Background: Elongation factor-Tu (EF-Tu) chaperones aminoacyl-tRNA (aa-tRNA) to the elongating ribosome as a ternary complex with GTP.
Results: EF-Ts facilitates EF-Tu·GTP binding to aa-tRNA through direct, transient interactions.
Conclusion: EF-Ts accelerates the formation and decay of ternary complex through the formation of a transient EF-Tu/Ts·GTP·aa-tRNA quaternary complex.
Significance: These newly described interactions of EF-Ts may serve to regulate ternary complex abundance in the cell.
Keywords: G Protein, Guanine Nucleotide Exchange Factor (GEF), Protein Synthesis, Translation, Translation Elongation Factor, Elongation Factor Ts, Elongation Factor Tu, Guanosine Nucleotide Exchange Factor, Ternary Complex
Abstract
During protein synthesis, elongation factor-Tu (EF-Tu) bound to GTP chaperones the entry of aminoacyl-tRNA (aa-tRNA) into actively translating ribosomes. In so doing, EF-Tu increases the rate and fidelity of the translation mechanism. Recent evidence suggests that EF-Ts, the guanosine nucleotide exchange factor for EF-Tu, directly accelerates both the formation and dissociation of the EF-Tu-GTP-Phe-tRNAPhe ternary complex (Burnett, B. J., Altman, R. B., Ferrao, R., Alejo, J. L., Kaur, N., Kanji, J., and Blanchard, S. C. (2013) J. Biol. Chem. 288, 13917–13928). A central feature of this model is the existence of a quaternary complex of EF-Tu/Ts·GTP·aa-tRNAaa. Here, through comparative investigations of phenylalanyl, methionyl, and arginyl ternary complexes, and the development of a strategy to monitor their formation and decay using fluorescence resonance energy transfer, we reveal the generality of this newly described EF-Ts function and the first direct evidence of the transient quaternary complex species. These findings suggest that EF-Ts may regulate ternary complex abundance in the cell through mechanisms that are distinct from its guanosine nucleotide exchange factor functions.
Introduction
Guanosine nucleotide-binding proteins (G-proteins) regulate a diverse range of essential cellular processes in all three kingdoms of life, including the universally conserved and energy-intensive process of protein synthesis. Elongation factor Tu (EF-Tu)2 in bacteria and elongation factor 1A (eEF1A) in eukaryotes are conserved members of the TRAFAC (translation factor) class of G-proteins comprising three globular domains (I–III). Both proteins play a critical role in translation by chaperoning messenger RNA (mRNA) codon-dependent incorporation of aminoacyl-tRNA (aa-tRNA) into the aminoacyl (A) site of the two-subunit ribosome during the substrate selection step of the elongation cycle (2–6).
EF-Tu, through its capacity to form a ternary complex with aa-tRNA in the presence of guanosine triphosphate (GTP) (EF-Tu·GTP·aa-tRNA), increases both the rate and fidelity of aa-tRNA selection on the ribosome (7–10) (see Fig. 1A). EF-Tu simultaneously promotes ribosome binding and couples the aa-tRNA selection mechanism to irreversible GTP hydrolysis (11). Following the release of inorganic phosphate (12), aa-tRNA dissociates from EF-Tu·guanosine diphosphate (GDP) to enter the ribosome where it undergoes irreversible peptide bond formation (13, 14).
FIGURE 1.
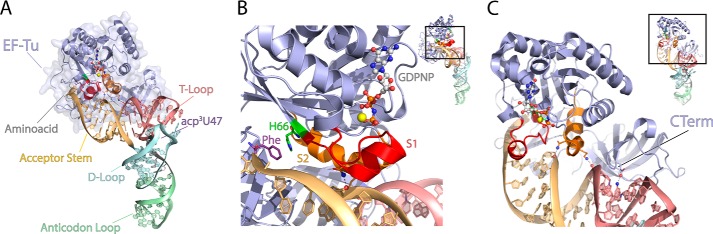
EF-Tu binds both the aminoacyl moiety and the tRNA body. A, E. coli EF-Tu (blue) bound in a ternary complex with GDPNP and Saccharomyces cerevisiae Phe-tRNAPhe (Protein Data Bank code 1OB2). The following functional domains of tRNA are highlighted: acceptor stem (orange), T-loop (red), D-loop (blue), and the anticodon loop (green) as well as GDPNP (carbon atoms in gray, nitrogen in blue, oxygen in red, and phosphorus in orange) and the acp3U47 residue, the site of fluorophore labeling used in this report. B, the aminoacyl binding pocket is stabilized by the GTP-dependent coordination of switch 1 (S1; red) and switch 2 (S2; orange) regions. C, conserved contacts between the acceptor stem of aa-tRNA and all three domains of EF-Tu. The approximate position of the C terminus (CTerm) of EF-Tu is shown.
The rates of protein synthesis and cellular growth critically depend on and are highly correlated with the availability of the complete complement of ternary complex species needed to translate the mRNA open reading frame (15–19). In Escherichia coli, there are ∼86 genes encoding for elongator tRNAs that exhibit a range of expression levels (19–21). Depending on the species, organism, and cellular growth rate, the total tRNA concentration in the cell ranges from 100 μm to 1 mm (22). Although a component of these fluctuations likely relates to the proximity of each gene to the origin of replication as postulated for ribosomal RNA (20, 24), changes in tRNA gene expression also reflect regulatory cues (21).
Ternary complex levels also depend on the extent to which tRNAs are aminoacylated, which is a prerequisite for EF-Tu·GTP binding (7, 25). Direct measurements suggest that ∼80% of expressed tRNAs are charged during logarithmic growth in nutrient-rich conditions (26, 27). Under nutrient-poor conditions, however, where amino acid reserves become limiting, this level drops substantially (27, 28). Such conditions may ultimately trigger the stringent response leading to the down-regulation of the translational machinery (27).
The abundance of distinct ternary complex species is also dictated by the affinity of EF-Tu for each aa-tRNA. EF-Tu exhibits tRNA-specific dissociation constants ranging from a few nanomolar to a few hundred nanomolar (8, 29, 30). A substantial proportion of the overall binding energy between EF-Tu·GTP and aa-tRNA arises from contacts between the aminoacyl moiety of tRNA and the amino acid binding pocket formed at the interface of domains I and II of EF-Tu (31–33) (see Fig. 1B). In the case of the phenylalanyl-tRNAPhe ternary complex (Phe-TC), the aromatic ring of the 2′-linked phenylalanyl moiety has been shown to directly interact with the conserved histidine 66 (His-66) residue of EF-Tu. The switch regions of EF-Tu (S1 and S2), which are present in all G-proteins and facilitate nucleotide interactions, help position His-66, thereby enabling EF-Tu to read out the aminoacyl state of the tRNA molecule in a manner that depends on the phosphorylation state of the guanosine nucleotide (32, 34) (see Fig. 1B).
The distinct chemical and physical properties of each amino acid predict significant differences in EF-Tu binding affinity to distinct tRNA species (31, 32). These disparities are thought to be dampened by a thermodynamic compensation mechanism based on unique EF-Tu interactions with sequence determinants in the tRNA acceptor stem and T-loop (Fig. 1C) that result in a relatively uniform EF-Tu·GTP affinity for each aa-tRNA species (29, 35, 36). In this model, the tRNA species bearing an aminoacyl moiety that contributes strongly to the free energy of ternary complex affinity have tRNA bodies that bind weakly to EF-Tu. Conversely, tRNAs bearing aminoacyl moieties that contribute relatively little to the free energy of ternary complex formation bind strongly to EF-Tu (31, 36). Taken together with structural and kinetic evidence that EF-Tu undergoes large scale conformational changes during aa-tRNA binding and release (34, 37–39), this model posits a degree of coupling between these two distinct determinants of EF-Tu affinity. Under steady-state, logarithmic growth conditions where aa-tRNA, EF-Tu, and GTP are at high concentrations, the vast majority of aa-tRNAs are bound to EF-Tu·GTP in ternary complex (40). At growth saturation or under nutrient-limited conditions, however, where the effective aa-tRNA concentrations are reduced, lower affinity species shift away from the EF-Tu·GTP-bound state, reducing their cellular concentrations.
