Background: TbBILBO1 is the only known protein component of the flagellar pocket collar, but its assembly remains unknown.
Results: Structural dissections of the three different domains of TbBILBO1 revealed their roles in protein assembly.
Conclusion: TbBILBO1 forms a linear filament that interacts laterally to form a fibrous bundle.
Significance: The data show how two types of coiled coil act together to assemble TbBILBO1 into long filaments.
Keywords: Calcium-binding Protein, Cytoskeleton, Electron Microscopy (EM), Protein Assembly, Trypanosoma brucei, EF-hand, Leucine Zipper, TbBILBO1, Coiled Coil, Flagellar Pocket Collar
Abstract
Trypanosoma brucei BILBO1 (TbBILBO1) is an essential component of the flagellar pocket collar of trypanosomes. We recently reported the high resolution structure of the N-terminal domain of TbBILBO1. Here, we provide further structural dissections of its other three constituent domains: EF-hand, coiled coil, and leucine zipper. We found that the EF-hand changes its conformation upon calcium binding, the central coiled coil forms an antiparallel dimer, and the C-terminal leucine zipper appears to contain targeting information. Furthermore, interdimer interactions between adjacent leucine zippers allow TbBILBO1 to form extended filaments in vitro. These filaments were additionally found to condense into fibers through lateral interactions. Based on these experimental data, we propose a mechanism for TbBILBO1 assembly at the flagellar pocket collar.
Introduction
Trypanosomes are unicellular protists belonging to the order Kinetoplastida, an early diverging eukaryotic lineage (1). They are obligate pathogens responsible for a number of serious diseases in both humans and livestock (2–4). Trypanosoma brucei, which is the best studied species, lives freely in the bloodstream of an infected mammalian host. Transmitted by tsetse flies between host mammals and alternating, respectively, between the procyclic and bloodstream forms in these two life cycle stages, T. brucei is the causative agent of sleeping sickness in Africa (2).
Trypanosome cells are enclosed in a subpellicular corset of microtubules, giving them their characteristic shape (5). The parasite has a single flagellum that adheres lengthwise to the cell body, and its root is present in an intracellular invagination of the plasma membrane termed the flagellar pocket (FP).5 The FP is the sole site for all endo- and exocytic activity in trypanosomes (6). This activity not only involves the uptake of nutrients but also the removal of antibodies bound to its glycosylphosphatidylinositol-anchored surface glycoprotein coat (7–9). The FP constitutes only around 5% of the total plasma membrane, and given its essential role in metabolism and immune evasion, trypanosomes unsurprisingly exhibit some of the highest rates of endocytic and secretory traffic of any eukaryote (6, 10).
At the neck of the FP is an electron-dense structure termed the flagellar pocket collar (FPC). The FPC encircles the top of the FP to clamp the neck to the flagellar membrane that encloses the axoneme. A recent electron tomography study of the region around the FP suggests that the electron-dense material comprising the FPC forms a horseshoe shape, thus leaving a gap for placement of a specialized microtubule quartet that extends from alongside the basal body down the longitudinal axis of the cell to the cell anterior (12).
Identified in a screen for novel cytoskeletal localizations using polyclonal mouse antisera, T. brucei BILBO1 (TbBILBO1) was the first characterized component of the FPC (13). Loss of TbBILBO1 is lethal in both the procyclic and bloodstream forms of T. brucei, suggesting an essential role in cell viability. Depletion of TbBILBO1 by RNAi causes a cell cycle defect, preventing FP biogenesis (13). The mechanistic explanation for this effect is not known, nor is it clear what function TbBILBO1 is performing in interphase cells.
Cloning of full-length TbBILBO1 (TbBILBO1-FL) and structure-based functional analysis of the N-terminal domain (NTD) have been described recently (14). Here, we report our follow-up structural dissection of the other domains of TbBILBO1 and their respective roles in protein oligomerization. The results provide insights into the self-assembly of TbBILBO1 and suggest a molecular mechanism for the filament-like assembly of the protein at the FPC.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were obtained from commercial sources: HRP-conjugated anti-mouse (Pierce), anti-GFP (Roche Applied Science), Alexa Fluor 488 goat anti-mouse (Invitrogen), Alexa Fluor 568 goat anti-rabbit (Invitrogen). Sypro® Orange dye (5000× concentration in dimethyl sulfoxide) for thermal shift assays was purchased from Invitrogen. 5-nm Ni-NTA-Nanogold® was purchased from Nanoprobes.
Cloning
Cloning of TbBILBO1-FL has been described previously (14). Briefly, this involved amplification of the ORF from T. brucei genomic DNA by PCR and ligation into the HM15b vector. This vector provides an N-terminal His6-maltose-binding protein (MBP) tag. The His6 part of the tag can be removed using thrombin protease.
N-terminal YFP-tagged TbBILBO1 truncation constructs were subcloned from HM15b-TbBILBO1-FL into the pXS2-YFP vector as described previously (14). All TbBILBO1 truncation constructs carrying an N-terminal His6-MBP or MBP-His10 tag were subcloned from HM15b-TbBILBO1-FL into either the HM15b or the custom expression vector MalpET. Construct sequences were verified by DNA sequencing.
For production of untagged or MBP-His10-tagged recombinant protein of individual domains, PCR products of the TbBILBO1 EF-hand (EFh) (residues 178–250) and coiled-coil domain (CCD) (residues 263–533) were ligated into the MalpET vector. This vector provides an N-terminal MBP-His10 tag, which is cleavable by the tobacco etch virus protease.
Cell Lines, Culture, and Generation
The procyclic T. brucei 427 Lister strain was used for localization tests of TbBILBO1 targeting constructs. Transfection of cells was carried out as described previously (14). Transfected cells were cultured overnight in SDM79 medium supplemented with 7.5 μg/ml hemin and 20% (w/v) heat-inactivated fetal calf serum (Sigma-Aldrich) at 27 °C.
Protein Expression and Purification
All recombinant proteins were expressed in Escherichia coli BL21(DE3) cells. Bacteria transformed with cloned constructs were grown at 37 °C to an A600 of ∼0.6–0.8 and then subjected to cold shock (on ice for 30 min). Protein expression was induced by the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside, and protein production was continued for 20–22 h at 16 °C. Cells were harvested and lysed as described previously for the TbBILBO1-NTD using lysis buffer containing 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20 mm imidazole, and 5% (v/v) glycerol (14).
Protein purification was carried out initially on a Ni-HiTrap column (GE Healthcare) as described previously for the TbBILBO1-NTD (14). For His6-MBP-TbBILBO1-FL and truncation constructs, eluted proteins from the Ni-HiTrap column were either used directly for negative staining EM or further purified on a Sephacryl S-400 16/60 column (GE Healthcare) pre-equilibrated with 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 5% (v/v) glycerol for rotary metal-shadowing EM analysis. For MBP-His10-tagged TbBILBO1-EFh and -CCD, the N-terminal MBP-His10 tag derived from the vector was removed by incubation with 2% (w/w) tobacco etch virus protease while dialyzing against the same lysis buffer to remove excessive imidazole (overnight, 4 °C). The sample was then loaded again onto a Ni-HiTrap column to remove the cleaved MBP-His10 tag, and the target protein was further purified on a Superdex S-200 16/60 column (GE Healthcare) using the same running buffer as for TbBILBO1-FL. For rotary metal-shadowing EM experiments on MBP-His10-TbBILBO1-CCD, the sample was purified directly on a Superdex S-200 16/60 column following the first Ni-HiTrap purification.
