Background: TAT-RasGAP317–326 is a Ras GTPase-activating protein (RasGAP)-derived peptide that requires deleted in liver cancer-1 (DLC1) for its antimetastatic activities.
Results: A WXW motif within TAT-RasGAP317–326 mediates RasGAP-DLC1 interaction.
Conclusion: The tryptophan residues of TAT-RasGAP317–326 are crucial for its activity
Significance: The WXW motif could be used to design anticancer small molecules bearing TAT-RasGAP317–326 activities.
Keywords: Cell Migration, Cell-penetrating peptide (CPP), Deleted in Liver Cancer 1 (DLC1), Docking, GTPase-activating Protein (GAP), Peptides, RasGAP, TAT-RasGAP317–326, Anticancer Peptide, Sensitizer
Abstract
TAT-RasGAP317–326, a cell-permeable 10-amino acid-long peptide derived from the N2 fragment of p120 Ras GTPase-activating protein (RasGAP), sensitizes tumor cells to apoptosis induced by various anticancer therapies. This RasGAP-derived peptide, by targeting the deleted in liver cancer-1 (DLC1) tumor suppressor, also hampers cell migration and invasion by promoting cell adherence and by inhibiting cell movement. Here, we systematically investigated the role of each amino acid within the RasGAP317–326 sequence for the anticancer activities of TAT-RasGAP317–326. We report here that the first three amino acids of this sequence, tryptophan, methionine, and tryptophan (WMW), are necessary and sufficient to sensitize cancer cells to cisplatin-induced apoptosis and to reduce cell migration. The WMW motif was found to be critical for the binding of fragment N2 to DLC1. These results define the interaction mode between the active anticancer sequence of RasGAP and DLC1. This knowledge will facilitate the design of small molecules bearing the tumor-sensitizing and antimetastatic activities of TAT-RasGAP317–326.
Introduction
Cancer is the second leading cause of death worldwide (1). Ninety percent of cancer-related death is attributed to metastases (1). Surgery, chemotherapy, and radiotherapy remain at present the main clinical therapeutic tools to treat cancer. These treatments are very efficacious against some cancers but are of limited long term efficacy against others. Besides developing new anticancer modalities, increasing the efficacy of current therapies is of clear and timely interest.
p120 Ras GTPase-activating protein (from now on referred to as RasGAP)2 is a multidomain protein. It was first reported as a negative modulator of the Ras signaling pathway (2). This activity requires the GAP domain of the protein (2). It was found later that other domains of the protein had signaling activities that were not related to its function as a GAP. For example, the N-terminal moiety of RasGAP can positively regulate MAP kinase kinases such as Ras and Mos (3). RasGAP is also a specific caspase-3 substrate (4). Because of the presence of two sites within RasGAP that have differential sensitivities toward caspase-3-mediated cleavage, the RasGAP/caspase-3 module acts as a stress sensor in cells (5). In the presence of low stresses, caspase-3 is mildly activated, and this leads to the partial cleavage of RasGAP into an N-terminal fragment that exerts potent, Akt-dependent, antiapoptotic activities (4, 6, 7). When cells are subjected to strong stresses, RasGAP is fully processed by caspase-3, further cleaving the N-terminal fragment and henceforth destroying its ability to protect cells.
One of the fragments generated by the full cleavage of RasGAP by caspase-3, fragment N2 (RasGAP158–455), has the capacity to increase the sensitivity to anticancer treatments of various tumor cell lines, both in vitro and in vivo (6, 8). Similarly to fragment N2, a cell-permeable 10-amino acid peptide contained within the SH3 domain of fragment N2, called TAT-RasGAP317–326, was found to efficiently sensitize cancer cells to anticancer agent-induced apoptosis (9) and to inhibit tumor growth when combined with chemotherapy (8). We recently reported that fragment N2 was an efficient inhibitor of the metastatic cascade (10). TAT-RasGAP317–326 also inhibited cell migration and invasion into basement membrane matrix by strengthening adhesiveness of the cells to their substratum (11). However, in an attempt to use TAT-RasGAP317–326 as an antimetastatic tool, we found, using mouse models, that this peptide was not always delivered efficiently to tumors (10). This delivery issue would call for the development of small molecules bearing the activity of RasGAP317–326. However, such development would greatly benefit from a better understanding of the mode of action of TAT-RasGAP317–326.
Actin cytoskeleton dynamics controls adhesion, migration, and invasion and is mainly regulated by the small GTPases of the Rho family (e.g. Rho itself, Rac, and Cdc42 (12, 13)). We found that the TAT-RasGAP317–326 molecular properties by which it induces adhesion and inhibits migration rely on modulation of the actin cytoskeleton and requires deleted in liver cancer-1 (DLC1), a RhoGAP that functions as a tumor and metastasis suppressor (11, 14). Therefore, understanding whether and how TAT-RasGAP317–326 engages DLC1 is of crucial interest.
Although peptide therapeutics are gathering increasing interest for the treatment of tumors (15), classical issues associated with peptide-based therapy are impeding their development. These issues consist of the rapid clearance from the body, the lack of targetable ability, their short half-lives, and their expensive production costs. Consistent with this, the Lipinski's rule-of-five, a model that predicts the likeliness of a compound to be translated into an orally active drug, is of bad prognosis for peptide development (16).
The goal of the present study was to characterize the importance of each of the RasGAP317–326 amino acids for its sensitizing activity and its ability to increase cell adhesiveness. This was performed to better understand the mode of action of the peptide and to gather structure-function information that could be used for pharmacological development to facilitate the development of a small molecule that mimics TAT-RasGAP317–326. Our recent finding that fragment N2 requires DLC1 for its antimetastatic activities prompted us to dissect how these two molecules interact. Here, we report the exact binding mode between DLC1 and TAT-RasGAP317–326 and we identify a short WXW motif located at the N-terminal end of the peptide that carries its anticancer activities.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The osteosarcoma cell line used in this study is U2OS (ATCC; HTB-96). HEK-293T cells were used for protein interaction studies as reported previously (11). The cells were cultured at 37 °C and 5% CO2 and were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco; 61965) supplemented with 10% fetal bovine serum (FBS) (Gibco; 10270) for not more than 2 months and were constantly checked for their morphology.
Peptide Synthesis
The peptides used in this study were made of d-amino acids, which provide resistance to proteases and therefore increase their stability. In order for the side chains to be similarly exposed as in l-form peptides (8), the d-form peptides were synthesized starting with the last C-terminal residue (retro-inverso peptides). The TAT-RasGAP317–326 (GRKKRRQRRRGGWMWVTNLRTD), the TAT (HIV-TAT48–57) (GRKKRRQRRR), and all the peptides mentioned in the figures were synthesized at the Department of Biochemistry, University of Lausanne, Switzerland using the Fmoc (N-(9-fluorenyl)methoxycarbonyl) technology, purified by HPLC, and tested by mass spectrometry as reported earlier (9). Two glycine residues were inserted as a linker between TAT and the peptides of interest.
Trypsin-mediated Detachment Assays, Transfection, Immunoprecipitation, and Western Blotting
Trypsin-mediated detachment assays, Giemsa staining, calcium-phosphate transfection, immunoprecipitation, and Western blotting were performed as reported previously (11). The primary antibodies used were anti-HA (Covance; MMS-101r), anti-V5 (Invitrogen; 46-1157) and anti-GFP (Clontech; 632380).
