Background: (+)-3-Carene/(−)-sabinene synthases are associated with insect resistance in Sitka spruce.
Results: Amino acid position 596 is critical for the monoterpene synthase product profile.
Conclusion: This work revealed mechanistic underpinnings for patterns of functional evolution of this monoterpene synthase family.
Significance: Catalytic plasticity of conifer monoterpene synthases is a major factor in the diversification of the TPS-d family and evolution of insect resistance.
Keywords: Enzyme Mechanism, Enzyme Mutation, Plant, Plant Biochemistry, Plant Defense, Terpenoid, Terpenoid Synthase
Abstract
The monoterpene (+)-3-carene is associated with resistance of Sitka spruce against white pine weevil, a major North American forest insect pest of pine and spruce. High and low levels of (+)-3-carene in, respectively, resistant and susceptible Sitka spruce genotypes are due to variation of (+)-3-carene synthase gene copy number, transcript and protein expression levels, enzyme product profiles, and enzyme catalytic efficiency. A family of multiproduct (+)-3-carene synthase-like genes of Sitka spruce include the three (+)-3-carene synthases, PsTPS-3car1, PsTPS-3car2, PsTPS-3car3, and the (−)-sabinene synthase PsTPS-sab. Of these, PsTPS-3car2 is responsible for the relatively higher levels of (+)-3-carene in weevil-resistant trees. Here, we identified features of the PsTPS-3car1, PsTPS-3car2, PsTPS-3car3, and PsTPS-sab proteins that determine different product profiles. A series of domain swap and site-directed mutations, supported by structural comparisons, identified the amino acid in position 596 as critical for product profiles dominated by (+)-3-carene in PsTPS-3car1, PsTPS-3car2, and PsTPS-3car3, or (−)-sabinene in PsTPS-sab. A leucine in this position promotes formation of (+)-3-carene, whereas phenylalanine promotes (−)-sabinene. Homology modeling predicts that position 596 directs product profiles through differential stabilization of the reaction intermediate. Kinetic analysis revealed position 596 also plays a role in catalytic efficiency. Mutations of position 596 with different side chain properties resulted in a series of enzymes with different product profiles, further highlighting the inherent plasticity and potential for evolution of alternative product profiles of these monoterpene synthases of conifer defense against insects.
Introduction
White pine weevil (Pissodes strobi) is one of the most devastating insect pests of spruce (Picea spp.) and pine (Pinus spp.). Sitka spruce (Picea sitchensis), a conifer species in which most genotypes are highly susceptible to weevils (1), is native to the temperate rainforest ecosystem of the North American Pacific coast, and is also an economically valuable forest tree in Europe. Susceptibility to weevils caused the nearly complete halt of commercial Sitka spruce reforestation in the Pacific Northwest. However, successful field trials identified a few highly resistant Sitka spruce genotypes; most notably genotype H898, which has become a focus for research and breeding of conifer resistance to stem boring insects (1).
One of the major defenses of conifers against insects is the chemically complex oleoresin, which includes dozens of different monoterpenes and diterpene resin acids (2–5). Previous work (6) explored the monoterpene and diterpene resin acid profiles of Sitka spruce from different geographic regions of the natural distribution where trees displayed strong, intermediate, or weak resistance. Resistance was positively associated with higher levels of the bicyclic monoterpene (+)-3-carene (6). Subsequently, Hall et al. (7) used a combination of genomic, target specific proteomic, and biochemical approaches to study the basis of variation of (+)-3-carene levels in two contrasting genotypes of Sitka spruce, resistant genotype H898 trees with relatively high levels of (+)-3-carene and susceptible genotype Q903 trees with trace levels of (+)-3-carene. This work identified a small family of (+)-3-carene synthase-like genes in Sitka spruce that contains the three (+)-3-carene synthases PsTPS-3car1,3 PsTPS-3car2, PsTPS-3car3, and the (−)-sabinene synthase PsTPS-sab. Genotype-specific variations of gene copy number, transcript and protein expression, and catalytic efficiencies of members of this family are responsible for the difference in (+)-3-carene levels (7). Specifically, the genomic presence, transcript and protein expression, and enzyme activity of PsTPS-3car2 accounted for much of the high levels of (+)-3-carene in the resistant genotype.
Members of the Sitka spruce (+)-3-carene synthase-like family showed between 82.5 and 95.7% pairwise amino acid sequence identity (Fig. 1). These four enzymes are multiproduct enzymes with the same overall product profile of monoterpenes, however, with different relative amounts of individual compounds (7). Most notably, PsTPS-3car1, PsTPS-3car2, and PsTPS-3car3 have (+)-3-carene as the predominant product, whereas PsTPS-sab forms (−)-sabinene as the predominant product. All four enzymes produce α-terpinolene as the second most abundant product plus a set of additional minor products (7) (Table 1, supplemental Table S1). These similar traits and the particular presence of a (−)-sabinene synthase as a closely related enzyme with a group of (+)-3-carene synthases suggested a pattern of divergent evolution in which PsTPS-sab arose from a PsTPS-3car ancestor through gene duplication and shift of function (7).
FIGURE 1.
Amino acid sequence alignment of the C-terminal α-domain of spruce TPS-3car and TPS-sab enzymes of a family of (+)-3-carene synthase-like monoterpene synthases. The alignment includes protein sequences of (+)-3-carene synthases and (−)-sabinene synthase from Sitka spruce (P. sitchensis; PsTPS-3car1, PsTPS-3car2, PsTPS-3car3, and PsTPS-sab (7)); as well as (+)-3-carene synthases from Norway spruce (P. abies; PaTPS-3car (29) and white spruce (P. glauca; PgTPS-3car (30)). Amino acids with highlighted blue background color are those different from the consensus. A diagrammatic representation of the secondary structures of C-terminal domain of the (+)-3-carene synthase-like enzymes is shown with cylinders representing α-helices and ribbons represent loops. The conserved DDXXD motif is identified by the red line. Positions 595, 596, and 599 in helix J are marked with asterisks.
TABLE 1.
