [PubMed] [Google Scholar]
- 12. Tomasz A., Waks S. (1975) Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc. Natl. Acad. Sci. U.S.A. 72, 4162–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanchez-Puelles J. M., Ronda C., Garcia J. L., Garcia P., Lopez R., Garcia E. (1986) Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158, 289–293 [DOI] [PubMed] [Google Scholar]
- 14. Fernández-Tornero C., López R., García E., Giménez-Gallego G., Romero A. (2001) A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 8, 1020–1024 [DOI] [PubMed] [Google Scholar]
- 15. Yuan Z. Q., Lv Z. Y., Gan H. Q., Xian M., Zhang K. X., Mai J. Y., Yu X. B., Wu Z. D. (2011) Intranasal immunization with autolysin (LytA) in mice model induced protection against five prevalent Streptococcus pneumoniae serotypes in China. Immunol. Res. 51, 108–115 [DOI] [PubMed] [Google Scholar]
- 16. Pérez-Dorado I., González A., Morales M., Sanles R., Striker W., Vollmer W., Mobashery S., García J. L., Martínez-Ripoll M., García P., Hermoso J. A. (2010) Insights into pneumococcal fratricide from the crystal structures of the modular killing factor LytC. Nat. Struct. Mol. Biol. 17, 576–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Claverys J. P., Håvarstein L. S. (2007) Cannibalism and fratricide: mechanisms and raisons d'etre. Nat. Rev. Microbiol. 5, 219–229 [DOI] [PubMed] [Google Scholar]
- 18. De Las Rivas B., García J. L., López R., García P. (2002) Purification and polar localization of pneumococcal LytB, a putative endo-β-n-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184, 4988–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gosink K. K., Mann E. R., Guglielmo C., Tuomanen E. I., Masure H. R. (2000) Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68, 5690–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramos-Sevillano E., Moscoso M., García P., García E., Yuste J. (2011) Nasopharyngeal colonization and invasive disease are enhanced by the cell wall hydrolases LytB and LytC of Streptococcus pneumoniae. PLoS ONE 6, e23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Atilano M. L., Pereira P. M., Vaz F., Catalão M. J., Reed P., Grilo I. R., Sobral R. G., Ligoxygakis P., Pinho M. G., Filipe S. R. (2014) Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. eLife 3, e02277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu L., Ma Y., Zhang J. R. (2006) Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J. Biol. Chem. 281, 15464–15474 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Brodersen D. E., de La Fortelle E., Vonrhein C., Bricogne G., Nyborg J., Kjeldgaard M. (2000) Applications of single-wavelength anomalous dispersion at high and atomic resolution. Acta Crystallogr. D. Biol. Crystallogr. 56, 431–441 [DOI] [PubMed] [Google Scholar]
- 25. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terwilliger T. C. (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D. Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vollmer W. (2007) Preparation and analysis of pneumococcal murein (peptidoglycan). In Molecular Biology of Streptococci, pp. 531–536, Horizon Bioscience, Norfolk, UK [Google Scholar]
- 36. Zhou R., Chen S., Recsei P. (1988) A dye release assay for determination of lysostaphin activity. Anal. Biochem. 171, 141–144 [DOI] [PubMed] [Google Scholar]
- 37. Bricker A. L., Camilli A. (1999) Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172, 131–135 [DOI] [PubMed] [Google Scholar]
- 38. Sung C. K., Li H., Claverys J. P., Morrison D. A. (2001) An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67, 5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heckman K. L., Pease L. R. (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 [DOI] [PubMed] [Google Scholar]
- 40. Zhang J. R., Mostov K. E., Lamm M. E., Nanno M., Shimida S., Ohwaki M., Tuomanen E. (2000) The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102, 827–837 [DOI] [PubMed] [Google Scholar]
- 41. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bublitz M., Polle L., Holland C., Heinz D. W., Nimtz M., Schubert W. D. (2009) Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes. Mol. Microbiol. 71, 1509–1522 [DOI] [PubMed] [Google Scholar]
- 43. Hashimoto W., Ochiai A., Momma K., Itoh T., Mikami B., Maruyama Y., Murata K. (2009) Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ. Biochem. Biophys. Res. Commun. 381, 16–21 [DOI] [PubMed] [Google Scholar]
- 44. Biswas R., Voggu L., Simon U. K., Hentschel P., Thumm G., Götz F. (2006) Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259, 260–268 [DOI] [PubMed] [Google Scholar]