Wound-healing Scratch Assay
Five hundred thousand U2OS were grown in 3.5-cm plates. The day after, the cells were pretreated as indicated in the figures for 3 h. A wound in the cell layer was made with a tip (0.4-mm diameter at its extremity). Five pictures per wound were taken just after wounding (0 h), and then 24 and 48 h after wounding (the same five fields per wound were photographed at each time point). Wound areas were measured using the ImageJ software. The migration area was calculated by subtracting the 48 h value by the 0 h value, and the data were normalized against the untreated condition. For time-course experiments, the data obtained after 24 h were treated similarly than for 48 h.
Apoptosis Measurements
One hundred and fifty thousand U2OS cells were grown in 3.5-cm plates. The day after, the cells were treated for 22 h with 20 μm TAT-coupled peptides in the presence or in the absence of 30 μm cisplatin (Sigma-Aldrich; P4394) as indicated in the figures. The cells were then fixed, and the nuclei were stained in 2% paraformaldehyde containing 10 μg/ml Hoechst 33342 (Molecular Probes; H1399) for 15 min. The pyknotic nuclei were visualized using the Nikon Eclipse TS100 microscope equipped with fluorescence; they were then scored and reported in percentage over total number of cells.
Greyscale Heat Map
Heat map was performed by semiquantitative evaluation of the effects of the alanine-substituted peptides as compared with wild-type 317–326 peptide. The results on which the heat map is based appear in Fig. 1B (adherence; 20 μm values), Fig. 2A (migration), and Fig. 2B (apoptosis). This heat map is a greyscaled representation of whether the alanine-substituted peptides recapitulate the effects of 317–326. Specifically for apoptosis, the minimal effect (in black) was set to the “cisplatin alone” condition, and the maximal effect (in white) was set to the “cisplatin + 317–326” condition; for adherence, the minimal effect (in black) was set to the “untreated + trypsin” condition, and the maximal effect (in white) was set to the “317–326 + trypsin” condition; for migration, the minimal effect (in white) was set to the “317–326” condition, and the maximal effect (in black) was set to the “untreated” condition.
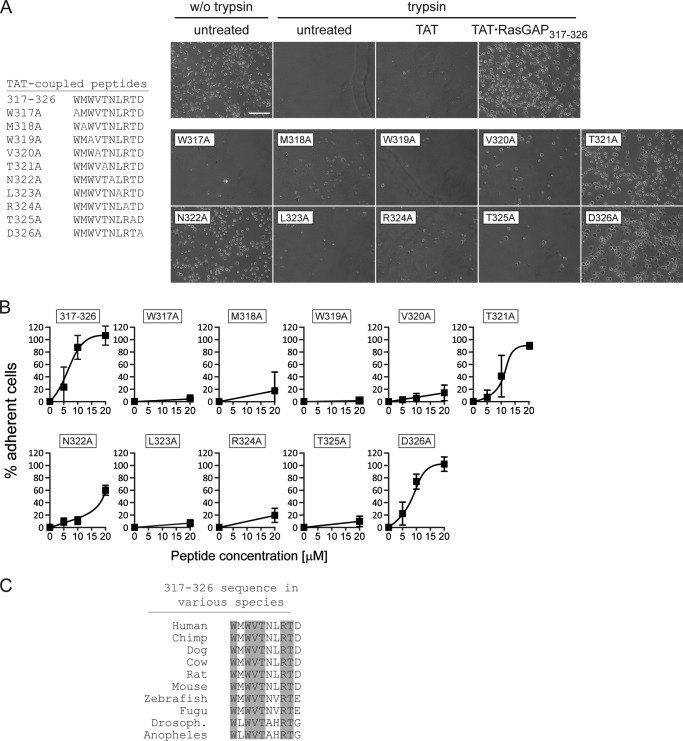
FIGURE 1.
Cell adherence induced by alanine-substituted RasGAP317–326 peptides. A and B, U2OS cells were cultured overnight and then treated for 8 h with 20 μm TAT or 20 μm of the indicated TAT-coupled peptides or left untreated. The cells were then subjected to trypsin-mediated detachment assays. The images shown in panel A correspond to 20 μm peptide-treated cells (n = 4 independent experiments). Scale bar: 200 μm for all images. w/o trypsin, without trypsin. B, dose-response quantitation (n = 4 independent experiments). C, alignment of the p120 RasGAP317–326 sequences from different species. The amino acids conserved during evolution are shaded in gray. Drosoph., Drosophila.
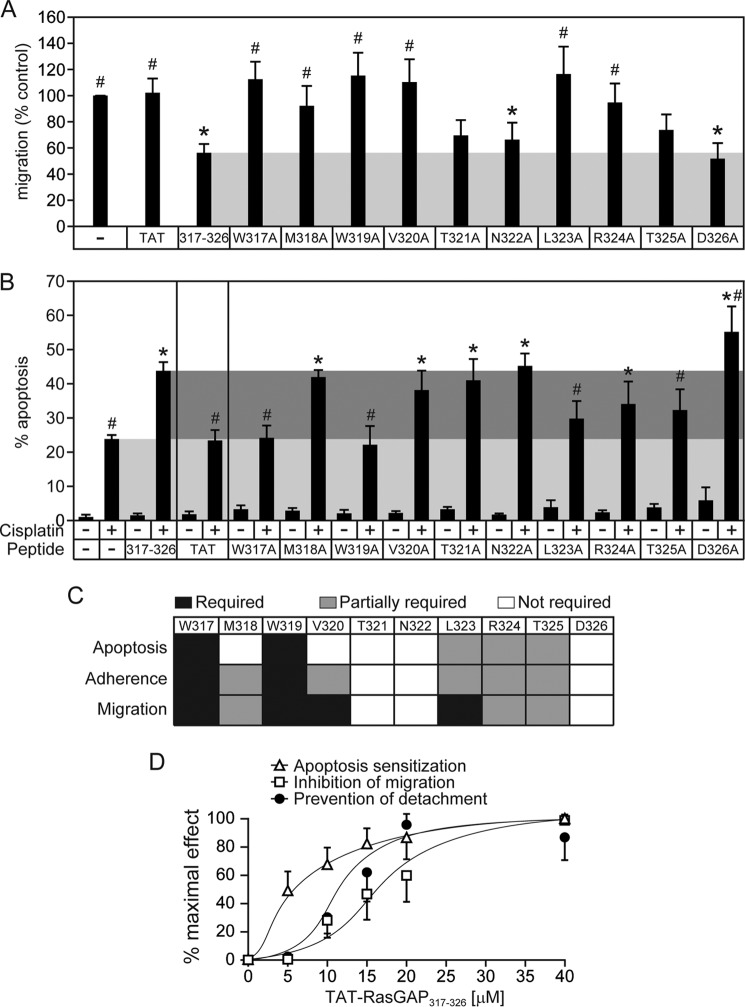
FIGURE 2.