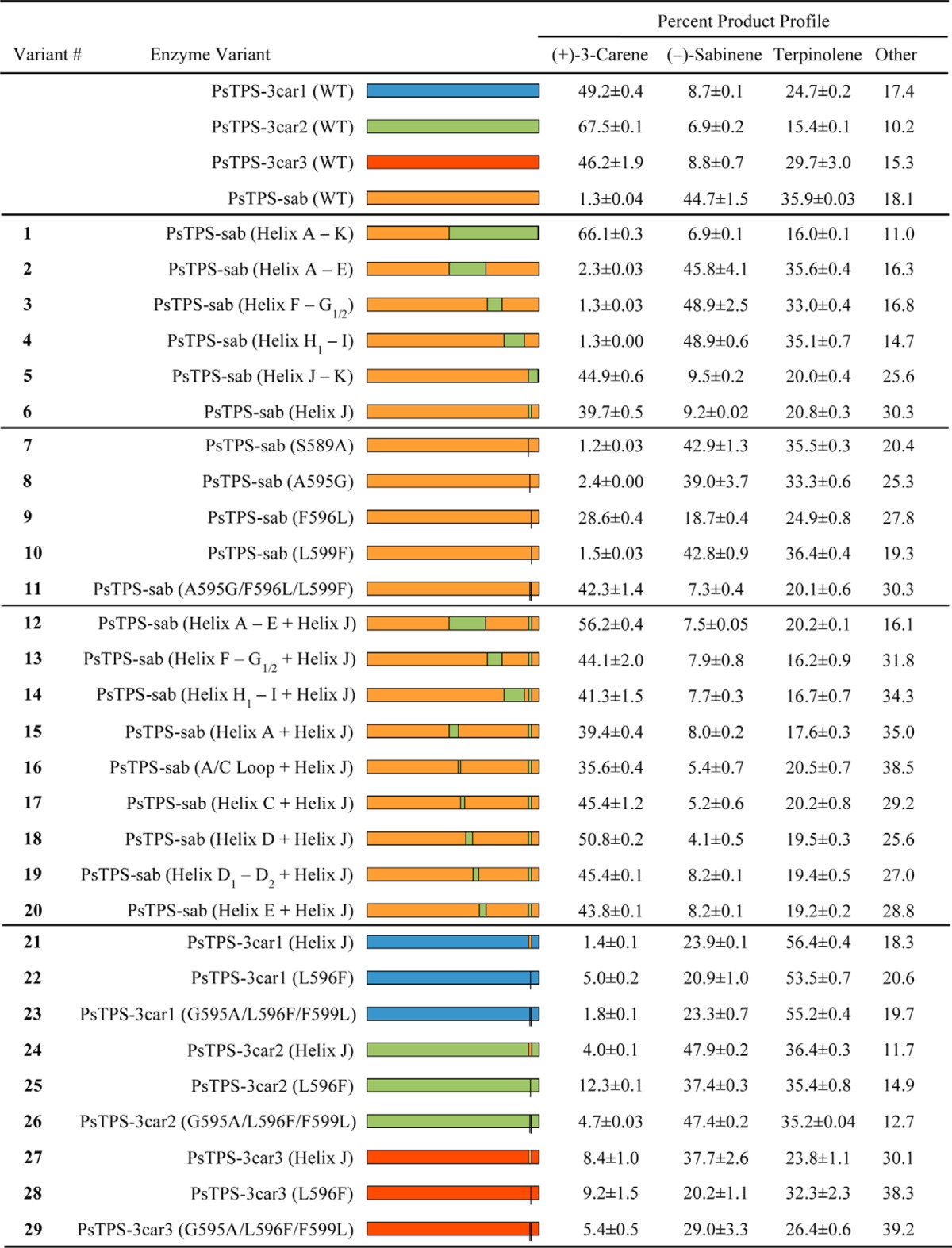
Product profiles of PsTPS-3car1, PsTPS-3car2, PsTPS-3car3, PsTPS-sab, and their variants
Based on general knowledge of monoterpene synthases (8), PsTPS-3car and PsTPS-sab enzymes are thought to employ divalent metal ion-dependent ionization/isomerization/cyclization reaction mechanisms (Fig. 2). Initial ionization of the substrate geranyl diphosphate allows the formation of linalyl diphosphate. Attack from the allylic double bond upon reionization of linalyl diphosphate results in the formation of the α-terpinyl cation, an important proposed carbocation intermediate for the formation of various cyclic monoterpenes found in the product profiles of PsTPS-3car and PsTPS-sab enzymes. This intermediate can undergo a series of hydride shifts and/or additional cyclizations until reactions are terminated by deprotonation or addition of a nucleophile. Previous work on angiosperm monoterpene synthases has shown how individual amino acids may interact with reaction intermediates and determine product profile. For example, substituting an asparagine to an isoleucine removed the ability of a Salvia fruticosa 1,8-cineole synthase to deprotonate a water molecule in the active site, preventing water capture by the reaction intermediate and altering the reaction pathway to produce sabinene instead of 1,8-cineole as the predominant product (9). Similarly, stereo-specificity of two Thymus vulgaris sabinene hydrate synthases was inter-converted by reciprocal substitution between a pair isoleucine and asparagine residues (10).
FIGURE 2.
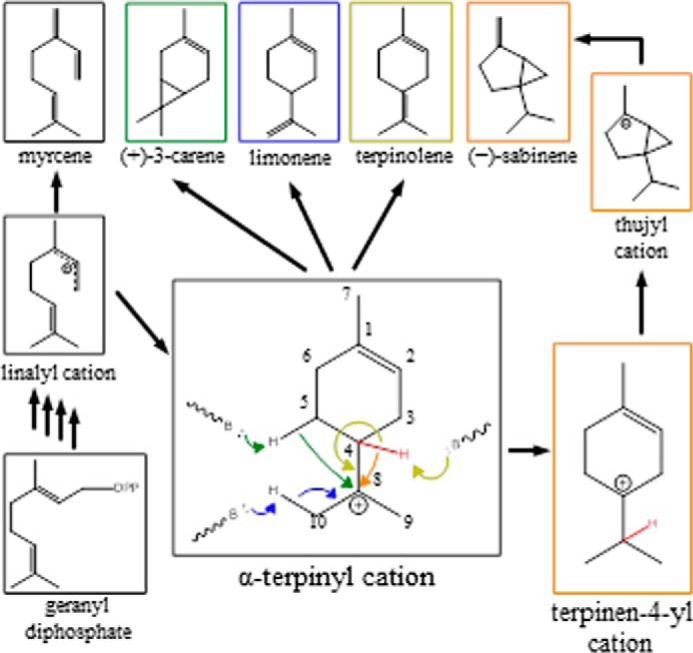
Proposed reaction mechanisms explaining monoterpenes of the product profiles of PsTPS-3car and PsTPS-sab enzymes and their variants. Cyclic monoterpene products, including the major products (+)-3-carene, (−)-sabinene, and α-terpinolene, are proposed to be derived from an α-terpinyl cation intermediate. Formation of (−)-sabinene is proposed to involve a terpinen-4-yl cation intermediate. Proposed hydride shifts, cyclizations, and termination reactions by proton loss are indicated with arrows color coded with the corresponding products.
The high sequence similarity yet different product profiles of the Sitka spruce PsTPS-3car and PsTPS-sab enzymes, and their different roles in contributing to insect resistance, has made them attractive targets for investigating which particular structural features of these enzymes affect their functions. Here, we used domain-swapping and site-directed mutagenesis, guided by sequence comparisons and supported by structural homology modeling, to test which specific domains and amino acids direct PsTPS-3car versus PsTPS-sab product profile and how these domains and amino acids might interact with the reaction intermediates. Our results indicate changes of sequence and functions that may have occurred in the natural evolution of the (+)-3-carene synthase-like family of spruce defense.
EXPERIMENTAL PROCEDURES
Mutagenesis
Mutagenesis of the cDNA clones PsTPS-3car1, PsTPS-3car2, PsTPS-3car3, and PsTPS-sab (7) was performed using Phusion Hot Start II DNA Polymerase (Thermo Scientific) following the manufacturer's instructions with 25 ng of template DNA per reaction. Primers are listed in supplemental Table S2. All mutations were verified by Sanger sequencing prior to expression.
