Background: Lgr5 was shown to mark proliferating stem cells in several adult organs.
Results: Lgr5 expression was detected in proliferating hematopoietic stem and progenitor cells (HSPCs) at the early stage of hematopoiesis.
Conclusion: Lgr5 marks short-term (ST) hematopoietic stem and progenitor cells during the embryonic and fetal hematopoiesis.
Significance: Extending Lgr5 as a HSPC marker into the hematopoietic system during development.
Keywords: Embryo, Hematopoiesis, Proliferation, Stem Cells, Transplantation, Lgr5, Hematopoietic Stem Cells, Progenitor Cells
Abstract
Lgr5 is a marker for proliferating stem cells in adult intestine, stomach, and hair follicle. However, Lgr5 is not expressed in adult hematopoietic stem and progenitor cells (HSPCs). Whether Lgr5 is expressed in the embryonic and fetal HSPCs that undergo rapid proliferation is unknown. Here we report the detection of Lgr5 expression in HSPCs in the aorta-gonad-mesonephros (AGM) and fetal liver. We also found that a portion of Lgr5+ cells expressed the Runx1 gene that is critical for the ontogeny of HSPCs. A small portion of Lgr5+ cells also expressed HSPC surface markers c-Kit and CD34 in AGM or CD41 in fetal liver. Furthermore, the majority of Lgr5+ cells expressed Ki67, indicating their proliferating state. Transplantation of fetal liver-derived Lgr5-GFP+ cells (E12.5) demonstrated that Lgr5-GFP+ cells were able to reconstitute myeloid and lymphoid lineages in adult recipients, but the engraftment was short-term (4–8 weeks) and 20-fold lower compared with the Lgr5-GFP− control. Our data show that Lgr5-expressing cells mark short-term hematopoietic stem and progenitor cells, consistent with the role of Lgr5 in supporting HSPCs rapid proliferation during embryonic and fetal development.
Introduction
In mammals, the ontogeny of hematopoiesis occurs sequentially in several anatomical sites during development, including yolk sac, aorta-gonad-mesonephros (AGM)3 region, placenta, and fetal liver; finally, hematopoietic stem cells (HSCs) migrate to the bone marrow (BM) around the time of birth and support hematopoiesis through the rest of adult life (1, 2). One of the distinguishing features between embryonic or fetal HSCs and adult HSCs is that the former are cycling and undergo rapid proliferation and the latter are normally maintained in the quiescent or slow cycling state (3, 4).
Recently Lgr5 was shown to mark cycling stem cells in intestine, stomach, hair follicle, liver, mammary gland, and inner ear (5–10). While Lgr5 was not, or only very weakly, expressed in the adult BM, we asked whether Lgr5 was expressed in rapidly proliferating HSPCs during mouse embryonic development.
To investigate whether Lgr5 is involved in the development of HSPCs during early hematopoiesis, we evaluated the pattern of Lgr5 expression in embryonic and fetal hematopoietic organs including AGM region and fetal liver (2, 11). In mouse embryos, the generation of HSCs initiates in AGM on E10.5, and continues very rapidly through E12 (12–15). A pool of HSCs is formed predominantly in fetal liver on E12 by recruiting definitive HSCs via the blood circulation from extra-hepatic sources (AGM region and/or placenta) (15, 16). The definitive HSCs gradually form a massive pool in the fetal liver, becoming the main source of HSCs, which subsequently colonize the bone marrow around the time of birth (17–20). It is within these limited time windows and in these sites (E11.5-E12.5 AGM region and E11.5-E13.5 fetal liver) that we found Lgr5 expressing cells coincided with the generation and proliferation of HSPCs. These data show that Lgr5 expresses in HSPCs in the early stage of embryonic and fetal hematopoietic system, suggesting its potential role in facilitating HSPC proliferation and expansion.
EXPERIMENTAL PROCEDURES
Embryo Generation
Timed matings were set up between males of transgenic mouse line Lgr5GFP/+ (C57BL/6 background) and wild-type C57BL6/Ly5.2 females. Vaginal plug day was E0.5. Lgr5GFP/+ heterozygous embryos were collected from E11.5 to E12.5. Lgr5GFP/+embryos were identified under the fluorescence dissection microscope. All mice used in this study were housed in the animal facility at Stowers Institute for Medical Research (SIMR) and handled according to Institute and National Institutes of Health (NIH) guidelines. All procedures were approved by the Institutional Animal Care and Use Committee of SIMR.
Hematopoietic Tissues Dissection and Hematopoietic Cells Preparation
The dissection of hematopoietic sites (AGM and fetal liver) and dissociation of single hematopoietic cells were performed as described previously (13, 18, 21). Slight modifications were done for the latter part. AoMs (≤10) were collected into a 1.5 ml Eppendorf tube and incubated 15–20 min at 37 °C in the 1 mg/ml collagenase 1A (Sigma) before passing through the tissues 3 times with a 20 gauge needle and a 1 ml syringe, and then incubated another 10–15 min, followed by passing through the tissues 3 times with 22 gauge and 25 gauge needles and a 1 ml syringe (for AoM dissociation) or a 3 ml syringe (for fetal liver) and dissociated into single cells.