Ability of alanine-substituted RasGAP317–326 peptides to inhibit cell migration and sensitize cancer cells against cisplatin-induced apoptosis. A, U2OS cells were grown to confluence and were pretreated for 3 h with 20 μm of the indicated TAT-coupled peptides. The cells were then subjected to wound healing scratch assays. The graph displays the percentage of migration over untreated cells. Asterisks and number signs denote statistical significant differences after one-way ANOVA (asterisk: different from untreated condition; number sign: different from 317–326 treatment; n = 7 independent experiments). Error bars indicate mean ± 95% confidence intervals. B, U2OS cells were cultured overnight and were subsequently treated for 22 h with 20 μm of the indicated TAT-coupled peptides in the presence or in the absence of 30 μm cisplatin. The number of pyknotic nuclei was then scored and reported as the percentage of apoptosis. The light gray region displays the level of apoptosis induced by cisplatin alone, and the dark gray region shows the apoptosis sensitization zone. Asterisks and number signs denote statistical significant differences after one-way ANOVA with the cisplatin-only treatment and with the cisplatin + 317–326 treatment, respectively (n = 6 independent experiments). Error bars indicate mean ± 95% confidence intervals, C, schematic representation of the importance of each amino acid within the wild-type sequence for the apoptosis sensitization, adhesion promotion, and migration inhibition processes. D, comparison of the dose-dependent effect of 317–326 on apoptosis sensitization, adhesion (prevention of detachment), and inhibition of migration. Each experiment was performed as described in earlier figures and panels except that here all the responses are reported as the percentage of the maximum observed effect. Error bars indicate mean ± 95% confidence intervals.
Plasmids
The extension .dn3 indicates that the backbone plasmid is pcDNA3 (plasmid 1) from Invitrogen. The Stag-HA-hRasGAP[158–455].dn3 (plasmid 644) (here called HA-N2), Stag-HA-hRasGAP[158–455](W317A).dn3 (plasmid 793) (here called HA-N2 (W317A)), and V5-mDLC1[1–1092].dn3 (plasmid 791) (here called V5-DLC1) plasmids were described earlier (11). The Stag-HA-hRasGAP[158–455](W319A).dn3 (plasmid 803) (here called HA-N2 (W319A)) plasmid encodes a Stag- and HA-tagged form of fragment N2 bearing a mutation of tryptophan 319 into an alanine residue (W319A). The template vector used for starting the mutagenesis is the HA-N2 plasmid. Mutagenesis was performed using the mega-primer procedure (17) as follows. (i) The W319A mutation was generated by PCR amplification of HA-N2 using oligonucleotide (oligo) 1019 (human RasGAP nucleotides 1052–1097 (NCBI entry M23379) except for nucleotides (underlined) that create a W319A mutation and a silent mutation generating a Bsu36I restriction site: (GAA TTA GAA GAT GGA TGG ATG GCG (W319A) GTT ACA AAC C (N1-N2 of Bsu36I) TA AGG (N7 of Bsu36I) ACAGATG)) and oligo 62 (TACCTAGCATGAACAGATTG (random sequence) AGGGGCAAACAACAGATG (pcDNA3 nucleotides 1080–1063)). (ii) The PCR product obtained in (i) was purified and elongated on the HA-N2 template. (iii) The PCR reaction was resumed after the addition of oligo 28 and oligo 70. The PCR in (iii) was digested with BsiWI and NotI and ligated into plasmid HA-N2 digested with the same enzymes. The DLC1 mutant plasmids were generated using the same methodology. The V5-mDLC1[1–1092] (R677A).dn3 (plasmid 804) (here called V5-DLC1 (R677A)) plasmid encodes a V5-tagged form of the Mus musculus DLC1 transcript variant 2 (NM_015802.3) bearing a mutation of arginine 677 into an alanine residue (R677A). The template vector used for starting the mutagenesis is the V5-DLC1 plasmid. Mutagenesis was performed as follows. (i) The R677A mutation was generated by PCR amplification of V5-DLC1 using oligo 1016 (mouse Dlc1 nucleotides 2368–2411 (NCBI entry NM_015802.3) except for nucleotides (underlined) that create a R677A mutation and a silent mutation generating an EcoRI restriction site: (GTC GGG CTC TTC GCG (R677A) AAG TCA GGT GTC AAA TCC CG A (N2 of EcoRI) AT T (N5 of EcoRI) CAGGCT) and oligo 62. (ii) The PCR product obtained in (i) was purified and elongated on the V5-DLC1 template. (iii) The PCR reaction was resumed after the addition of oligo 28 (TAATACGACTCACTATAGGGAGA (pcDNA3 sequence 863–885)) and oligo 70 (TACCTAGCATGAACAGATTG (same random sequence as in nucleotide oligo 62)). The PCR in (iii) was digested with EcoRV and XhoI and ligated into plasmid V5-DLC1 digested with the same enzymes. The V5-mDLC1[1–1092](N829A,L830A).dn3 (plasmid 805) (here called V5-DLC1 (N829A,L830A)) plasmid encodes a V5-tagged form of the M. musculus DLC1 transcript variant 2 (NM_015802.3) bearing a mutation of asparagine 829 and leucine 830 into alanine residues (N829A,L830A). The template vector used for starting the mutagenesis is the V5-DLC1. Mutagenesis was performed as follows. (i) The N829A and L830A mutations were generated by PCR amplification of V5-DLC1 using oligo 1017 (mouse Dlc1 nucleotides 2821–2859 (NCBI entry NM_015802.3) except for nucleotides (underlined) that create the N829A and L830A mutations and a new NotI restriction site: (AAA GAC CTG AAT GAA GCGGCC (N829A,L830A and (N1–6) of NotI) GCG GCG ACT CAA GGG CTG) and oligo 62). (ii) The PCR product obtained in (i) was purified and elongated on the V5-DLC1 template. (iii) The PCR reaction was resumed after the addition of oligo 28 and oligo 70. The PCR in (iii) was digested with EcoRI and XhoI and ligated into plasmid V5-DLC1 digested with the same enzymes. The extension .gfp indicates that the backbone plasmid is pEGFP-C1 (plasmid 6) from Clontech and encodes the green fluorescent protein. The pEGFP-hDLC1.gfp (plasmid 811) (here called GFP-DLC1), pEGFP-hDLC2.gfp (plasmid 812) (here called GFP-DLC2) and pEGFP-hDLC3.gfp (plasmid 813) (here called GFP-DLC3) plasmids encode GFP-tagged versions of human DLC1, DLC2α, and DLC3α, respectively, and were described earlier (18, 19).
The extension .pgx indicates that the backbone plasmid is pGEX-4T-1 from GE Healthcare. GST-hDLC1[625–878].pgx (plasmid 816) (here called GST-DLC1-GAP) is an expression vector encoding the GST fusion human DLC1 GAP domain corresponding to nucleotide 1873–2634 of the human DLC1 gene (GenBank: AAK97501.1).
In Silico Prediction of Protein Interaction
The rigid docking of p120 RasGAP to DLC1 was performed using the PatchDock web server (20, 21), version beta 1.3, with the default complex type and a 4.0 Å clustering root mean square deviation. The DLC1 GAP domain (Protein Data Bank (PDB) ID: 3KUQ (22, 23)) was used as the receptor and the RasGAP SH3 domain (PDB ID: 2J05 (24)) as the ligand. The receptor and ligand binding sites were not imposed, and no additional constraint was applied during the docking. The best-scored calculated binding mode was retained for structural analysis.