Protein Expression and Purification
Recombinant plasmids were transformed into E. coli C41 containing the pRARE 2 plasmid isolated from Rosetta 2 cells (Novagen) to negate codon bias. Individual colonies were inoculated into 50 ml of Terrific Broth containing kanamycin (50 mg/liter) and chloramphenicol (50 mg/liter) and cultured at 37 °C and 180 rpm until A600 = 1.0. Cultures were then cooled to 16 °C, induced by addition of isopropyl β-d-1-thiogalactopyranoside (final concentration 0.1 mm), and grown for 16 h at 180 rpm before harvesting. Recombinant protein was extracted and nickel affinity purified as previously described (7, 11). Protein was quantified by BCA assay (Thermo Scientific) and SDS-PAGE with measurement of protein band intensity performed with the program ImageJ.
Enzyme Assays
Monoterpene synthase activities were assayed in triplicate as previously described with minor modifications (7, 11, 12). 500-μl reactions containing 25 mm HEPES, 100 mm KCl, 10 mm MgCl2, 5 mm dithiothreitol, 10% glycerol, 61.6 μm geranyl diphosphate (Echelon Biosciences Inc.), and affinity-purified protein extract were overlaid with 500 μl of pentane containing 2.5 μm isobutylbenzene as an internal standard and incubated at 30 °C for either 1 (all enzymes derived from PsTPS-3car1, PsTPS-3car2, and PsTPS-sab) or 4 h (all enzymes derived from PsTPS-3car3). Reaction products were extracted into pentane by vortexing for 30 s followed by phase separation through centrifugation at 1000 × g for 30 min at 4 °C.
To determine enzyme kinetic parameters, assays were performed with nine different concentrations of geranyl diphosphate ranging from 1 to 60 μm. PsTPS-3car2 (WT) was assayed for 20 min at 30 °C; all other enzymes were assayed for 10 min at 30 °C. Enzyme concentrations in each assay were 19.9–26.9 pm for PsTPS-3car2 (WT), 12.9–19.4 pm for variant 24, 4.6–15.9 pm for variant 25, 10.0–11.7 pm for variant 26, 22.3–24.3 pm for PsTPS-sab (WT), 60.1–62.7 pm for variant 6, 23.9–38.0 pm for variant 9, and 60.3–60.7 pm for variant 11. Kinetic analysis was performed by non-linear regression using the EXCEL template ANEMONA.
Gas Chromatography/Mass Spectrometry (GC/MS) Analysis
Assay products were identified by GC (Agilent 6890A series)/MSD (5973N mass selective detector, quadropole analyzer, electron ionization, 70 eV) by comparison of retention times and mass spectra with authentic standards and by comparison with mass spectral libraries (Wiley7Nist05). Monoterpene synthase assay products were analyzed on a DB-WAX capillary column (J&W 122–7032; 250-μm internal diameter, 30 m length, 0.25-μm film thickness) with an initial temperature of 40 °C (4 min), increasing by 3 °C min−1 to 85 °C, then by 30 °C min−1 to 250 °C (held for 2.5 min), injector temperature was 250 °C, flow rate was 1.4 ml of He min−1, and run time was 27 min. Compounds were quantified using response factors calculated by comparison to a known concentration of isobutylbenzene.
Homology Modeling and Ligand Docking
Homology models for the (+)-3-carene synthase-like enzymes and their variants were produced using the SWISS-MODEL server (13, 14) and underwent energy minimization using the YASARA force field (15). Models were based on the structure of Salvia officinalis (+)-bornyl diphosphate synthase (PDB code 1N22) containing the substrate analog (4R)-7-aza-7,8-dihydrolimonene (16). Ramachandran plots of all models verified high stereochemical quality having greater than 90% of residues in most favored regions. Energy-minimized ligands for docking were produced using the PRODRG server (17). Docking studies with the α-terpinyl cation and the protein models were performed using Molegro Virtual Docking. The substrate analog (4R)-7-aza-7,8-dihydrolimonene was used as a positional template for docking. Because this analog is inverted in the active site of the S. officinalis (+)-bornyl diphosphate synthase crystal structure, similarity measurements used in the template docking parameters were relaxed to allow for increased flexibility in the positioning of the α-terpinyl cation. This resulted in two to three of the top five most energetically favorable positions of the α-terpinyl cation oriented in the appropriate direction. Of these, the most energetically favorable position was used. The results were visualized in PyMOL.
RESULTS
Exchange of the Helix J Region Shifts the (−)-Sabinene Synthase Product Profile of PsTPS-sab to a (+)-3-Carene Synthase Profile Resembling PsTPS-3car
We performed a series of domain swaps and site-directed substitutions between PsTPS-sab and PsTPS-3car to explore which regions and specific amino acids of these enzymes affect the differences of their product profiles. Conifer monoterpene synthases of the TPS-d1 group possess a βα-domain structure (9, 16, 18). The α-domain of these enzymes adopts an α-α barrel structure comprised of 14 helixes and two loops, which harbors the class I active site (Fig. 1). To test if product profiles could be altered through mutation of the α-domain as seen in other TPS-d1 monoterpene synthases (19–21), an initial domain swap was performed on PsTPS-sab wild type (WT) enzyme so its helix A-helix K region would be identical to that of PsTPS-3car2 (WT) (Table 1). The resulting enzyme (variant 1) showed a product profile nearly identical to that of PsTPS-3car2 (WT), producing 66.1% (+)-3-carene and 6.9% (−)-sabinene plus other additional monoterpenes (Table 1, supplemental Table S1), indicating successful conversion into a PsTPS-3car type (+)-3-carene synthase. To identify the specific regions that caused this change in product profile, four additional domain swaps were performed on PsTPS-sab (WT): helix A-helix E (variant 2), helix F-helix G1/2 (variant 3), helix H1-helix I (variant 4), and helix J-helix K (variant 5). Of these, variants 2, 3, and 4 showed no substantial change in product profile compared with PsTPS-sab (WT); however, variant 5 displayed a product profile containing 44.9% (+)-3-carene and 9.5% (−)-sabinene (Table 1). To narrow down which parts of the helix J-helix K region caused this change, we performed separate substitutions of helix J (variant 6), J/K loop, and helix K on PsTPS-sab (WT). Changes of the J/K loop and helix K regions had no effect on product profile compared with PsTPS-sab (WT). In contrast, variant 6 produced a profile similar to that of PsTPS-3car of 39.7% (+)-3-carene and 9.2% (−)-sabinene (Table 1). In summary, these results indicated that sequence variation in the 11-amino acid long helix J region were responsible for much of the difference of PsTPS-sab and PsTPS-3car product profiles.
Mutation of Three Amino Acids of the Helix J Region Had Major Effects on Shifting the Product Profile of PsTPS-sab to a Profile Resembling PsTPS-3car
Of the four amino acids in the helix J region that differ between PsTPS-sab and PsTPS-3car2 (Fig. 1), individual site-directed substitution in positions 589 (variant 7), 595 (variant 8), and 599 (variant 10) produced no change in product profile relative to PsTPS-sab (WT). However, substitution of the Leu in position 596 (variant 9), an amino acid conserved across all known conifer TPS-3car enzymes, to Phe produced an enzyme with a product profile of 28.6% (+)-3-carene and 18.7% (−)-sabinene (Table 1, Fig. 3d).
FIGURE 3.