Whole-mount Immunostaining and Imaging
Whole-mount immunostaining was performed as described previously (22). Briefly, embryos were fixed for 2–3 h in 2% paraformaldehyde/PBS (6 ml) at 4 °C, and dehydrated in graded concentrations of methanol/PBS (50%, 75%, 100%; 10 min each). The yolk sac, rostral half of the body, limb buds, and lateral body wall were removed. Samples were incubated in Dent's bleach for 2 h at room temperature before being rehydrated with 50% methanol/PBS and several ice-cold PBS washes lasting 10 min each. Endogenous biotin activity was blocked with 3% BSA. All subsequent treatment was performed as reported (23). Goat anti-mouse was the primary antibody for GFP. Secondary antibodies were donkey anti-rat IgG-Alexa546 (Invitrogen) and donkey anti-goat IgG-Alexa488 (Invitrogen). Immunostained embryos were analyzed with a confocal microscope Zeiss LSM 5 PASCAL. Three-dimensional reconstructions were generated from z-stacks with Imaris X64 7.6.1.
Immunohistochemistry and Immunofluorescence Assay
The embryos (E7.5-E12.5) were fixed in Zn+ formalin from 30 min to 3 h at room temperature, according to the embryo size, and processed for paraffin sections. The volume of the formalin was at least 10 times that of the embryos. For immunostaining, we followed procedures reported previously (23, 24). Sections were incubated; Anti-GFP and anti-Ki67 were added to the tissue section slides.
Flow Analysis and Cell Sorting
We followed procedures reported previously (23, 24). For cell surface phenotyping, we used lineage markers including CD3, CD4, CD8, Mac-1, Gr1, B220, IgM, and Ter119 (eBioscience), CD45.1 and CD45.2. Monoclonal antibodies against CD45, C-Kit, CD41, and CD34 were also used. Dead cells were gated out by high 7AAD staining and forward light scatter. Cell sorting and analysis were performed using a MoFlo (Dako) and/or CyAn ADP (Dako). Data analysis was performed using FlowJo software.
Analysis of Multi-lineage Repopulating Activity
Total Lgr5-GFP+ cells (2000–3000) and Lgr5-GFP− cells were sorted from 4 E12.5 fetal liver derived from Lgr5GFP/+ embryos (CD45.2), and transplanted per irradiated (9Gy) adult recipient mouse (CD45.1) with 2 X 10e5 adult recipient spleen cells for rescue. We would like to point out that the sorting procedure could significantly reduce the engraftment. For example, we could use as low as 1 × 105 fetal liver cells without sorting to reach 50–60% of engraftment. Repopulation assays were carried out at 4, 12, and 16 weeks post transplantation by measuring the percentage of donor-derived CD45.2 cells in peripheral blood (engraftment), and lineage potentials were measured for their myeloid and lymphoid percentage.
Quantitative Real-Time RT-PCR (Q-RT-PCR)
We followed procedures reported previously (25). For each reaction, RNA from each type of sorted cell was divided into two or three reactions equivalent to 200 cells. Q-RT-PCR reactions were performed using an iCycler (Bio-Rad iQ5) according to the manufacturer's instructions. Amplification of GAPDH was used to normalize for sample RNA content. Specificity of products was confirmed by assessing band size in 2% agarose gels. The oligonucleotides used in Q-RT-PCR: Lgr5 (GPR49, 110bp), Forward-AGGCTGCCAAAAACTTCAGA, Reverse-TCCATGCTAAGTTCAGAGATCG; Runx1b (119bp), Forward-CCTCCGGTAGTAATAAAGGCTTCTG, Reverse-CCGATTGAGTAAGGACCCTGAA; Gata2 (297bp), Forward-GACTATGGCAGCAGTCTCTTCC, Reverse-GGTGGTTGTCGTCTGACAATT; Lgr4 (111bp), Forward-GGCTCAGCGCCTTCACCCAA, Reverse-ACCAGCCAGTTGTAGCTCCTCT; Gapdh (156bp), Forward-TGGCAAAGTGGAGATTGTTGCC, Reverse-AAGATGGTGATGGGCTTCCCG.
Statistics
Statistics were analyzed with Student's t test. The results are shown with S.D.