Recombinant Protein Production and Isothermal Titration Calorimetry
Recombinant GST-DLC1-GAP domain was produced as reported previously using BL21 transformed with the GST-GAP-DLC1.pgx construct (25). Elution of the GST-DLC1-GAP domain in 30 mm reduced l-glutathione (Sigma-Aldrich, G4251, in 50 mm Tris (pH:7.4), 100 mm NaCl) was followed by 2 h of dialysis in 4 liters of PBS. The dialysis buffer was then used as titration buffer and to resuspend TAT-RasGAP317–326 and the corresponding tryptophan mutants. Isothermal titration calorimetry was performed as reported previously using the ITC200 system (GE Healthcare Life Sciences) unless otherwise mentioned (26). The sample cell (200 μl) was loaded with 50 μm GST-DLC1-GAP, whereas the TAT-RasGAP317–326 and the tryptophan mutant concentrations in the syringe were 500 μm. The titration experiment started with an injection of 0.5 μl followed by 15 injections of 2.49 μl, each lasting 5 s, with a 3-min interval between each injection. Controls and measurements were performed as reported earlier (26).
Statistical Analysis
All experiments were performed independently. The results were expressed as mean ± 95% confidence intervals. One-way ANOVAs and repeated measurement ANOVAs were performed using the R software (version 2.11.0) and were followed by a Tukey test for multiple comparisons. Asterisks denote statistically significant differences (p value < 0.05).
RESULTS
The individual contribution of each amino acid of the RasGAP 317–326 sequence was evaluated through an alanine-scanning approach. This approach has been successfully used to identify several protein-protein interactions (27). For example, it has been employed to map the binding between the human immunodeficiency virus (HIV) GP120 glycoprotein and the human CD4 receptor (28). This methodology consists of substituting amino acids of interest into alanine residues (27). Alanine that has a methyl side chain is considered as neutral and, in contrast to glycine that is also a small amino acid with only a hydrogen side chain, prevents introduction of conformational flexibility. Alanine substitutions therefore tend to keep intact the overall secondary structure of the investigated protein (27).
We first evaluated the consequence of these mutations on adhesion induced by TAT-RasGAP317–326 (from now on referred to as 317–326). Alanine substitutions of the tryptophan residues at positions 317 and 319 abrogated the capacity of the peptide to increase cell adherence (Fig. 1, A and B). Substitutions at positions 318, 320, 323, 324, and 325 strongly reduced the pro-adherence activity of 317–326, but some residual adherence was nevertheless observed. Replacement of amino acids 321, 322, and 326 had no or only limited effect on the ability of the peptide to increase cell adherence. These results indicate that the tryptophan and leucine residues of 317–326 are critically required for the pro-adhesion activity of the peptide. Residues 318, 320, and 324–325 are also important for this activity but do not appear as indispensable as the tryptophan residues because their replacement does not totally abolish the activity of 317–326. One could therefore generate a consensus sequence for RasGAP317–326-mediated adhesion increase, WMWVXXLRTX, where the X represents residues that are not required for this activity and where the bold residues are those that seem the most important for the pro-adhesive activity of the peptide. Interestingly, this consensus sequence is almost identical to the amino acids that are conserved between vertebrates and insects in the RasGAP 317–326 sequence (WXWVTXXRTX) (Fig. 1C). Hence the data presented in Fig. 1 indicate that most evolutionary conserved amino acids are important for the increase cell adherence mediated by the 317–326 sequence of RasGAP.
We next assessed whether the amino acids identified to be required for the pro-adhesion activity of 317–326 were also playing a role in the capacity of the peptide to inhibit migration and to sensitize tumor cells to genotoxins. Similarly to the adhesion increase, alanine substitutions of the Trp-317, Trp-319, Leu-323, Arg-324, and Thr-325 amino acids completely or partially abrogated the capacity of 317–326 to inhibit cell migration (Fig. 2A) or to favor cell death induced by cisplatin, a commonly used chemotherapeutic agent (Fig. 2B). The Met-318 and Val-320 residues seem, however, to be differentially required for 317–326-mediated effects on apoptosis, adhesion, and migration (Fig. 2C). This differential requirement of Met-318 and Val-320 could be the consequence of 317–326 mediating its effects on apoptosis, adhesion, and migration via different signaling pathways. Alternatively, this could be explained by a difference in the sensitivity of the assays used to measure apoptosis, adhesion, and migration and/or a difference in the sensitivity of these biological processes themselves. We therefore performed a dose response with 317–326 and measured apoptosis sensitization, adhesion increase, and migration inhibition. Fig. 2D shows that these three assays or biological processes do not display similar sensitivities to the RasGAP-derived peptide. Indeed, the EC50 for apoptosis sensitization (5 μm) was lower than the EC50 for adhesion promotion (11.5 μm), which was lower than the EC50 for migration (16.5 μm). In other words, less 317–326 is required to render tumor cells more sensitive to cisplatin-induced apoptosis than is required to mediate increased adhesiveness and even less so to inhibit cell migration. This indicates that a decrease in the activity of the peptide will first impact on its capacity to inhibit migration, then on its ability to increase cell adherence, and lastly on its capacity to sensitize cells to anticancer treatments. Hence a mutation that partially affects the activity of 317–326 should have a stronger impact on migration inhibition than on cell adherence promotion and tumor sensitization. This is particularly obvious with the V320A mutation. This can also be seen somewhat with the M318A and L323A mutations. Therefore, these results are not contradictory with the possibility that modulation of apoptosis, adhesion, and migration by 317–326 is the consequence of one protein or molecule being targeted by the peptide.
