Background: The capsule of Group B Streptococcus is an important virulence factor and vaccine target.
Results: Analysis of type IX polysaccharide revealed a structure similar to type V and VII structures.
Conclusion: The structural and phylogenetic basis for the differentiation between types V, VII, and IX was elucidated.
Significance: Determination of the type IX structure is instrumental for the development of a carbohydrate-based vaccine.
Keywords: Bacterial Genetics, Nuclear Magnetic Resonance, Polysaccharide, Streptococcus, Vaccine, Group B Streptococcus, Capsular Polysaccharide
Abstract
The Group B Streptococcus capsular polysaccharide type IX was isolated and purified, and the structure of its repeating unit was determined. Type IX capsule →4)[NeupNAc-α-(2→3)-Galp-β-(1→4)-GlcpNAc-β-(1→6)]-β-GlcpNAc-(1→4)-β-Galp-(1→4)-β-Glcp-(1→ appears most similar to types VII and V, although it contains two GlcpNAc residues. Genetic analysis identified differences in cpsM, cpsO, and cpsI gene sequences as responsible for the differentiation between the three capsular polysaccharide types, leading us to hypothesize that type V emerged from a recombination event in a type IX background.
Introduction
Streptococcus agalactiae (Group B Streptococcus) is a β-hemolytic, encapsulated Gram-positive microorganism that colonizes the anogenital tract of 25–30% of healthy women. It is a major cause of neonatal sepsis and meningitis, particularly in infants born to mothers carrying the bacteria (1). The pathogen can also infect adults with underlying disease, particularly the elderly, and cause bovine mastitis.
The GBS2 capsule is a major virulence factor enabling the bacterium to evade human innate immune defenses (2, 3). It is constituted by high molecular weight polysaccharides of which 10 variants exist (Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX) differing in chemical composition. As for other Gram-positive and Gram-negative bacteria, individual polysaccharide repeating units are assembled on a carrier lipid (undecaprenyl phosphate) by the sequential activities of glycosyltransferase enzymes. Repeat units are transferred across the plasma membrane by a “flippase” protein. Polymerization occurs at the periplasmic face of the plasma membrane and is catalyzed by a polymerase enzyme.
All the genes responsible for the synthesis and cell wall attachment of the GBS capsular polysaccharides (CPS) are clustered in the cps operon. This operon is composed of 16–18 genes, the sequences of which differ among serotypes (4).
High titers of maternal antibodies to each CPS variant correlate with serotype-specific reduction of disease risk in the newborn (5). Hence, maternal vaccination with CPS conjugates shows great promise for the prevention of GBS neonatal disease (6).
The most common serotypes among GBS infective isolates are Ia, Ib, II, III, and V, although there are some geographical and historical variations (7). For instance, the frequency of type V has increased in the last few years, whereas serotype IV has recently emerged as a cause of adult and neonatal invasive disease (8). Therefore, accurately monitoring the variation of this important target antigen is essential for predicting vaccine coverage.
The serotype IX was identified only in 2007 during the analysis of three human isolates that failed to react with antisera against the nine GBS types described at that time. Preparation of specific antisera allowed the identification of additional type IX strains among formerly non-typeable isolates (9). The identity of GBS type IX was also confirmed by a multiplex PCR assay containing a mix of primers specific for the 10 different allelic variants of the cps genes (10). Interestingly, type IX strains can also cause neonatal infections (11, 12).
The precise chemical structures of GBS type Ia, Ib, II, III, IV, V, VI, VII, and VIII CPSs are well described (13–20). They are composed of repeating units of four to seven monosaccharides with a backbone and one or two side chains. Four monosaccharides (i.e. Glcp, Galp, GlcpNAc, and NeupNAc) are present in all nine described serotypes, and the NeupNAc residue is always found at the terminus of one of their side chains. However, the pattern of glycosidic linkages is unique to each serotype. Structural comparison of the nine GBS polysaccharide variants and alignment of their cps gene sequences indicated that their differential evolution proceeded through en bloc replacement of individual glycosyltransferase genes with DNA sequences encoding enzymes with new linkage specificities (22).
The chemical structure of CPS type IX and the genetic composition of its cps operon have not yet been delineated. Here we report the isolation, purification, structural characterization, and immunochemical analysis of type IX CPS and the genetic analysis of the cps genes encoding the enzymes responsible for its synthesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains
GBS type IX strains IT-NI-016 (isolated from a neonatal early onset disease case), IT-PW-62, and IT-PW-64 (from colonized pregnant women) were kindly provided by Alberto Berardi (Policlinico di Modena, Italy), Roberta Creti, Lucilla Baldassarri, and Graziella Orefici (Istituto Superiore di Sanità, Rome, Italy). The three strains as well as CZ-PW-119 (VI), CZ-PW-045 (VII), and CZ-NI-016 (serotype IV) were isolated and typed by latex and molecular approaches in the frame of the DEVANI study (10, 11, 23). The capsular genotype of type IX isolates was confirmed by genome analysis (see below).
Strains 2603 V/R (serotype V), COH1 (serotype III), 18RS21 (serotype II), and JM9130013 (serotype VIII) were obtained from Dennis Kasper (Harvard Medical School, Boston, MA). Strains 515 (serotype Ia) and H36B (serotype Ib) were obtained from Carol Baker (Baylor College of Medicine, Houston, TX).
Isolation and Purification of the Type IX Capsular Polysaccharide
The GBS strain IT-NI-016 was used for preparation of CPS IX from 1 liter of bacterial culture grown to exponential phase in Todd Hewitt broth. The purification process was based on previously described procedures (24). Briefly, the bacterial pellet was recovered by centrifugation at 4,000 rpm for 20 min and incubated with 0.8 n NaOH at 37 °C for 36 h. After centrifugation at 4,000 rpm for 20 min, 1 m Tris buffer (1:9, v/v) was added to the supernatant and diluted with 1:1 (v/v) HCl to reach a neutral pH.
To further purify type IX CPS, 2 m CaCl2 (0.1 m final concentration) and ethanol (30% (v/v) final concentration) were added to the solution. After centrifugation at 4,000 × g for 20 min, the supernatant was subjected to tangential flow filtration with a 10,000-molecular weight cutoff (Hydrosart Sartorius; 0.1-m2 surface) against 14 volumes of 50 mm Tris, 500 mm NaCl, pH 8.8 and 7 volumes of 10 mm sodium phosphate, pH 7.2.
To reconstitute full N-acetylation of possibly present GlcpNAc and NeupNAc residues, 1:5 (v/v) of 400 mm sodium acetate buffer, pH 4.4 (final concentration, 80 mm) was added. The material was stored at room temperature for 15 min and centrifuged at 4,000 × g for 20 min to remove any minor particles. The supernatant was subjected to filtration with a 10,000-molecular weight cutoff (Vivaspin Sartorius; 20-ml device) against 300 mm Na2CO3, 300 mm NaCl, pH 8.8. A 1:1 diluted solution of 4.15 μl/ml acetic anhydride in ethanol was added, and the reaction was incubated at room temperature for 2 h. The sample was subjected to filtration with a 10,000-molecular weight cutoff (Vivaspin Sartorius; 20-ml device) against Milli-Q water, CaCl2 (0.1 m final concentration) and ethanol (30% (v/v) final concentration) were added, and the sample was centrifuged at 4,000 × g for 20 min. The purified CPS IX was finally recovered by precipitation with ethanol (80%, v/v), and the pellet was dried under vacuum (SpeedVac, Thermo).
The purity of the polysaccharide preparation was assessed by colorimetric assays, which indicated a content of residual proteins and nucleic acids below 1% (w/w). Endotoxin content measured by Limulus amebocyte lysate was <10 endotoxin units/μg of saccharide. Highly purified GBS type Ia, Ib, II, III, V, and VII CPSs were obtained by applying the same purification process as described above for type IX.