RESULTS
Detection of Lgr5-expressing Cells in AGM and Fetal Liver between E11.5 and E12.5
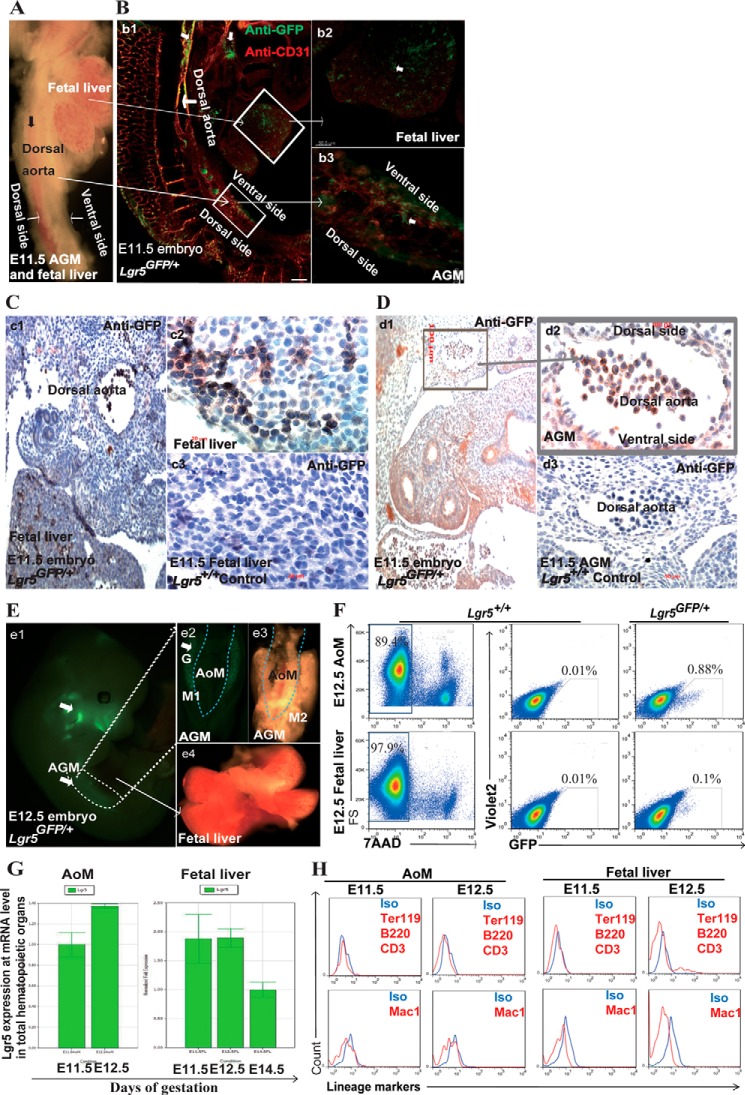
Using Lgr5-EGFP knock-in mice, we compared Lgr5GFP/+ and Lgr5+/+ embryos at the same developmental stage as control. We first conducted a whole-mount immunostaining of embryos at E11.5 and observed the Lgr5-GFP labeled cells in the fetal liver and dorsal aorta (major blood vessel), including intra-aortic clusters, and the ventral side of AGM region (22) (Fig. 1, A and B). We then conducted immunostaining and further confirmed that Lgr5-GFP+ cells were in the fetal liver sinusoidal lumens and within primitive hepatic cell cords (Fig. 1C) as well as in the dorsal aorta and mesenchyme tissue surrounding it (Fig. 1, C and D). We dissected and isolated fetal liver and the aorta and surrounding mesenchyme tissue (AoM) by removing the gonad bridges from AGM. Using flow cytometry, we examined Lgr5-GFP cells during E11.5–12.5, when definitive HSCs were generated in AGM region and recruited to fetal liver in the mouse embryos (Fig. 1, E and F). We then determined the Lgr5 expression at the mRNA level using Q-RT-PCR, and found that its relative expression level changed dynamically in accordance with the migration of HSCs from AGM to fetal liver during E11.5–12.5 (Fig. 1G). We further asked whether those Lgr5-GFP cells were hematopoietic cells and found that they did not express any mature hematopoietic markers including Ter119, B220, CD3, and Mac1 (Fig. 1H), suggesting that the Lgr5-GFP cells were either HSPCs or non-hematopoietic cells.
FIGURE 1.
Detection of Lgr5-GFP expression in AGM and fetal liver in Lgr5-GFP mouse embryos. A, a light image of live Lgr5GFP/+ AGM region and fetal liver at E11.5. B, confocal image of Lgr5-GFP and CD31 expression in Lgr5GFP/+ embryo at E11.5 after whole-mount embryo fluorescence staining: Lgr5-GFP expression in the major blood vessels including the wall of dorsal aorta and hematopoietic organs (b1, Scale bar, 300 μm); The boxed areas in (b1) are shown at high magnification (Scale bar, 50 μm) in b2, (fetal liver) and in b3 (AGM region). GFP signal was detected in the fetal liver (b2) and in the ventral side of the aorta and its surrounding mesenchyme tissue (b3). C, Lgr5-GFP+ cells were detected in the fetal liver in the cross section of Lgr5GFP/+ embryo at E11.5 (c1, scale bar, 100 μm), and a high magnification image of the boxed area is shown in c2 (scale bar, 10 μm). Lgr5-GFP+ cells were found in the fetal liver sinusoidal lumens and within primitive hepatic cell cords (c2). c3 shows a negative control from the Lgr5+/+ fetal liver (scale bar, 10 μm). D, Lgr5 expressed in the ventral side of aorta and its surrounding mesenchyme tissue in the cross section of Lgr5GFP/+ embryo at E11.5 (d1, Scale bar, 100 μm). A high magnification image of the boxed area is shown in d2 (Scale bar, 10 μm) and the negative control (Lgr5+/+ AGM at E11.5) is shown in d3 (scale bar, 10 μm). E, a live image of an Lgr5GFP/+ embryo and embryonic hematopoietic organs showing Lgr5-GFP+ signal in the head and AGM region (white arrow) (e1), Fluorescent image of the ventral side AGM region from E12.5 Lgr5GFP/+ embryo, strong GFP signal was observed in the urogenital ridges (white arrow) (e2). Bright field image of the dorsal side of E12.5 AGM showing the separation of AoM from the urogenital ridges containing the developing gonads (G) and mesonephro (M) along the dotted lines (e3). No visible GFP signal was observed in the live unstained E12.5 Lgr5GFP/+ fetal liver (e4). F, representative flow cytometry plots showing Lgr5 expression based on GFP fluorescence in Lgr5GFP/+ E12.5 AoM and fetal liver. All GFP signal was gated from 7AAD− population. The negative controls with the same gating were from Lgr5+/+ AoM and fetal liver cells. G, Q-RT-PCR analysis of Lgr5 expression in the developing hematopoietic organs: AoM (AGM region) at E11.5-E12.5 and fetal liver at E11.5-E14.5. H, representative histograms showing that lineage markers were not detected in Lgr5-GFP+ cells from AoM and fetal liver at E11.5-E12.5. Isotype antibody staining was used as negative control.