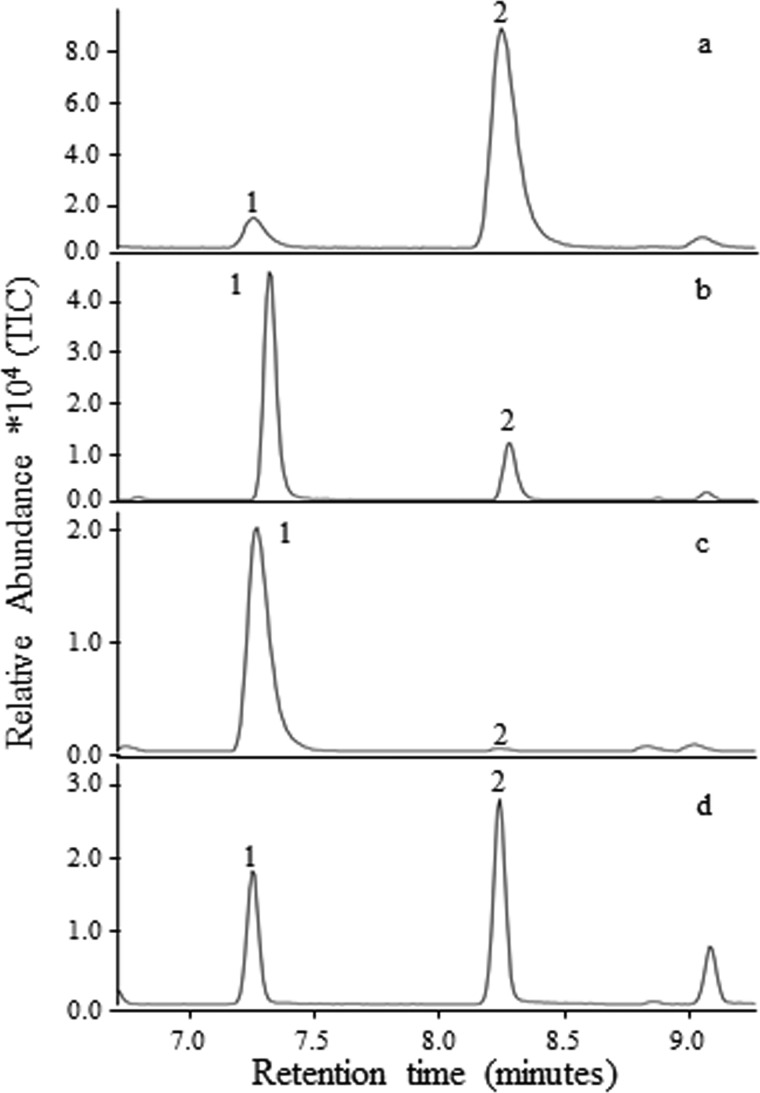
Select regions of total ion GC/MS traces of products formed by PsTPS-3car and PsTPS-sab and their variants in position 596. Traces a and b show shifts in the abundance of (−)-sabinene (1) and (+)-3-carene (2) in the product profiles of PsTPS-3car2 (WT) and PsTPS-3car2 (L596F) variant 25, respectively. Traces c and d show shifts in the abundance of (−)-sabinene (1) and (+)-3-carene (2) in the product profiles of PsTPS-sab (WT) and PsTPS-sab (F596L) variant 9, respectively. Products were confirmed by comparison of mass spectra retention times with those of authentic standards.
The proportion of (+)-3-carene in the product profile of PsTPS-sab variant 9 was less compared with the product profiles of variants 5 and 6, suggesting that at least one of the conserved Ala589, Gly595, and Phe599 of the PsTPS-3car enzymes has a synergistic effect with Leu596 on (+)-3-carene formation. To test this hypothesis, we assessed the product profiles of all six possible PsTPS-sab variants that were produced from combinations of variant 9 with additional substitutions in positions 589, 595, and/or 599. Of these, variant 11 produced the highest levels of (+)-3-carene and a product profile closest to variants 5 and 6 with 42.3% (+)-3-carene and 7.3% (−)-sabinene (Table 1).
The observed effects these substitutions had on product profile identified Phe596 as critical in PsTPS-sab for determining (−)-sabinene as the major product, and Leu596 as critical for (+)-3-carene formation in the mutated PsTPS-sab enzyme with positions Gly595 and Phe599 providing synergistic effects. In turn, it can be proposed these three amino acids play an important role in (+)-3-carene formation in PsTPS-3car.
Mutations in the Helix A-E Region Synergistically Affect the Shift of (−)-Sabinene Synthase Product Profile to a (+)-3-Carene Synthase Product Profile
The three helix J amino acid substitutions Ala595-Gly, Phe596-Leu, and Leu599-Phe of PsTPS-sab variant 11 explained the product profile changes observed in variants 5 and 6 relative to PsTPS-sab (WT); however, the (+)-3-carene biosynthesis levels of variant 11 were only two-thirds of that observed in PsTPS-sab variant 1 and PsTPS-3car2 (WT). To elucidate which additional amino acids promote (+)-3-carene biosynthesis, we performed further domain swaps to variant 6 with the helix A-helix E, helix F-helix G1/2, and helix H-helix I regions of PsTPS-3car2 (variants 12, 13, and 14, respectively). Of the resulting enzymes, variants 13 and 14 showed no increase in (+)-3-carene formation, whereas variant 12 produced a product profile containing 56.2% (+)-3-carene and 7.5% (−)-sabinene. Next, we divided the helix A-helix E region of PsTPS-3car2 into six smaller regions based on individual helices and loops for the design of additional domain swaps in the background of PsTPS-sab variant 6. Of these, the additional swap of helix A (variant 15) and the A/C loop (variant 16) showed no increase in (+)-3-carene formation compared with variant 6. Additional swaps of helix C (variant 17), helix D (variant 18), helix D1-D2 (variant 19), and helix E (variant 20) all showed slight increases in (+)-3-carene resulting in product profiles that contained, respectively, 45.4, 50.8, 45.5, and 43.8% (+)-3-carene and 5.2, 4.1, 8.2, and 8.2% (−)-sabinene. These results suggest that some or all of the 13 amino acids that differ between PsTPS-3car2 (WT) and PsTPS-sab (WT) within the helix C-E region provide additional synergistic effects to (+)-3-carene formation.
Reciprocal Mutations in Positions 595, 596, and 599 Result in Conversion of PsTPS-3car (+)-3-Carene Synthases to (−)-Sabinene Synthases Resembling PsTPS-sab
To substantiate results obtained with substitutions in PsTPS-sab according to which positions 595, 596, and 599 are critical for determining the predominant (−)-sabinene or (+)-3-carene product profiles of PsTPS-sab and PsTPS-3car, respectively, we made the reciprocal substitutions corresponding to PsTPS-sab variants 6, 9, and 11 in each of the three different PsTPS-3car enzymes, PsTPS-3car1, PsTPS-3car2, and PsTPS-3car3 (Table 1).
These substitutions in the PsTPS-3car1 (WT) background resulted in variants 21, 22, and 23 and reduced the formation of (+)-3-carene to 1.4 (variant 21), 5.0 (variant 22), and 1.8% (variant 23) of the overall product profile compared with 49.2% in PsTPS-3car1 (WT) (Table 1). These variant PsTPS-3car1 enzymes produced (−)-sabinene at 23.0, 20.9, and 23.3% of total product profile, respectively; compared with 8.7% in PsTPS-3car1 (WT). Although α-terpinolene was the major product at 56.4, 53.5, and 55.2% of product profile, respectively, these substitutions all caused a large increase in (−)-sabinene product levels. The same three substitutions in PsTPS-3car2 (WT) resulted in variants 24, 25, and 26 with (−)-sabinene as the major product at 47.9, 37.4, and 47.4% of total product profile, respectively (Table 1, Fig. 3). In PsTPS-3car3 (WT), the same three substitutions resulted in variants 27, 28, and 29, again with (−)-sabinene as the major product in the helix J substitution (variant 27; 37.7%) and in the triple amino acid substitution variant 29 (29.0%). (−)-Sabinene was also the second most abundant product in the position 596 variant 28 with 20.2% of product profile, respectively. These results demonstrated that Gly595, Leu596, and Phe599 are critical in PsTPS-3car for determining (+)-3-carene as the major product, and that Phe596 is critical for (−)-sabinene biosynthesis with Ala595 and Leu599 providing synergistic effects.