Immunofluorescence Microscopy
Immunofluorescence microscopic analysis of transiently expressed YFP-TbBILBO1-FL, -NTD-EFh, -LZ, and -ΔLZ was carried out as described previously (14). Briefly, cells were attached to coverslips by centrifugation and fixed using 4% (w/v) paraformaldehyde. After permeabilization with Triton X-100, the fixed cells were washed with PBS and then blocked using BSA. Finally, sequential incubations with primary and secondary antibodies were carried out, and cells were imaged using an inverted microscope. Images were processed using ImageJ and Adobe Photoshop CS3 software (Adobe Systems, Inc., San Jose, CA).
Rotary Metal-shadowing Electron Microscopy
For rotary metal-shadowing EM analysis, purified MBP-His10-TbBILBO1-CCD, His6-MBP-TbBILBO1-FL, -ΔEF, and -ΔLZ, and TbBILBO1-ΔNTD were prepared at 0.05–0.1 mg/ml in EM buffer (100 mm (NH4)HCO3-H2CO3, pH 7.5, and 30% (v/v) glycerol) and sprayed onto freshly cleaved mica chips. The specimens were dried in a BAF400 high vacuum coater (Oerlikon Balzers, Balzers, Liechtenstein) for at least 6 h and then rotary-shadowed with 0.7-nm platinum/carbon at an elevation angle of 5° and backed with 10 nm carbon at 90 °C. Electron micrographs were taken on an FEI Morgagni 268D transmission electron microscope operated at 80 kV and equipped with an Olympus-SIS charge-coupled device camera. Images were examined and analyzed using ImageJ software.
Negative Staining and Nanogold-labeling Electron Microscopy
For negative staining EM analysis, His6-MBP-TbBILBO1-FL purified by Ni-HiTrap column (Novagen) was used. For Nanogold-labeling EM, full-length TbBILBO1 purified by Ni-HiTrap column was further dialyzed against the labeling buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 5% (v/v) glycerol) to remove the excess imidazole. Samples were incubated with 5-nm Ni-NTA-Nanogold (Nanoprobes) with a 1:50 molar ratio in the labeling buffer (30 min at room temperature) and loaded onto a Sephacryl S-400 16/60 gel filtration column to remove the unbound gold particles. Protein samples were adsorbed onto glow-discharged Formvar- and carbon-coated copper grids (30 s at room temperature) and stained in 2% (w/v) uranyl acetate (2 min at room temperature). The samples were viewed, and images were recorded either on an FEI Morgagni 80-kV electron microscope equipped with an Olympus-SIS CCD camera (for direct negative staining) or on a JEOL 1210 TEM with a Morada digital camera (for Nanogold labeling).
Circular Dichroism (CD)
CD measurements were performed using a Chirascan-plus CD spectrometer (Applied Photophysics). 150 μl of TbBILBO1-EFh (residues 178–250, 50 μg/ml) was incubated with either 2 mm CaCl2 or 2 mm EDTA (1 h at room temperature). Three independent CD spectra were collected (each in duplicate) between 190 and 260 nm, using a 0.5-cm path-length cuvette. A sampling time per point of 1 s and a bandwidth of 1 nm were used. CD signals of buffer alone were subtracted from all sample measurements.
Static Light Scattering (SLS)
SLS measurements were carried out on a mini-DAWNTM Treos® instrument (Wyatt Technology Corp.) connected in-line to an AKTA purifier equipped with a Superdex S-200 10/300 GL size exclusion chromatography (SEC) column. For molecular mass determination of the TbBILBO1-EFh, the column was pre-equilibrated overnight with SLS buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 2 mm CaCl2 or 2 mm EDTA). 100 μl of TbBILBO1-EFh (10 mg/ml) was incubated with either 2 mm CaCl2 or 2 mm EDTA (1 h at room temperature) and then injected to the column and run with a flow rate 0.5 ml/min. All measurements were taken at room temperature. The results were analyzed using the ASTRA program. For molecular mass determination of the TbBILBO1-CCD, the column was equilibrated with SLS buffer without CaCl2 or EDTA.
Thermal Shift Assay
The assay was carried out based on a previously published protocol (15). Solutions of 7.5 ml of 17× Sypro Orange (Molecular Probes) and 12.5 ml of protein buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 5% (v/v) glycerol) containing either 5 mm CaCl2 or 5 mm EDTA were used. 5 ml of TbBILBO1-EFh (2.5 mg/ml) was added onto a 96-well thin-wall PCR plate (Bio-Rad), which was then sealed with optical-quality sealing tape (Bio-Rad) and heated in an iCycler iQ real-time PCR detection system (Bio-Rad) from 20 to 95 °C with increments of 0.5 °C.
Chemical Cross-linking by Glutaraldehyde
The TbBILBO1-CCD (10 μl, 0.1 mg/ml) was incubated in cross-linking buffer (20 mm HEPES-NaOH, pH 8.0, and 100 mm NaCl) in the absence or presence of 0.01, 0.001, or 0.0001% (v/v) glutaraldehyde (room temperature for 10 min). Reactions were terminated by adding 1 μl of 1 m Tris-HCl, pH 8.0, to the 10-μl reaction solution. Results were analyzed by SDS-PAGE.
RESULTS
Protein Domain Organization
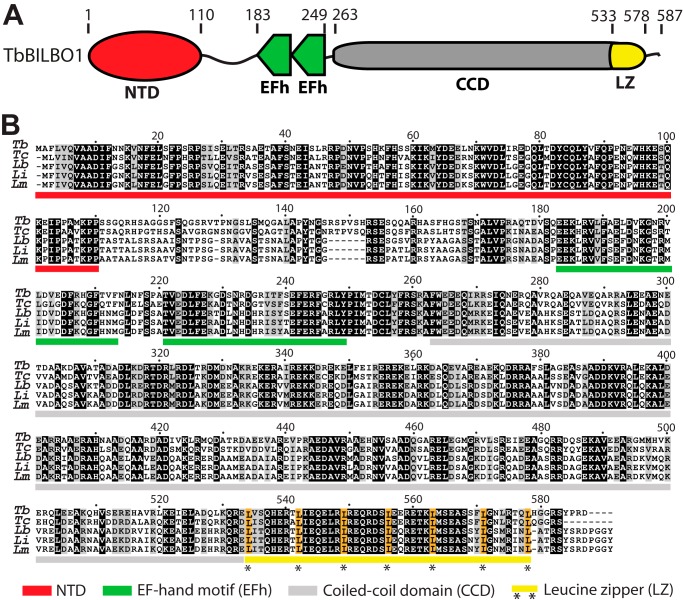
TbBILBO1 is a modular protein with three structured domains, comprising the NTD (residues 1–110), two tandem EFh motifs (residues 183–249), and the coiled-coil region (residues 263–578). Regularly spaced repetitive leucines in a predicted coiled coil are the characteristic feature of a leucine zipper (LZ) (16). Closer examination of the coiled-coil region by the 2ZIP-Server (17) revealed a putative LZ (residues 534–578) at its C-terminal end (Fig. 1A). The primary sequences of these structured domains are all highly conserved in the Trypanosomatida, with the exception of a linker region that lies between the NTD and the EFh (Fig. 1B). T. brucei and Trypanosoma cruzi exhibit a short (6 amino acids) insert in this linker region, which is not found in Leishmania species. Sequence conservation in the linker region is also lower than in the structured domains (Fig. 1B).
FIGURE 1.