Proliferating Lgr5-GFP+ Cells Were Detected in AGM and Fetal Liver
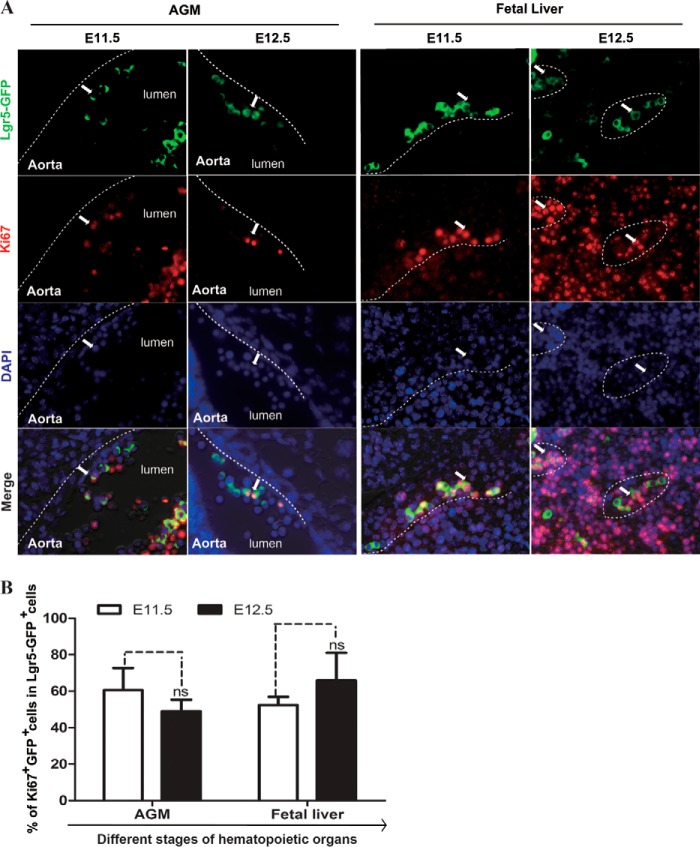
To assess the proliferation status of Lgr5-GFP cells in the hematopoietic system of the embryo, we stained embryos at E11.5 and E12.5 with an antibody recognizing the proliferation marker Ki67 (Fig. 2A). We found that Lgr5-GFP was co-expressed broadly with Ki67 in both AGM and fetal liver (Fig. 2B). In E11.5 AGMs, Lgr5, and Ki67 double positive cells were observed mainly within the intra-aortic clusters (Fig. 2A), where HSCs emerged and matured (13, 14). In E12.5 embryos, the main HSC expansion sites switched to the fetal liver, where proliferative Lgr5-GFP cells increased from 52% at E11.5 to 65.8% at E12.5 (Fig. 2B). Our data showed that in the mouse embryo, Lgr5-GFP cells appeared in the AGM region on E11.5, but their numbers declined in parallel with an increase from E11.5 to E12.5 in the fetal liver. Hence, the location and the number of Lgr5-GFP cells correlated well with the ontogeny of HSPCs.
FIGURE 2.
The majority of Lgr5-expressing cells are proliferating. A, co-staining of Ki67+ (red) with Lgr5-GFP+ cells (green) in the ventral side of intra-aortic clusters in E11.5 and E12.5 AGM, and in the sinusoid of E11.5 and E12.5 fetal liver. (Scale bar, 10 μm). The wall of the dorsal aorta (AGM region) and the outline of sinusoid (fetal liver) are shown along the dotted lines. B, bar graphs showing the percentage of Lgr5-GFP+ cells that are Ki67+ in different hematopoietic sites. * indicates significance and ns indicates no significant difference.
Lgr5-expressing Cells Express Genes That Are Critical for HSPCs Generation and Proliferation
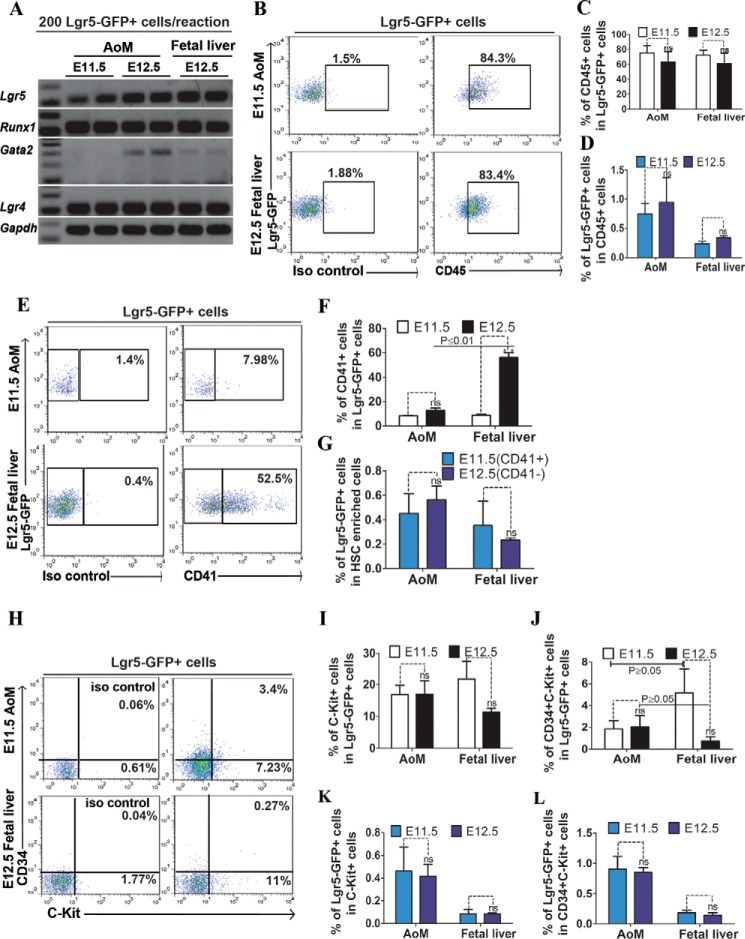
Transcriptional regulation is a key in controlling the emergence of HSPCs and their subsequent proliferation and maturation. Both gain and loss of function studies have demonstrated that Runx1 and Gata2 are critical for the specification, self-renewal, and differentiation of HSCs (26, 27). Using Q-RT-PCR, we examined the relative expression levels of the Runx1 and Gata2 genes. We detected their expression in, at least a portion of, Lgr5-expressing cells isolated from AoM and fetal liver (Fig. 3A). We also detected Lgr4 expression (Fig. 3A), which was reported to be often co-expressed with Lgr5 in Wnt-driven proliferative cells in adult tissue (8), and in Lgr5+ cells from AoM and fetal liver (Fig. 3A).