Analysis of Monosaccharide Composition in CPS IX
The monosaccharides constituting the GBS type IX repeating unit were obtained by acidic hydrolysis. Two different series of standard samples with five increasing concentrations, the first containing Glcp, Galp, and GlcpNAc (Sigma) ranging between 0.2 and 4.0 μg/ml (as saccharide powder/ml) and the second containing NeupNAc (Sigma) between 1.0 and 10.0 μg/ml (as saccharide powder/ml), were prepared for building individual calibration curves for each monosaccharide. Two CPS IX samples targeting final concentrations in the calibration curve range were prepared. All the reference and analytical samples for Glcp/Galp/GlcpNAc were prepared in 2 m trifluoroacetic acid (Sigma), incubated at 100 °C for 2 h, dried under vacuum (SpeedVac Thermo), and suspended in water. Reference and analytical samples for NeupNAc were prepared in 0.05 m HCl (Merck), incubated at 75 °C for 1 h, and neutralized by addition of NaOH.
All analytical samples were filtered with 0.45-μm Acrodisc (Pall) filters before analysis. High performance anionic exchange chromatography-pulsed amperometric detection analysis was performed with a Dionex ICS3000 equipped with a CarboPac PA1 column (4 × 250 mm; Dionex) coupled with a PA1 guard column (4 × 50 mm; Dionex). Samples (20-μl injection volume) were run at 1 ml/min using an isocratic elution with either 24 mm NaOH (Carlo Erba) followed by a washing step with 0.5 m NaOH or 30 mm sodium acetate (Sigma) in 100 mm NaOH followed by a washing step with NaNO3 (gradient from 50 to 200 mm) in 100 mm NaOH. The effluent was monitored using an electrochemical detector in the pulse amperometric mode with a gold working electrode and an Ag/AgCl reference electrode. A quadruple potential waveform for carbohydrates was applied. The resulting chromatographic data were processed using Chromeleon software 6.8 (Dionex).
Detection of Monosaccharide Configurations
The configurations of the monosaccharides were determined by methylation according to the procedure of Ciucanu and Kerek (25) followed by hydrolysis, reduction, acetylation, and GC analysis.
A 1-mg solution of polysaccharide sample in 95:5 DMSO/water (0.3–0.5 ml) was added to 3–5 mg of finely powdered NaOH, and after 10 min of incubation at room temperature, 0.1 ml of methyl iodide was added. The reaction was stirred at room temperature for 3 h. Water (2 ml) and dichloromethane (10 ml) were then added, and the organic layer was washed with water (2 ml for three times) and then dried with Na2SO4. The dried sample was then hydrolyzed by the addition of 0.8 ml of 4 m trifluoroacetic acid and incubation at 100 °C for 2 h with stirring. After complete evaporation under vacuum (Thermo SpeedVac at 45 °C for 4 h), the dried sample was reduced with 1 m aqueous sodium borohydride (0.4 ml) and stirred for 5 h at room temperature. The sample was then acetylated with a mixture of pyridine (2 ml) and acetic anhydride (1 ml), and the reaction was stirred overnight. The excess reagent was removed by evaporation, and alditol acetates were extracted with a dichloromethane/water mixture. The generated monosaccharides were analyzed by capillary gas-liquid chromatography using a Thermo Scientific Trace GC Ultra/TSQ Quantum XLS equipped with a Restek Rxi-17Sil MS column (30 m, 0.25-mm inner diameter, 0.25 μm) in the isothermal program at 190 °C for 80 min.
NMR Spectroscopy
1H and 13C NMR experiments were recorded on a Bruker Avance III 400- or 500-MHz spectrometer equipped with a high precision temperature controller using a 5-mm broadband probe (Bruker). TopSpin version 2.6 software (Bruker) was used for data acquisition and processing.
1H NMR spectra were collected at 25 or 35 ± 0.1 °C with 32,000 data points over a 10-ppm spectral width, accumulating 128 scans. The spectra were weighted with 0.2-Hz line broadening and Fourier-transformed. The transmitter was set at the water frequency, which was used as the reference signal (4.79 ppm).
13C NMR spectra were recorded at 100.6 or 125.7 MHz and 25 ± 0.1 °C with 32,000 data points over a 200-ppm spectral width, accumulating 4,000 scans. The spectra were weighted with 0.2-Hz line broadening and Fourier-transformed. The transmitter was set at the ethanol frequency, which was used as the reference signal (17.47 ppm). All monodimensional proton NMR spectra were obtained in a quantitative manner using a total recycle time to ensure full recovery of each signal (5 × longitudinal relaxation time T1).
Bidimensional 1H-1H homo- and 1H-13C heterocorrelated experiments (correlation spectroscopy, total correlation spectroscopy, NOESY, heteronuclear single quantum correlation, heteronuclear multiple quantum correlation, and heteronuclear multiple bond correlation) were acquired with standard pulse programs. Typically, 2,048 and 256 data points were collected in F2 and F1 dimensions, respectively, and 128 scans were accumulated prior to Fourier transformation to yield a digital resolution of 0.2 and 3.0 Hz per point in F2 and F1, respectively.
Selective 1H monodimensional total correlation spectroscopy and NOESY experiments were collected at 35 ± 0.1 °C with 32,000 data points over a 10-ppm spectral width, accumulating 1,024 scans. A Gaussian shape was used for selective excitation. For NOESY, a mixing time of 300 ms was used. The spectra were weighted with 0.2-Hz line broadening and Fourier-transformed.
The NMR analytical samples of type Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX CPSs were prepared by solubilizing ∼0.5–2 mg of dried saccharide in 0.75 ml of deuterium oxide (99.9 atom % deuterium; Aldrich). The solutions were mixed to obtain a uniform concentration and subsequently transferred to a 5-mm NMR tube (Wilmad).
Estimation of the Molecular Weight of CPS IX by Size Exclusion Chromatography
The average molecular weight of CPS IX was estimated by an Ultimate 3000 system (Dionex) on PolySep GFC-P 6000 and PolySep GFC-P 5000 analytical columns with a GFC-Guard column (Phenomenex) connected in series and calibrated with a series of defined pullulan standards (Polymer) of average molecular weights ranging from 20,000 to 800,000. The void and total volumes were determined with dextran and sodium azide, respectively. The polysaccharide samples were analyzed at a concentration of 1 mg/ml using 10 mm sodium phosphate buffer, pH 7.2 as the mobile phase at a flow rate of 0.5 ml/min.
Sera and Monoclonal Antibody Reagents
Polyclonal rabbit sera were obtained from the Statens Serum Institute (Copenhagen, Denmark). CPS type IX-specific mouse antiserum was obtained by immunizing CD1 mice with purified CRM197-CPS type IX conjugate prepared as described previously (25). Animal treatments were performed in compliance with the Italian laws and approved by the institutional review board (Animal Ethics Committee) of Novartis Vaccines and Diagnostics, Siena, Italy.
CPS Serotyping and Flow Cytometry Analysis
GBS serotyping by latex agglutination assay was conducted using the Strep-B-Latex kit (Statens Serum Institute) according to the manufacturer's instructions.
For flow cytometry analysis using lectins, bacteria were grown overnight on tryptic soy agar plates at 37 °C, harvested using a sterile loop, and diluted in phosphate-buffered saline (PBS) to an A600 of 0.3. The bacterial suspension (400 μl) was centrifuged at 3,000 × g, suspended in 200 μl of PBS containing 1% BSA, and incubated for 20 min at 25 °C with shaking (blocking). Bacteria were then diluted 1:10 in PBS containing 0.1% BSA, 0.05% Tween 20 (PBSAT), and 5 μg/ml biotinylated Maackia amurensis lectin (MAL) I (Vector Laboratories) specific for NeupNAc-α-(2→3)-Galp-β-(1→4)-GlcpNAc-β-(1→trisaccharide). The solution was incubated at 4 °C for 1 h. Samples were washed twice in PBSAT, suspended in phycoerythrin-conjugated streptavidin (Southern Biotech) diluted 1:200 in PBSAT, and incubated for 30 min at 4 °C. Bacteria were washed twice in PBS, fixed in PBS containing 2% (w/v) paraformaldehyde for 20 min at room temperature, centrifuged, and suspended in 150 μl of PBS.