Finally, the total ternary complex concentration in the cell is proportional to the concentration of GTP and the activated EF-Tu·GTP pool. The concentration of this latter complex in turn depends on the function of elongation factor Ts (EF-Ts), the guanosine nucleotide exchange factor for EF-Tu. The guanosine nucleotide exchange factor functions of EF-Ts facilitate the conversion of EF-Tu·GDP to EF-Tu·GTP following each round of tRNA selection. Given that the amount of EF-Tu·GDP is proportional to the concentration of ribosomes in the cell (10–100 μm) and the rate of protein synthesis (approximately 2–20 amino acids polymerized per second), high concentrations of EF-Ts promote a rapid rate of cell growth (41). In E. coli, the concentration of EF-Ts is tightly coupled with ribosome biogenesis and thus the ribosome concentration (15, 41–43, 45–47). Hence, a stoichiometry of EF-Tu/Ts·GDP in the cell that approaches 1:1 with ribosomes would facilitate prompt nucleotide exchange on EF-Tu and the maintenance of the highest concentration of EF-Tu·GTP possible.
Our recent efforts, which established a kinetic framework for examining ternary complex stability using fluorescence-based methods (1), provided evidence that EF-Ts directly regulates conformational changes in EF-Tu that facilitate the formation and disassembly of the Phe-TC. These observations suggested that EF-Ts has a capacity to directly regulate ternary complex concentration via the formation of an EF-Tu/Ts·GTP·aa-tRNA quaternary complex (1). Here, we directly examined whether such findings are general in nature by measuring the impact of EF-Ts on the kinetic properties of distinct ternary complex species that exhibit distinct apparent KD values (29). These investigations, combined with fluorescence resonance energy transfer (FRET) methods that facilitated the direct monitoring of EF-Tu/Ts interactions, yielded a general kinetic framework that explains how EF-Ts may influence the thermodynamic compensation model underpinning ternary complex stability, why quaternary complex has escaped detection previously, and how cellular interactions between EF-Ts and ternary complex can influence the regulation of protein synthesis in the cell.
EXPERIMENTAL PROCEDURES
Purification of Native Elongation Factors
Cleavable His6-EF-Tu (tufB) and His6-EF-Ts (tsf) were expressed recombinantly in DH5α cells and purified by nickel-nitrilotriacetic acid affinity chromatography in the absence of magnesium as described previously (48). After removal of His6, the elongation factors were purified further using Superdex 75 gel filtration chromatography as described previously (1).
Purification and Labeling of Elongation Factors
Labeled EF-Tu was constructed and purified using the expression system described for native EF-Tu with the modification of a C-terminal acyl-carrier protein recognition epitope (AcpS). Labeling was achieved by incubating purified EF-Tu (C-terminal AcpS) with a 10-fold excess of CoA-linked cyanine fluorophore (either Cy3 or Cy5Q) and a 5-fold excess of AcpS phosphopantetheinyltransferase enzyme in buffer A (50 mm HEPES, pH 7.5, 10 mm Mg(OAc)2) for 2 h. EF-Ts (ΔC, A160C) was generated by site-directed mutagenesis of its two native cysteines (at positions 23 and 78) to serines and Ala-160 to cysteine. EF-Ts (ΔC, A160C) was labeled by incubation with a 2-fold excess of Cy5 maleimide for 90 min at room temperature in buffer B (100 mm phosphate-buffered saline, pH 6.5, 100 μm tris(2-carboxyethyl)phosphine, 100 mm NH4Cl).
Purification and Labeling of tRNA
Wild-type E. coli tRNAPhe was expressed and purified from the bacterial strain MRE600 as described previously (49). tRNAArg and tRNAMet were purchased from Chemical Block and further purified by hydrophobic interaction chromatography (using a Tosoh Corp. Phenyl-5PW column) by applying a shallow gradient from buffer C (10 mm NH4OAc, pH 5.8, 1.7 m (NH4)2SO4) to buffer D (10 mm NH4OAc, pH 5.8, 10% methanol) over 20 column volumes. tRNA molecules were site-specifically labeled with either Cy3 or Cy3B via the naturally occurring modified nucleotide acp3U at position 47 (48, 50). This labeling strategy has been shown previously to not inhibit aminoacylation or translation reactions (48, 50). Each tRNA was aminoacylated by adding a 0.1× molar ratio of tRNA synthetase and a 1000-fold excess of amino acid in a volume of 10 μl in buffer E (50 mm Tris-HCl, pH 8, 20 mm KCl, 100 mm NH4Cl, 1 mm DTT, 2.5 mm ATP, 0.5 mm EDTA, 10 mm MgCl2) and incubating at 37 °C for 20 min. The extent of aminoacylation was verified by analytical hydrophobic interaction chromatography.
Fluorometer Experiments
The fluorescence measurements shown in Figs. 2, 4, and 5 were obtained using a Photon Technology International fluorescence spectrometer with a 532-nm long pass emission filter (LP03-532RS-25 RazorEdge by Semrock). Reactions were analyzed in buffer F (100 mm HEPES, pH 7, 20 mm KCl, 100 mm NH4Cl, 1 mm DTT, 0.5 mm EDTA, 2.5 mm MgCl2, 10 μm GTP) in a 3-ml quartz cuvette with constant mixing at room temperature. Measurements were made by manually adding EF-Tu or an EF-Tu/Ts complex (preincubated in buffer F with 10 μm GTP for 10 min at room temperature) to a reaction of 5 nm fluorescently labeled aa-tRNA while exciting at 532 nm and monitoring at 565 nm. The dissociation constant (KD) was determined as described previously (1). Apparent rates (kapp) were determined from pre-steady-state measurements where the time-dependent changes in fluorescence observed were fit to a single exponential function (51).
FIGURE 2.
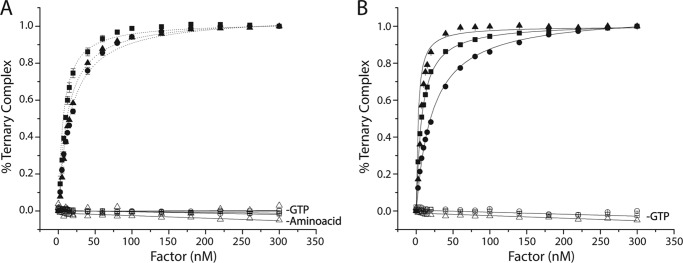
EF-Tu binds ternary complex with nanomolar affinity. Titration of EF-Tu·GTP (A) or EF-Tu/Ts·GTP (B) into a solution of Cy3-labeled Phe-tRNAPhe (solid triangles), Met-tRNAMet (solid squares), or Arginyl-tRNAArg (solid circles) in the presence of 10 μm GTP. The apparent dissociation constant (KD) for each tRNA species was estimated by fitting (see “Experimental Procedures”). Identical experiments performed in the absence of GTP or the tRNA synthetase (open symbols). Error bars represent the S.D. of three separate experiments.
FIGURE 4.
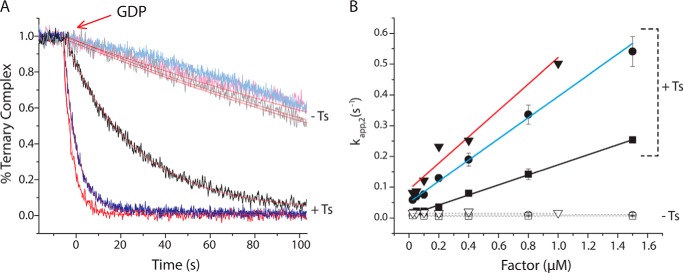
EF-Ts accelerates ternary complex decay in response to GDP. A, addition of a 10-fold molar excess of GDP to Arg-TC (blue) or Met-TC (black) preformed with EF-Tu (light colored curves) or EF-Tu/Ts (dark colored curves) in the presence of 10 μm GTP. B, the apparent rate of ternary complex dissociation (kapp,2) was estimated by fitting each decay process to a single exponential function. The relationship between kapp,2 and the concentration of EF-Tu (open symbols) or EF-Tu/Ts (solid symbols) is shown for both Met-TC (squares) and Arg-TC (circles). Error bars represent the S.D. of three separate experiments. For comparison, kapp,2 (acquired previously) is shown for Phe-TC (red triangles) (1).