FIGURE 3.
Phenotypic analyses of Lgr5+ cells isolated from AoM and fetal liver at E11.5 and E12.5. A, RT-PCR analysis of Runx1 and Gata2 expression in 200 Lgr5-GFP+ cells isolated from AoM and fetal liver; Gapdh was as an internal control. We used flow cytometry and analyzed hematopoietic cell or HSC markers in Lgr5-GFP+ cells: CD45 (B), CD41 (E), and c-Kit and CD34 (H) in Lgr5-GFP+ cells isolated from E11.5 AoM and E12.5 fetal liver. Isotype antibody staining was used as control. Quantification of the percentage of selected markers expressing cells in Lgr5-GFP+ cells in AoM and fetal liver is shown in C, D, F, G, I, J, K, L. All error bars indicate S.D. * indicates significance, and ns indicates no significant difference.
HSPCs Related Surface Markers Are Expressed in Lgr5-expressing Cells, and Their Expression Changes Dynamically during Development
We stained single cells from Lgr5GFP/+ AoM and fetal liver at E11.5 and E12.5 with various surface markers and analyzed their expression in Lgr5-GFP cells using flow cytometry. We found Lgr5-GFP cells from these organs strongly expressed the pan-hematopoietic marker CD45 (27) (Fig. 3B). However, a fetal HSC marker CD41 was expressed primarily in fetal liver derived cells (28)(Fig. 3E). In contrast, c-Kit and CD34 were expressed at low levels in both populations (16, 29)(Fig. 3H). In the AoM, about 17% of Lgr5-GFP cells expressed c-Kit (Fig. 3I), and the percentage of CD45+ cells declined from 75% at E11.5 to 63% at E12.5 (Fig. 3C). Conversely, in the fetal liver, Lgr5-GFP+CD41+ cells increased from 9% to 56%. (Fig. 3F), while the percentage of c-Kit and CD34 marked Lgr5-GFP cells declined from 5% at E11.5 to 0.7% at E12.5 (Fig. 3, J–L). Hence, the data suggest that the population of Lgr5-GFP cells, which also express c-Kit and CD41, exhibits the phenotypic characteristics of HSPCs, and their location varies spatiotemporally during early embryogenesis.
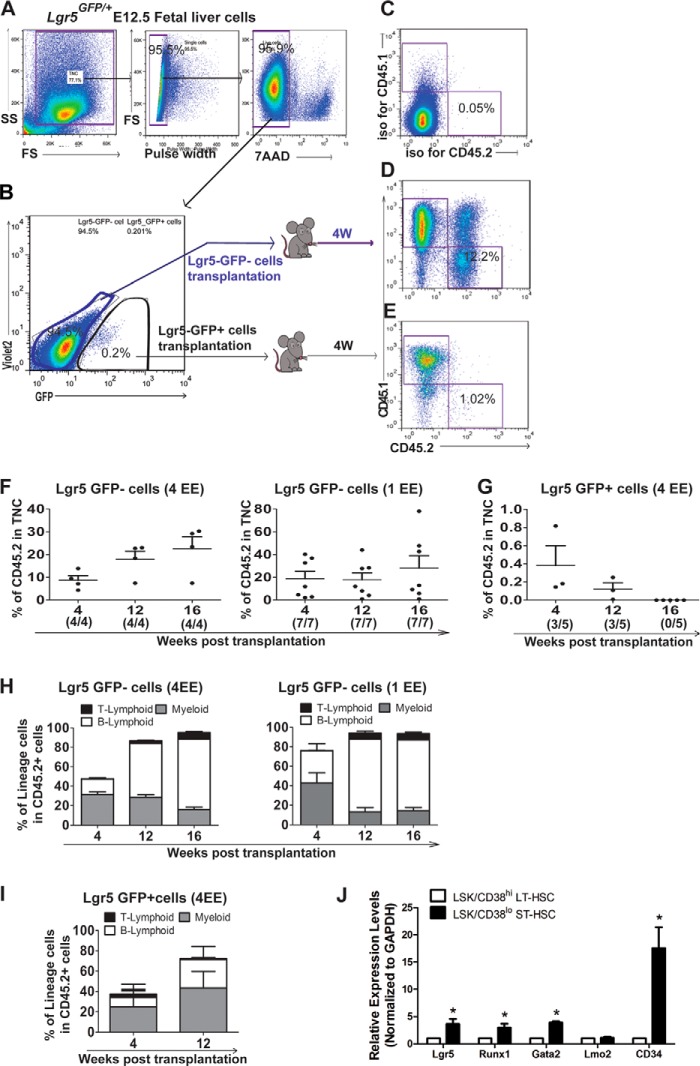
Lgr5-GFP Cells from Fetal Liver at E12.5 Can Give Rise to Hematopoietic Lineages with Low and Short-term Repopulation Activity
To test whether Lgr5-GFP cells could reconstitute hematopoietic lineages in vivo, we separated live (7AAD−) Lgr5-GFP+ cells and Lgr5-GFP− cells from Lgr5GFP/+ fetal livers (CD45.2) at E12.5 using fluorescent-activated cell sorting (FACS), and transplanted them respectively into irradiated adult recipients (CD45.1) (Fig. 4, A and B) together with 2 × 105 adult spleen cells (CD45.1) as rescue cells. While the control group from transplantation of Lgr5-GFP− cells showed a robust long-term reconstitution (20-fold higher at 4 weeks compared with the Lgr-5GFP+ group) (Fig. 4, D and F), we detected only a very low percentage of engraftment (on average 0.4%) with multilineage at 4 weeks post-transplantation from Lgr5-GFP+ donor cells (Fig. 4, E and G). Furthermore, the engraftment decreased to the baseline levels at 16 weeks (Fig. 4G). Taken together, our data show that Lgr5-GFP+ cells are short-term hematopoietic stem and progenitor cells with multilineage potential (Fig. 4, H and I) (30). This conclusion was further verified by our RT-PCR analysis of FACS sorted ST-HSCs (CD38loLSK) from fetal liver (31) (Fig. 4J), and the Lgr5, together with Runx1 and Gata2 were expressed 3–4-fold higher than that in LT-HSCs, while Lgr4 was higher in LT-HSCs, consistent with Lgr4 predominant expression in quiescent and lower expression in cycling intestinal stem cells (32).