- 45. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Q., Sudek S., McMullan D., Miller M. D., Geierstanger B., Jones D. H., Krishna S. S., Spraggon G., Bursalay B., Abdubek P., Acosta C., Ambing E., Astakhova T., Axelrod H. L., Carlton D., Caruthers J., Chiu H. J., Clayton T., Deller M. C., Duan L., Elias Y., Elsliger M. A., Feuerhelm J., Grzechnik S. K., Hale J., Han G. W., Haugen J., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kozbial P., Kumar A., Marciano D., Morse A. T., Nigoghossian E., Okach L., Oommachen S., Paulsen J., Reyes R., Rife C. L., Trout C. V., van den Bedem H., Weekes D., White A., Wolf G., Zubieta C., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2009) Structural basis of murein peptide specificity of a γ-d-glutamyl-l-diamino acid endopeptidase. Structure 17, 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu J. Z., Fujiwara T., Komatsuzawa H., Sugai M., Sakon J. (2006) Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 281, 549–558 [DOI] [PubMed] [Google Scholar]
- 48. Bateman A., Rawlings N. D. (2003) The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28, 234–237 [DOI] [PubMed] [Google Scholar]
- 49. Hanawa-Suetsugu K., Sekine S., Sakai H., Hori-Takemoto C., Terada T., Unzai S., Tame J. R., Kuramitsu S., Shirouzu M., Yokoyama S. (2004) Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. U.S.A. 101, 9595–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Aalten D. M., Synstad B., Brurberg M. B., Hough E., Riise B. W., Eijsink V. G., Wierenga R. K. (2000) Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 97, 5842–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 52. Bogaert D., De Groot R., Hermans P. W. (2004) Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154 [DOI] [PubMed] [Google Scholar]
- 53. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cabanes D., Dussurget O., Dehoux P., Cossart P. (2004) Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51, 1601–1614 [DOI] [PubMed] [Google Scholar]
- 55. Camiade E., Peltier J., Bourgeois I., Couture-Tosi E., Courtin P., Antunes A., Chapot-Chartier M. P., Dupuy B., Pons J. L. (2010) Characterization of Acp, a peptidoglycan hydrolase of Clostridium perfringens with N-acetylglucosaminidase activity that is implicated in cell separation and stress-induced autolysis. J. Bacteriol. 192, 2373–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jonquières R., Bierne H., Fiedler F., Gounon P., Cossart P. (1999) Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34, 902–914 [DOI] [PubMed] [Google Scholar]
- 57. Eckert C., Lecerf M., Dubost L., Arthur M., Mesnage S. (2006) Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J. Bacteriol. 188, 8513–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vos P. D., Garrity G. M., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H., Whitman W. B. (2009) The Firmicutes, Springer-Verlag, Dordrecht, The Netherlands [Google Scholar]
- 59. Mellroth P., Sandalova T., Kikhney A., Vilaplana F., Hesek D., Lee M., Mobashery S., Normark S., Svergun D., Henriques-Normark B., Achour A. (2014) Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae. mBio 5, e01120–01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Varea J., Saiz J. L., López-Zumel C., Monterroso B., Medrano F. J., Arrondo J. L., Iloro I., Laynez J., Garcia J. L., Menéndez M. (2000) Do sequence repeats play an equivalent role in the choline-binding module of pneumococcal LytA amidase? J. Biol. Chem. 275, 26842–26855 [DOI] [PubMed] [Google Scholar]
- 61. Molina R., González A., Stelter M., Pérez-Dorado I., Kahn R., Morales M., Moscoso M., Campuzano S., Campillo N. E., Mobashery S., García J. L., García P., Hermoso J. A. (2009) Crystal structure of CbpF, a bifunctional choline-binding protein and autolysis regulator from Streptococcus pneumoniae. EMBO Rep. 10, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Romero P., López R., García E. (2007) Key role of amino acid residues in the dimerization and catalytic activation of the autolysin LytA, an important virulence factor in Streptococcus pneumoniae. J. Biol. Chem. 282, 17729–17737 [DOI] [PubMed] [Google Scholar]
- 63. Fernández-Tornero C., García E., López R., Giménez-Gallego G., Romero A. (2002) Two new crystal forms of the choline-binding domain of the major pneumococcal autolysin: insights into the dynamics of the active homodimer. J. Mol. Biol. 321, 163–173 [DOI] [PubMed] [Google Scholar]
- 64. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]