FIGURE 5.
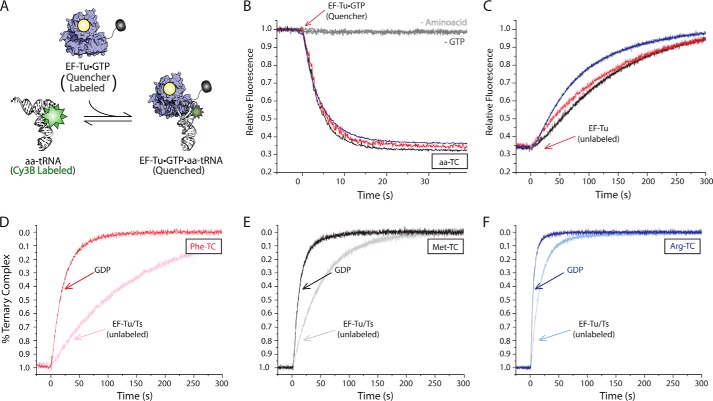
Measuring ternary complex formation and dissociation via FRET. A, schematic illustration of the TC quenching assay showing Cy5Q-labeled EF-Tu (blue) bound to GTP (yellow) quenches the fluorescence of Cy3B-labeled aa-tRNA (white) upon ternary complex formation. B, addition of 0.4 μm EF-Tu·GTP (Cy5Q) to 5 nm Cy3B-labeled Phe-tRNAPhe (red), Met-tRNAMet (black), or Arg-tRNAArg (blue) in the presence of 10 μm GTP. Omission of the tRNA synthetase (−Amino acid) or GTP (−GTP) from the ternary complex formation assay resulted in no change in Cy3B fluorescence intensity. C, addition of excess unlabeled EF-Tu to the ternary complexes formed in B. Addition of excess GDP (bold colors) or unlabeled EF-Tu/Ts (light colors) to Phe-TC (D), Met-TC (E), and Arg-TC (F) preformed with EF-Tu/Ts (Cy5Q). Apparent rates of ternary complex decay were estimated by fitting to a single exponential function.
Rapid Stopped-flow Experiments
The fluorescence measurements shown in Figs. 3, 6, and 7 were obtained in buffer F at room temperature using an SX20 stopped-flow spectrometer from Applied Photophysics equipped with either a 550-nm long pass emission filter (OG550 by Schott) or a 633-nm long pass emission filter (LP01-633RS-25 RazorEdge by Semrock) for monitoring Cy3 and Cy5 fluorescence, respectively. All concentrations referenced herein are stated as final concentrations after mixing. Error bars represent S.E. of three separate reactions.
FIGURE 3.
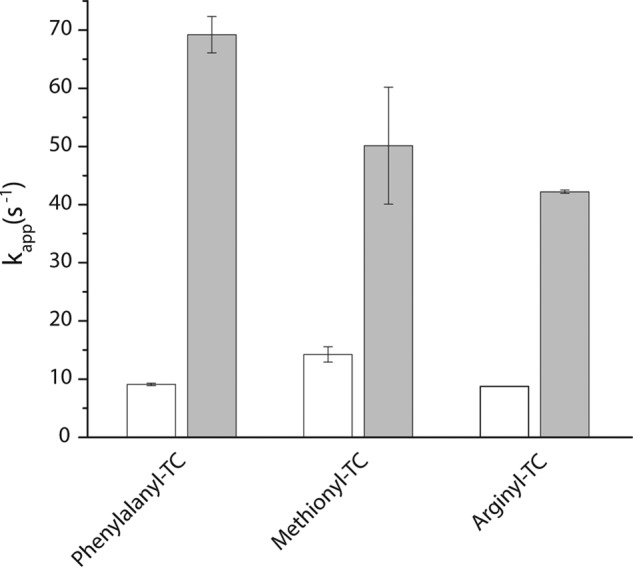
EF-Ts directly facilitates ternary complex formation. The estimated rate of ternary complex formation (kapp,1) measured by a change in fluorescence intensity upon stopped-flow delivery of 5 μm EF-Tu·GTP (open bars) or 5 μm EF-Tu/Ts (solid bars) to 100 nm Cy3-labeled phenylalanyl-tRNAPhe, arginyl-tRNAArg, or methionyl-tRNAMet. Error bars represent the S.D. of three separate experiments.
FIGURE 6.
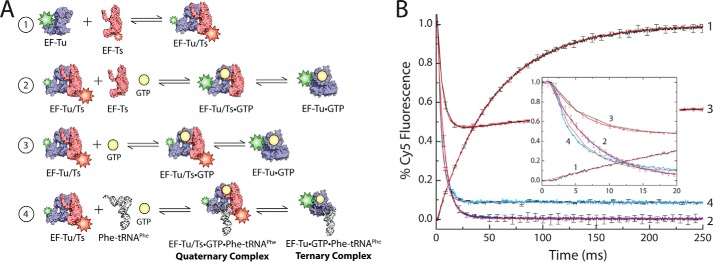
Significant amounts of EF-Tu/Ts remain bound to GTP under physiological nucleotide concentrations. A, schematic illustrating the EF-Tu/Ts FRET assay. 1, Cy3-labeled EF-Tu (blue) is added to Cy5-labeled EF-Ts (red). 2, EF-Tu(Cy3)/Ts(Cy5) mixed with unlabeled EF-Ts and GTP. 3, EF-Tu(Cy3)/Ts(Cy5) added to GTP. 4, EF-Tu(Cy3)/Ts(Cy5) mixed with unlabeled Phe-tRNAPhe and GTP. B, 2 μm EF-Tu(Cy3) mixed with 2 μm EF-Ts(Cy5) while monitoring Cy5 fluorescence (curve 1), 2 μm EF-Tu(Cy3)/Ts(Cy5) added to 20 μm unlabeled EF-Ts and 1 mm GTP (curve 2), 2 μm EF-Tu(Cy3)/Ts(Cy5) mixed with 1 mm GTP (curve 3), and 2 μm EF-Tu(Cy3)/Ts(Cy5) mixed with 0.5 μm Phe-tRNAPhe and 1 mm GTP (curve 4). A plot focusing on the early time points in the reactions is shown for clarity (inset). Apparent rates were estimated by fitting to either a single exponential function (curves 1, 2, and 4) or a double exponential function (curve 3). Error bars represent the S.D. of three separate experiments.
FIGURE 7.
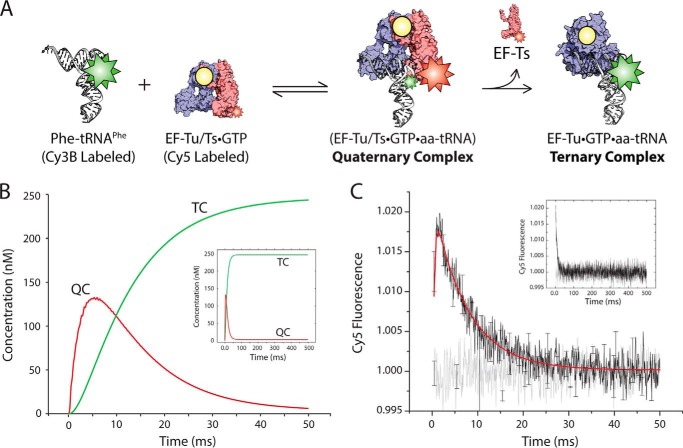
Direct evidence of the transient quaternary complex. A, schematic illustrating the FRET assay used to detect quaternary complex. Cy3B-labeled Phe-tRNAPhe (white) is mixed with EF-Tu/Ts·GTP harboring a Cy5 fluorophore linked to EF-Ts. B, simulation of the time-dependent formation of quaternary complex (red curve; QC) and ternary complex (green curve; TC) based on the schematic presented in A and outlined under “Experimental Procedures.” C, transient FRET observed upon mixing 2 μm EF-Tu/Ts(Cy5) and 250 nm Phe-tRNAPhe (Cy3B) in the presence of 1 mm GTP (black). These data were fit to a double exponential function (red). This experiment was repeated with deacylated tRNAPhe (Cy3B) (gray). Error bars represent the S.D. of three separate experiments.