Position 596 Is Important for Functional Plasticity
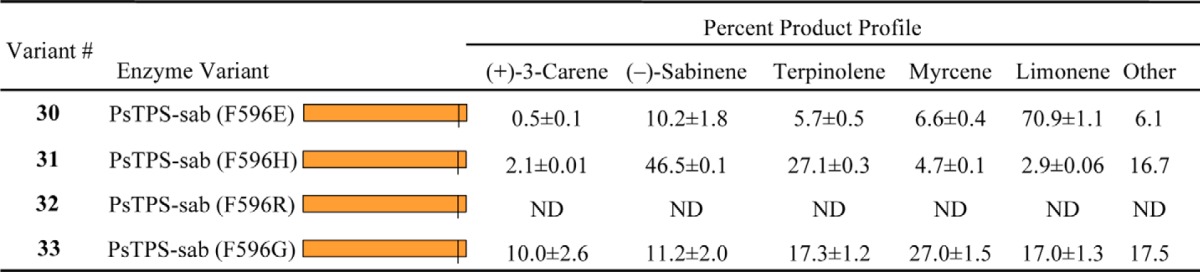
Reciprocal Phe-Leu substitutions between PsTPS-sab and PsTPS-3car1, PsTPS-3car2, and PsTPS-3car3 (variants 9, 22, 25, and 28) showed the importance of position 596 in specifying (−)-sabinene or (+)-3-carene as the major product. To further explore the effect of variations in position 596, we tested four additional substitutions in the PsTPS-sab background sequence that introduced side chains with different electrostatic and steric properties (Table 2). Substitution of Phe to Glu (variant 30) produced an enzyme with limonene as the major product at 70.9% of product profile. Substitution to His (variant 31) produced an enzyme with a product profile nearly identical to PsTPS-sab (WT) with (−)-sabinene and terpinolene being 46.5 and 27.1% of the product profile, respectively. Substitution to Arg (variant 32) produced an enzyme with no detectable activity. Finally, substitution to Gly (variant 33) produced an enzyme displaying a product profile with major products myrcene (27%), α-terpinolene (17.3%), limonene (17.0%), (−)-sabinene (11.2%), and (+)-3-carene (10.0%). The range of changes in product profile seen in these variants highlights the inherent potential for plasticity of this monoterpene synthase and suggests the 596 amino acid plays an important role in directing product outcome and evolution of enzyme function.
TABLE 2.
Product profiles of PsTPS-sab variants
Homology Models Place Position 596 in the Active Site of PsTPS-sab and PsTPS-3car
To assess if the amino acid in position 596 interacts with the α-terpinyl cation reaction intermediate or has a catalytic role in directing the predominantly (−)-sabinene or (+)-3-carene product profiles in the PsTPS-sab and PsTPS-3car enzymes, we performed substrate docking experiments with homology models of these proteins and the proposed α-terpinyl cation reaction intermediate. As tertiary structures of plant monoterpene synthases are well conserved, we produced homology models on the structure of (+)-bornyl synthase from S. officinalis (PDB code 1N22) (16) (Fig. 4). Due to the large active site cavity volume and consistent with the multiproduct nature of the PsTPS-sab and PsTPS-3car enzymes, several possible ligand positions were obtained in docking experiments with the α-terpinyl cation. To mitigate this problem, docking of the α-terpinyl cation was performed using the position of the substrate analog (4R)-7-aza-7,8-dihydrolimonene within the S. officinalis (+)-bornyl diphosphate synthase x-ray crystal structure (PDB code 1N22) as a positional template.
FIGURE 4.
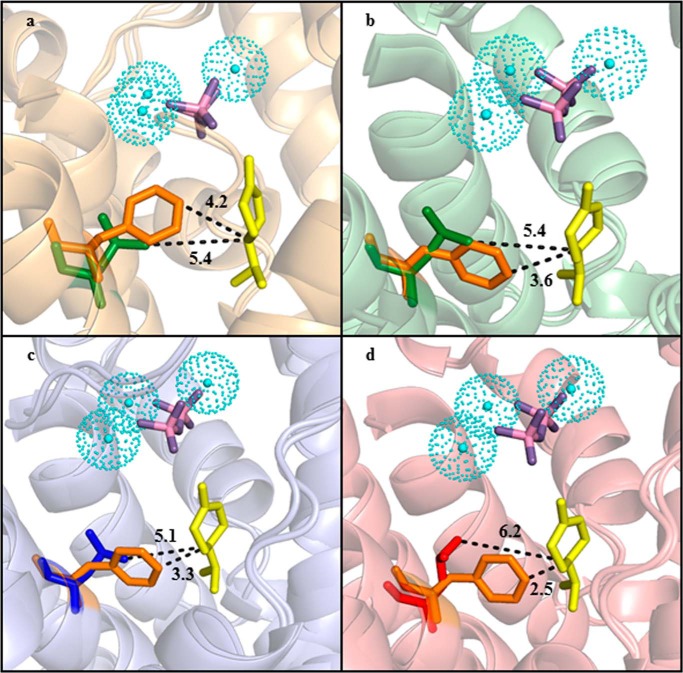
Homology models of the active sites of PsTPS-sab (WT), PsTPS-sab (F596L), PsTPS-3car2 (WT), PsTPS-3car2 (L596F), PsTPS-3car1 (WT), PsTPS-3car1 (L596F), PsTPS-3car3 (WT), and PsTPS-3car3 (L596F). Superimposition of the PsTPS-sab (WT) and PsTPS-sab (F596L) enzymes (a), superimposition of the PsTPS-3car2 (WT) and PsTPS-3car2 (L596F) enzymes (b), superimposition of the PsTPS-3car1 (WT) and PsTPS-3car1 (L596F) enzymes (c), and superimposition of the PsTPS-3car3 (WT) and PsTPS-3car3 (L596F) enzymes (d). Helices, loops, and individual amino acids shown in orange denote those found in PsTPS-sab (WT); green denotes those found in PsTPS-3car2 (WT); blue denotes those found in PsTPS-3car1 (WT); red denotes those found in PsTPS-3car3 (WT). The Phe or Leu side chains found in position 596 are shown. The trinuclear magnesium cluster is shown in cyan and the diphosphate ion is shown in pink and purple. Dotted lines mark the shortest distance between the amino acid side chain in position 596 and the C4 carbon (Fig. 2) of the α-terpinyl cation, which is shown in yellow.
