Background: Hepcidin controls systemic iron balance and is regulated by bone morphogenetic protein (BMP) signaling.
Results: MicroRNA miR-130a is up-regulated by iron deficiency and targets BMP type I receptor ALK2 to suppress BMP signaling and hepcidin.
Conclusion: miR-130a is a novel regulator of BMP signaling and hepcidin.
Significance: This pathway may increase iron availability in the setting of iron deficiency.
Keywords: Activin A Receptor, Type I (ACVR1); Bone Morphogenetic Protein (BMP); Iron Metabolism; MicroRNA (miRNA); Signal Transduction; SMAD Transcription Factor; ALK2; SMAD4; Hepcidin; miR-130a
Abstract
Systemic iron balance is controlled by the liver peptide hormone hepcidin, which is transcriptionally regulated by the bone morphogenetic protein (BMP)-SMAD pathway. In iron deficiency, liver BMP-SMAD signaling and hepcidin are suppressed as a compensatory mechanism to increase iron availability. MicroRNAs are small regulatory RNAs that have an increasingly recognized role in many biologic processes but are only recently implicated in iron homeostasis regulation. Here, we demonstrate that liver expression of the microRNA miR-130a is up-regulated by iron deficiency in mice. We identify the BMP6-SMAD signaling pathway as a functional target of miR-130a in hepatoma-derived Hep3B cells. Although the TGF-β/BMP common mediator SMAD4 was previously reported to be an miR-130a target to inhibit TGF-β signaling, we do not confirm SMAD4 as an miR-130a target in our biologic system. Instead, we determine that the BMP type I receptor ALK2 is a novel target of miR-130a and that miR-130a binds to two specific sites in the 3′-untranslated region to reduce ALK2 mRNA stability. Moreover, we show in mice that the increased liver miR-130a during iron deficiency is associated with reduced liver Alk2 mRNA levels. Finally, we demonstrate that down-regulation of ALK2 by miR-130a has a functional effect to inhibit BMP6-induced hepcidin transcription in Hep3B cells. Our data suggest that iron deficiency increases liver miR-130a, which, by targeting ALK2, may contribute to reduce BMP-SMAD signaling, suppress hepcidin synthesis, and thereby promote iron availability.
Introduction
The liver orchestrates systemic iron homeostasis by synthesizing the peptide hormone hepcidin, which controls iron entry into the circulation by regulating degradation of the iron exporter ferroportin (1). Iron administration stimulates hepcidin expression as a negative feedback mechanism to prevent further iron entry into plasma, whereas iron deficiency inhibits hepcidin expression to increase iron availability. Dysregulated hepcidin expression leads to many diseases, including the iron overload disorder hereditary hemochromatosis and the anemia of chronic disease (1).
The bone morphogenetic protein (BMP)3-hemojuvelin (HJV)-SMAD signaling pathway is central to hepcidin transcriptional regulation by iron (2–6). Iron stimulates hepatic expression of BMP6 ligand (6–8), which binds to a complex of BMP type I and type II serine threonine kinase receptors and the co-receptor HJV to induce phosphorylation of intracellular SMAD1, SMAD5, and SMAD8 proteins (2, 3). These phosphorylated SMAD1, SMAD5, and SMAD8 proteins (p-SMAD1/5/8) complex with SMAD4 and translocate to the nucleus to induce hepcidin transcription (2). ALK2 (also known as ACVR1) and ALK3 (also known as BMPR1A) appear to be the primary endogenous BMP type I receptors involved in hepcidin regulation and systemic iron balance, because liver-specific deletion of either ALK2 or ALK3 in mice results in hepcidin deficiency and iron overload (9). ACTRIIA (also known as ACVR2A) has been proposed as the main BMP type II receptor involved in hepcidin regulation, because ACTRIIA is the predominant BMP type II receptor endogenously expressed in the liver, and because HJV induces preferential binding of BMP ligands to ACTRIIA versus other BMP type II receptors (10).
MicroRNAs (miRNAs) are short, single-stranded, noncoding RNAs that control gene expression by binding to complementary sequences in target mRNA 3′ untranslated regions (UTRs) to inhibit translation or induce degradation (11). Estimated to target more than 60% of all genes, miRNAs have an increasingly recognized role in many physiologic and pathologic processes, including development, differentiation, metabolism, hepatitis C, and cancer (11). However, a role for miRNAs in iron homeostasis is only recently emerging (12). One group reported that miR-122 may regulate hepcidin expression and systemic iron balance by inhibiting expression of the hemochromatosis protein HFE and HJV (13). Others have proposed a role for heme and iron in miRNA processing from primary transcripts to their precursor and mature forms (14, 15).