TbBILBO1 domain organization and sequence alignment. A, schematic depicting the arrangement of the four domains of TbBILBO1: NTD, EF, CCD, and LZ. Amino acid numbers are indicated above the schematic. B, sequence alignment of TbBILBO1 and homologs from related trypanosomatids. Identical (black) and similar (gray) residues are shaded. Domains are indicated by bars under the aligned sequences using the color scheme detailed at the bottom. Conserved leucines in the C-terminal LZ are highlighted in gold. Tb, T. brucei; Tc, T. cruzi; Lb, Leishmania braziliensis; Li, Leishmania infantum; Lm, Leishmania major.
Targeting of TbBILBO1 to the FPC
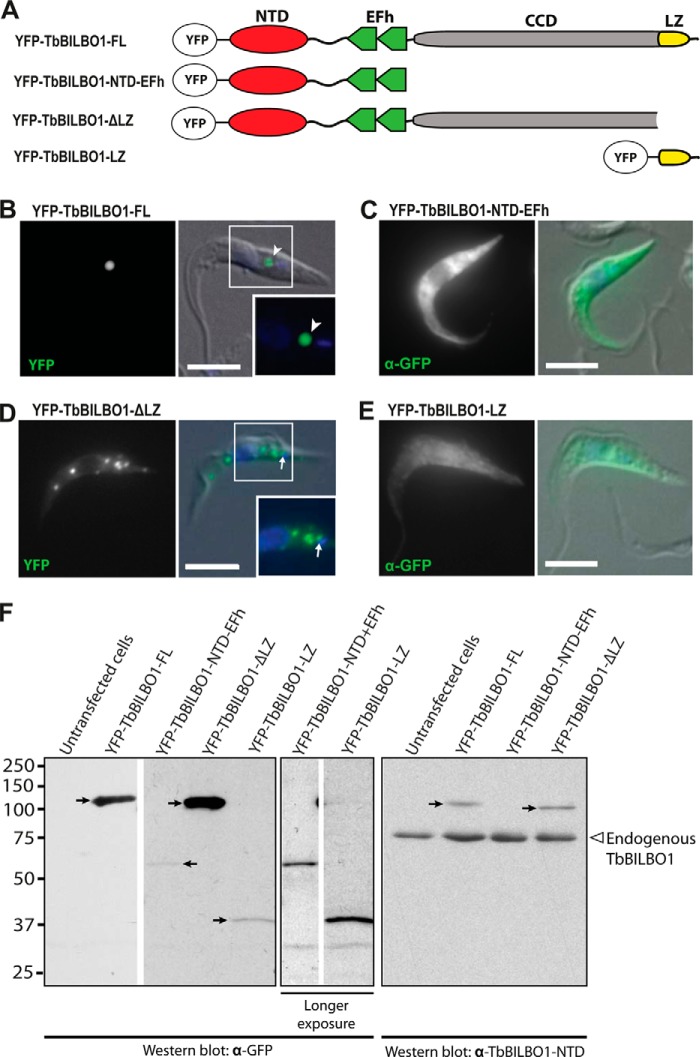
We recently reported that the NTD is not required for TbBILBO1 targeting to the FPC (14). To determine whether the putative LZ played a role in targeting, we tested three more YFP-tagged truncation constructs (Fig. 2A). These constructs were transiently expressed in T. brucei and their localization analyzed by immunofluorescence microscopy. YFP-TbBILBO1-FL localized to the FPC region (Fig. 2B, arrowhead), which matches the distribution seen for the endogenous protein (13). Similar to what was seen for the NTD alone (14), a construct containing the NTD and EFh domains localized to the cytoplasm and not to the FPC (Fig. 2C). Furthermore, a second, longer construct encoding the NTD, EFh, and CCD but not the LZ still did not target correctly. This construct (YFP-TbBILBO1-ΔLZ) instead formed numerous cytoplasmic punctae. Although there was a concentration of these punctae in the region of the FPC, they did not correctly localize to the FPC (Fig. 2D, arrow). This suggested that the LZ was required for correct targeting. The LZ by itself was cytoplasmic (Fig. 2E). It was concluded that the LZ is necessary but not sufficient for targeting to the FPC. Expression of the truncation constructs was confirmed by immunoblots of whole-cell lysates from transfected cells using anti-GFP antibodies (Fig. 2F, left and middle panels). All proteins were expressed, although the expression levels of the two short constructs, YFP-TbBILBO1-NTD-EFh and -LZ, were considerably lower than the others. Blotting with anti-TbBILBO1 antibodies showed that the total amount of any given truncation construct was significantly less than the total amount of endogenous TbBILBO1 in the population (Fig. 2F, right panel).
FIGURE 2.
The TbBILBO1-LZ is necessary but not sufficient for targeting to the FPC. A, TbBILBO1 truncation constructs used for the experiment. B–E, localization of YFP-tagged TbBILBO1 truncations in transiently transfected T. brucei cells. Fluorescence microscopy was used to visualize protein localizations. For YFP-TbBILBO1-NTD-EFh and -LZ, low expression levels precluded observation of YFP directly, and anti-GFP antibodies were used to visualize the protein instead. DAPI (blue) was used to stain DNA. The insets in B and D, indicated by white-outlined boxes, are enlargements of the smaller boxed areas. Scale bars: 5 μm. F, immunoblots of whole-cell lysates from transiently transfected T. brucei cells probed using anti-GFP antibodies or anti-TbBILBO1-NTD. The YFP-tagged proteins are indicated by arrows. Longer exposures were used for YFP-TbBILBO1-NTD-EFh and -LZ to better visualize the target proteins. Endogenous TbBILBO1 proteins are marked by an empty arrowhead. The YFP-TbBILBO1-LZ construct is not detectable with the anti-TbIBLBO1-NTD antibodies, as it does not contain the NTD.
The EFh Motifs Change Their Conformation upon Binding Calcium
Previously, we had determined the structure of the NTD and its essential role in cell viability but found that it is not required for TbBILBO1 localization (14). Given their predicted structural properties and/or role in FPC targeting (Figs. 1 and 2), it was hypothesized that the other domains of TbBILBO1 are important for its assembly at the FPC.
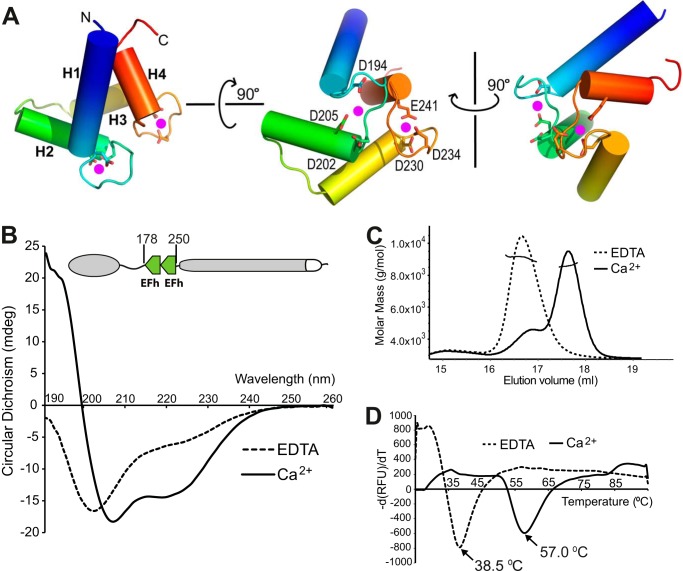
TbBILBO1 has two predicted tandem EFh motifs between residues 183 and 249. EFh motifs have been found in a large number of protein families, with most of them possessing the ability to bind calcium (18). The TbBILBO1-EFh is homologous to a human calmodulin-like protein, hCLP, with a sequence identity of 29.4% (19). Taking the hCLP structure as the template model, we carried out homology modeling of the TbBILBO1-EFh using the (PS)2-v2 protein structure prediction server (20). The results showed that the TbBILBO1-EFh has several conserved acidic residues in the loops, which are likely to coordinate calcium binding (Fig. 3A) (20).