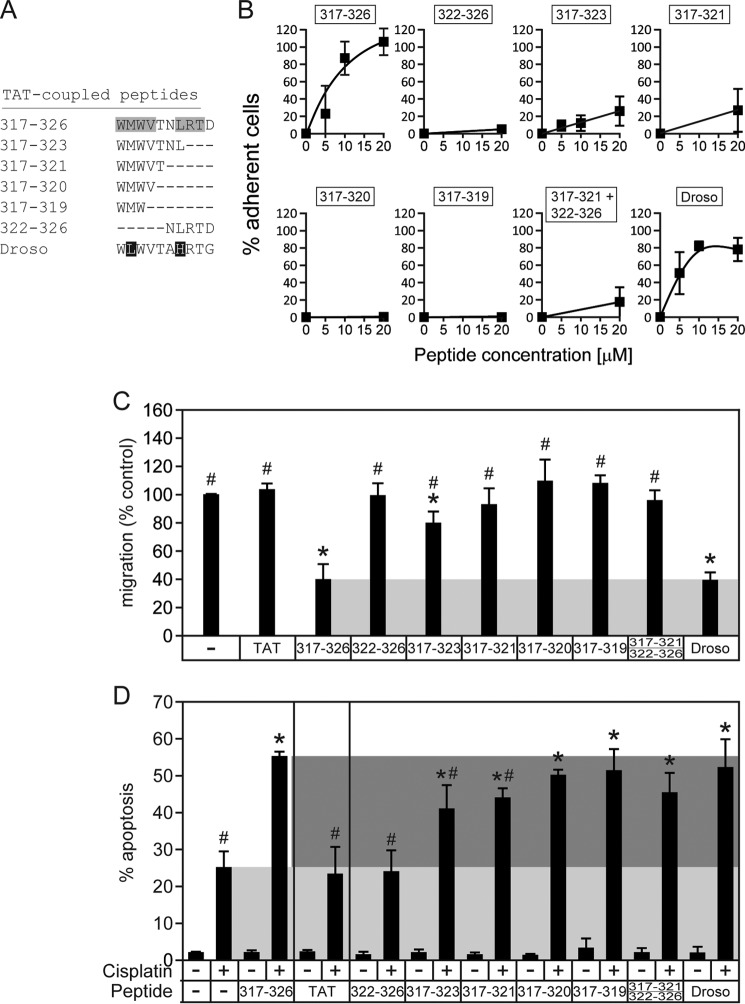
If the two N-terminal tryptophan residues of 317–326 are the only crucial residues for the activity of the peptide, it should be possible to shorten the 317–326 sequence without losing its activity. Hence, truncated versions of 317–326 were analyzed for their effects on adhesion, migration, and sensitization to apoptosis. Fig. 3, A and B, show that 317–323 and 317–321 still induce increased adherence, although to a lower extent than the parental 317–326 peptide. In contrast, 322–326 failed to augment cell adhesiveness. This indicates that the 317–321 sequence is sufficient to increase cell adherence. Although the 322–326 sequence lacked activity, it still may be required for optimal 317–326 activity as revealed by the point mutations within this sequence (Figs. 1 and 2). It was reported that the 322–326 sequence participates in the formation of a β-sheet required for dimerization of the RasGAP SH3 domains. One could therefore hypothesize that the 322–326 fulfills structural functions to constrain the 317–321 stretch in a conformation that is optimal for its activity. Co-incubation of 317–321 and 322–326 failed to potentiate 317–321-induced adhesion, indicating that both domains have to be carried by the same entity to be physiologically fully active. When the active peptides are narrowed down to four (317–320) and three (317–319) amino acids, their pro-adhesive property is even more decreased (Fig. 3B). We also tested the ability of the truncated peptides to inhibit migration (Fig. 3C) and to sensitize to cisplatin-induced apoptosis (Fig. 3D). The slight 317–323-mediated increase of adherence was translated into a weak but statistically significant inhibition of migration, whereas smaller peptides did not affect migration or affected it to a much lesser extent (Fig. 3C). On the other hand, the apoptosis-sensitizing effect of the truncated versions, including the 317–319 tripeptide, was similar to the one induced by the full-length peptide (Fig. 3D). Of note, the Drosophila peptide that carries a leucine residue at position 318 was highly efficient in recapitulating the effects of the human version (Fig. 3, A–D). Altogether, these results indicate that the WXW motif within the 317–326 sequence carries the anticancer activities of the peptide.
FIGURE 3.
Ability of truncated and variant RasGAP317–326 peptides to increase adherence, inhibit cell migration, and sensitize cancer cells to cisplatin-induced apoptosis. A, sequences of the peptides tested. Residues shaded in gray are those that are found required for adhesion based on the alanine-scanning performed in Fig. 1. The residues in white on a black background are those that differ from the residues shaded in gray. Droso, Drosophila. B, U2OS cells were cultured overnight and were subsequently treated for 8 h with 20 μm TAT or 20 μm of the indicated TAT-coupled peptides or left untreated. The cells were then subjected to trypsin-mediated detachment assays. Error bars indicate mean ± 95% confidence intervals, C, U2OS cells were subjected to wound-healing scratch assays in the presence of 20 μm of the indicated TAT-coupled peptides. Asterisks and number signs denote statistical significant differences after one-way ANOVA from the untreated condition and from the 317–326 treatment, respectively (n = 7 independent experiments). Error bars indicate mean ± 95% confidence intervals, D, U2OS cells were cultured overnight and were subsequently treated for 22 h with 20 μm of the indicated TAT-coupled peptides in the presence or in the absence of 30 μm cisplatin. The number of pyknotic nuclei was then scored and reported as the percentage of apoptosis. The light gray region displays the level of apoptosis induced by cisplatin alone, and the dark gray region shows the zone where sensitization occurs. Asterisks and number signs denote statistical significant differences after one-way ANOVA with the cisplatin-only treatment and with the cisplatin + 317–326 treatment, respectively (n = 6 independent experiments). Error bars indicate mean ± 95% confidence intervals.
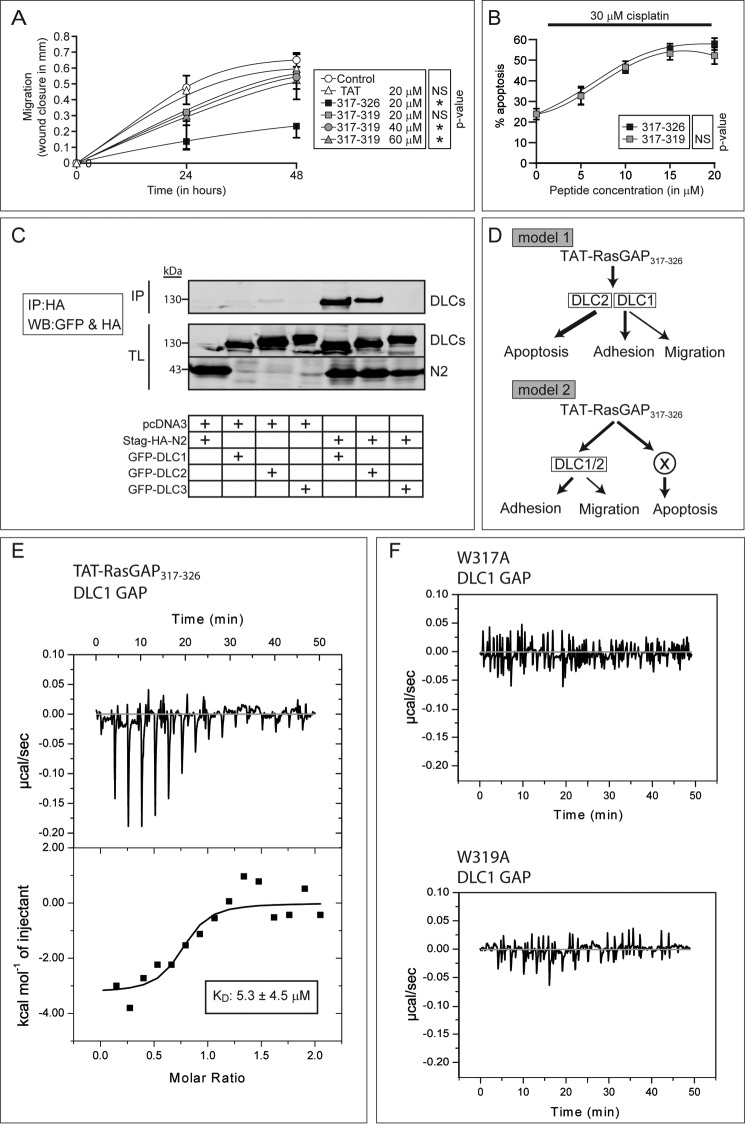
To better understand why the 317–319 tripeptide sensitizes tumor cells to apoptosis but does not recapitulate the effects on adhesion and more importantly on migration, we evaluated the effects of increased dose of 317–319 on migration. Wound closure was measured over a 48-h period to have a longer window for the evaluation of migration dynamics. At a 40 μm concentration, the 317–319 tripeptide significantly inhibited migration (Fig. 4A). When the 317–319 concentration increased to 60 μm, the effect on migration was not stronger than with 40 μm, indicating that 40 μm is already a saturating dose and that 317–319 cannot reach the inhibitory effect obtained with the full-length peptide. In contrast, the dose-response experiment depicted in Fig. 4B shows that 317–319 is as efficient as 317–326 in sensitizing tumor cells to cisplatin-induced apoptosis. This finding may indicate that two different molecules are targeted by 317–326 to mediate inhibition of migration and to sensitize tumor cells to death.