Nucleotide Purification
GTP and GDP were purchased from Sigma and further purified on a Tricorn Mono Q 5/50 GL ion exchange column as described previously (1).
Simulation of Ternary Complex Formation
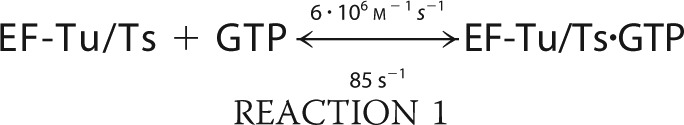
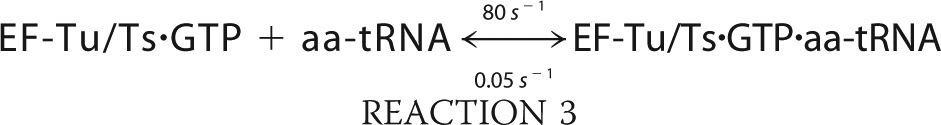
We simulated the formation of ternary complex using MATLAB (R2010b) built on the following six reactions.
 |
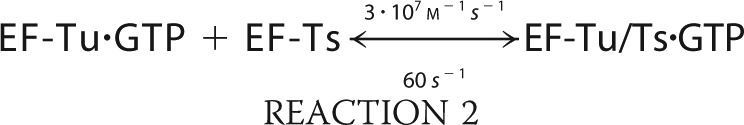 |
 |
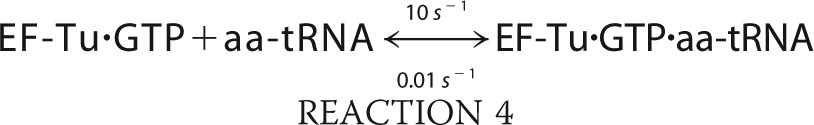 |
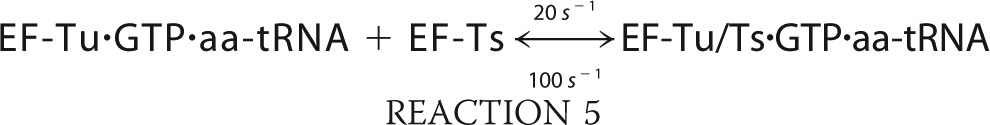 |
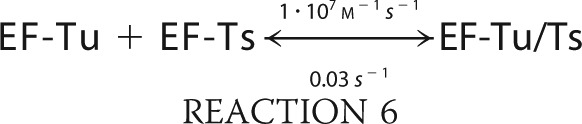 |
The simulation was conducted with the ode45 (Dormand-Prince) solver with an absolute tolerance setting of 1.0·10−6 using the starting conditions of 2 μm EF-Tu/Ts, 1 mm GTP, and 250 nm aa-tRNA.
RESULTS
EF-Tu Binds Distinct aa-tRNAs with High Affinity
To monitor ternary complex formation, we used a steady-state, fluorescence-based approach that yields an increase in relative intensity of Cy3 fluorescence when EF-Tu binds to site-specifically labeled aa-tRNA (1). We measured the apparent affinity of highly purified, nucleotide-free EF-Tu or an EF-Tu/Ts complex for three distinct elongator tRNA species, each containing the same acp3U post-transcriptional modification at position 47 used for Cy3 labeling: Phe-tRNAPhe, Met-tRNAMet, and Arg-tRNAArg. For these experiments, EF-Tu and EF-Tu/Ts were preincubated with an excess of GTP (10 μm) to ensure complex formation (“Experimental Procedures”). In each case, EF-Tu·GTP or EF-Tu/Ts·GTP was titrated into a 1.5-ml reaction containing 5 nm aa-tRNA (Cy3-acp3U) and 10 μm GTP (“Experimental Procedures”). As anticipated from previous studies on Phe-tRNAPhe (Cy3-acp3U) (1), each aa-tRNA species exhibited a marked increase in Cy3 relative fluorescence as a function of increasing factor concentration (Fig. 2). Under the current experimental conditions (“Experimental Procedures”), Cy3 fluorescence increased ∼32, 45, and 30% for Phe-tRNAPhe, Met-tRNAMet, and Arg-tRNAArg, respectively. As expected, no change in fluorescence intensity was observed when identical experiments were performed either in the absence of GTP or when the tRNA was not aminoacylated. To provide an estimate of the affinity of each factor for aa-tRNA from these experiments, each titration was fit to an equation for a bimolecular interaction (1). As expected from prior investigations (7, 29), all three aa-tRNA species exhibited an apparent KD in the nanomolar range both in the presence and absence of EF-Ts (Fig. 2 and Table 1). The lowest apparent KD was for Phe-tRNAPhe (6 nm), whereas Arg-tRNAArg was the highest (22 nm) (Table 1). Compared with the relatively large impact on Phe-TC affinity (a 3-fold increase) in the presence of EF-Ts (1), the apparent KD values of Met-tRNAMet and Arg-tRNAArg were only modestly affected (approximately ±20–30%; Table 1).
TABLE 1.
Kinetic parameters measured in the presence and absence of EF-Ts
| Kinetic parameters |
||||||
|---|---|---|---|---|---|---|
|
KD (Fig. 2) |
kapp,1 (Fig. 3) |
kapp,2 (Fig. 4) |
||||
| −EF-Ts | +EF-Ts | −EF-Ts | +EF-Ts | −EF-Ts | +EF-Ts | |
| nm | s−1 | s−1 | ||||
| Phe-TC | 18.2 ± 1.2 | 6.1 ± 0.8 | 9.01 ± 0.2 | 69.2 ± 3.1 | 0.015 ± 1E−3a | 0.25 ± 0.04a |
| Met-TC | 10.8 ± 1.5 | 7.5 ± 1.2 | 14.2 ± 1.3 | 50.1 ± 10.0 | 0.01 ± 1.7E−3 | 0.08 ± 0.01 |
| Arg-TC | 19.6 ± 0.8 | 22 ± 0.3 | 8.73 ± 0.04 | 42.19 ± 0.3 | 0.01 ± 1.6E−4 | 0.19 ± 0.02 |
a Data obtained from Burnett et al. (1) at 0.4 μm EF-Tu or EF-Tu/Ts.
EF-Ts Accelerates the Rate of Ternary Complex Formation
To delineate the kinetic features governing ternary complex formation, pre-steady-state, rapid mixing experiments were performed using the same Cy3-labeled Phe-tRNAPhe, Met-tRNAMet, and Arg-tRNAArg reagents under conditions estimating the physiological concentrations of each component (5 μm factor, 1 mm GTP, and 100 nm aa-tRNA). Purified EF-Tu and EF-Tu/Ts complex were pre-incubated with saturating concentrations of GTP (1 mm) to ensure complete nucleotide binding. For all three aa-tRNAs, the apparent rate of ternary complex formation using EF-Tu was between ∼8 and ∼14 s−1 (Fig. 3, open bars). Identical experiments conducted using the EF-Tu/Ts complex resulted in an increase in the apparent rate of ternary complex formation for each tRNA species to ∼40 to ∼70 s−1 (Fig. 3, solid bars). As suggested for Phe-TC formation (1), these data argue that EF-Ts also facilitates rate-determining conformational events in EF-Tu that are required for nucleotide-dependent binding to both Met-tRNAMet and Arg-tRNAArg.