Models of all four PsTPS-sab and PsTPS-3car enzymes show amino acid 596 positioned in the active site opposite the DDXXD motif and beside the J/K loop believed to partake in the conformational change that promotes closure of the active site during catalysis (16, 22). Of the 36 to 38 amino acids predicted to have side chains within 7 Å of the most energetically favorable conformation of the α-terpinyl cation reaction intermediate, only three amino acids were consistently distinct between PsTPS-3car and PsTPS-sab: amino acids in the 595, 596, and 599 positions. In this position of the reaction intermediate, Leu596 was consistently at a distance of more than 5 Å from the intermediate with no obvious impact on catalysis (Fig. 4). In contrast, all models with Phe596 showed this amino acid was consistently within 4 Å from the C4 carbon of the intermediate (Fig. 4). Taking the conformational freedom of the carbocation into account, the proximity of the Phe aromatic ring could facilitate steric or van der Waals interactions capable of stabilizing the intermediate in such a manner that promotes (−)-sabinene biosynthesis (Fig. 2).
Kinetic Properties of PsTPS-sab, PsTPS-3car2, and Selected Variants
To investigate if positions that affect contrasting (−)-sabinene or (+)-3-carene product profiles of PsTPS-sab and PsTPS-3car, respectively, also affect differences in other properties of enzyme activity, we determined kinetic parameters for PsTPS-sab (WT), PsTPS-3car2 (WT), and three variants of each of these enzymes with reciprocal substitutions of either helix J, the 596 amino acid, or the 595, 596, and 599 amino acids (Table 3). These enzymes that produced (+)-3-carene as the major product, namely PsTPS-3car2 (WT) and PsTPS-sab variants 6, 9, and 11, had kcat and apparent Km values in the same order of magnitude. The catalytic efficiencies (kcat/Km) of these enzymes were similar at 0.37, 0.04, 0.16, and 0.10 min−1 μm−1, respectively. Likewise, all enzymes that produced (−)-sabinene as the major product, PsTPS-sab (WT) and PsTPS-3car2 variants 24, 25, and 26, had apparent Km values in the same order of magnitude and similar catalytic efficiencies with kcat/Km values of 0.92, 1.09, 2.2, and 1.22 min−1 μm−1, respectively.
TABLE 3.
Kinetic properties of PsTPS-3car2, PsTPS-sab, and their variants
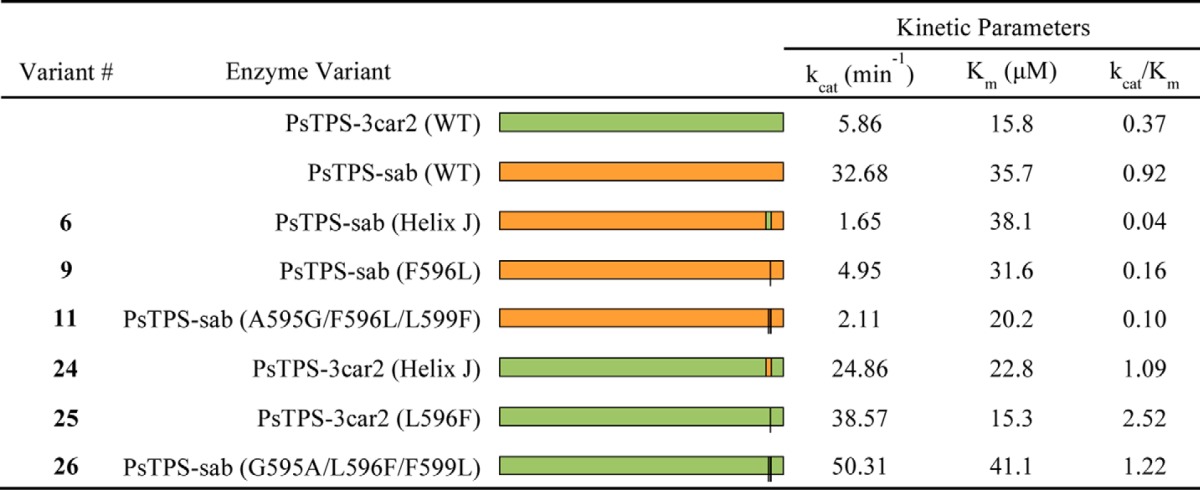
In conclusion, the results obtained with the reciprocal single residue substitutions in position 596 (variants 9 and 25) showed this position consistently affects both the contrasting product profiles and catalytic efficiencies of enzymes that produce alternatively (−)-sabinene or (+)-3-carene as one of their major products. The single amino acid Leu to Phe substitution in position 596 in the PsTPS-3car2 background (variant 25) increased kcat by more than 6-fold similar to PsTPS-sab (WT), whereas the opposite effect was observed with the Phe to Leu substitution of the 596 amino acid in the PsTPS-sab background (variant 9).
DISCUSSION
Using a mutational approach we investigated the effects naturally occurring amino acid variations have on functions of monoterpene synthases of the PsTPS-3car/PsTPS-sab family, a group of enzymes that contribute to phenotypic variation of weevil resistance in Sitka spruce (7). Through a progressive series of domain swaps and site-directed mutations, we identified residues at position 596 as critical for enzyme product profile and kinetic properties of the multiproduct PsTPS-3car and PsTPS-sab enzymes. This position appears to be a site of functional plasticity as different substitutions in this position gave rise to enzyme variants with a range of different product profiles. This position had the largest single effect on controlling alternatively (−)-sabinene or (+)-3-carene as major products.
Analysis of the PsTPS-3car (WT) and PsTPS-sab (WT) homology models revealed predicted active sites of similar size and contour. As product selectivity in reactions with multiple outcomes is predominantly determined by energies of the different transition-state intermediate structures leading to each product (23), we suggest the different product profiles of PsTPS-3car and PsTPS-sab are a result of unique active site residues that stabilize a common α-terpinyl cation reaction intermediate in different ways. The changes in (−)-sabinene product levels seen in amino acid substitutions in position 596 of all enzymes (variants 9, 22, 25, and 28) supported this amino acid having a critical role in directing product outcome (Table 1, Fig. 4). Molecular docking in the class I active site of PsTPS-sab (WT) and PsTPS-3car (L596F) predicted the side chain of Phe596 to be close enough to the α-terpinyl cation for steric blocking or van der Waals forces to mold the energy landscape of the reaction and promote (−)-sabinene formation (Fig. 4). Similar to what has been described for other terpene synthases, these effects could help stabilize a carbocation on the C4 carbon, promoting formation of the terpinen-4-yl cation, through a cation-π interaction (22, 24, 25) or hyperconjugation promoted through a C-H/π interaction (26, 27). Phe596 could also affect the conformation of the cyclohexene ring of the α-terpinyl cation by promoting an equatorial position for the C8 carbon, preventing the 5,8-cyclication needed to form (+)-3-carene (23).
Additional substitutions at positions 595 and 599 in the background of the PsTPS-sab (F596L), variant 9, and in the backgrounds of the three PsTPS-3car (L596F), variants 22, 25, and 28, resulted in an increased formation of the predominant monoterpene. Proximity of these two synergistic positions to the 596 position suggest they either contribute to optimal side chain orientation of the residue in the 596 position or alter the active site contour toward stabilizing the conformation of the α-terpinyl cation reaction intermediate within the active site as opposed to directly interacting with the carbocation intermediate. The additional 13-amino acid substitutions within helix C, D, D1-D2, and E (variants 17, 18, 19, and 20, respectively) cause a subtle increase in (+)-3-carene biosynthesis levels. As none of these amino acids were found to be positioned in the active site cavity, these substitutions likely influence product specificity by contributing to the overall barrel structure and/or contour of the class I active site as has been demonstrated in other terpene synthases (11). Quantum chemical calculations have shown that changes of the carbocation structure in response to the distribution of energy in its vibrational nodes could play a substantial role in terpene synthase reaction selectivity (28), suggesting that Phe596 or these synergistic amino acids might play a role in modulating the reaction energy landscape of the intermediate to promote dynamic tendencies that lead to the formation of (−)-sabinene as opposed to directly stabilizing the intermediate.