Flow cytometry using specific anti-capsular polysaccharide antibodies was performed as described elsewhere (26) with minor differences. Briefly, bacteria grown in Todd Hewitt broth to exponential phase were harvested and fixed in PBS containing 0.1% (w/v) paraformaldehyde. The fixed cells were washed with PBS and incubated for 1 h at room temperature with immune mouse sera raised against type V or type IX purified polysaccharides diluted 1:200 in PBS containing 0.1% BSA. The cells were incubated for 1 h at 23 °C with R-phycoerythrin-conjugated F(ab)2 goat anti-mouse immunoglobulin G diluted 1:100 in PBS containing 0.1% BSA. All data were collected using a FACSCalibur and a BD FACS CANTO II (BD Biosciences) by acquiring 10,000 events, and data analysis was performed with FlowJo software (v.8.6, TreeStar Inc.).
DNA Sequence Analysis of cps Capsule Biosynthetic Clusters
For high throughput full genome deep sequencing of type IX strains IX IT-NI-016, IT-PW-62, and IT-PW-64, 5 μg of genomic DNA was randomly sheared by a Covaris S2 instrument with set target size of 300 bp. DNA libraries were automatically generated by mixing 2.5 μg of sheared DNA with 2.5 μl of indexed Illumina adapter from TruSeq DNA Sample Preparation kits (Illumina). SPRIworks Fragment Library kit I cartridges (Beckman Coulter) and size selection of 300–600 bp were used for library preparation. Library enrichment was performed using a TruSeq Sample Preparation kit (Illumina), and amplified products were purified by Ampure XP Magnetic Beads (Beckman Coulter) according to the manufacturer's protocol. Adapted libraries were quantified for optimal cluster density with the 7900 HT Fast RT-PCR System (Invitrogen) using the KAPA SYBR FAST ABI Prism qPCR kit (Kapa Biosystems). Libraries were then pooled, denatured, diluted to a final concentration of 6 pm, and loaded on the cBOTTM System (Illumina) for the cluster generation step. Sequencing was performed on a HiSeq2000 in a 100-bp paired-end run using TruSeq SBS chemistry (Illumina).
For alignment comparisons, nucleotide sequences of the cps serotype-specific regions from reference GBS strains were retrieved from the NCBI database (type V reference strain 2603V/R, accession number NC_004116; type VII reference, accession number AY376403). Multiple and pairwise sequence alignments were performed with MUSCLE using Geneious version 7.05 (Biomatters).
Genetic Engineering of GBS Types IX, V, and VII
Two different plasmids were designed to obtain the chimeric CPS-expressing strains. First, DNA fragments consisting of the cps type V-specific (cps5M, cps5O, and cps5I) or the type IX-specific genes (cps9M and cps9I) were amplified by PCR from S. agalactiae CJB111 or from IT-NI-016 genomic DNA, respectively, using specifically designed primers (Table 1) and the following reaction cycle: 1 min at 98 °C; 10 s at 98 °C, 20 s at 55 °C, and 3 min at 72 °C (30 cycles); and 7 min at 72 °C. Then the resulting fragments were alternatively cloned into the expression vector pAM-p80 (26) to obtain plasmids pAM-cps5MOI (containing cps5M, cps5O, and cps5I; pAM-V) and pAM-cps9MI (cps9M and cps9I; pAM-IX). Plasmids were purified and used to transform GBS by electroporation. Strain IT-NI-016 (serotype IX) was transformed with pAM-cps5MOI, and pAM-cps9MI was used to transform strain 2603 V/R. Both plasmids were also used to transform serotype VII strain CZ-PW-045.
TABLE 1.
List of oligonucleotides used for plasmid construction
| Primer name | Sequence (5′ → 3′) | Length |
|---|---|---|
| bp | ||
| pAM-IXMI-F | GCGGCGCGGCCGCGACATATTTGCTCTGATATGGCAG | 37 |
| pAM-IXMI-R | GCGGCAGATCTGGGATAATGATACTAATCATCTTC | 35 |
| pAM-VMOI-F | GCGGCGCGGCCGCGCTCTGATATGGCAGGAGGTAAGG | 37 |
| pAM-VMOI-R | GCGGCAGATCTGGGATAATGATACTAACTTTATCC | 35 |
RESULTS
The Type IX CPS Chemical Structure Is Unique among GBS and Contains Two GlcpNAc Residues per Hexasaccharide Repeating Unit
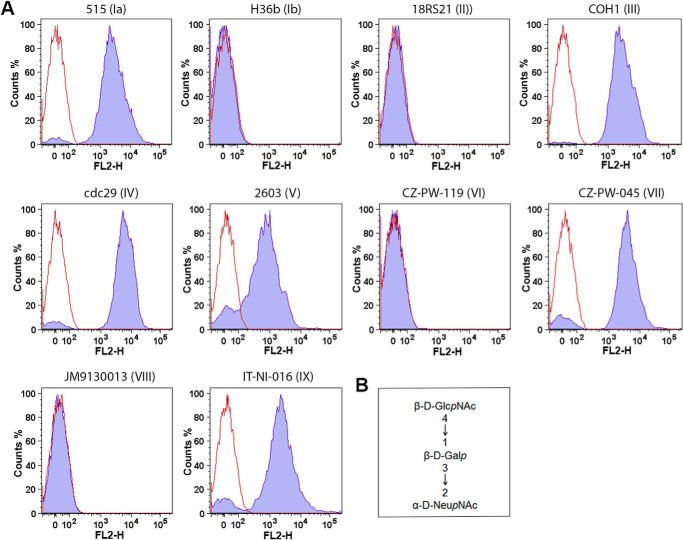
In a first attempt to investigate the chemical structure of the type IX capsular polysaccharide, we performed flow cytometry on different GBS strains representing the 10 serotypes. Bacteria were labeled with biotinylated MAL I specific for NeupNAc-α-(2→3)-Galp-β-(1→4)-GlcpNAc-β-(1→. As expected, MAL I reacted with serotype Ia, III, IV, V, and VII bacteria that contain the trisaccharide in their side chain but not with types Ib, II, VI, and VIII (Fig. 1). The type IX strain IT-NI-016 was clearly labeled by the lectin, indicating that this trisaccharide is also present on the surface of type IX bacteria.
FIGURE 1.
A, flow cytometry analysis of strains belonging to the 10 GBS serotypes after incubation with MAL I specific for B, NeupNAc-α-(2→3)-Galp-β-(1→4)-GlcpNAc-β-(1→.
Type IX CPS was then isolated by alkaline treatment of GBS IT-NI-016 and highly purified following the procedures described under “Experimental Procedures.” The purified material was spotted onto nitrocellulose and showed good reactivity with a commercial rabbit type IX antiserum in dot blot analysis (data not shown). The CPS IX preparation was structurally characterized by evaluating its molecular size, monosaccharide composition, and structural identity.
The average molecular weight estimated by size exclusion chromatography-HPLC was ∼100,000. High performance anionic exchange chromatography-pulsed amperometric detection compositional analysis revealed Glc, Gal, GlcNAc, and NeuNAc in a molar ratio of 1.0:1.9:2.0:1.3. The data indicated that CPS IX is composed of the same monosaccharide residues as Ia, Ib, II, III, IV, V, V, and VII, although, differently from the other CPS types, it contains 2 mol of GlcNAc per repeating unit.
To preliminarily assess the configuration of the monosaccharides composing a type IX repeating unit, a sugar linkage-type analysis was performed by GC-MS. Several variably linked monosaccharide units were revealed, including 3-monosubstituted Galp ((→3)-Galp-(1→)) and 4-monosubstituted Galp ((→4)-Galp-(1→)), Glcp ((→4)-Glcp-(1→)), and GlcpNAc ((→4)-GlcpNAc-(1→)).