EF-Ts Facilitates Ternary Complex Dissociation in the Presence of GDP
To examine whether EF-Ts influences the dissociation pathways of these two distinct ternary complex species as has been demonstrated for the Phe-TC (1), we performed GDP chase experiments on Met-TC and Arg-TC preformed with either EF-Tu or EF-Tu/Ts. As noted previously (1), a hallmark kinetic signature of the EF-Tu/Ts·GTP·aa-tRNA quaternary complex is the factor concentration dependence of the ternary complex dissociation rate in the presence of GDP. In such experiments, ternary complex is formed by adding 0.4 μm factor (preincubated with 10 μm GTP) to 5 nm Cy3-labeled Phe-tRNAPhe, Met-tRNAMet, or Arg-tRNAArg. A 10-fold excess of GDP (100 μm) is then added, and the rate of fluorescence decay is then tracked over time.
In the absence of EF-Ts, all three ternary complexes were observed to decay at a slow rate of ∼0.01 s−1, consistent with ternary complex being a stable, high affinity species (Fig. 4A). As shown previously for Phe-TC (1), the measured decay rate was independent of EF-Tu concentration as expected for a spontaneous dissociation process followed by GDP binding, an event that prevents ternary complex from re-forming (Fig. 4B). When the same experiment was performed with EF-Tu/Ts, however, the rate of fluorescence intensity decay was ∼25-, ∼13-, and ∼20-fold faster for Phe-TC, Met-TC, and Arg-TC, respectively (Fig. 4A). The observed rate of decay was also linearly dependent on the concentration of EF-Tu/Ts (Fig. 4B). This trend is inconsistent with a simple unimolecular dissociation pathway, suggesting that EF-Ts facilitates ternary complex dissociation through a bimolecular interaction that influences the nucleotide binding pocket of EF-Tu. Here, the apparent EF-Ts on-rate estimated from the slope of the concentration-dependent process ranged from ∼0.3 to 1.0 μm−1 s−1 depending on the tRNA species and taking into account that the concentration of free EF-Ts is roughly 50% of the total factor concentration. As this rate is more than 2 orders of magnitude slower than a diffusion-limited process as well as the established rate of nucleotide binding to EF-Tu (52), we conclude that one or more rate-determining events in EF-Tu, likely related to GTP release, precede the EF-Ts-facilitated ternary complex dissociation process leading to the loss of fluorescence.
EF-Ts Increases the Rate of Ternary Complex Turnover under Steady State Conditions
To probe whether EF-Ts influences rate-determining step that precede the dissociation process observed in the presence of GDP, we next sought to ascertain whether the rate of ternary complex turnover was influenced by EF-Ts under steady-state conditions. To do so, we developed a FRET-based approach to directly examine the rate of ternary complex turnover by site-specifically labeling EF-Tu with a quencher fluorophore at its C terminus (“Experimental Procedures”). The C terminus of EF-Tu and the acp3U47 residue in tRNA are in close proximity (approximately 30 Å) (Fig. 1). To dampen the environment-dependent increase in Cy3 fluorescence that accompanies ternary complex formation (Fig. 2), Phe-tRNAPhe, Met-tRNAMet, and Arg-tRNAArg were labeled at their acp3U47 residues using Cy3B, a fluorophore less prone to cis-trans isomerization and environment-dependent changes in quantum yield (53–55). Hence, the formation of ternary complex is anticipated to result in a substantial decrease in Cy3B fluorescence intensity as a result of energy transfer to the nearby quencher (Fig. 5A).
Using this approach, ternary complex formation resulted in an ∼65% decrease in Cy3B fluorescence intensity for all three aa-tRNAs examined (Fig. 5B). As expected, this change in signal was strictly dependent on GTP and the aminoacyl moiety of aa-tRNA (Fig. 5B). Consistent with the findings presented for unlabeled EF-Tu (Fig. 4), this reduction in Cy3B fluorescence intensity could be fully recovered by the addition of a 10-fold molar excess of GDP (data not shown). These data convincingly revealed that quencher-labeled EF-Tu is fully competent to form ternary complex.
To directly examine ternary complex turnover under steady-state conditions, a competition experiment was performed in which a 10-fold molar excess of unlabeled EF-Tu or EF-Tu/Ts was added to a preformed, quencher-labeled EF-Tu·GTP·Phe-tRNAPhe (Cy3B-acp3U47) ternary complex. As the vast majority of ternary complex re-formed after dissociation will not be quenched, the intrinsic rate of ternary complex turnover in this assay is revealed by the rate of increase in Cy3B fluorescence intensity.
In both the absence and presence of EF-Ts, Cy3B fluorescence recovered at a rate that was tRNA-specific (Fig. 5, C–F). For the unlabeled EF-Tu chase experiment, the rates of steady-state ternary complex turnover approximated those observed upon quenching with GDP in the absence of EF-Ts (Fig. 5C and compare Table 2 koff with Table 1 kapp,2). However, distinct responses were evidenced in each EF-Tu/Ts chase experiment (Fig. 5, D–F). Here, the steady-state turnover rates were approximately an order of magnitude slower than those observed in the presence of GDP (compare Table 1 kapp,2 with Table 2 koff).
TABLE 2.
Apparent rates of ternary complex dissociation
|
koff (Fig. 5) |
||
|---|---|---|
| −EF-Ts | +EF-Ts | |
| s−1 | ||
| Phe-TC | 0.007 ± 0.001 | 0.014 ± 0.001 |
| Met-TC | 0.007 ± 0.001 | 0.022 ± 0.002 |
| Arg-TC | 0.011 ± 0.001 | 0.053 ± 0.004 |
These findings suggest that EF-Ts has the propensity to destabilize each ternary complex and to specifically promote ternary complex dissociation in the presence of GDP. Such trends are consistent with the notion that nucleotide exchange occurs at a slow rate within ternary complex and that EF-Ts increases the rate of such processes via direct interactions with EF-Tu while it remains bound to aa-tRNA (1). We speculate that, when only GTP is present in the reaction, nucleotide can reload onto EF-Tu prior to its dissociation from aa-tRNA. When GDP is present in excess of GTP, however, GDP binding to EF-Tu leads to ternary complex dissociation. We therefore infer that EF-Ts can act as a guanosine nucleotide exchange factor for EF-Tu while it is bound to aa-tRNA via a quaternary complex where the formation of an EF-Tu·GDP·aa-tRNA ternary complex stimulates EF-Tu release from aa-tRNA, which is accelerated by EF-Ts.
EF-Tu/Ts·GTP Is an Abundant Species under Cellular Conditions
The notion that EF-Ts can interact with EF-Tu while it is bound to aa-tRNA provides a plausible model for the observation that EF-Ts accelerates the process of ternary complex formation (Fig. 3). As speculated previously (1), EF-Ts may increase the rate of ternary complex formation via a transient EF-Tu/Ts·GTP·aa-tRNA quaternary complex. A key issue with this model is that the EF-Tu/Ts·GTP species is expected to dissociate at a rate of ∼60 s−1 at physiological GTP concentrations (approximately 1 mm) (52). Meaningful concentrations of the EF-Tu/Ts·GTP complex, however, are expected at EF-Ts concentrations above 1 μm as the bimolecular rate constant between EF-Ts and EF-Tu·GTP is 3·107 m−1 s−1 (52). Indeed, such considerations predict that ∼50% of EF-Tu will be in an EF-Tu/Ts·GTP complex under the conditions where EF-Ts is observed to accelerate ternary complex formation (Fig. 3). At cellular EF-Ts concentrations, which under nutrient-rich conditions are estimated to be ≥10 μm (41, 42), the proportion of EF-Tu/Ts·GTP complex is expected to be even higher as the apparent rate of EF-Tu/Ts·GTP re-formation will be as much as ∼5-fold faster than the rate of EF-Ts dissociation from EF-Tu·GTP.