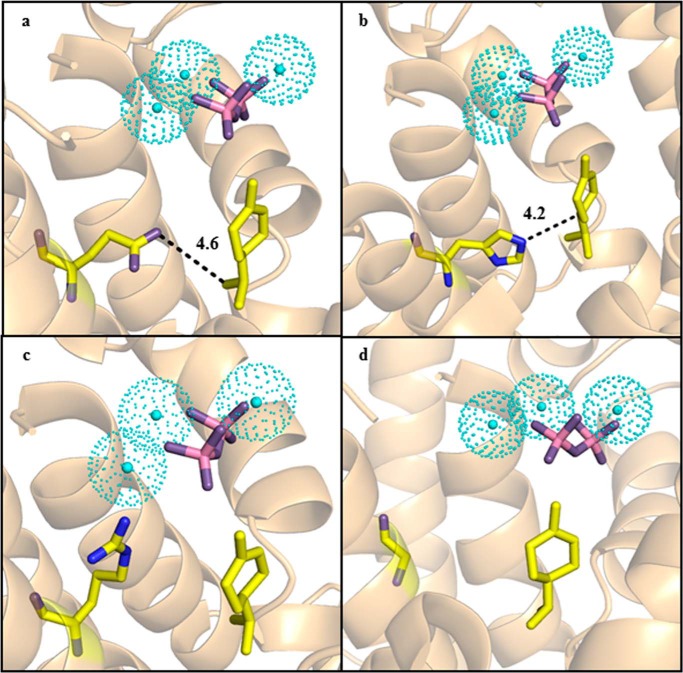
The distinct product profiles of PsTPS-sab (F596E) (variant 30), PsTPS-sab (F596H) (variant 31), PsTPS-sab (F596R) (variant 32), and PsTPS-sab (F596G) (variant 33) also support the conclusion of the amino acid in position 596 playing a role in modulating carbocation stabilization. Substitution to a glutamic acid (variant 30) introduces a negative charge poised to interact with the reaction intermediate. The observed high levels of limonene formation (Table 2) and homology modeling suggest this residue plays a role in promoting the deprotonation of the C9 or C10 carbons of the α-terpinyl cation to terminate the reaction (Fig. 2; Fig. 5a). Substitution to a histidine (variant 31) resulted in a product profile nearly identical to PsTPS-sab (WT). Protein modeling positions the histidine residue similarly to the phenylalanine in PsTPS-sab (WT) (Figs. 4a and 5b), suggesting the specific steric and aromatic effects histidine has on the reaction intermediate are similar to those of phenylalanine and promote (−)-sabinene biosynthesis. Substitution to an arginine (variant 32) introduces a positive charge and increases the steric constraints of the intermediate (Fig. 5c). It is not clear how exactly this affects enzyme functioning, but the absence of enzyme activity suggests it disrupted either the trinuclear magnesium ion configuration or reaction intermediate formation. Perhaps most interesting, substitution to a glycine (variant 33) shows that the absence of a side chain in the 596 amino acid position results in a very broad monoterpene profile with no major predominant product. In addition to removing a side chain that could direct carbocation stabilization, the glycine residue likely allows the reaction intermediate a larger degree of freedom by expanding the active site pocket (Fig. 5d). The result is less specificity in carbocation stabilization and a more general product profile. These results further highlighted the importance of the amino acid in the 596 position promoting specific stabilization of the reaction intermediate to produce a unique product profile.
FIGURE 5.
Homology models of the active sites of PsTPS-sab (F596E), PsTPS-sab (F596H), PsTPS-sab (F596R), and PsTPS-sab (F596G) active sites. Helices and loops shown in orange are those of the PsTPS-sab (WT) background structure. The modified 596 amino acid in each enzyme is shown: Glu in PsTPS-sab (F596E) (a); His in PsTPS-sab (F596H) (b); Arg in PsTPS-sab (F596R) (c); and Gly in PsTPS-sab (F596G) (d). The trinuclear magnesium cluster is shown in cyan and the diphosphate ion is shown in pink and purple. Where applicable, dotted lines mark the shortest distance between the amino acid side chain in position 596 and the C4 carbon (Fig. 2) of the α-terpinyl cation, which is shown in yellow.
As this study centered around those residues of PsTPS-car and PsTPS-sab that appear to influence the fate of product profile once the α-terpinyl cation is formed, it is likely that differences in the kinetic properties are due to final reaction steps of product formation and subsequent product release from the active site rather than geranyl diphosphate ionization and the formation of the α-terpinyl cation. Although the formation of (−)-sabinene requires intramolecular proton transfer and cyclization, followed by deprotonation, those enzymes with a phenylalanine in the 596 position had significantly higher catalytic rates and catalytic efficiencies than the enzymes that produced higher levels of (+)-3-carene synthase, which only requires a 5,8-ring closure to form (+)-3-carene and terminate the reaction. Although it is possible that the amino acids in the active site are poised to act on the intermediate leading to (−)-sabinene, our current structural/functional understanding of these enzymes is insufficient to explain the difference in kinetic ability.
Previous work proposed that gene duplications and divergent functional evolution lead to predominance of either (+)-3-carene or (−)-sabinene as alternative primary products of closely related members of the Sitka spruce (+)-3-carene synthase-like family, the four members of which generally share the same products in varying quantities (7). The mutational work presented here supports this hypothesis and provides mechanistic insights into how functional variations in this family may have evolved. The higher sequence identity of PsTPS-3car2 and PsTPS-3car3 to each other (97.2% nucleotide identity) and their higher sequence relatedness to PsTPS-sab (91.8 and 92.6% nucleotide identity, respectively), compared with the sequence relatedness between PsTPS-3car1 and PsTPS-sab (89.3% nucleotide identity) support a phylogeny of this gene family according to which PsTPS-3car1 has diverged the most from a common ancestor of these four genes (7). The present work supports this pattern, given the relative ease with which PsTPS-3car2 and PsTPS-3car3 could be converted to (−)-sabinene synthases compared with PsTPS-3car1. Mutagenesis studies here show that as little as one base pair substitution could convert a more ancestral (+)-3-carene synthase to a (−)-sabinene synthase highlighting how PsTPS-sab gene function could have evolved. Together, these results provide evidence of active site plasticity and underscore that subtle alterations of the active site contour, such as shifting a backbone atom by a fraction of an Angstrom or the addition or removal of a methyl group can have a large effect on enzyme activity. In the context of the evolution of a family of defense and resistance related conifer TPS-d genes, we highlight how gene duplications and conspicuously minor sequence variation may lead to diversification of terpenoid profiles as is seen with the complex mixtures of oleoresin monoterpenes in general, and with the intra-specific variation of (+)-3-carene profiles in Sitka spruce in particular.