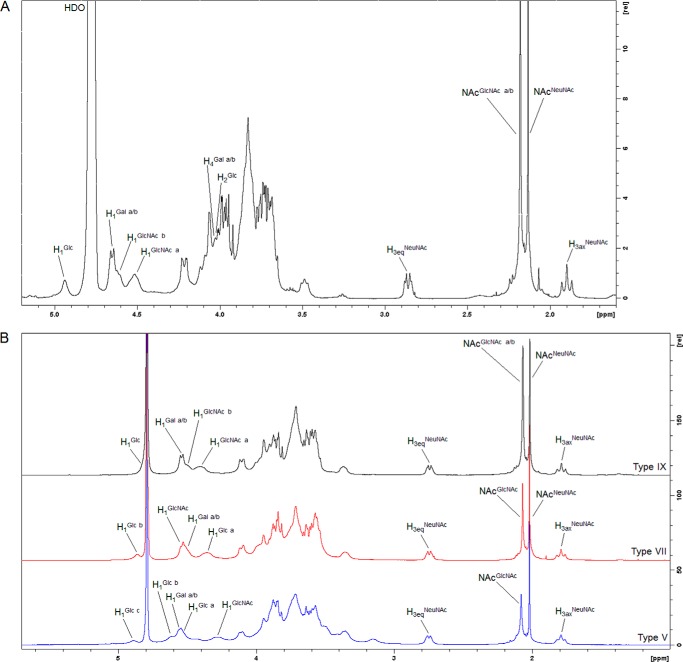
Further structural evaluation was achieved by comparing the 1H monodimensional NMR spectrum of CPS IX with those of other CPS types for which peak assignments have already been reported (13–21) (Fig. 2A). Comparison of the less crowded regions (anomeric protons of Glcp/Galp/GlcpNAc, H1; equatorial and axial protons at C2 of NeupNAc, H3eq and H3ax; N-acetyl groups of GlcpNAc and NeupNAc, CH3NAc; Fig. 2A) suggested the type IX structure by NMR chemical shift analogies. The integration of their relative peak areas confirmed the molar ratio determined by high performance anionic exchange chromatography-pulsed amperometric detection, i.e. H1Glc/Gal/GlcNAc:H3eqNeuNAc:NAcGlcNAc/NeuNAc:H3eqNeuNAc in a ratio of ∼5:1:6:1. As an additional confirmation of CPS IX compositional analysis, the anomeric region of the 1H monodimensional spectrum collected on the fully hydrolyzed polysaccharide showed the H1α and H1β peaks of Glcp, Galp, and GlcpN (generated from GlcpNAc by the strong acid treatment) in a ratio of ∼1:2:2 (sum of H1α and H1β integral value for each monosaccharide).
FIGURE 2.
A, 1H NMR spectrum of GBS type IX capsular polysaccharide recorded at 35 ± 0. 1 °C. B, comparison of 1H NMR spectra obtained for capsular polysaccharides IX, VII, and V recorded at 25 ± 0.1 °C. For type V, H1Glc a, anomeric proton of Glcp residues in the branch; H1Glc b and H1Glc c, anomeric protons of Glcp residue in the backbone; H1Gal a/b, anomeric protons of Galp residues in the branch and backbone; H1GlcNAc, anomeric proton of GlcpNAc residue in the branch; H3eqNeuNAc and H3axNeuNAc, protons at position C3 equatorial and axial of NeupNAc residue in the branch. For type VII, H1Glc a and H1Glc b, anomeric protons of Glcp residue in the backbone; H1Gal a/b, anomeric protons of Galp residues in the branch and backbone; H1GlcNAc, anomeric proton of GlcpNAc residue in the branch; H3eqNeuNAc and H3axNeuNAc, protons at position C3 equatorial and axial of NeupNAc residue in the branch. For type IX, H1Glc, anomeric proton of Glcp residues in the backbone; H1Gal a/b, anomeric protons of Galp residues in the branch and backbone; H1GlcNAc a, anomeric proton of GlcpNAc residue in the branch; H1GlcNAc a, anomeric proton of GlcpNAc residue in the backbone; H4Gal a/b, protons at position C4 of Galp residues in the branch and backbone; H2Glc, proton at position C2 of Glcp residue in the backbone; H3eqNeuNAc and H3axNeuNAc, protons at position C3 equatorial and axial of NeupNAc residue in the branch.
The 1H monodimensional and homocorrelated two-dimensional correlation spectroscopy NMR spectra collected at 35 °C allowed assigning five anomeric signals at 4.938 (H1Glc), 4.649 (both H1Gal), 4.612 (H1GlcNAc), and 4.514 (H1GlcNAc) ppm as well as signals at 2.858 and 1.897 ppm of the equatorial and axial H3 of α-linked NeupNAc, respectively (Table 2 and Fig. 2A). One signal (H1Glc) was assigned to an α-glycosidic configuration (3J1,2 ∼ 4.2 Hz), whereas the other four signals showed a larger 3J coupling constant (e.g. H1Gal, 3J1,2 = 7.5 Hz) and thus belonged to β-anomeric protons.
TABLE 2.
NMR chemical shift of protons at C1 and C2 for GBS type IX CPS recorded at 35 ± 0.1 °C
Other signals are as follows: H3eqNeuNAc, 2.858 ppm; H3axNeuNAc, 1.897 ppm; NAcGlcNAc a/b, 2.179 ppm; and NAcNeuNAc, 2.132 ppm.
| Sugar residue | H1 | H2 |
|---|---|---|
| Glcp | 4.938 | 4.009 |
| Galp a | 4.649 | 3.668 |
| Galp b | 4.649 | 3.862 |
| GlcpNAc a | 4.514 | 3.479 |
| GlcpNAc b | 4.612 | 3.722 |
The location of the H2Glc signal at 4.009 ppm (unresolved doublet with small diaxial coupling, 3J1,2 ∼ 4.2 Hz) unambiguously characterized this unit as being β-Glcp (Fig. 2A). The very low field resonance for H4Gal (4.067 ppm) together with the large value of 3J1,2 coupling for H1 (7.5 Hz) characterized both Galp units as β-Galp (Fig. 2A).
The monodimensional 13C and the heterocorrelated two-dimensional (1H-13C) heteronuclear multiple quantum correlation NMR spectra showed five signals in the anomeric region at 104.04, 103.71, 103.28 (2C), 101.70, and 100.50 ppm in addition to the signal at 40.33 ppm corresponding to the C3 of the NeupNAc residue, confirming that the type IX CPS is composed of a hexasaccharide repeating unit. Additional signals were observed at 52.38 ppm (3C), which corresponded to the C2 of the two GlcpNAc residues and C5 of NeupNAc residue, and at 22.57, 23.32, and 22.74 ppm, corresponding to the methyl of N-acetyl groups of both GlcpNAc residues and NeupNAc residues, respectively. The 1H NMR signals of these methyl groups fell at 2.066 (GlcpNAc, 6H) and 2.015 ppm (NeupNAc, 3H).
All protons at C2 of Glcp, Galp, and GlcpNAc residues were assigned using correlation spectroscopy as well as the total correlation spectroscopy method. The two GlcpNAc and the two Galp residues present in the CPS IX repeating unit were designated “a” and “b” to facilitate their differentiation.
To identify the O-glycosidic linkages, selective total correlation spectroscopy and NOESY experiments were collected by irradiating the four anomeric protons (H1Glc, H1Gal a, and H1Gal b together and H1GlcNAc a and H1GlcNAc b) and the equatorial/axial protons at C3 of the NeupNAc residue (H3eqNeuNAc and H3axNeuNAc). The same chemical shift and J coupling values revealed for both H1Gal signals, which fell at the same resonance of other already assigned GBS CPS types, unambiguously characterized these units as being Galp β-linked to the position C4 of GlcpNAc residues (Galp-(1→4)-GlcpNAc).
In accordance with flow cytometry results using MAL I, interglycosidic NOEs from H3eqNeuNAc and H3axNeuNAc to H3Gal a and from H1Gal a to H4GlcNAc a were revealed, confirming the presence of the NeupNAc-α-(2→3)-Galp-β-(1→4)-GlcpNAc-β-(1→) trisaccharide branch. In this regard, the NMR represented a validation and confirmation of the previous data and inferences. Therefore, by the time we carried out NMR experiments, only the configuration of a second β-GlcpNAc residue, named GlcNAc b, and the anchoring site of branched trisaccharide to the backbone were unknown. Inter-residual NOE couplings H1Glc-H4GlcNAc b, H1GlcNAc a-H6GlcNAc b, and H1GlcNAc b-H4Gal b were revealed by selective irradiation of the anomeric protons of Glc, GlcNAc a, and GlcNAc b residues, indicating that the (→4)-β-GlcpNAc-(1→4)-β-Galp-(1→4)-α-Glcp-(1→) trisaccharide constitutes the backbone of the CPS IX repeating unit. Low intensity NOE correlations were also revealed between H1Glc, H1GlcNAc, and H1Gal b and protons of their own saccharide ring and the vicinal residue.