To directly test these predictions, we set out to measure the interactions between EF-Tu and EF-Ts through a FRET-based approach (Fig. 6A). To do so, we used enzymatic tagging procedures to site-specifically label EF-Tu with a Cy3 fluorophore at its C terminus and EF-Ts with a Cy5 fluorophore through maleimide chemistry at a position engineered to yield high FRET (A160C) (38) (“Experimental Procedures”). When 2 μm Cy3-labeled EF-Tu was rapidly mixed with 2 μm Cy5-labeled EF-Ts under stopped flow (“Experimental Procedures”), a rapid increase in Cy5 emission was observed (kapp = 18.5 s−1), which as expected from the kinetics and high affinity nature of the EF-Tu/Ts interaction (52) remained stable over time (Fig. 6, A and B, black curve, and Table 3).
TABLE 3.
Apparent rates of EF-Tu and EF-Ts interactions
Rxn, reaction.
|
k (Fig. 6) | |||
|---|---|---|---|
| Rxn 1 | Rxn 2 | Rxn 3a | Rxn 4 |
| s−1 | |||
| 18.5 ± 0.5 | 148 ± 5 | 177 ± 0.8 (decay) | 242 ± 10 |
| 5.8 ± 0.04 (rise) | |||
a Double exponential fit.
To determine the dissociation rate of the EF-Tu/Ts complex in the presence of saturating concentrations of GTP, a preformed EF-Tu(Cy3)/Ts(Cy5) complex was rapidly mixed with 1 mm GTP in the presence of a 10-fold molar excess of unlabeled EF-Ts to prevent the complex from re-forming (Fig. 6, A and B, blue curve). As expected (52), a rapid loss of fluorescence was observed (∼148 s−1) (Table 3). This value is ∼2-fold faster that the EF-Tu/Ts·GTP dissociation rate estimated using native components (52), suggesting that the modifications to EF-Tu and EF-Ts may modestly destabilize the EF-Tu/Ts complex.
We next investigated the extent to which GTP disrupts the EF-Tu/Ts complex under steady state conditions in the absence of unlabeled EF-Ts where a dynamic equilibrium between dissociation and association is expected. In line with known kinetic parameters of this system (52), mixing the preformed EF-Tu(Cy3)/Ts(Cy5) complex (2 μm) with GTP (1 mm) led to a rapid loss of FRET, which stabilized at approximately ∼55% of the starting intensity (Fig. 6, A and B, red curve, and Table 3). These data argue that the EF-Tu/Ts·GTP complex is an abundant species both in our experiments and the cellular milieu.
To test the hypothesis that EF-Ts accelerates the binding of aa-tRNA to EF-Tu·GTP, a preformed EF-Tu(Cy3)/Ts(Cy5) complex was rapidly mixed with 1 mm GTP and 500 nm Phe-tRNAPhe (Fig. 6, A and B, cyan curve). Here, Cy5 fluorescence was observed to decay at a rate of ∼242 s−1 (Table 3), a rate significantly faster than when only GTP or unlabeled EF-Ts was added (Fig. 6B, inset). This finding suggests that aa-tRNA contributes to the apparent rate of EF-Tu(Cy3)/Ts(Cy5) dissociation as would be expected if ternary complex is formed rapidly (approximately ≥140 s−1).
Quaternary Complex Is Transient by Nature
To directly observe the EF-Tu/Ts·GTP·aa-tRNA quaternary complex, we designed an experiment to detect FRET between EF-Ts and aa-tRNA during ternary complex formation where the hypothesized architecture of quaternary complex predicts a physical proximity between the Cy3B fluorophore at position U47 of aa-tRNA and Cy5 at position 160 of EF-Ts (1) (Fig. 7A). The conditions of this experiment were derived by simulating the expected rate of quaternary complex formation and decay that should accompany ternary complex formation based on a framework that included the known kinetic features of the interactions among EF-Tu, EF-Ts, guanosine nucleotide, and Phe-tRNAPhe (1, 52) (Fig. 7B, red curve). Using this approach, the optimal conditions for detecting quaternary complex were determined to be a concentration of 2 μm EF-Tu/Ts, 250 nm Phe-tRNAPhe, and 1 mm GTP.
As anticipated by these simulations, when 2 μm EF-Tu/Ts(Cy5) was rapidly mixed with 250 nm Phe-tRNAPhe (Cy3B-acp3U47) and 1 mm GTP, FRET was observed between the excited Cy3B fluorophore and the Cy5-labeled EF-Ts (Fig. 7C, black). No evidence of FRET was observed when the exact same experiment was performed using deacylated tRNAPhe (Fig. 7C, gray).
The biphasic nature of the transient FRET signal observed was fit to a double exponential process to estimate the apparent rates of quaternary complex formation and decay, respectively. Here, the rise and decay portions were estimated to occur at rates of ∼870 ± 23 and 143 ± 7 s−1, respectively (Table 4). These apparent rates suggest that the appearance of FRET is diffusion-limited and that the rate of quaternary complex decay and thus ternary complex formation may be as fast as 140 s−1. Notably, this rate is strikingly consistent with our stopped-flow measurements based on changes in Cy3 relative fluorescence intensity where ternary complex formation occurs at a rate of ∼70 s−1 (Fig. 3) under conditions in which ∼50% of EF-Tu is expected to be in an EF-Tu/Ts·GTP complex (Fig. 6). Such findings provide compelling, direct evidence that ternary complex formation occurs via a highly transient quaternary complex species under cellular conditions.
TABLE 4.
Apparent rates of quaternary complex formation and decay
|
k (Fig. 7) | |
|---|---|
| Rise | Decay |
| s−1 | |
| 870 ± 23 | 143 ± 7 |
DISCUSSION
Contemporary models posit that the EF-Tu life cycle parallels the quintessential model of G-protein function (56–58). After aa-tRNA is delivered to the actively translating ribosome, EF-Tu·GDP is released as an inactive species whereupon the guanosine nucleotide exchange factor, EF-Ts, operates on it to exchange GDP for GTP. Thereafter, EF-Tu·GTP is released as an activated species that is again competent to bind free aa-tRNA with uniformly high affinity (13, 14, 36, 52, 57–60).
The data presented here and elsewhere (1, 61) argue that refinements to this model are needed. At cellular EF-Ts concentrations (approximately >1 μm), our findings suggest that ternary complex preferentially forms via a transient EF-Tu/Ts·GTP·aa-tRNA quaternary complex (Fig. 8). This pathway is kinetically preferred because EF-Ts lowers the activation barrier for rate-determining conformational processes in EF-Tu that accompany the formation of a high affinity complex with aa-tRNA. Given what is presently known, the nature of these conformational processes likely surrounds the allosteric linkage between the aminoacyl moiety linked to the terminus of aa-tRNA and the switch regions surrounding the GTP nucleotide that coordinate magnesium and the γ-phosphate group (Fig. 1).
FIGURE 8.
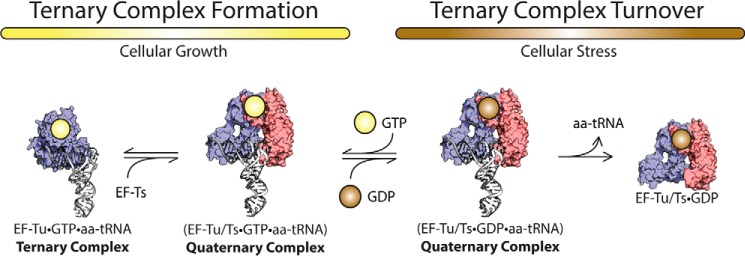
EF-Ts regulates ternary complex abundance. During growth-promoting conditions, EF-Ts (red) facilitates the formation of GTP (yellow)-bound EF-Tu (blue) to aa-tRNA (white) by accelerating rate-determining conformational processes in EF-Tu. The guanosine nucleotide exchange function of EF-Ts also serves to preferentially load GDP during conditions of cellular stress.