FIGURE 3.
Calcium binding induces structural changes in the TbBILBO1-EFh motifs. A, three orthogonal views of the TbBILBO1-EFh (residues 178–250) derived from homology-based modeling. The schematic is color-ramped from blue at the N terminus to red at the C terminus. The four α-helices (H1–H4) in the two EFh motifs are shown as cylinders. Conserved acidic residues in the loops, which are likely to coordinate calcium binding, are shown as sticks. The two predicted calcium-binding sites are shown as magenta dots. B, far-UV CD spectra of recombinant untagged TbBILBO1-EFh in the presence of either 2 mm CaCl2 or 2 mm EDTA. Complete removal of calcium by EDTA resulted in the apo-form of the protein with a classical random coil structure, showing a maximum below 0 millidegrees and a minimum at ∼200 nm. Conversely, calcium binding led to a characteristic α-helical spectrum with minima at 209 and 220 nm. C, SLS results of the two forms of the TbBILBO1-EFh. Although both were monomeric, the apo-form was eluted much earlier than the calcium-loaded form, suggesting that in the absence of calcium, the EFh is improperly folded and therefore more loosely packed. It therefore behaves like a larger protein. D, melting curves of the TbBILBO1-EFh with and without loaded calcium measured by ThermoFluor assays. The melting temperatures for the apo- and calcium-bound forms were 38.5 and 57.0 °C, respectively. This further demonstrates that binding to calcium makes the EFh undergo a conformational switch from unfolded random coils to a well folded compact structure, conferring greater thermal stability.
It is known that EFh motifs often undergo structural rearrangements upon binding calcium (18, 21). To verify whether the putative EFh motifs in TbBILBO1 indeed bind calcium and whether calcium binding induces any conformational change to the protein, we performed biophysical analysis using CD, SLS, and temperature-based ThermoFluor methods. The experiments were carried out on untagged recombinant TbBILBO1-EFh (residues 178–250). The far-UV CD analysis showed that removal of calcium by 2 mm EDTA (apo-form) resulted in a classical random coil structure with a maximum below 0 millidegrees and a minimum at ∼200 nm. In contrast, calcium binding led to a characteristic spectrum for α-helices with two minima at 209 and 220 nm and a positive signal at ≤200 nm (Fig. 3B). The SLS results indicated that the apo-form EFh was eluted much earlier than the calcium-loaded form even though both proteins were monomeric and had the same molecular mass (Fig. 3C). This suggested that in the absence of calcium, the EFh motifs are improperly folded and therefore more loosely packed, allowing them to be eluted earlier from the gel filtration column. Furthermore, ThermoFluor assays showed that the melting temperatures for the calcium-bound and the apo forms were 57.0 and 38.5 °C, respectively (Fig. 3D). This further demonstrated that binding to calcium makes the EFh motifs become well folded, with a more compact structure that resists induced unfolding at high temperatures.
The TbBILBO1 Coiled-coil Domain Forms an Antiparallel Homodimer
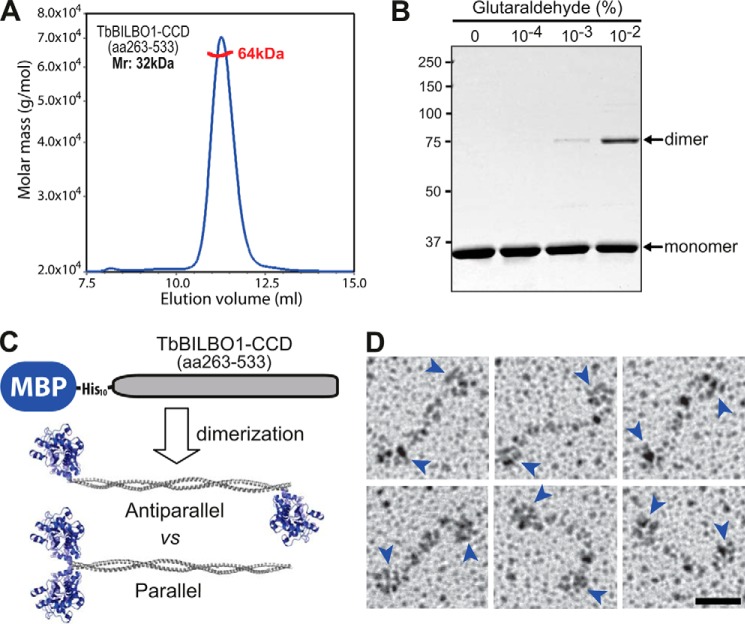
Given that the CCD (residues 263–533) represents nearly half of the full-length protein, it is necessary to know the structural arrangement of this domain in order to find out how TbBILBO1 assembles. Using SLS it was found that untagged recombinant CCD forms a dimer in solution (Fig. 4A). The dimeric conformation of the CCD was further confirmed by glutaraldehyde-mediated cross-linking, which showed the formation of homodimers after incubation of the protein with 0.001–0.01% (v/v) glutaraldehyde (Fig. 4B). Initial crystallization experiments using the CCD did not yield any crystals that diffracted well, so low resolution EM methods were employed instead to elucidate the relative orientation of the two coiled coils in the dimer. For these experiments, recombinant TbBILBO1-CCD was expressed with an N-terminal MBP-His10 tag. The 42-kDa MBP has a globular structure of ∼5 nm in diameter, which is significantly larger than the ∼1.5-nm width of a typical two-stranded coiled coil. If the CCD forms an antiparallel dimer, one would expect to see a dumbbell-like structure with two globules at opposite ends of the coiled-coil rod. Conversely, a parallel dimer would show a hammer-like structure with both MBP moieties spatially close to each other (Fig. 4C). Using rotary metal-shadowing EM, the fusion protein was found to appear in a dumbbell-like configuration, suggesting that the TbBILBO1-CCD forms an antiparallel dimer (Fig. 4D).
FIGURE 4.
The TbBILBO1-CCD forms an antiparallel dimer. A, SLS data from recombinant untagged TbBILBO1-CCD (residues 263–533) showed an average molecular mass of 64 kDa (red line), suggesting that it forms a dimer. B, cross-linking results of untagged TbBILBO1-CCD (residues 263–533) with different concentrations of glutaraldehyde. C, schematic showing the predicted distributions of the N-terminal MBP tag (42 kDa) in parallel and antiparallel dimers. D, gallery of rotary metal-shadowing EM images of MBP-His10-TbBILBO1-CCD. Blue arrowheads indicate the globular MBP tags. Scale bar: 20 nm.