Overall, NOE data were consistent with the structure of the CPS type IX repeating unit illustrated in Fig. 3 that was assembled by combining the NMR results with data obtained from other methodologies described previously. To further confirm the uniqueness of the type IX CPS structure, its collected 1H NMR profile was compared with those obtained for its most similar polysaccharides belonging to types V and VII (Fig. 2B). The different fingerprints of the anomeric region supported the previously elucidated structural diversity. Closer scrutiny of the N-acetyl group region (NeupNAc and GlcpNAc signals) showed a higher intensity of the GlcpNAc peak in CPS IX due to the presence of two GlcpNAc residues compared with the other two capsular polysaccharides.
FIGURE 3.
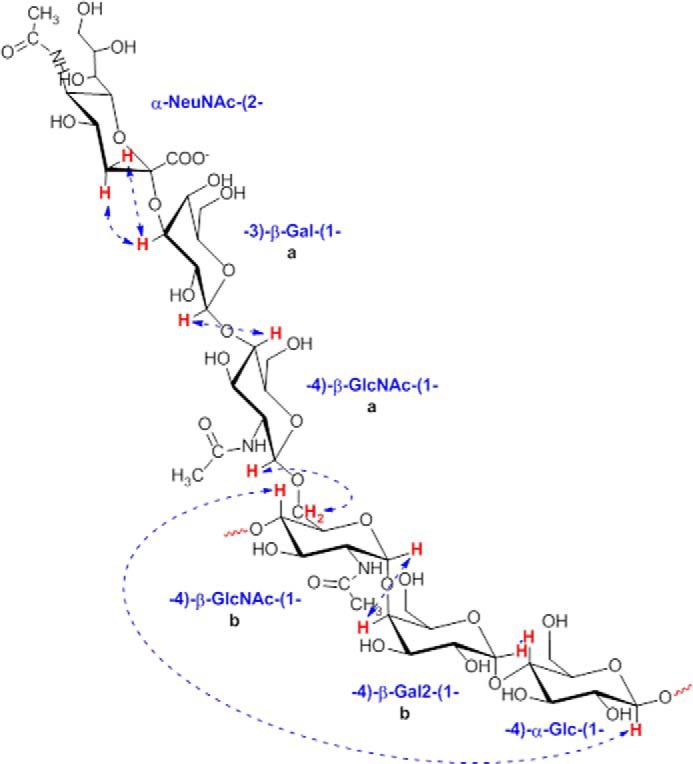
Repeating unit structure of CPS IX with inter-residue NOEs revealed by selective NOESY experiments on anomeric protons and H3eqNeuNAc. The double GlcpNAc and Galp residues are labeled a and b.
CPS Type IX Mostly Resembles Types V and VII Both at Structural and Genetic Levels
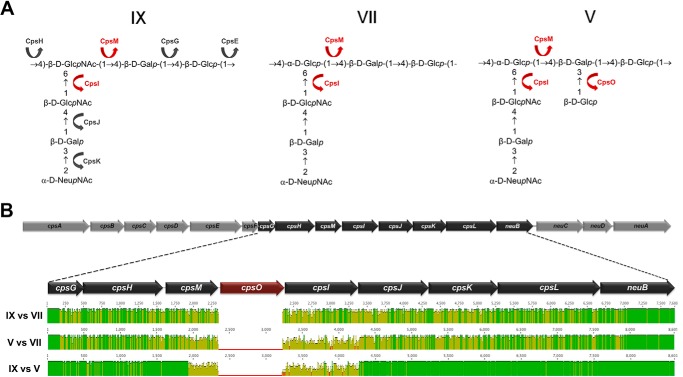
As indicated above and illustrated in Fig. 4A, the type IX polysaccharide repeating unit most closely resembles that of type VII (19) and of type V (17). The type V structure is identical to type VII except for the presence of a second side chain containing a Glcp residue β-linked to the C3 of the backbone Galp residue. Type IX contains only one lateral chain, just like type VII, but the Glcp residue present in the backbone of types V and VII is here replaced by GlcpNAc.
FIGURE 4.
Comparison of CPS type IX with types VII and V. A, chemical structure of the repeating units of type V, VII, and IX CPSs. The putative function of the enzymes involved in the assembly of the type IX CPS is indicated by arrows. cpsM, cpsO, and cpsI glycosyltransferases, likely catalyzing the addition of the differing monosaccharides, are labeled in red for the three CPS types. B, schematic illustration of the cps9 operon and pairwise alignments between the central regions comprising genes cpsG to neuB of cps9, cps7, and cps5. The cps5O gene present only in type V is indicated in red. A gap was introduced between cpsM and cpsI in the alignment of types IX and VII (top) to compensate for the lack of cpsO. Green areas indicate regions with 100% identity, and variable regions are indicated in yellow.
The machinery responsible for the synthesis of GBS capsular polysaccharides is encoded in the cps operon. The 5′ cpsABCD genes, possibly involved in the regulation of capsular synthesis, and the 3′ neuBCDA, responsible for the synthesis of sialic acid, are highly conserved (22). The remaining cpsE–L genes are more variable between the different serotypes and encode the enzymes responsible for the synthesis of the repeating units, transport to the outside leaflet of the cell membrane, and repeating unit polymerization. To investigate the evolutionary relationship between the synthesis machineries of type IX, V, and VII capsular polysaccharides, we sought to compare the DNA sequences of their corresponding cps operons (herein cps9, cps5, and cps7).
First, three independent type IX isolates were subjected to deep sequencing, and their cps9 sequences were compared. The overall DNA sequence identity exceeded 99% in pairwise alignment, similar to the high intraserotype homology observed for the cps operons of all other GBS serotypes downloaded from public databases.
Second, cps9 genes were aligned to their homologs in all other serotypes (Table 3). This analysis revealed two well conserved regions spanning from cpsA to cpsF and from neuB to neuA, flanking a more variable central region from cpsG to cpsL. In this central region, type IX showed a higher similarity to types V and VII compared with the other serotypes.
TABLE 3.
Nucleotide sequence identity between type IX cps genes (strain IT-NI-016) and types V (2603/VR), VII (7271; AY376403), Ia (A909), Ib (H36B), II (18RS21), III (NEM316), IV (CNTC 1/82; AF355776.1), and VI (NT6 type VI; AF337958)
Identity levels are indicated by a gray color scale. (n.a., not available; −, lack of gene).
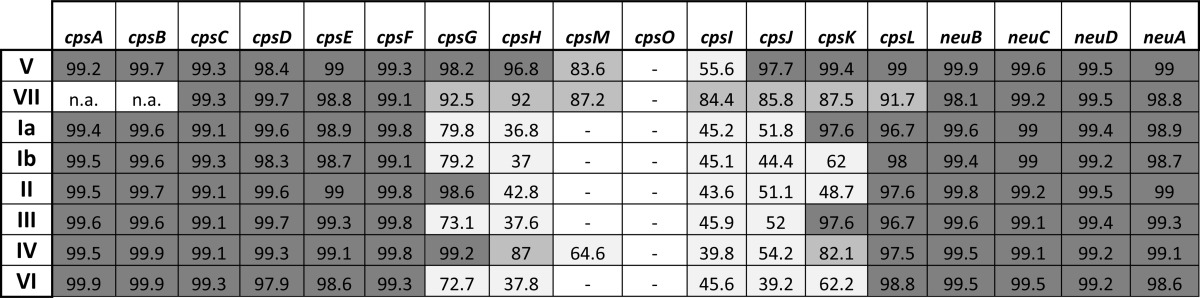
The putative function of each of the glycosyltransferases and polymerases involved in the assembly of the type IX, V, and VII hexasaccharide repeating unit was predicted based on literature data and on comparisons with other serotypes (Fig. 4A). A schematic pairwise alignment of the DNA segment comprising genes cpsF to cpsL from serotypes IX, V, and VII is illustrated in Fig. 4B. The highest sequence identity in this region (>95%) was observed between types V and IX from the full cpsF up to nucleotide 326 of cpsM and from cpsJ to cpsL. Sequence identity decreased dramatically (<60%) for the second half of cpsM and the full cpsI gene sequence, a region that also included the unique type V cpsO gene (see also Table 3). The data suggested a recombination event as the possible cause for the differentiation between the two serotypes.