FIGURE 4.
Direct interaction of 317–326 with DLC1 requires a WXW motif that is sufficient to mediate the tumor sensitization effect of 317–326. A and B, effect of increasing doses of 317–319 on U2OS cell migration over time (A) and on sensitization to apoptosis induced by 30 μm cisplatin (B) (n = 3 independent experiments). Asterisks denote statistical significance after repeated measurement ANOVAs (A). Error bars indicate mean ± 95% confidence intervals, C, HEK-293T cells were transfected with the indicated combinations of plasmids. Two milligrams of cell lysates were immunoprecipitated (IP) using an anti-HA antibody. Forty μg of total lysates (TL) were also loaded. Western blotting (WB) against the HA tag (N2) and GFP (DLC1–3) was performed. The cropped blot is representative of three independent experiments. D, working models. Model 1 suggests that DLC1 and DLC2 mediate every 317–326 anticancer effects, whereas model 2 includes a third player that would mediate the effects of 317–326 on apoptosis. E, isothermal titration calorimetry was used to investigate the binding between 317–326 and the GAP domain of DLC1. This panel displays the raw data of the heat pulses resulting from titration of the recombinant GST-DLC1-GAP domain (50 μm) in the calorimetric cell with serial injections of 317–326 at a 500 μm concentration. The lower panel displays the integrated heat pulses (in kcal), normalized per mole of 317–326, as a function of the molar ratio (317–326 concentration/DLC1-GAP concentration). The resulting curve was fitted to a model equation allowing the determination of the KD. The dissociation constant and the molar ratio displayed here are the mean of three experiments. F, as controls, the recombinant GST-DLC1-GAP domain was titrated with the RasGAP W317A mutant peptide (upper right) and the RasGAP W319A mutant peptide (lower right) (n = 3 independent experiments).
The DLC family consists of three isoforms (DLC1, DLC2, and DLC3) with nonredundant activity (14). DLC1 regulates cell migration (29). DLC2 could modulate apoptosis as it can be localized at the mitochondria level (30). Fragment N2, the parental polypeptide from which 317–326 is derived, can bind DLC1 (11) but whether fragment N2 also binds the other DLC isoforms was not investigated. Fig. 4C shows that DLC2, but not DLC3, interacts with fragment N2. As fragment N2 binds both DLC1 and DLC2, this raises the possibility that 317–326 modulates adhesion/migration through DLC1 and apoptosis through DLC2 (model 1 in Fig. 4D). Another possibility would be that DLC1 and DLC2 are both required for the effects on adhesion/migration and that a still unidentified protein mediates the apoptotic response (model 2 in Fig. 4D). Jaiswal et al. (31) have shown that the SH3 domain of RasGAP binds to the DLC1 GAP domain using isothermal titration calorimetry (ITC). However, whether 317–326 directly binds DLC proteins has not been reported. We used ITC to assess this point. ITC records temperature changes generated when two molecules interact (32). Repeated injection of 317–326 into the cell containing the DLC1 GAP domain led to a marked variation in temperature (endergonic reaction) that was progressively reduced with subsequent 317–326 injections (indicative of binding site saturation and hence the specificity of the interaction) (Fig. 4E, upper panel). In contrast, injections of the two tryptophan peptide mutants (W317A and W319A) in the incubation cell of the ITC device containing the DLC1 GAP domain induced minimal temperature changes (these changes are caused by the injection of a given volume in the cell) (Fig. 4F). The dissociation constant (KD) of the interaction between the RasGAP 317–326 sequence and the DLC1 GAP domain was calculated to be ∼5 μm (Fig. 4E, lower panel). This KD is in agreement with the functional TAT-RasGAP317–326 EC50 values obtained for apoptosis (5 μm), adhesion (11.5 μm) and migration (16.5 μm) (Fig. 2D).
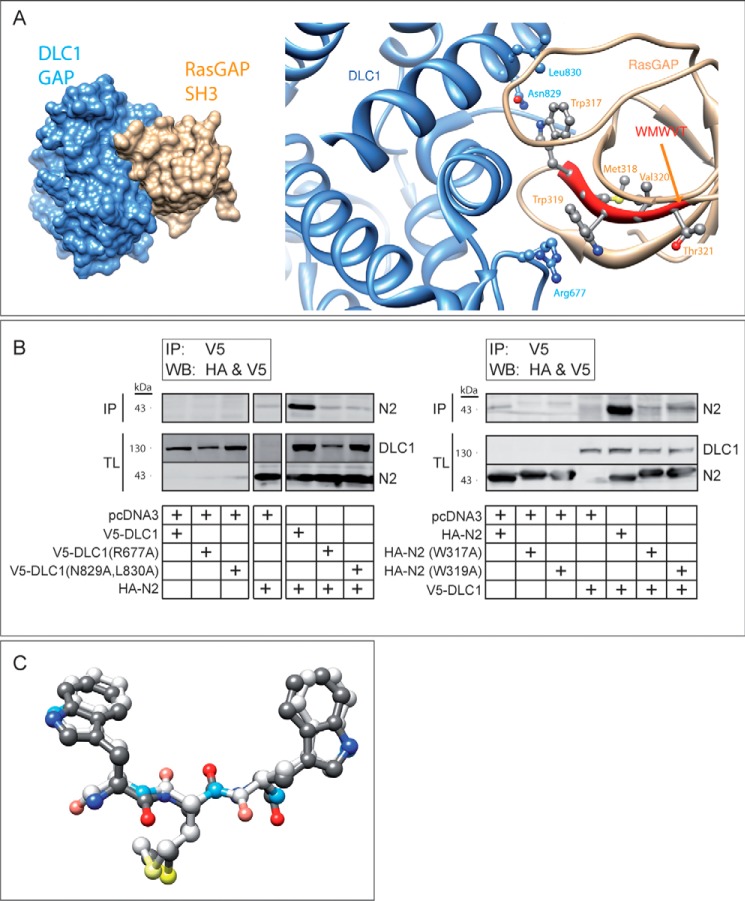
We next wanted to determine the binding mode of 317–326 to DLC1. In silico docking experiments of the RasGAP SH3 domain (PDB: 2JO5) to the DLC1 GAP domain (PDB: 3KUQ) revealed a preferred binding mode involving polar and nonpolar interactions of RasGAP Trp-317 to DLC1 Leu-830 and Asn-829, as well as a cation-π interaction between RasGAP Trp-319 and DLC1 Arg-677 (Fig. 5A). This in silico prediction was in agreement with the immunoprecipitation assays using DLC1 mutants bearing point mutations at position 677 and at positions 829 and 830. Fig. 5B (left blot) shows that these mutated DLC1 proteins have impaired fragment N2 binding abilities. Conversely, alanine substitutions of Trp-317 and Trp-319 in fragment N2 greatly reduced its capacity to bind wild-type DLC1 (Fig. 5B, right blot).
FIGURE 5.