The TbBILBO1 Leucine Zipper Mediates Protein Oligomerization
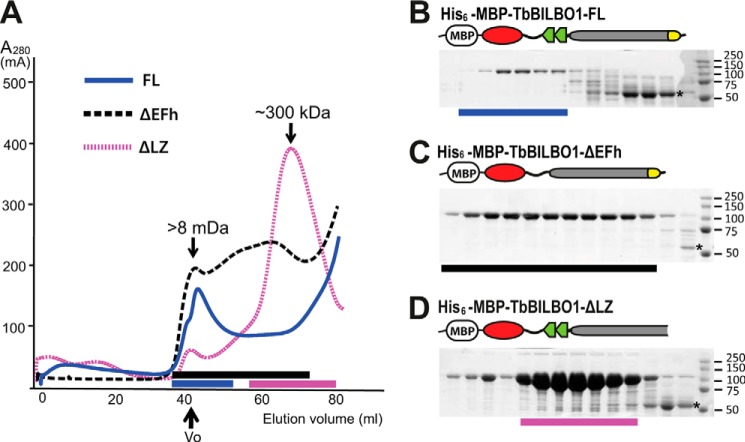
Recombinant His6-TbBILBO1-FL was expressed in bacteria, but no soluble protein could be purified by affinity chromatography.6 His6-MBP-TbBILBO1-FL was however soluble when expressed and could be purified by affinity chromatography. The purified protein was checked by SEC (Fig. 5). Curiously, a large fraction of His6-MBP-TbBILBO1-FL failed to pass through the column. The intact protein that did pass through was almost exclusively present in the void volume (V0) of the column, suggesting that the protein either aggregated or formed very large oligomers (Fig. 5, A and B). Deletion of the EFh motifs resulted in more protein being eluted, but the majority remained at V0 (Fig. 5, A and C). Interestingly, however, deletion of the C-terminal LZ not only allowed almost all protein to pass through the column but also shifted the elution peak to a position corresponding to a molecular mass of 300 kDa (Fig. 5, A and D). The gel filtration column separates proteins based on their Stokes radius, which is determined by both the molecular weight and the shape (23). An elongated molecule typically elutes much faster than a globular protein on SEC (24). Therefore, the 300-kDa peak is consistent with the ∼200-kDa dimeric His6-MBP-TbBILBO1-ΔLZ containing an extended coiled coil. This implied that the LZ mediates oligomerization of TbBILBO1.
FIGURE 5.
The TbBILBO1-LZ is essential for protein oligomerization. A, SEC elution profiles of three different His6-MBP-tagged constructs of TbBILBO1. The full-length protein was predominantly in the void volume (V0). Note that although deletion of the EFh motifs (ΔEFh) did not significantly alter its elution, removal of the leucine zipper (ΔLZ) caused a nearly complete shift to the lower molecular weight fractions. B–D, Coomassie-stained SDS-polyacrylamide gels showing proteins in the fractions indicated with colored bars as in A. The degradation products that contain both the His6-MBP tag and the TbBILBO1-NTD are starred.
The TbBILBO1 Leucine Zipper Mediates Filament Assembly
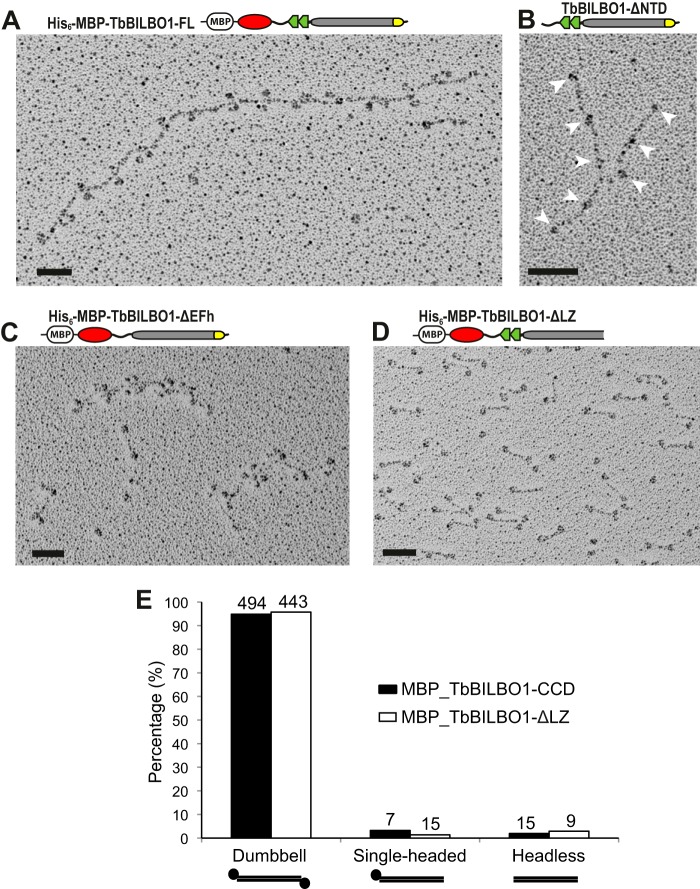
The behavior of His6-MBP-TbBILBO1-FL on SEC suggested that it might form large oligomers. To examine this possibility, rotary metal-shadowing EM experiments were carried out using the purified His6-MBP-TbBILBO1-FL eluted in the V0 of the SEC column. The EM images revealed a filament-like structure decorated with paired globules along the filament (Fig. 6A). The lengths of the interval rods were measured at 40–45 nm, in good agreement with the length of the TbBILBO1-CCD rod (Fig. 4D). The globular structures, 5–7 nm in diameter, matched the size of the MBP moiety together with the fused TbBILBO1-NTD. This assembly mode suggests that the oligomerization of TbBILBO1 is mediated by one or more of the structural elements flanking the CCD: the NTD and the EFh on the N-terminal side and the C-terminal LZ (Fig. 1A). This hypothesis was systematically tested by removing each domain in turn and then examining the purified protein by rotary metal-shadowing EM.
FIGURE 6.
The TbBILBO1-LZ mediates the formation of linear filaments. A, rotary metal-shadowing EM image of His6-MBP-tagged TbBILBO1-FL. The protein formed a linear structure with globular MBP moieties regularly spaced along the filament. B, deletion of the NTD did not preclude filament formation. Note that the untagged version of this construct is readily soluble and does not require an MBP tag; the smaller bead-like structures of the EFhs are still visible (arrowheads). C, deletion of the EFh motifs did not preclude filament formation, although the average length was decreased. D, deletion of the LZ completely disrupted filament assembly. Only dumbbell-like dimer structures were observed. Scale bars: 50 nm. E, the frequencies of each observed structure for both the coiled coil alone (see Fig. 4D) and LZ-deletion constructs were quantified and compared. Numbers above the bars are the number of structures present in the rotary metal-shadowing micrographs. The structures were counted in eight separate images obtained from two independent experiments.
Removal of the NTD did not significantly affect filament formation, although it led to a reduction in the average filament length (Fig. 6B). Deletion of the EFh motifs produced a similar effect (Fig. 6C). However, deletion of the LZ completely abolished filament assembly, resulting in only dumbbell-like structures as seen for the MBP-His10-tagged CCD alone (compare Fig. 6D with Fig. 4D). The frequency of the dumbbell-like structures was quantified for the TbBILBO1-CCD (Fig. 4D) and compared with that seen for TbBILBO1-ΔLZ (Fig. 6D). For both constructs, the frequency of occurrence in the rotary metal-shadowing micrographs was around 95% (Fig. 6E). This emphasizes that in the absence of the LZ, TbBILBO1 preferentially forms antiparallel dimers. These results are consistent with the SEC data, which also suggested that deleting the LZ favored dimer formation (Fig. 5, A and D).
TbBILBO1 Assembles into Fibrous Bundles
Given that the His6-MBP-TbBILBO1-FL forms filament-like oligomers (Fig. 6A) and the majority of it was unable to pass through the SEC column (Fig. 5, A and B), it seemed likely that TbBILBO1 might form even larger and higher order assemblies. Therefore, His6-MBP-TbBILBO1-FL was examined directly after affinity purification without doing SEC. It was found that the linear filaments were capable of forming much larger assemblies. Negative staining EM images showed that two TbBILBO1 filaments could laterally interact with each other (Fig. 7, A and B). Furthermore, multiple filaments were frequently observed to interact laterally to assemble into a highly ordered fiber (Fig. 7, C and D). These bundled fibers could condense and extend for a few micrometers, suggesting that it is a specific interaction-mediated higher order assembly. Furthermore, the examination of ∼100 of these fibrous structures in our collected EM images indicated that, although the single and double filaments appeared in similar frequencies, the highly ordered fibrous bundle was much more abundant (Fig. 7E). This suggested that the bundled structure could be stably maintained after formation.