FIGURE 4.
Functional characterization of Lgr5-GFP+ cells using repopulation assay. A, single live (7AAD−) cells were gated from Lgr5GFP/+E12.5 fetal liver cells (CD45.2). B, Lgr5-GFP+ and Lgr5-GFP− cells derived from a whole liver were further sorted and injected into irradiated (9Gy) adult recipient mice with rescue cells (CD45.1), respectively. The peripheral blood was collected for repopulation assay 4 weeks post-transplantation. C, base level of engraftment was set up using an isotype control antibody staining. D, representative flow cytometry plots showing the population of Lgr5-GFP−-derived cells (CD45.2). E, representative flow cytometry plots showing the population of Lgr5-GFP+ -derived cells (CD45.2). F, bar graphs showing the engraftment of donor cells (Lgr5-GFP− cells) at 4, 12, and 16 weeks post-transplantation (4/4 and 7/7 indicate how many mice out of transplanted recipients showed engraftment after 4 and 1 EE of donor cells injection). G, bar graphs showing the engraftment of donor cells (Lgr5-GFP+ cells) at 4, 12, and 16 weeks post transplantation (3/5 and 0/5 indicate how many mice out of transplanted recipients showed engraftment). H, bar graphs showing the percentage of myeloid and lymphoid lineage cells in Lgr5-GFP−-derived cells. I, bar graphs showing the percentage of myeloid and lymphoid lineage cells in Lgr5-GFP+-derived cells. J, bar graphs showing relative expression level of Lgr5, Runx1,Gata2, Lgr4, Lmo2, and CD34 genes in sorted ST-HSCs (CD38loLSK) from fetal liver.
DISCUSSION
It was recently reported that Lgr5 marks proliferating adult stem cells in several tissues. Although Lgr5 was not expressed in adult BM, here we showed that Lgr5 was expressed and potentially functions in the rapid proliferation of HSPCs during embryonic and fetal development.
Lgr5 Expression Is Correlated with Ontogeny of Hematopoiesis during Early Development
In this study, we found that Lgr5 was expressed dynamically in AGM region and fetal liver at E11.5-E12.5, when HSPCs underwent rapid expansion. Furthermore, the expression level of Lgr5 changed dynamically, implying that different niches may modulate HSPCs at various developmental stages. Our immunostaining of E11.5 and E12.5 embryos further confirmed that Lgr5 was expressed in the dorsal aorta (major blood vessel), including intra-aortic clusters, where pre-definitive HSCs underwent maturation (28) as well as in the mesenchyme surrounding the ventral side of aorta (AoM) region, where HSCs emerged and matured (33), and also in the fetal liver. Consistent with the cycling feature of Lgr5+ stem cells in other tissues, we showed that the majority of Lgr5-GFP+ cells were proliferating.