Structural model of RasGAP binding to DLC1. A, in silico docking of DLC1 GAP domain (PDB ID: 3KUQ; in blue) and RasGAP SH3 domain (PDB ID: 2JO5; in orange) interaction. The right panel displays the interaction between the SH3 domain of RasGAP and the GAP domain of DLC1. This interaction involves the Trp-317 and Trp-319 residues of RasGAP and the Arg-677, Asn-829, and Leu-830 residues of DLC1. The left panel shows a lower magnification image of this interaction. This binding mode had a geometric shape complementarity score of 10024. See Refs. 20 and 21 for score interpretation. The next best binding mode (not shown) had a score of 9804. The spatially nearest calculated pose (not shown) had a root mean square deviation of 15.1 Å as compared with the binding mode displayed in the figure and had a geometric shape complementarity score of 9488. B, immunoprecipitation assays displaying the interaction between wild-type fragment N2 and mutants of DLC1 (left blot) and between wild-type DLC1 and mutants of fragment N2 (right blot). HEK 293T cells were transfected with the indicated combinations of plasmids. Two milligrams of cell lysates were immunoprecipitated (IP) using an anti-V5 antibody. Forty μg of total lysates (TL) were also loaded. Western blotting (WB) against the HA tag (N2) and the V5 tag (DLC1) was performed. The left blot was cropped, but all the lanes originate from the same exposure of one given blot. Cropped blots are representative of three independent experiments. C, retro-inverso WMW peptide (d-amino acids) superimposed on the conformation of the WMW residues of RasGAP (l-amino acids). The two peptides are colored according to the atom types. Lighter colors have been applied to the retro-inverso peptide.
Fig. 5A was derived from the structure of the RasGAP sequence in its natural l-form. One could question whether the TAT-RasGAP317–326 peptide, synthesized with d-amino acids, adopts a similar configuration. To assess this point, we performed an in silico superimposition between the WMW sequences in d-amino acids and the one derived from the SH3 domain of RasGAP (l-form, PDB ID: 2J05). Fig. 5C shows a virtual identical structure for both conformations, indicating that the d-amino acid version of 317–326 is likely to engage DLC1 as the corresponding natural l-form does.
DISCUSSION
Increasing the rate of death induced by chemotherapy specifically in cancer cells and impairing their invasiveness are prime strategies to fight cancer. We previously reported that a cell-permeable peptide derived from RasGAP, 317–326, had the ability to act as a dual anticancer peptide by sensitizing tumor cells to anticancer therapy-induced cell death and by hampering invasiveness (8, 9, 11). The mode of action of this peptide is still imprecisely defined. However, recent data demonstrated that the DLC1 tumor suppressor binds to the parental molecule (fragment N2) that contains 317–326 and that DLC1 is required for some of the anticancer activities of 317–326 (11). This new information allowed us to evaluate whether 317–326 is able to bind the DLC1 GAP domain using ITC and their mode of binding using site-directed mutagenesis and in silico modeling approaches. The KD of this interaction is about 10-fold higher than the KD calculated for the interaction between the SH3 domain of RasGAP with the DLC1 GAP domain (0.6 μm) (31). The weaker binding affinity of 317–326 with the DLC1 GAP domain could be explained by the number of amino acids involved in the interaction. Indeed, it appears that only two amino acids are necessary for this binding, whereas it has been reported that three amino acids are involved in the interaction between the RasGAP SH3 domain and the DLC1 GAP domain (31).
In this study, we show that sensitization to apoptosis, migration, and adhesion was differentially modulated by 317–326 (Fig. 2D) but that this globally required the same crucial amino acids (Fig. 2C). The full-length RasGAP peptide could be narrowed down to a three-amino acid peptide, 317–319, that still bears most of the anticancer activities of the longer construct (Fig. 4, A and B). However, in contrast to its effect on apoptosis sensitization, the ability of 317–319 to inhibit migration was reduced as compared with that of the 10 amino acid 317–326 sequence. This suggests that two targets are involved in transducing the RasGAP peptide anticancer properties. Here, we show that another DLC isoform, DLC2, is also engaged by fragment N2 (Fig. 4E), raising the possibility that the second target is DLC2. However, this remains to be demonstrated. It has been previously reported that the SH3 of RasGAP inhibits the GAP activity of DLC1 (33). These results have been confirmed very recently by the observation that RasGAP SH3 domain inhibits the GAP activity of DLC1 in vitro (31). Therefore, a reasonable assumption is that 317–319, by engaging the Arg-677 finger of DLC1, also inhibits the GAP activity of DLC1. However, DLC1 exerts some of its functions in a GAP-independent manner (29), and it remains possible that 317–326 mediates its effects by modulating the GAP-independent activities of DLC1. In favor of this second possibility is our recent observation that Rho inhibition using the C3 exoenzyme does not prevent 317–326 from increasing adhesion (11).
We have demonstrated here that the Trp-317 and Trp-319 residues are required for 317–326 to mediate its anticancer effects. The importance of these residues for protein-protein interaction is consistent with previously published structural data showing that Trp-317 and Trp-319 establish a hydrophobic pocket allowing the dimerization of the SH3 domain of RasGAP (24). Data collected in the present study suggest that other amino acids, although not strictly required for the biological activity of the RasGAP-derived peptide, are nevertheless of importance, in particular as structural scaffolding residues (amino acids 323–325). Interestingly, the hydrophilic residues of 317–326 (Thr-321, Asn-322, and Asp-326), known to be exposed at the surface of the RasGAP SH3 crystal (24), are dispensable for 317–326-mediated anticancer effects. This suggests that hydrophobicity is a key chemical property through which 317–326 (and likely the endogenous p120 RasGAP SH3 domain) functions. Met-318, a residue that is not strictly conserved during evolution but whose hydrophobicity is however evolutionary conserved (leucine in insects), could be indeed replaced by a hydrophobic leucine residue without any loss in activity of 317–326. Another important question is whether a WaW motif (where a represents an aliphatic residue) can be found elsewhere within the human proteome and be a putative target of DLC1/2. Not surprisingly, hundreds of proteins bear such a motif, but none of them correspond to known DLC-interacting proteins or are predicted to be involved in biological functions potentially modulated by the DLC proteins (data not shown) (29).
The discovery that a very short peptidic motif, consisting of only two tryptophan amino acids separated by a spacer residue (WXW), still bears, at least to some extent, the anticancer activities of the parental molecule provides the basis to undertake the rational design of small molecules with similar activities. Such drugs, by potentiating chemotherapy and impinging on metastatic progression, may be of considerable benefit to cancer patients.
This work was supported by a grant from Oncosuisse (KFS-02543-02-2010). C. W. is a co-inventor of the TAT-RasGAP317–326 compound as an antitumor agent (patent owned by the University of Lausanne) and may receive royalties from patent licensing if the compound is commercialized.