Comparison between the variable region of types IX and VII revealed 99% identity for cpsG up to nucleotide 59 and lower sequence identity (between 84 and 92%) from that position up to nucleotide 246 of neuB. The degree of similarity between types VII and IX matches that observed when comparing type VII with type V if one excludes the hypervariable cps5MOI region. Also, in this case, the sequence identity decreased from nucleotide 178 of cpsG up to nucleotide 82 of neuB. This observation is in contrast with the previously reported high conservation of cpsL among all GBS serotypes (22). Overall, these results indicate that, at the genetic level, type IX is more closely related to type V than to type VII.
Sequence Polymorphisms in cpsMOI Genes Determine Capsular Polysaccharide Specificity among Serotypes V, VII, and IX
As reported above, the cpsMOI genes showed the highest degree of variability in the otherwise highly similar cps5, cps7, and cps9 operons. This observation led us to hypothesize that differences in these three genes could account for the structural diversity among serotypes IX, V, and VII.
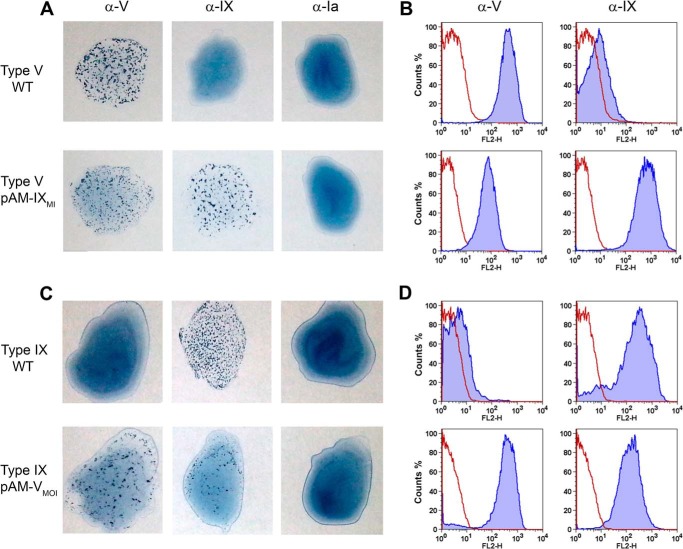
To test this hypothesis, we investigated whether episomal transfer of cpsMOI genes between the three serotypes could be sufficient to drive the synthesis of heterologous GBS polysaccharides. To this end, we first designed the plasmid construct pAM-IX containing cps9MI genes under the GBS p80 promoter (27). The plasmid was subsequently used to transform a type V strain. Expression of different types of CPS in the recipient and recombinant strains was analyzed by latex agglutination and flow cytometry using mouse sera raised against type V and rabbit sera against type IX polysaccharides.
As shown in Fig. 5A, the recipient strain showed positive latex agglutination only with anti-type V, whereas recombinant GBS V harboring pAM-IX showed double agglutination with both type V and type IX antibodies. Similar results were obtained by flow cytometry experiments (Fig. 5B) where the recipient strain showed a fluorescence shift only with anti-CPS V, whereas positive shifts were observed in the recombinant strain for both anti-CPS V and anti-CPS IX antibodies.
FIGURE 5.
Concomitant synthesis of CPS IX and CPS V in recombinant type V and type IX strains transformed with pAM-cps5MOI and pAM-cps9MI, respectively. A and C, serotyping of wild-type and recombinant bacteria by latex agglutination. B and D, flow cytometry analysis of the same strains using anti-CPS V and anti-CPS IX mouse polyclonal antibodies. The red peaks represent negative controls only labeled with secondary antibodies; the blue peaks represent bacteria labeled with the anti-CPS antibodies.
The data indicated that anti-type IX antibodies are highly specific for the type IX CPS and do not cross-react with the similar but not identical type V structure. Furthermore, we confirmed that transfer of type IX cps9MI genes in a type V background is sufficient to drive the synthesis of both type V and IX CPSs. Fluorescence intensities with anti-CPS V antibodies decreased in the recombinant strain compared with the wild-type recipient strain, indicating decreased synthesis of CPS V when cps5MOI and cps9MI were co-expressed by recombinant bacteria.
We then constructed the plasmid pAM-V containing the cps5MOI genes under the GBS p80 promoter, and this construct was transferred into a GBS type IX isolate. As shown in Fig. 5, C and D, latex serotyping and flow cytometry using anti-CPS V and anti-CPS IX rabbit and mouse sera, respectively, revealed the presence of polysaccharide IX only in the recipient strain and of both type IX and type V CPSs in recombinant bacteria. Again, type IX CPS decreased in GBS co-synthesizing type V. Once more the data confirmed specificity of anti-type V antibodies and the possibility of co-expressing type V and IX CPSs in a type IX background by transfer of the cps5MOI genes.
Finally, we introduced in a GBS type VII strain either pAM-IX or pAM-V plasmids. Latex serotyping showed type VII reactivity in the recipient strain, double type VII and IX reactivity for the clone harboring pAM-IX, and double type VII and V reactivity for pAM-V-transformed bacteria (Fig. 6). As in former experiments, latex agglutination for the endogenous type VII decreased in transformed recipient strains also expressing heterologous polysaccharides. The obtained results confirmed that differences in cpsMOI gene can fully explain the diversity in the polysaccharide structures and serological characteristics among CPS types IX, V, and VII.
FIGURE 6.
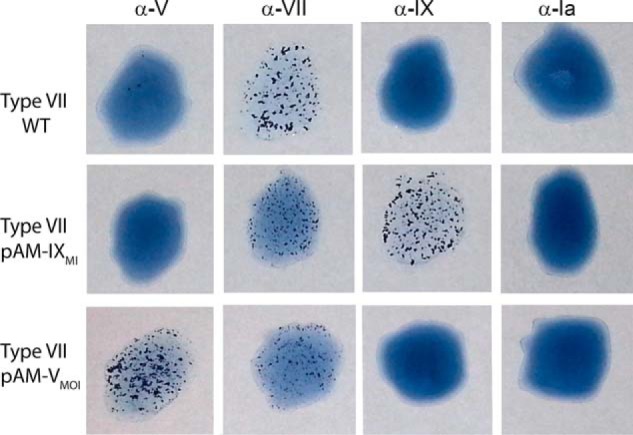
Concomitant synthesis of CPS VII and CPS IX and of CPS VII and CPSV in recombinant type VII strains transformed with plasmids harboring type IX cps9MI or type V cps5MOI genes. Latex agglutination of GBS wild-type and recombinant bacteria using anti-type V, -type VII, and -type IX antisera as well as anti-type Ia as a negative control.
DISCUSSION
The CPS repeating unit of the recently described GBS serotype IX was confirmed to contain a unique structural motif differing from all previously identified GBS CPSs. Similar to CPS types Ia, Ib, II, III, IV, V, VI, and VII, type IX CPS is exclusively composed of Glcp, Galp, GlcpNAc, and NeupNAc residues. The NeupNAc is always positioned at the terminal end of the repeating unit side chain, and its involvement in the immunodominant CPS epitope has been demonstrated for type III, Ia, II, V, and VI antigens (28–34).
The overall CPS type IX structure most closely resembles that of type VII (19) except that the trilinked Glcp residue in its backbone is replaced by GlcpNAc, resulting in 2 mol of GlcNAc/mol of repeating unit. In addition to this monosaccharide substitution, type IX differs from type V by the presence in the latter of a second side chain of Glcp linked β-(1→3) to the backbone Galp residue.
Recent comparisons of GBS latex agglutination serotyping results and PCR-based capsular gene typing revealed some inconsistencies between the two methods mostly associated with type IX (23). Serotyping difficulties could in part be due to weak serum cross-reactivity, requiring preabsorption with the different antigens for correct serological discrimination between the different types.