The Phenotypic Characteristics of Lgr5-expressing Cells Vary According to Tissue and Time during Early Embryogenesis
The expression of several HSPC-related genes and markers in Lgr5-GFP+ cells revealed their HSPC characteristics. At the mRNA level, Runx1 expressed in some of the Lgr5-GFP+ cells in the hematopoietic sites. Gata2 was weakly expressed in Lgr5-GFP+ cells. Moreover, at E11.5 AGM Gata2 was not expressed in Lgr5-GFP+ cells, consistent with the nature of Lgr5-GFP+ cells as ST-HSCs and progenitor cells and Gata2 expressed in the aortic endothelium and neighboring mesenchymal cells at E11.5 (34, 35). The majority of Lgr5-GFP+ cells did not express lineage markers, and a fraction of Lgr5-GFP+ cells expressed markers CD45, CD41, C-Kit, and CD34, with varying percentages and at different developmental stages. These observations confirmed that at least a fraction of Lgr5-expressing cells were hematopoietic cells with HSPCs phenotypic characteristics.
Lgr5-expressing Cells Reconstituted in Short-term Hematopoietic Lineages with Very Low Efficiency in Adult Recipient Mice
Given that only a fraction of Lgr5-expressing cells co-expressed stem cell signature genes or markers, the majority of Lgr5-GFP+ cells most likely enriched progenitor cells. However, our functional study revealed that a small number of Lgr5-GFP+ cells isolated from fetal liver at E12.5 were able to reconstitute hematopoietic lineages in adult recipient mice, albeit with very low efficiency. Taking into account an average 25% mosaic expression of Lgr5-GFP in the knock-in mice (6), the actual engraftment could be up to 4 times higher (on average 1.6% in theory at 4 weeks post-transplantation) in our experimental setting. Thus, while the majority of Lgr5-GFP cells were most likely proliferating hematopoietic progenitor cells, our data indicate that a small population of short-term Lgr5 expressing HSCs exists, consistent with the role of Lgr5 in supporting rapid proliferation of hematopoietic (stem) and progenitor cells during early stage embryo and fetal development. Compared with the single cell lineage tracing approach widely used in solid tissues, transplantation experiments offer the advantage of being able to test the behavior and properties of a population of candidate cells in terms of their robustness and duration in supporting tissue homeostasis and/or regeneration. Our observations here suggest a role for the Lgr5 and the associated Wnt-R-spondin signaling (32), in the biology of these proliferative, early hematopoietic progenitors.
Acknowledgments
We thank Drs. E. Dzierzak, N. Speck, and E. Yoder for scientific discussion and J. Haug and L. M. Wiedemann for technical training and scientific discussion. We appreciate technical support from the core facilities of Histology, Cytometry and Microscopy Center at the Stowers Institute for Medical Research. We thank all the members of the Li laboratory for scientific discussion.
This work was supported by the China Scholarship Council and the Stowers Institute for Medical Research.
- AGM
- aorta-gonad-mesonephros
- HSCs
- hematopoietic stem cells
- Q-RT-PCR
- quantitative real-time-PCR.
REFERENCES
- 1. Yoder M. C. (2004) Generation of HSCs in the embryo and assays to detect them. Oncogene 23, 7161–7163 [DOI] [PubMed] [Google Scholar]
- 2. Mikkola H. K., Orkin S. H. (2006) The journey of developing hematopoietic stem cells. Development 133, 3733–3744 [DOI] [PubMed] [Google Scholar]
- 3. Pietras E. M., Warr M. R., Passegué E. (2011) Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pietras E. M., Passegué E. (2013) Linking HSCs to their youth. Nature Cell Biol. 15, 885–887 [DOI] [PubMed] [Google Scholar]
- 5. Chai R., Kuo B., Wang T., Liaw E. J., Xia A., Jan T. A., Liu Z., Taketo M. M., Oghalai J. S., Nusse R., Zuo J., Cheng A. G. (2012) Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc. Natl. Acad. Sci. U. S. A. 109, 8167–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., Clevers H. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 7. Jaks V., Barker N., Kasper M., van Es J. H., Snippert H. J., Clevers H., Toftgård R. (2008) Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nature Genetics 40, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 8. Barker N., Huch M., Kujala P., van de Wetering M., Snippert H. J., van Es J. H., Sato T., Stange D. E., Begthel H., van den Born M., Danenberg E., van den Brink S., Korving J., Abo A., Peters P. J., Wright N., Poulsom R., Clevers H. (2010) Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 [DOI] [PubMed] [Google Scholar]
- 9. Huch M., Dorrell C., Boj S. F., van Es J. H., Li V. S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M. J., Haft A., Vries R. G., Grompe M., Clevers H. (2013) In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Plaks V., Brenot A., Lawson D. A., Linnemann J. R., Van Kappel E. C., Wong K. C., de Sauvage F., Klein O. D., Werb Z. (2013) Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 3, 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsu S. Y., Liang S. G., Hsueh A. J. (1998) Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol. Endocrinol. 12, 1830–1845 [DOI] [PubMed] [Google Scholar]