- RasGAP
- Ras GTPase-activating protein
- DLC
- deleted in liver cancer protein
- ANOVA
- analysis of variance
- ITC
- isothermal titration calorimetry
- oligo
- oligonucleotide
- h
- human
- TAT
- transactivator of transcription.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Trahey M., McCormick F. (1987) A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238, 542–545 [DOI] [PubMed] [Google Scholar]
- 3. Pomerance M., Thang M. N., Tocque B., Pierre M. (1996) The Ras-GTPase-activating protein SH3 domain is required for Cdc2 activation and Mos induction by oncogenic Ras in Xenopus oocytes independently of mitogen-activated protein kinase activation. Mol. Cell. Biol. 16, 3179–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang J. Y., Michod D., Walicki J., Murphy B. M., Kasibhatla S., Martin S. J., Widmann C. (2004) Partial cleavage of RasGAP by caspases is required for cell survival in mild stress conditions. Mol. Cell. Biol. 24, 10425–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khalil H., Bertrand M. J., Vandenabeele P., Widmann C. (2014) Caspase-3 and RasGAP: a stress-sensing survival/demise switch. Trends Cell Biol. 24, 83–89 [DOI] [PubMed] [Google Scholar]
- 6. Yang J. Y., Widmann C. (2001) Antiapoptotic signaling generated by caspase-induced cleavage of RasGAP. Mol. Cell. Biol. 21, 5346–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang J. Y., Widmann C. (2002) The RasGAP N-terminal fragment generated by caspase cleavage protects cells in a Ras/PI3K/Akt-dependent manner that does not rely on NF-κB activation. J. Biol. Chem. 277, 14641–14646 [DOI] [PubMed] [Google Scholar]
- 8. Michod D., Annibaldi A., Schaefer S., Dapples C., Rochat B., Widmann C. (2009) Effect of RasGAP N2 fragment-derived peptide on tumor growth in mice. J. Natl. Cancer Inst. 101, 828–832 [DOI] [PubMed] [Google Scholar]
- 9. Michod D., Yang J. Y., Chen J., Bonny C., Widmann C. (2004) A RasGAP-derived cell permeable peptide potently enhances genotoxin-induced cytotoxicity in tumor cells. Oncogene 23, 8971–8978 [DOI] [PubMed] [Google Scholar]
- 10. Barras D., Lorusso G., Lhermitte B., Viertl D., Rüegg C., Widmann C. (2014) Fragment N2, a caspase-3 generated RasGAP fragment, inhibits breast cancer metastatic progression. Int. J. Cancer 135, 242–247 [DOI] [PubMed] [Google Scholar]
- 11. Barras D., Lorusso G., Ruegg C., Widmann C. (2014) Inhibition of cell migration and invasion mediated by the TAT-RasGAP317–326 peptide requires the DLC1 tumor suppressor. Oncogene 10.1038/onc.2013.465 [DOI] [PubMed] [Google Scholar]
- 12. Raftopoulou M., Hall A. (2004) Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 13. Nürnberg A., Kitzing T., Grosse R. (2011) Nucleating actin for invasion. Nat. Rev. Cancer 11, 177–187 [DOI] [PubMed] [Google Scholar]
- 14. Durkin M. E., Yuan B. Z., Zhou X., Zimonjic D. B., Lowy D. R., Thorgeirsson S. S., Popescu N. C. (2007) DLC-1:a Rho GTPase-activating protein and tumour suppressor. J. Cell Mol. Med. 11, 1185–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saladin P. M., Zhang B. D., Reichert J. M. (2009) Current trends in the clinical development of peptide therapeutics. IDrugs. 12, 779–784 [PubMed] [Google Scholar]
- 16. Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 [DOI] [PubMed] [Google Scholar]
- 17. Nelson R. M., Long G. L. (1989) A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal. Biochem. 180, 147–151 [DOI] [PubMed] [Google Scholar]
- 18. Holeiter G., Heering J., Erlmann P., Schmid S., Jähne R., Olayioye M. A. (2008) Deleted in liver cancer 1 controls cell migration through a Dia1-dependent signaling pathway. Cancer Res. 68, 8743–8751 [DOI] [PubMed] [Google Scholar]
- 19. Holeiter G., Bischoff A., Braun A. C., Huck B., Erlmann P., Schmid S., Herr R., Brummer T., Olayioye M. A. (2012) The RhoGAP protein Deleted in Liver Cancer 3 (DLC3) is essential for adherens junctions integrity. Oncogenesis. 1, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–WW367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duhovny D., Nussinov R., Wolfson H. J. (2002) Efficient unbound docking of rigid molecules. in Algorithms in Bioinformatics, Lecture Notes in Computer Science, Vol. 2452, pp. 185–200, Springer-Verlag, Berlin Heidelberg [Google Scholar]
- 22. Rose P. W., Beran B., Bi C., Bluhm W. F., Dimitropoulos D., Goodsell D. S., Prlic A., Quesada M., Quinn G. B., Westbrook J. D., Young J., Yukich B., Zardecki C., Berman H. M., Bourne P. E. (2011) The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 39, D392–D401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rose P. W., Bi C., Bluhm W. F., Christie C. H., Dimitropoulos D., Dutta S., Green R. K., Goodsell D. S., Prlic A., Quesada M., Quinn G. B., Ramos A. G., Westbrook J. D., Young J., Zardecki C., Berman H. M., Bourne P. E. (2013) The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 41, D475–D482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ross B., Kristensen O., Favre D., Walicki J., Kastrup J. S., Widmann C., Gajhede M. (2007) High resolution crystal structures of the p120 RasGAP SH3 domain. Biochem. Biophys. Res. Commun. 353, 463–468 [DOI] [PubMed] [Google Scholar]
- 25. Erlmann P., Schmid S., Horenkamp F. A., Geyer M., Pomorski T. G., Olayioye M. A. (2009) DLC1 activation requires lipid interaction through a polybasic region preceding the RhoGAP domain. Mol. Biol. Cell 20, 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lapouge K., Perozzo R., Iwaszkiewicz J., Bertelli C., Zoete V., Michielin O., Scapozza L., Haas D. (2013) RNA pentaloop structures as effective targets of regulators belonging to the RsmA/CsrA protein family. RNA. Biol. 10, 1031–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morrison K. L., Weiss G. A. (2001) Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 5, 302–307 [DOI] [PubMed] [Google Scholar]
- 28. Ashkenazi A., Presta L. G., Marsters S. A., Camerato T. R., Rosenthal K. A., Fendly B. M., Capon D. J. (1990) Mapping the CD4 binding site for human immunodeficiency virus by alanine-scanning mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 87, 7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barras D., Widmann C. (2014) GAP-independent functions of DLC1 in metastasis. Cancer Metastasis Rev. 33, 87–100 [DOI] [PubMed] [Google Scholar]
- 30. Ng D. C., Chan S. F., Kok K. H., Yam J. W., Ching Y. P., Ng I. O., Jin D. Y. (2006) Mitochondrial targeting of growth suppressor protein DLC2 through the START domain. FEBS Lett. 580, 191–198 [DOI] [PubMed] [Google Scholar]
- 31. Jaiswal M., Dvorsky R., Amin E., Risse S. L., Fansa E. K., Zhang S. C., Taha M. S., Gauhar A. R., Nakhaei-Rad S., Kordes C., Koessmeier K. T., Cirstea I. C., Olayioye M. A., Häussinger D., Ahmadian M. R. (2014) Functional cross-talk between Ras and Rho pathways: a Ras-specific GTPase-activating protein (p120RasGAP) competitively inhibits the RhoGAP activity of deleted in liver cancer (DLC) tumor suppressor by masking the catalytic arginine finger. J. Biol. Chem. 289, 6839–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Neill M. A., Gaisford S. (2011) Application and use of isothermal calorimetry in pharmaceutical development. Int. J. Pharm. 417, 83–93 [DOI] [PubMed] [Google Scholar]
- 33. Yang X. Y., Guan M., Vigil D., Der C. J., Lowy D. R., Popescu N. C. (2009) p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. Oncogene 28, 1401–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]