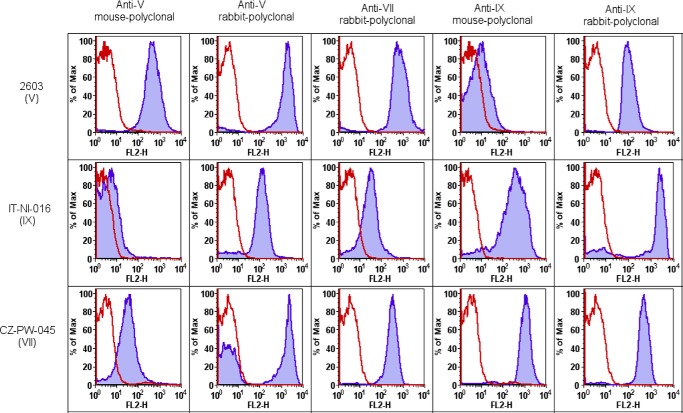
We further investigated immunological cross-reactivity among types IX, V, and VII by FACS analysis of GBS belonging to the three related serotypes using both commercial rabbit sera and mouse sera obtained by immunizing animals with purified CPS V and CPS IX conjugated to CRM197. The obtained data indicate higher cross-reactivity between types VII and IX compared with type V and confirmed a higher specificity of mouse antibodies raised against purified polysaccharides compared with commercial rabbit antisera in the highly sensitive FACS setting (Fig. 7). The difficulties in obtaining highly specific antisera confirm the strong similarity between immunodominant epitopes of these structurally similar polysaccharides and could also explain why type IX was identified only recently. One consequence of this similarity is the possibility that the three polysaccharide types could induce cross-reactive functional antibodies. The high sensitivity of the available type VII and IX strains to complement did not allow testing this hypothesis by functional opsonophagocytic or in vivo mouse protection assays.
FIGURE 7.
Flow cytometry analysis of type V, VII, and IX strains using anti-CPS V, -CPS VII, and -CPS IX rabbit commercial sera (r; Statens Serum Institute) and anti-CPS V and -CPS IX mouse sera (m) obtained by immunizing animals with the purified polysaccharides conjugated to CRM197. The red peaks represent negative controls labeled with secondary antibodies; the blue peaks represent bacteria incubated with the anti-CPS sera and labeled secondary antibodies.
The observation that type IX is genetically closer to type V than to type VII was unexpected given the closer structural similarity and higher serological cross-reactivity between types IX and VII compared with type V. However, genetic evidence seems to indicate that the type V cps operon originated from a pre-existing type IX cps by a horizontal transfer-recombination event targeting positions 326 of cps9M and 10 of cps9J. This event resulted in the formation of a functional cpsM chimera, catalyzing the incorporation of a Glcp residue in position 3 in place of GlcpNAc, and in a new variant of cpsI, attaching the first GlcpNAc of the lateral chain to this third backbone residue. Finally, the generation of a new Glcp lateral branch resulted from the acquisition of the cps5O gene.
A proposed phylogenetic model for the evolution of capsular types VII, IX, and V in the GBS population is shown in Fig. 8. According to this model, CPS VII and IX were derived from a common ancestor by sequential addition of point mutations, leading to divergent structures containing either Glcp or GlcpNAc in position 3 of the backbone of the repeating unit. A subsequent recombination event in a type IX background generated the type V CPS where the third backbone residue is again Glcp. Unfortunately, the mining of publicly available genome databases has not yet provided any clues on the exact identity of the donor strain/species. However, we were able to identify cpsO homologs in Streptococcus suis and Streptococcus infantarius, thus suggesting the possible involvement of these related streptococci in the origin of type V CPS.
FIGURE 8.
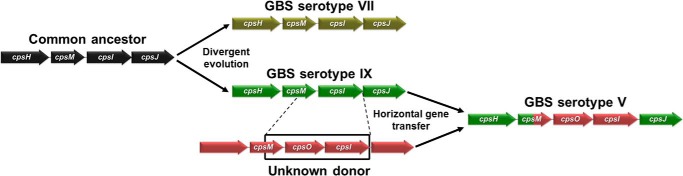
Proposed model for the genetic relationship between serotypes IX, VII, and V. According to this model, types VII and IX were derived from a common ancestor by sequential addition of point mutations, whereas type V emerged from a subsequent recombination event in a type IX background. The boxed region highlights the genetic segment involved in the recombination event resulting in a type V CPS.
The phylogenetic relationship between serotype V and serotype IX GBS strains remains to be investigated in detail. Type V strains are highly diverse in that, based on multilocus sequence typing analysis, they have been assigned to different GBS clonal complexes, including the prevalent CC1 but also CC19, CC23, and CC10 (35). The three type IX isolates sequenced in this study belong to CC130, which to our knowledge has not been ascribed to any type V strain. The genetic distance between the two serotypes will need to be confirmed by genome wide analyses on multiple strains and would imply that in addition to the proposed lateral transfer involving part of the cps operon that led to the appearance of CPS type V capsular switching events have occurred in ancestor strains belonging to one of the two serotypes. Interestingly, although the relative frequency of type IX isolates in the GBS population seems to be very low and more comprehensive molecular or seroepidemiological studies are needed, serotype V recently emerged as one of the major serotypes causing adult disease and is now increasingly causing neonatal infections (7).
The data showing that an anti-CPS V reactive polysaccharide could be synthesized by a recombinant type IX strain expressing cps5MOI in trans support the proposed model for the emergence of type V from a genetic exchange limited to the three genes. Similarly, an anti-CPS IX-reactive polysaccharide was synthesized in a type V strain carrying cps9MI in a plasmid vector. Interestingly, the sole addition of the same plasmid to a type VII strain was sufficient to drive the synthesis of an anti-IX-reactive CPS. The data indicate that the cps7H polymerase, which differs from cps9H by 29 amino acids, can still operate on a repeating unit substrate containing a terminal GlcpNAc instead of Glcp.
The above results are in agreement with previous data from Chaffin et al. (36), who reported that a single gene can confer GBS serotype specificity. In that case, the authors reported that cpsH differences were solely responsible for the structural diversity between types Ia and III. In the present study, differences in the cpsMOI genes account for the diversity among types IX, V, and VII, whereas the cpsH enzyme shows more relaxed substrate specificity among the three serotypes. Similar to what observed by Chaffin et al. (36), synthesis of a heterologous CPS reduced the synthesis of the homologous immunoreactive polysaccharide. This is possibly due to competition between the two alternative repeating units as substrates for the enzymes catalyzing the downstream steps of CPS production, also taking into account that multicopy expression from a strong promoter might favor the synthesis of the heterologous repeating unit.
As for other pathogens, the CPSs are excellent antigen targets for the prevention of GBS infections, and conjugate vaccines have been developed against the major serotypes Ia, Ib, II, III, and V. Elucidation of the CPS type IX motif provides additional knowledge that can be instrumental for the development of carbohydrate-based vaccines covering all GBS serotypes.
Acknowledgments
We are grateful to Alberto Berardi, Roberta Creti, Lucilla Baldassarri, Graziella Orefici, and the DEVANI consortium for providing type IX GBS strains. We thank Stefano Censini and Nicola Pacchiani of Novartis Vaccines (Siena, Italy) for support with genome sequence analysis.
Footnotes
- GBS
- Group B Streptococcus
- Glcp
- glucose
- CPS
- capsular polysaccharide
- Galp
- galactose
- GlcpNAc
- N-acetylglucosamine
- GlcpN
- glucosamine
- NeupNAc
- N-acetylneuraminic acid
- MAL
- M. amurensis lectin.