- 12. de Bruijn M. F., Ma X., Robin C., Ottersbach K., Sanchez M. J., Dzierzak E. (2002) Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 16, 673–683 [DOI] [PubMed] [Google Scholar]
- 13. Ma X., de Bruijn M., Robin C., Peeters M., Kong-A-San J., de Wit T., Snoijs C., Dzierzak E. (2002) Expression of the Ly-6A (Sca-1) lacZ transgene in mouse haematopoietic stem cells and embryos. Br. J. Haematol. 116, 401–408 [DOI] [PubMed] [Google Scholar]
- 14. Mascarenhas M. I., Parker A., Dzierzak E., Ottersbach K. (2009) Identification of novel regulators of hematopoietic stem cell development through refinement of stem cell localization and expression profiling. Blood 114, 4645–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Medvinsky A. L., Samoylina N. L., Müller A. M., Dzierzak E. A. (1993) An early pre-liver intraembryonic source of CFU-S in the developing mouse. Nature 364, 64–67 [DOI] [PubMed] [Google Scholar]
- 16. Ohneda O., Fennie C., Zheng Z., Donahue C., La H., Villacorta R., Cairns B., Lasky L. A. (1998) Hematopoietic stem cell maintenance and differentiation are supported by embryonic aorta-gonad-mesonephros region-derived endothelium. Blood 92, 908–919 [PubMed] [Google Scholar]
- 17. Dzierzak E. (2003) Ontogenic emergence of definitive hematopoietic stem cells. Curr. Opin. Hematol. 10, 229–234 [DOI] [PubMed] [Google Scholar]
- 18. Jordan C. T., McKearn J. P., Lemischka I. R. (1990) Cellular and developmental properties of fetal hematopoietic stem cells. Cell 61, 953–963 [DOI] [PubMed] [Google Scholar]
- 19. Galloway J. L., Zon L. I. (2003) Ontogeny of hematopoiesis: examining the emergence of hematopoietic cells in the vertebrate embryo. Curr. Topics Dev. Biol. 53, 139–158 [DOI] [PubMed] [Google Scholar]
- 20. Li L. (2005) Finding the hematopoietic stem cell niche in the placenta. Dev. Cell 8, 297–298 [DOI] [PubMed] [Google Scholar]
- 21. Bravo J., Li Z., Speck N. A., Warren A. J. (2001) The leukemia-associated AML1 (Runx1)–CBF beta complex functions as a DNA-induced molecular clamp. Nat. Struct. Biol. 8, 371–378 [DOI] [PubMed] [Google Scholar]
- 22. Yokomizo T., Yamada-Inagawa T., Yzaguirre A. D., Chen M. J., Speck N. A., Dzierzak E. (2012) Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nature Protocols 7, 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He X. C., Zhang J., Tong W. G., Tawfik O., Ross J., Scoville D. H., Tian Q., Zeng X., He X., Wiedemann L. M., Mishina Y., Li L. (2004) BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nature Genetics 36, 1117–1121 [DOI] [PubMed] [Google Scholar]
- 24. Sugimura R., He X. C., Venkatraman A., Arai F., Box A., Semerad C., Haug J. S., Peng L., Zhong X. B., Suda T., Li L. (2012) Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell 150, 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haug J. S., He X. C., Grindley J. C., Wunderlich J. P., Gaudenz K., Ross J. T., Paulson A., Wagner K. P., Xie Y., Zhu R., Yin T., Perry J. M., Hembree M. J., Redenbaugh E. P., Radice G. L., Seidel C., Li L. (2008) N-cadherin expression level distinguishes reserved versus primed states of hematopoietic stem cells. Cell Stem Cell 2, 367–379 [DOI] [PubMed] [Google Scholar]
- 26. North T. E., de Bruijn M. F., Stacy T., Talebian L., Lind E., Robin C., Binder M., Dzierzak E., Speck N. A. (2002) Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672 [DOI] [PubMed] [Google Scholar]
- 27. Taoudi S., Gonneau C., Moore K., Sheridan J. M., Blackburn C. C., Taylor E., Medvinsky A. (2008) Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell 3, 99–108 [DOI] [PubMed] [Google Scholar]
- 28. Ferkowicz M. J., Starr M., Xie X., Li W., Johnson S. A., Shelley W. C., Morrison P. R., Yoder M. C. (2003) CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development 130, 4393–4403 [DOI] [PubMed] [Google Scholar]
- 29. Delassus S., Titley I., Enver T. (1999) Functional and molecular analysis of hematopoietic progenitors derived from the aorta-gonad-mesonephros region of the mouse embryo. Blood 94, 1495–1503 [PubMed] [Google Scholar]
- 30. Morrison S. J., Uchida N., Weissman I. L. (1995) The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 11, 35–71 [DOI] [PubMed] [Google Scholar]
- 31. Randall T. D., Lund F. E., Howard M. C., Weissman I. L. (1996) Expression of murine CD38 defines a population of long-term reconstituting hematopoietic stem cells. Blood 87, 4057–4067 [PubMed] [Google Scholar]
- 32. de Lau W., Barker N., Low T. Y., Koo B. K., Li V. S., Teunissen H., Kujala P., Haegebarth A., Peters P. J., van de Wetering M., Stange D. E., van Es J. E., Guardavaccaro D., Schasfoort R. B., Mohri Y., Nishimori K., Mohammed S., Heck A. J., Clevers H. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 [DOI] [PubMed] [Google Scholar]
- 33. Boisset J. C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. (2010) In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 [DOI] [PubMed] [Google Scholar]
- 34. Ling K. W., Ottersbach K., van Hamburg J. P., Oziemlak A., Tsai F. Y., Orkin S. H., Ploemacher R., Hendriks R. W., Dzierzak E. (2004) GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodrigues N. P., Tipping A. J., Wang Z., Enver T. (2012) GATA-2 mediated regulation of normal hematopoietic stem/progenitor cell function, myelodysplasia and myeloid leukemia. Int. J. Biochem. Cell Biol. 44, 457–460 [DOI] [PMC free article] [PubMed] [Google Scholar]