In conclusion, we provide a mechanistic underpinning for apparent patterns of the functional evolution of the Sitka spruce (+)-3-carene synthase/(−)-sabinene synthase gene family associated with white pine weevil resistance in Sitka spruce. With as few as one amino acid substitution, we observed large changes in both product profile and kinetic capabilities of the enzymes. These results underscore the large catalytic plasticity of conifer monoterpene synthases as a major factor in the expansive evolutionary diversification of the TPS-d family, which in the case of PsTPS-3car and PsTPS-sab explains formation of either (+)-3-carene or (−)-sabinene as the major product with a single residue switch.
Acknowledgments
We thank Karen Reid for excellent lab management support and Lina Madilao for expert assistance with the GC/MS analyses.
This work was supported in part by grants from the Natural Sciences and Engineering Research Council of Canada (to J. B.), Genome Canada, Genome BC, and Genome Quebec in support of the SMarTForests Project LSARP Program (to Project Leader J. B.).

This article contains supplemental Tables S1 and S2.
- Ps
- P. sitchensis
- TPS
- terpene synthase
- TPS-car
- (+)-3-carene synthase
- TPS-sab
- (−)-sabinene synthase.
REFERENCES
- 1. King J. N., Alfaro R. I., Cartwright C. (2004) Genetic resistance of Sitka spruce (Picea sitchensis) populations to the white pine weevil (Pissodes strobi): distribution of resistance. Forestry 77, 269–278 [Google Scholar]
- 2. Zulak K. G., Lippert D. N., Kuzyk M. A., Domanski D., Chou T., Borchers C. H., Bohlmann J. (2009) Targeted proteomics using selected reaction monitoring reveals the induction of specific terpene synthases in a multi-level study of methyl jasmonate-treated Norway spruce (Picea abies). Plant J. 60, 1015–1030 [DOI] [PubMed] [Google Scholar]
- 3. Keeling C. I., Bohlmann J. (2006) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 170, 657–675 [DOI] [PubMed] [Google Scholar]
- 4. Keeling C. I., Bohlmann J. (2006) Diterpene resin acids in conifers. Phytochemistry 67, 2415–2423 [DOI] [PubMed] [Google Scholar]
- 5. Phillips M. A., Croteau R. B. (1999) Resin-based defenses in conifers. Trends Plant Sci. 4, 184–190 [DOI] [PubMed] [Google Scholar]
- 6. Robert J. A., Madilao L. L., White R., Yanchuk A., King J., Bohlmann J. (2010) Terpenoid metabolite profiling in Sitka spruce identifies association of dehydroabietic acid, (+)-3-carene, and terpinolene with resistance against white pine weevil. Botany 88, 810–820 [Google Scholar]
- 7. Hall D. E., Robert J. A., Keeling C. I., Domanski D., Quesada A. L., Jancsik S., Kuzyk M. A., Hamberger B., Borchers C. H., Bohlmann J. (2011) An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J. 65, 936–948 [DOI] [PubMed] [Google Scholar]
- 8. Davis E. M., Croteau R. (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Top. Curr. Chem. 209, 53–95 [Google Scholar]
- 9. Kampranis S. C., Ioannidis D., Purvis A., Mahrez W., Ninga E., Katerelos N. A., Anssour S., Dunwell J. M., Degenhardt J., Makris A. M. (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell 19, 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krause S. T., Köllner T. G., Asbach J., Degenhardt J. (2013) Stereochemical mechanism of two sabinene hydrate synthases forming antipodal monoterpenes in thyme (Thymus vulgaris). Arch. Biochem. Biophys. 529, 112–121 [DOI] [PubMed] [Google Scholar]
- 11. Keeling C. I., Weisshaar S., Lin R. P., Bohlmann J. (2008) Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc. Natl. Acad. Sci. U.S.A. 105, 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Maille P. E., Chappell J., Noel J. P. (2004) A single-vial analytical and quantitative gas chromatography-mass spectrometry assay for terpene synthases. Anal. Biochem. 335, 210–217 [DOI] [PubMed] [Google Scholar]
- 13. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 14. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whittington D. A., Wise M. L., Urbansky M., Coates R. M., Croteau R. B., Christianson D. W. (2002) Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc. Natl. Acad. Sci. U.S.A. 99, 15375–15380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 18. Hyatt D. C., Youn B., Zhao Y., Santhamma B., Coates R. M., Croteau R. B., Kang C. (2007) Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc. Natl. Acad. Sci. U.S.A. 104, 5360–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katoh S., Hyatt D., Croteau R. (2004) Altering product outcome in Abies grandis (−)-limonene synthase and (−)-limonene/(−)-α-pinene synthase by domain swapping and directed mutagenesis. Arch. Biochem. Biophys. 425, 65–76 [DOI] [PubMed] [Google Scholar]
- 20. Hyatt D. C., Croteau R. (2005) Mutational analysis of a monoterpene synthase reaction: altered catalysis through directed mutagenesis of α-pinene synthase from Abies grandis. Arch. Biochem. Biophys. 439, 222–233 [DOI] [PubMed] [Google Scholar]
- 21. Croteau R. (1987) Biosynthesis and catabolism of monoterpenoids. Chem. Rev. 87, 929–954 [Google Scholar]
- 22. Starks C. M., Back K., Chappell J., Noel J. P. (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820 [DOI] [PubMed] [Google Scholar]
- 23. Weitman M., Major D. T. (2010) Challenges posed to bornyl diphosphate synthase: diverging reaction mechanisms in monoterpenes. J. Am. Chem. Soc. 132, 6349–6360 [DOI] [PubMed] [Google Scholar]
- 24. Caruthers J. M., Kang I., Rynkiewicz M. J., Cane D. E., Christianson D. W. (2000) Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti. J. Biol. Chem. 275, 25533–25539 [DOI] [PubMed] [Google Scholar]
- 25. Thoma R., Schulz-Gasch T., D'Arcy B., Benz J., Aebi J., Dehmlow H., Hennig M., Stihle M., Ruf A. (2004) Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432, 118–122 [DOI] [PubMed] [Google Scholar]
- 26. Faraldos J. A., Antonczak A. K., González V., Fullerton R., Tippmann E. M., Allemann R. K. (2011) Probing eudesmane cation-π interactions in catalysis by aristolochene synthase with non-canonical amino acids. J. Am. Chem. Soc. 133, 13906–13909 [DOI] [PubMed] [Google Scholar]
- 27. Tantillo D. J. (2010) The carbocation continuum in terpene biosynthesis: where are the secondary cations?. Chem. Soc. Rev. 39, 2847–2854 [DOI] [PubMed] [Google Scholar]
- 28. Hong Y. J., Tantillo D. J. (2014) Biosynthetic consequences of multiple sequential post-transition-state bifurcations. Nat. Chem. 6, 104–111 [DOI] [PubMed] [Google Scholar]
- 29. Fäldt J., Martin D., Miller B., Rawat S., Bohlmann J. (2003) Traumatic resin defense in Norway spruce (Picea abies): methyl jasmonate-induced terpene synthase gene expression, and cDNA cloning and functional characterization of (+)-3-carene synthase. Plant Mol. Biol. 51, 119–133 [DOI] [PubMed] [Google Scholar]
- 30. Keeling C. I., Weisshaar S., Ralph S. G., Jancsik S., Hamberger B., Dullat H. K., Bohlmann J. (2011) Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol. 11, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]