FIGURE 7.
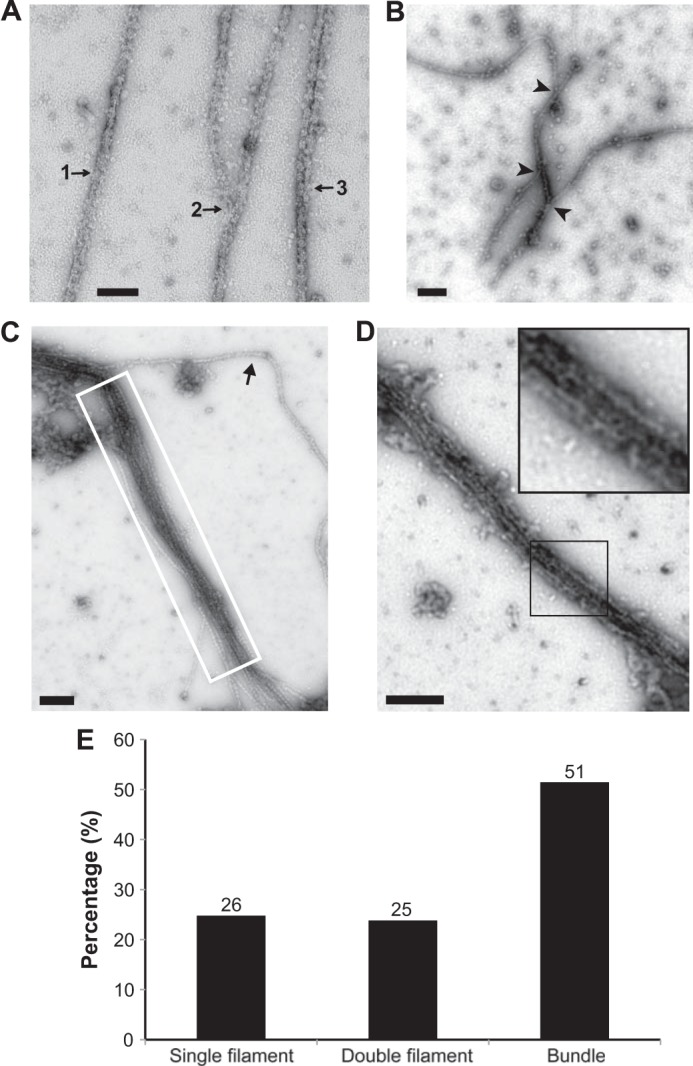
Lateral assemblies of TbBILBO1 linear oligomers analyzed by negative staining EM. A, lateral interaction of His6-MBP-TbBILBO1-FL filaments at three different stages: 1) single filament; 2) two semi-aligned filaments; 3) an assembled two-filament structure. B, lateral association of three filaments. Arrowheads indicate the junctions of the interactions. C, multiple laterally associated filaments of TbBILBO1 forming a bundle (white-boxed). The arrow indicates a filament that is only partially associated to one end of the multifilament bundle. D, a condensed bundle formed by multiple tightly associated TbBILBO1 filaments. All images were acquired using negative staining EM. The inset is an enlarged view of a segment of the structure. The inset is an enlarged region within the black-outlined box. Scale bars: 100 nm in A; 200 nm in B–D. E, the frequency of single, double, and bundled filamentous structures was quantified. Numbers above the bars are the number of structures found in the negative staining micrographs. The structures were counted in 22 separate images from two independent experiments.
To reveal the substructures of the lateral associations, we further labeled the N-terminal polyhistidine tag with 5-nm Ni-NTA-Nanogold (Fig. 8A). The labeled sample was then examined by negative staining EM (Fig. 8, B–E). The micrographs showed similar linear structures marked by gold particles ∼45 nm apart (Fig. 8B). These filaments were found to interact bilaterally with each other (Fig. 8C) and form two-stranded structures (Fig. 8D). Frequently, bundled structures formed by multiple filaments were observed (Fig. 8E). Furthermore, we found that the Nanogold particles were clustered regularly along the assembled bundles with intercluster distances of ∼45 nm (Fig. 8E, white arrowheads). It suggests that the TbBILBO1-NTDs in the fibrous bundles are in register and spatially close to one another.
FIGURE 8.
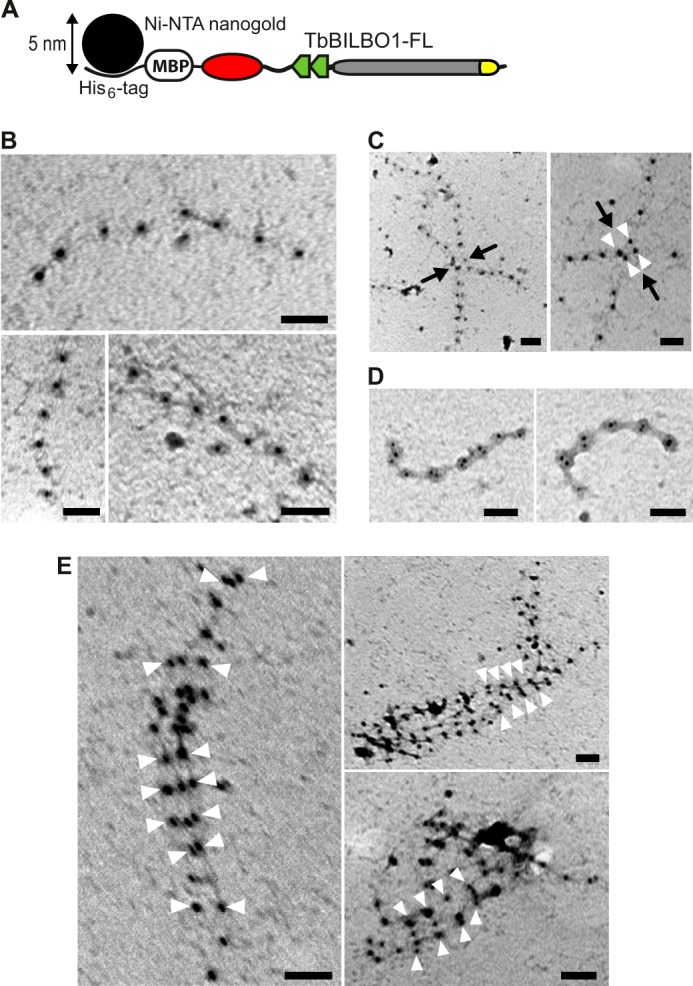
Linear and lateral assemblies of full-length TbBILBO1 revealed by Nanogold labeling-coupled negative staining EM. A, schematic showing the binding of the Ni-NTA-Nanogold particle to the His6 tag at the N terminus of MBP in the fusion protein. B, negative staining EM image of His6-MBP-TbBILBO1-FL labeled with 5-nm Ni-NTA-Nanogold. The oligomers are single linear filaments. C, lateral interactions of two filaments. Black arrows indicate the junctions of the interactions. White arrowheads mark the paired Nanogold particles. D, laterally associated two-filament structures. E, lateral assembly of multiple filaments. White arrowheads mark the clustered Nanogold particles in register. Scale bars: 50 nm.