Our direct measurements suggest that the lifetime quaternary complex is on the order of ∼10 ms (Fig. 7). As the predominant means for investigating ternary complex formation have entailed size exclusion chromatography, filter binding, and gel mobility shift assays (8, 23, 62) where rapid kinetic studies have been specifically lacking, its inherent instability is likely the key reason why quaternary complex has escaped detection previously. Although the existence of a quaternary encounter complex species is an expected result based on purely physical considerations, the capacity of EF-Ts to increase the rate of ternary complex formation substantially above that of protein synthesis (approximately 100 versus 10–20 s−1; Figs. 3 and 7) argues that it serves a critical biological role related to replenishing the supply of ternary complexes as they are consumed by the process of protein synthesis.
The data presented also suggest that EF-Ts destabilizes ternary complex species to make their abundance sensitive to the nucleotide status of the cell (Fig. 5). Such tendencies are consistent with EF-Ts remodeling the switch regions in EF-Tu while it is bound to aa-tRNA (as for the free protein (44)) to influence the rate of nucleotide exchange. Two notable corollaries to this observation are that 1) the nucleotide exchange rate may not be the same for all tRNA species, and 2) the activated EF-Tu molecule in ternary complex may be deactivated by energy-neutral processes. Evidence supporting the first corollary is shown for the three tRNA species investigated in this study (Phe-tRNAPhe, Met-tRNAMet, and Arg-tRNAArg) where the effects of EF-Ts on ternary complex differ by as much as an order of magnitude (Figs. 3, 4, and 5).
As the effective concentration of quaternary complex will be strongly dependent on EF-Ts concentration, such effects are also expected to be highly responsive to the active EF-Ts pool. Although the second corollary is likely irrelevant under nutrient-rich conditions where the intracellular concentration of GTP is much higher than GDP, it may play a significant role in regulating ternary complex abundance upon a change in the GTP/GDP ratio. EF-Ts rapidly dissociates ternary complex when GDP loads onto EF-Tu while it is bound to TC (Figs. 4, 5, and 8). Correspondingly, changes in EF-Ts concentration as well as the GDP/GTP ratio are likely to contribute differentially to the relative stabilities and effective concentrations of specific ternary complex species in interdependent ways. Although it is not presently clear whether EF-Ts contributes evenly or differentially to the stabilities of distinct ternary complex species, the data obtained for the three ternary complex species assessed here suggest that ternary complex affinities become somewhat more dispersed in the presence of EF-Ts (Table 1). Such differences likely arise from a balance of the distinct contributions to ternary complex affinity, including contacts between aa-tRNA and EF-Tu as well as the nucleotide exchange rate. A key prediction of these considerations is that the abundance and lifetime of each ternary complex type will be sensitive to the metabolic state of the cell where EF-Ts can serve to rapidly dissipate the ternary complex pool through an energy-neutral mechanism in the event of cellular stress. Such a capacity would provide a means for the cell to immediately cease the energy-expensive process of translation without waiting for ternary complex to be consumed and allow for the repurposing of GTP.
The demonstration that EF-Ts actively resolves quaternary complex species that contain GDP (Fig. 5) also has potentially important implications for the mechanism of tRNA selection as an EF-Tu·GDP·aa-tRNA ternary complex is formed each time an actively translating ribosome decodes mRNA (59). A key open question that must be resolved through future investigations is whether EF-Ts is able to gain access to the EF-Tu·GDP·aa-tRNA ternary complex while engaged at the A site of the ribosome. If so, EF-Ts may accelerate the rate of EF-Tu·GDP release from the ribosome following aa-tRNA entry into the peptidyltransferase center, a step reported to be rate-limiting to the process (13, 14). To our knowledge such questions have yet to be explored.
Acknowledgment
We are grateful to Dr. Chris Walsh for providing the AcpS expression vector.
This work was supported, in whole or in part, by National Institutes of Health Grants GM079238, GM098859, and MH099491 (to S. C. B.).
- EF
- elongation factor
- aa-tRNA
- aminoacyl-tRNA
- TC
- ternary complex
- AcpS
- acyl-carrier protein recognition epitope
- acp3U
- 3-(3-amino-3-carboxypropyl)uridine
- GDPNP
- guanosine 5′-(β,γ-imido)triphosphate.
REFERENCES
- 1. Burnett B. J., Altman R. B., Ferrao R., Alejo J. L., Kaur N., Kanji J., Blanchard S. C. (2013) Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu·GTP·aminoacyl-tRNA ternary complex. J. Biol. Chem. 288, 13917–13928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 3. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 4. Thomposon R. C., Dix D. B. (1982) Accuracy of protein biosynthesis. A kinetic study of the reaction of poly(U)-programmed ribosomes with a leucyl-tRNA2-elongation factor Tu-GTP complex. J. Biol. Chem. 257, 6677–6682 [PubMed] [Google Scholar]
- 5. Rodnina M. V., Pape T., Fricke R., Wintermeyer W. (1995) Elongation factor Tu, a GTPase triggered by codon recognition on the ribosome: mechanism and GTP consumption. Biochem. Cell Biol. 73, 1221–1227 [DOI] [PubMed] [Google Scholar]
- 6. Rodnina M. V., Savelsbergh A., Wintermeyer W. (1999) Dynamics of translation on the ribosome: molecular mechanics of translocation. FEMS Microbiol. Rev. 23, 317–333 [DOI] [PubMed] [Google Scholar]
- 7. Abrahamson J. K., Laue T. M., Miller D. L., Johnson A. E. (1985) Direct determination of the association constant between elongation factor Tu and GTP and aminoacyl-tRNA using fluorescence. Biochemistry 24, 692–700 [DOI] [PubMed] [Google Scholar]
- 8. Louie A., Jurnak F. (1985) Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5-triphophate-aminoacyl-tRNA complexes. Biochemistry 24, 6433–6439 [DOI] [PubMed] [Google Scholar]
- 9. Rodnina M. V., Fricke R., Wintermeyer W. (1994) Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33, 12267–12275 [DOI] [PubMed] [Google Scholar]
- 10. Blanchard S. C., Gonzalez R. L., Kim H. D., Chu S., Puglisi J. D. (2004) tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11, 1008–1014 [DOI] [PubMed] [Google Scholar]
- 11. Geggier P., Dave R., Feldman M. B., Terry D. S., Altman R. B., Munro J. B., Blanchard S. C. (2010) Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 399, 576–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kothe U., Rodnina M. V. (2006) Delayed release of inorganic phosphate from elongation factor Tu following GTP hydrolysis on the ribosome. Biochemistry 45, 12767–12774 [DOI] [PubMed] [Google Scholar]
- 13. Pape T., Wintermeyer W., Rodnina M. V. (1998) Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pape T., Wintermeyer W., Rodnina M. (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C. (1978) Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell 14, 179–190 [DOI] [PubMed] [Google Scholar]
- 16. Johansson M., Lovmar M., Ehrenberg M. (2008) Rate and accuracy of bacterial protein synthesis revisited. Curr. Opin. Microbiol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 17. Young R., Bremer H. (1976) Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem. J. 160, 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Varenne S., Buc J., Lloubes R., Lazdunski C. (1984) Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 180, 549–576 [DOI] [PubMed] [Google Scholar]