REFERENCES
- 1. Schuchat A. (1998) Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11, 497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rubens C. E., Wessels M. R., Heggen L. M., Kasper D. L. (1987) Transposon mutagenesis of type III group B Streptococcus: correlation of capsule expression with virulence. Proc. Natl. Acad. Sci. U.S.A. 84, 7208–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wessels M. R., Rubens C. E., Benedí V. J., Kasper D. L. (1989) Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc. Natl. Acad. Sci. U.S.A. 86, 8983–8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cieslewicz M. J., Kasper D. L., Wang Y., Wessels M. R. (2001) Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276, 139–146 [DOI] [PubMed] [Google Scholar]
- 5. Baker C. J., Kasper D. L. (1976) Correlation of maternal antibody deficiency with susceptibility to neonatal group B streptococcal infection. N. Engl. J. Med. 294, 753–756 [DOI] [PubMed] [Google Scholar]
- 6. Edwards M. S. (2008) Group B streptococcal conjugate vaccine. A timely concept for which the time has come. Hum. Vaccin. 4, 444–448 [DOI] [PubMed] [Google Scholar]
- 7. Le Doare K., Heath P. T. (2013) An overview of global GBS epidemiology. Vaccine 31, Suppl. 4, D7–D12 [DOI] [PubMed] [Google Scholar]
- 8. Ferrieri P., Lynfield R., Creti R., Flores A. E. (2013) Serotype IV and invasive group B Streptococcus disease in neonates, Minnesota, U.S.A., 2000–2010. Emerg. Infect. Dis. 19, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slotved H. C., Kong F., Lambertsen L., Sauer S., Gilbert G. L. (2007) Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45, 2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imperi M., Pataracchia M., Alfarone G., Baldassarri L., Orefici G., Creti R. (2010) A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 80, 212–214 [DOI] [PubMed] [Google Scholar]
- 11. Melin P., Efstratiou A., (2013) Group B streptococcal epidemiology and vaccine needs in developed countries. Vaccine 31, Suppl. 4, D31–D42 [DOI] [PubMed] [Google Scholar]
- 12. Lamagni T. L., Keshishian C., Efstratiou A., Guy R., Henderson K. L., Broughton K., Sheridan E. (2013) Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991–2010. Clin. Infect. Dis. 57, 682–688 [DOI] [PubMed] [Google Scholar]
- 13. Jennings H. J., Katzenellenbogen E., Lugowski C., Kasper D. L. (1983) Structure of native polysaccharide antigens of type Ia and type Ib group B Streptococcus. Biochemistry 22, 1258–1264 [DOI] [PubMed] [Google Scholar]
- 14. Jennings H. J., Rosell K.-G., Katzenellenbogen E., Kasper D. L. (1983) Structural determination of the capsular polysaccharide antigen of type II group B streptococci. J. Biol. Chem. 258, 1793–1798 [PubMed] [Google Scholar]
- 15. Jennings H. J., Lugowski C., Kasper D. L. (1981) Conformational aspects critical to the immunospecificity of the type III group B streptococcal polysaccharide. Biochemistry 20, 4511–4518 [DOI] [PubMed] [Google Scholar]
- 16. Di Fabio J. L., Michon F., Brisson J.-R., Jennings H. J., Wessels M. R., Benedì V.-J., Kasper D. L. (1989) Structure of the capsular polysaccharide antigen of type IV group B Streptococcus. Can. J. Chem. 67, 877–882 [Google Scholar]
- 17. Wessels M. R., DiFabio J. L., Benedì V.-J., Kasper D. L., Michon F., Brisson J.-R., Jelínková J., Jennings H. J. (1991) Structural determination and immunochemical characterization of the type V group B Streptococcus agalactiae capsular polysaccharide. J. Biol. Chem. 266, 6714–6719 [PubMed] [Google Scholar]
- 18. von Hunolstein C., D'Ascenzi S., Wagner B., Jelínková J., Alfarone G., Recchia S., Wagner M., Orefici G. (1993) Immunochemistry of capsular type polysaccharide and virulence properties of type VI Streptococcus agalactiae (group B Streptococcus). Infect. Immun. 61, 1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kogan G., Brisson J.-R., Kasper D. L., von Hunolstein C., Orefici G., Jennings H. J. (1995) Structural elucidation of the novel type VII group B Streptococcus capsular polysaccharide by high resolution NMR spectroscopy. Carbohydr. Res. 277, 1–9 [DOI] [PubMed] [Google Scholar]
- 20. Kogan G., Uhrín D., Brisson J.-R., Paoletti L. C., Blodgett A. E., Kasper D. L., Jennings H. J. (1996) Structural and immunochemical characterization of the type VIII group B Streptococcus capsular polysaccharide. J. Biol. Chem. 271, 8786–8790 [DOI] [PubMed] [Google Scholar]
- 21. Pinto V., Berti F. (2014) Exploring the Group B Streptococcus capsular polysaccharides: the structural diversity provides the basis for development of NMR-based identity assays. J. Pharm. Biomed. Anal. 98C, 9–15 [DOI] [PubMed] [Google Scholar]
- 22. Cieslewicz M. J., Chaffin D., Glusman G., Kasper D., Madan A., Rodrigues S., Fahey J., Wessels M. R., Rubens C. E. (2005) Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73, 3096–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao K., Poulsen K., Maione D., Rinaudo C. D., Baldassarri L., Telford J. L., Sørensen U. B., Members of the DEVANI Study Group, and Kilian M. (2013) Capsular gene typing of Streptococcus agalactiae compared to serotyping by latex agglutination. J. Clin. Microbiol. 51, 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wessels M. R., Paoletti L. C., Kasper D. L., DiFabio J. L., Michon F., Holme K., Jennings H. J. (1990) Immunogenicity in animals of a polysaccharide-protein conjugate vaccine against type III group B Streptococcus. J. Clin. Investig. 86, 1428–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciucanu I., Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 [Google Scholar]
- 26. Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E. T., Rosini R., D'Agostino N., Miorin L., Buccato S., Mariani M., Galli G., Nogarotto R., Nardi-Dei V., Vegni F., Fraser C., Mancuso G., Teti G., Madoff L. C., Paoletti L. C., Rappuoli R., Kasper D. L., Telford J. L., Grandi G. (2005) Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309, 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buccato S., Maione D., Rinaudo C. D., Volpini G., Taddei A. R., Rosini R., Telford J. L., Grandi G., Margarit I. (2006) Use of Lactococcus lactis expressing pili from group B Streptococcus as a broad-coverage vaccine against streptococcal disease. J. Infect. Dis. 194, 331–340 [DOI] [PubMed] [Google Scholar]
- 28. Brisson J. R., Uhrinova S., Woods R. J., van der Zwan M., Jarrell H. C., Paoletti L. C., Kasper D. L., Jennings H. J. (1997) NMR and molecular dynamics studies of the conformational epitope of the type III group B Streptococcus capsular polysaccharide and derivatives. Biochemistry 36, 3278–3292 [DOI] [PubMed] [Google Scholar]
- 29. González-Outeiriño J., Kadirvelraj R., Woods R. J. (2005) Structural elucidation of type III group B Streptococcus capsular polysaccharide using molecular dynamics simulations: the role of sialic acid. Carbohydr. Res. 340, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 30. Jennings H. J., Katzenellenbogen E., Lugowski C., Michon F., Roy R., Kasper D. L. (1984) Structure, conformation, and immunology of sialic acid-containing polysaccharides of human pathogenic bacteria. Pure Appl. Chem. 56, 893–905 [Google Scholar]
- 31. Zou W., Mackenzie R., Thérien L., Hirama T., Yang Q., Gidney M. A., Jennings H. J. (1999) Conformational epitope of the type III group B Streptococcus capsular polysaccharide. J. Immunol. 163, 820–825 [PubMed] [Google Scholar]
- 32. Zou W., Jennings H. J. (2001) The conformational epitope of type III group B Streptococcus capsular polysaccharide. Adv. Exp. Med. Biol. 491, 473–484 [DOI] [PubMed] [Google Scholar]
- 33. Safari D., Dekker H. A., Rijkers G. T., van der Ende A., Kamerling J. P., Snippe H. (2011) The immune response to group B streptococcus type III capsular polysaccharide is directed to the -Glc-GlcNAc-Gal- backbone epitope. Glycoconj. J. 28, 557–562 [DOI] [PubMed] [Google Scholar]
- 34. Guttormsen H. K., Paoletti L. C., Mansfield K. G., Jachymek W., Jennings H. J., Kasper D. L. (2008) Rational chemical design of the carbohydrate in a glycoconjugate vaccine enhances IgM-to-IgG switching. Proc. Natl. Acad. Sci. U.S.A. 105, 5903–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Honsa E., Fricke T., Stephens A. J., Ko D., Kong F., Gilbert G. L., Huygens F., Giffard P. M. (2008) Assignment of Streptococcus agalactiae isolates to clonal complexes using a small set of single nucleotide polymorphisms. BMC Microbiol. 8, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaffin D. O., Beres S. B., Yim H. H., Rubens C. E. (2000) The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182, 4466–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]