DISCUSSION
We recently reported the high resolution structure of the TbBILBO1-NTD and demonstrated its essential requirement for cell viability (14). Here, we report the results of our investigation on the assembly mechanism of TbBILBO1. Bioinformatic analysis suggested that TbBILBO1 consists of four structural domains and not three as originally annotated (Fig. 1). The C-terminal end of the CCD was found to encode a putative LZ element. Immunofluorescence analysis using various truncations of the protein revealed that this C-terminal LZ is necessary but not sufficient for targeting (Fig. 2). The EFh motifs were found to bind calcium, which induced dramatic conformational changes to the protein (Fig. 3). To understand the architecture of the CCD, we employed rotary metal-shadowing EM to demonstrate that it forms an antiparallel dimer (Fig. 4). SEC results showed that purified recombinant TbBILBO1 forms large assemblies (Fig. 5). Further experiments were then carried out on the purified recombinant full-length protein and various truncations. Results from rotary metal-shadowing EM demonstrated that TbBILBO1-FL forms linear oligomers and that deletion of either the NTD or EFh domains does not significantly impair the oligomerization capacity. Removal of the C-terminal LZ, however, prevented the formation of oligomers and led to a predominance of dimeric forms (Fig. 6). When purified recombinant full-length TbBILBO1 was analyzed by EM without first separating the preparations by SEC, it was found that the protein formed large fibrous bundles with multiple strands associated together laterally (Fig. 7). Both the linear and the lateral assemblies of TbBILBO1 were also confirmed by Nanogold labeling EM studies (Fig. 8).
Drawing from all these observations, the following model for the structure and assembly of TbBILBO1 is proposed (Fig. 9). TbBILBO1 forms an antiparallel dimer via its central CCD, with the NTD and EFh domains projecting outward from the dimer in opposing directions. During purification of the various constructs of TbBILBO1, it was observed that the variable loop region between the NTD and EFh appears to be more prone to degradation than the rest of the protein. The ∼55-kDa degradation bands (Fig. 5, B–D, stars) were confirmed to contain both the MBP-His10 tag (42 kDa) and the TbBILBO1-NTD (∼13 kDa).6 This finding offers some support to the theory that the linker acts to sustain the NTD at a distance from the plane of the dimer.
FIGURE 9.
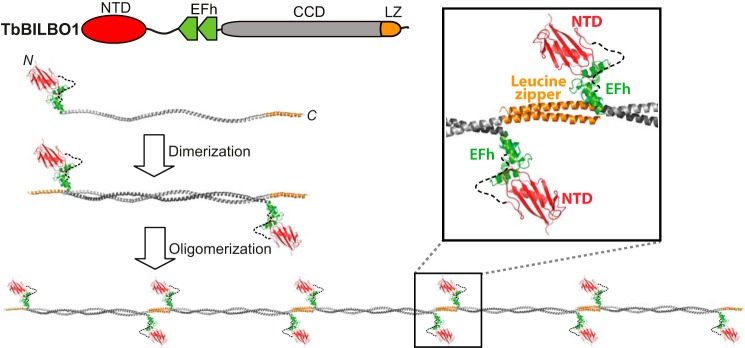
Putative assembly mechanism of TbBILBO1. This is a schematic of the inferred assembly mechanism of the TbBILBO1 filament. Two molecules form an antiparallel dimer via their CCDs, which then assembles into a filament through the interactions between the C-terminal LZs. Inset, an enlarged close-up view of the predicted interdimer junction is shown. The sizes of the CCD and LZ/NTD are not shown to scale.
The formation of interdimer contacts via the projecting C-terminal LZs then leads to the formation of a long linear filament (Fig. 9). The localization experiments showed that the LZ is necessary but not sufficient for correct targeting (Fig. 2). However, it is unclear whether the LZ specifically contains the targeting information or whether assembly of TbBILBO1 into oligomeric forms is a prerequisite for retention at the FPC. In the absence of the LZs, the truncated TbBILBO1 molecules were observed to form cytoplasmic punctae that clustered in the vicinity of the FPC, providing some tentative support for the latter hypothesis (Fig. 2D). Another open question is whether the position of the LZs makes the dimers radially symmetric; this has implications for the pitch of the filament and the relative orientation of the dimers within it.
Current knowledge about long homodimeric antiparallel coiled coils is very limited (25). Although the structures of many antiparallel coiled coils have been reported, most of them are intramolecular structures folding at a flexible hinge (26–28). Those that are formed intermolecularly are mostly heterodimers (29), and homodimeric antiparallel coiled coils are usually very short (30, 31). One exception is Rad50, which was found to form an 80-nm-long homodimeric antiparallel coiled coil upon ATP binding (32); but the Rad50 dimer does not assemble into filamentous polymers.
The morphology of the TbBILBO1 filament is reminiscent of that of previously reported lamin polymers (33, 34). However, in contrast to the antiparallel arrangement of the coiled-coil domain of TbBILBO1, lamin proteins form a parallel hammer-like homodimer, which further assembles linearly by head-to-tail interactions of neighboring dimers (33).
Most leucine zippers form parallel dimers (35–40). Nevertheless, both naturally occurring and designed antiparallel leucine zippers have been reported (11, 25, 41). Observation of the antiparallel homodimer of the LZ deletion construct (Fig. 6D) and the long filament of full-length TbBILBO1 (Fig. 6A) strongly suggests that the LZ is in an antiparallel configuration so as to mediate the interaction of two neighboring homodimers (Fig. 9).
Therefore, to the best of our knowledge, TbBILBO1 appears to be the first protein that not only contains a stably assembled long antiparallel coiled coil but is also interconnected by antiparallel LZs at the C termini of the helices to form a filament of potentially infinite length. The filament of TbBILBO1 is decorated with calcium-binding EF-hand motifs and functional globular domains (the NTDs), which together make TbBILBO1 a highly unusual protein oligomer. The positioning of the NTDs and EFh motifs in the filaments makes it possible that these domains have a role in coordinating the oligomeric interactions (Fig. 9, inset). This is partially supported by the observation that although deletion of these two regions does not affect filament assembly, it does appear to shorten the total length of the oligomers, implying a stabilizing function (Fig. 6, B and C). In the case of the EFh deletion, nonspecific lateral interactions seemed to occur, as demonstrated by multiple clustered bead-like structures along the filament (Fig. 6C). This offers further support for the hypothesis that the EFh might regulate the lateral associations of neighboring filaments. In the future, further examination of the EFh-regulated fibrous assembly of TbBILBO1 should be carried out in vitro and/or in vivo.
Previous studies showed that although overexpression of untagged and epitope-tagged TbBILBO1-FL is nontoxic, overexpression of either N- or C-terminally EGFP-tagged TbBILBO1 results in a growth arrest and a dominant negative phenotype similar to that seen following RNAi depletion of the protein (13). Our proposed assembly model of TbBILBO1 could provide a mechanistic explanation for these phenomena. In the model, both the N and C termini of TbBILBO1 are adjacent to the filament junction (Fig. 9, inset). It is tempting to speculate that the large size of the EGFP tag (37 kDa) might either sterically hinder the stable interaction at the junction or interfere with the lateral interactions between neighboring filaments (Fig. 9, inset).
The creation of the observed fibrous bundles of TbBILBO1 (Fig. 7) seems likely to occur by means of lateral interactions between filaments. The data obtained using Nanogold labeling of the filaments offer some support for the idea that the filaments are in register, with the NTDs in alignment (Fig. 8). This is in contrast to the spontaneous half-staggered lateral associations of vertebrate intermediate filament proteins (22).
In summary, our studies provide a structural view and shed light on the assembly mechanism for TbBILBO1 at the T. brucei FPC. Future studies will aim to provide a high resolution structural view of the interdimer junction of the filament and to investigate how TbBILBO1 regulates FPC biogenesis in vivo.