- 19. Wohlgemuth S. E., Gorochowski T. E., Roubos J. A. (2013) Translational sensitivity of the Escherichia coli genome to fluctuating tRNA availability. Nucleic Acids Res. 41, 8021–8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blattner F. R. (1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 21. Jinks-Robertson S., Gourse R. L., Nomura M. (1983) Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell 33, 865–876 [DOI] [PubMed] [Google Scholar]
- 22. Dong H., Nilsson L., Kurland C. G. (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 [DOI] [PubMed] [Google Scholar]
- 23. Ribeiro S., Nock S., Sprinzl M. (1995) Purification of aminoacyl-tRNA by affinity chromatography on immobilized Thermus thermophilus EF-TuGTP. Anal. Biochem. 228, 330–335 [DOI] [PubMed] [Google Scholar]
- 24. Condon C., Philips J., Fu Z.-Y., Squires C., Squires C. L. (1992) Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 11, 4175–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ehrenberg M., Bremer H., Dennis P. P. (2013) Medium-dependent control of the bacterial growth rate. Biochimie 95, 643–658 [DOI] [PubMed] [Google Scholar]
- 26. Yegian C. D., Stent G. S., Martin E. M. (1966) Intracellular condition of Escherichia coli transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 55, 839–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sørensen M. A. (2001) Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 307, 785–798 [DOI] [PubMed] [Google Scholar]
- 28. Dittmar K. A., Sørensen M. A., Elf J., Ehrenberg M., Pan T. (2005) Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6, 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Louie A., Ribeiro N. S., Reid B. R., Jurnak F. (1984) Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 259, 5010–5016 [PubMed] [Google Scholar]
- 30. Sanderson L. E., Uhlenbeck O. C. (2007) Exploring the specificity of bacterial elongation factor Tu for different tRNAs. Biochemistry 46, 6194–6200 [DOI] [PubMed] [Google Scholar]
- 31. Dale T., Sanderson L. E., Uhlenbeck O. C. (2004) The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry 43, 6159–6166 [DOI] [PubMed] [Google Scholar]
- 32. Chapman S. J., Schrader J. M., Uhlenbeck O. C. (2012) Histidine 66 in Escherichia coli elongation factor Tu selectively stabilizes aminoacyl-tRNAs. J. Biol. Chem. 287, 1229–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., Nyborg J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 [DOI] [PubMed] [Google Scholar]
- 34. Abel K., Yoder M. D., Hilgenfeld R., Jurnak F. (1996) An α to β conformational switch in EF-Tu. Structure 4, 1153–1159 [DOI] [PubMed] [Google Scholar]
- 35. Ott G., Schiesswohl M., Kiesewetter S., Förster C., Arnold L., Erdmann V. A., Sprinzl M. (1990) Ternary complexes of Escherichia coli aminoacyl-tRNAs with the elongation factor Tu and GTP: thermodynamic and structural studies. Biochim. Biophys. Acta 1050, 222–225 [DOI] [PubMed] [Google Scholar]
- 36. LaRiviere F. J., Wolfson A. D., Uhlenbeck O. C. (2001) Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294, 165–168 [DOI] [PubMed] [Google Scholar]
- 37. Nissen P., Kjeldgaard M., Thirup S., Clark B. F., Nyborg J. (1996) The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie 78, 921–933 [DOI] [PubMed] [Google Scholar]
- 38. Kawashima T., Berthet-Colominas C., Wulff M., Cusack S., Leberman R. (1996) The structure of the Escherichia coli EF-TuEF-Ts complex at 2.5 Å resolution. Nature 379, 511–518 [DOI] [PubMed] [Google Scholar]
- 39. Polekhina G., Thirup S., Kjeldgaard M., Nissen P., Lippmann C., Nyborg J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure 4, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 40. Cai Y. C., Bullard J. M., Thompson N. L., Spremulli L. L. (2000) Interaction of mitochondrial elongation factor Tu with aminoacyl-tRNA and elongation factor Ts. J. Biol. Chem. 275, 20308–20314 [DOI] [PubMed] [Google Scholar]
- 41. Bremer H., Dennis P. P. (1996) in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology (Neidhardt F. C., ed) 2nd Ed., p. 1559, American Society for Microbiology, Washington, D. C [Google Scholar]
- 42. Furano A. V. (1975) Content of elongation factor Tu in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 72, 4780–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miyajima A., Kaziro Y. (1978) Coordination of levels of elongation factors Tu, Ts, and G, and ribosomal protein S1 in Escherichia coli. J. Biochem. 83, 453–462 [DOI] [PubMed] [Google Scholar]
- 44. Schümmer T., Gromadski K. B., Rodnina M. V. (2007) Mechanism of EF-Ts-catalyzed guanine nucleotide exchange in EF-Tu: contribution of interactions mediated by helix B of EF-Tu. Biochemistry 46, 4977–4984 [DOI] [PubMed] [Google Scholar]
- 45. Gordon J. (1970) Regulation of the in Vivo synthesis of the polypeptide chain elongation factors in Escherichia coli. Biochemistry 9, 912–917 [DOI] [PubMed] [Google Scholar]
- 46. Gordon J., Weissbach H. (1970) Immunochemical distinction between the Escherichia coli polypeptide chain elongation factors Tu and Ts. Biochemistry 9, 4233–4236 [DOI] [PubMed] [Google Scholar]
- 47. Yamamoto M., Strycharz W. A., Nomura M. (1976) Identification of genes for elongation factor Ts and ribosomal protein S2 in E. coli. Cell 8, 129–138 [DOI] [PubMed] [Google Scholar]
- 48. Blanchard S. C., Kim H. D., Gonzalez R. L., Jr., Puglisi J. D., Chu S. (2004) tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. U.S.A. 101, 12893–12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dunkle J. A., Wang L., Feldman M. B., Pulk A., Chen V. B., Kapral G. J., Noeske J., Richardson J. S., Blanchard S. C., Cate J. H. (2011) Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Plumbridge J. A., Bäumert H. G., Ehrenberg M., Rigler R. (1980) Characterization of a new, fully active fluorescent derivative of E. coli tRNAPhe. Nucleic Acids Res. 8, 827–843 [PMC free article] [PubMed] [Google Scholar]
- 51. Fersht A. (1985) Enzyme Structure and Mechanism, 2nd Ed., pp. 128–133, W. H. Freeman and Co [Google Scholar]
- 52. Gromadski K. B., Wieden H. J., Rodnina M. V. (2002) Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41, 162–169 [DOI] [PubMed] [Google Scholar]
- 53. Luo G., Wang M., Konigsberg W. H., Xie X. S. (2007) Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 104, 12610–12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zechmeister L., Pinckard J. H. (1953) On stereoisomerism in the cyanine dye series. Experientia 9, 16–17 [DOI] [PubMed] [Google Scholar]
- 55. Hwang H., Kim H., Myong S. (2011) Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc. Natl. Acad. Sci. U.S.A. 108, 7414–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wittinghofer A., Vetter I. R. (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 [DOI] [PubMed] [Google Scholar]
- 57. Miller D. L., Weissbach H. (1970) Interactions between the elongation factors: the displacement of GDP from the Tu-GDP complex by factor Ts. Biochem. Biophys. Res. Commun. 38, 1016–1022 [DOI] [PubMed] [Google Scholar]
- 58. Weissbach H., Miller D. L., Hachmann J. (1970) Studies on the role of factor Ts in polypeptide synthesis. Arch. Biochem. Biophys. 137, 262–269 [DOI] [PubMed] [Google Scholar]
- 59. Voorhees R. M., Ramakrishnan V. (2013) Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236 [DOI] [PubMed] [Google Scholar]
- 60. Schuette J.-C., Murphy F. V., 4th, Kelley A. C., Weir J. R., Giesebrecht J., Connell S. R., Loerke J., Mielke T., Zhang W., Penczek P. A., Ramakrishnan V., Spahn C. M. (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 28, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Romero G., Chau V., Biltonen R. L. (1985) Kinetics and thermodynamics of the interaction of elongation factor Tu with Ts. J. Biol. Chem. 280, 6167–6174 [PubMed] [Google Scholar]
- 62. Pingoud A., Urbanke C., Krauss G., Peters F., Maass G. (1977) Ternary complex formation between elongation factor Tu, GTP and aminoacyl-tRNA: an equilibrium study. Eur. J. Biochem. 78, 403–409 [DOI] [PubMed] [Google Scholar]