Acknowledgments
We are grateful to G. Warren for insightful input and constructive criticism during the course of the project. We thank J. Lesigang for technical assistance in the laboratory and G. Resch and M. Brandstetter for helping with our EM analysis.
This work was supported by funding from the Max F. Perutz Laboratories and by Grant P24383-B21 from the Austrian Science Fund (to G. D.).
K. Vidilaseris, E. Shimanovskaya, H. J. Esson, B. Morriswood, and G. Dong, unpublished data.
- FP
- flagellar pocket
- FPC
- flagellar pocket collar
- TbBILBO1
- Trypanosoma brucei BILBO1
- CCD
- coiled-coil domain
- EFh
- EF-hand
- FL
- full-length
- LZ
- leucine zipper
- MBP
- maltose-binding protein
- NTD
- N-terminal domain
- SEC
- size exclusion chromatography
- SLS
- static light scattering
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Simpson A. G., Stevens J. R., Lukes J. (2006) The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 22, 168–174 [DOI] [PubMed] [Google Scholar]
- 2. Welburn S. C., Maudlin I. (2012) Priorities for the elimination of sleeping sickness. Adv. Parasitol. 79, 299–337 [DOI] [PubMed] [Google Scholar]
- 3. Schmunis G. A., Yadon Z. E. (2010) Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop. 115, 14–21 [DOI] [PubMed] [Google Scholar]
- 4. Alvar J., Vélez I. D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M., and WHO Leishmaniasis Control Team (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7, e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gull K. (1999) The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 53, 629–655 [DOI] [PubMed] [Google Scholar]
- 6. Field M. C., Carrington M. (2009) The trypanosome flagellar pocket. Nat. Rev. Microbiol. 7, 775–786 [DOI] [PubMed] [Google Scholar]
- 7. Engstler M., Pfohl T., Herminghaus S., Boshart M., Wiegertjes G., Heddergott N., Overath P. (2007) Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131, 505–515 [DOI] [PubMed] [Google Scholar]
- 8. Barry J. D. (1979) Capping of variable antigen on Trypanosoma brucei and its immunological and biological significance. J. Cell Sci. 37, 287–302 [DOI] [PubMed] [Google Scholar]
- 9. Pal A., Hall B. S., Jeffries T. R., Field M. C. (2003) Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. Biochem. J. 374, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engstler M., Thilo L., Weise F., Grünfelder C. G., Schwarz H., Boshart M., Overath P. (2004) Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J. Cell Sci. 117, 1105–1115 [DOI] [PubMed] [Google Scholar]
- 11. Bryson J. W., Betz S. F., Lu H. S., Suich D. J., Zhou H. X., O'Neil K. T., DeGrado W. F. (1995) Protein design: a hierarchic approach. Science 270, 935–941 [DOI] [PubMed] [Google Scholar]
- 12. Lacomble S., Vaughan S., Gadelha C., Morphew M. K., Shaw M. K., McIntosh J. R., Gull K. (2009) Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 122, 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonhivers M., Nowacki S., Landrein N., Robinson D. R. (2008) Biogenesis of the trypanosome endo-exocytotic organelle is cytoskeleton mediated. PLoS Biol. 6, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vidilaseris K., Morriswood B., Kontaxis G., Dong G. (2014) Structure of the TbBILBO1 N-terminal domain from Trypanosoma brucei reveals an essential requirement for a conserved surface patch. J. Biol. Chem. 289, 3724–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 16. Landschulz W. H., Johnson P. F., McKnight S. L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA-binding proteins. Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 17. Bornberg-Bauer E., Rivals E., Vingron M. (1998) Computational approaches to identify leucine zippers. Nucleic Acids Res. 26, 2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewit-Bentley A., Réty S. (2000) EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 10, 637–643 [DOI] [PubMed] [Google Scholar]
- 19. Han B. G., Han M., Sui H., Yaswen P., Walian P. J., Jap B. K. (2002) Crystal structure of human calmodulin-like protein: insights into its functional role. FEBS Lett. 521, 24–30 [DOI] [PubMed] [Google Scholar]
- 20. Chen C. C., Hwang J. K., Yang J. M. (2009) (PS)2-v2: template-based protein structure prediction server. BMC Bioinformatics 10, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng C., Liu H. H., Zhou J., Zhang B. (2010) EF-hand domains of MCFD2 mediate interactions with both LMAN1 and coagulation factor V or VIII. Blood 115, 1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herrmann H., Aebi U. (2004) Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 73, 749–789 [DOI] [PubMed] [Google Scholar]
- 23. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dubin P. L., Principi J. M. (1989) Optimization of size-exclusion separation of proteins on a Superose column. J. Chromatogr. 479, 159–164 [DOI] [PubMed] [Google Scholar]
- 25. Oakley M. G., Hollenbeck J. J. (2001) The design of antiparallel coiled coils. Curr. Opin. Struct. Biol. 11, 450–457 [DOI] [PubMed] [Google Scholar]
- 26. Biou V., Yaremchuk A., Tukalo M., Cusack S. (1994) The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science 263, 1404–1410 [DOI] [PubMed] [Google Scholar]
- 27. Stebbins C. E., Borukhov S., Orlova M., Polyakov A., Goldfarb A., Darst S. A. (1995) Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature 373, 636–640 [DOI] [PubMed] [Google Scholar]
- 28. Melby T. E., Ciampaglio C. N., Briscoe G., Erickson H. P. (1998) The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142, 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Passon D. M., Lee M., Rackham O., Stanley W. A., Sadowska A., Filipovska A., Fox A. H., Bond C. S. (2012) Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. USA 109, 4846–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuccola H. J., Rozzelle J. E., Lemon S. M., Erickson B. W., Hogle J. M. (1998) Structural basis of the oligomerization of hepatitis delta antigen. Structure 6, 821–830 [DOI] [PubMed] [Google Scholar]
- 31. Lu Q., Ye F., Wei Z., Wen Z., Zhang M. (2012) Antiparallel coiled-coil-mediated dimerization of myosin X. Proc. Natl. Acad. Sci. USA 109, 17388–17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 33. Dittmer T. A., Misteli T. (2011) The lamin protein family. Genome Biol. 12, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gerace L., Huber M. D. (2012) Nuclear lamina at the crossroads of the cytoplasm and nucleus. J. Struct. Biol. 177, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diao J. (2010) Crystal structure of a super leucine zipper, an extended two-stranded super long coiled coil. Protein Sci. 19, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lavigne P., Crump M. P., Gagné S. M., Hodges R. S., Kay C. M., Sykes B. D. (1998) Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper. J. Mol. Biol. 281, 165–181 [DOI] [PubMed] [Google Scholar]
- 37. Junius F. K., O'Donoghue S. I., Nilges M., Weiss A. S., King G. F. (1996) High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J. Biol. Chem. 271, 13663–13667 [DOI] [PubMed] [Google Scholar]
- 38. Alber T. (1992) Structure of the leucine zipper, Curr. Opin. Genet. Dev. 2, 205–210 [DOI] [PubMed] [Google Scholar]
- 39. O'Shea E. K., Klemm J. D., Kim P. S., Alber T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 [DOI] [PubMed] [Google Scholar]
- 40. Song X., Li B., Xiao Y., Chen C., Wang Q., Liu Y., Berezov A., Xu C., Gao Y., Li Z., Wu S. L., Cai Z., Zhang H., Karger B. L., Hancock W. W., Wells A. D., Zhou Z., Greene M. I. (2012) Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep. 1, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oakley M. G., Kim P. S. (1997) Protein dissection of the antiparallel coiled coil from Escherichia coli seryl tRNA synthetase. Biochemistry 36, 2544–2549 [DOI] [PubMed] [Google Scholar]