Here, we identify miR-130a as a novel regulator of BMP-SMAD signaling and hepcidin expression. We demonstrate that liver miR-130a is up-regulated by iron deficiency in mice. We also show that miR-130a targets the 3′UTR of ALK2 to inhibit BMP-SMAD signaling and hepcidin induction by BMP6 ligand in liver cells.
EXPERIMENTAL PROCEDURES
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee at Massachusetts General Hospital. miRNA microarray analysis and miR-130a validation were performed on livers from 129S6/SvEvTac mice fed a control diet (three females used for microarray; three females and two males used for validation) or iron-deficient diet (two females and one male used for microarray; two females and four males used for validation) for 5 weeks until 12 weeks of age as described previously (8). Additional validation studies were performed on female C57BL/6 mice (Taconic) fed a control diet (Lab Diets, 380 ppm iron, n = 8) or an iron-deficient diet (Harlan Teklad, 2–6 ppm iron, n = 8) for 5 weeks until 9 weeks of age.
Hematologic and Iron Analysis
Hematologic parameters were assessed in whole blood using the ADVIA 2120i and analyzed with multispecies software at Children's Hospital Boston. Serum iron and unsaturated iron binding capacity were measured by colorimetric assay (Pointe Scientific Inc.), and transferrin saturation was calculated as described previously (16). Quantitative measurement of non-heme iron in the liver was performed as described previously (16).
MicroRNA Array Analysis
Total RNA was prepared as described for miRNA extraction, and the quality was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies). MicroRNA array was performed by Exiqon Services, Denmark. Briefly, 10 μg of total RNA from liver were labeled with Hy3TM, and an RNA reference pool from each organ was labeled with Hy5TM fluorescent label, respectively, using the miRCURY LNATM microRNA power labeling kit, Hy3TMHy5TM. The Hy3TM- and Hy5TM-labeled pool reference RNA samples were mixed pairwise and hybridized to the miRCURY LNATM microRNA array (5th generation-hsa, mmu, and rno), which contains capture probes targeting all miRNAs for human, mouse, or rat registered in the miRBASE 16.0. The quantified signals (background-corrected) were normalized using the global Lowess (locally weighted scatterplot smoothing) regression algorithm (see ArrayExpress accession number E-MTAB-1520).
miRNA and mRNA Purification, Reverse Transcription, and Quantitative Real Time PCR (qRT-PCR)
For miRNA analysis, RNA was extracted from cells or liver using TRIzol® (Invitrogen). Following TRIzol® RNA preparation, 350 μl of RLT buffer from RNeasy mini kit (Qiagen) was added to dissolved RNA. Three and one-third volumes of 100% ethanol was added, and the samples were applied to an RNeasy spin column. Samples were washed twice with RPE buffer and eluted in RNase-free water. qRT-PCR was performed according to the manufacturer's directions using the Universal cDNA synthesis kit, SYBR Green master mix (Exiqon) supplemented with ROX Passive Reference Dye (Bio-Rad) and miRCURY LNATM Universal RT miRNA PCR primer sets hsa-miR-130a, hsa-miR-103, and RNU5G (Exiqon).
For mRNA analysis, RNA was isolated using the QIAshredder and RNeasy purification kits (Qiagen), and cDNA was synthesized with 1000 ng of RNA using the iScript cDNA synthesis kit (Bio-Rad) as described (17). qRT-PCR was performed using the Power SYBR Green PCR Master Mix as described previously (17) using primers listed in Table 1.
TABLE 1.
Primer sequences used for qRT-PCR analysis, cloning, or site-directed mutagenesis
F is forward, and R is reverse; h is human; m is mouse; and mut is mutant.
| Primer | Sequence (5′ to 3′) | Experimental method |
|---|---|---|
| hACVR1 F | tgccaaggggactggtgtaa | qRT-PCR |
| hACVR1 R | actgcgaacactacagagagaa | qRT-PCR |
| hHAMP F | cacaacagacgggacaac | qRT-PCR |
| hHAMP R | cgcagcagaaaatgcaga | qRT-PCR |
| hID1 F | gggattccactcgtgtgttt | qRT-PCR |
| hID1 R | ctgagaagcaccaaacgtga | qRT-PCR |
| hSMAD4 F | ctcatgtgatctatgcccgtc | qRT-PCR |
| hSMAD4 R | aggtgatacaactcgttcgtagt | qRT-PCR |
| hRPL19 F | tccgctgtggcaagaagaaggt | qRT-PCR |
| hRPPL9 R | accgtcacaggcttgcggat | qRT-PCR |
| mAcvr1 F | gtggaagattacaagccacca | qRT-PCR |
| mAcvr1 R | gggtctgagaaccatctgttagg | qRT-PCR |
| mBmp6 F | atggcaggactggatcattgc | qRT-PCR |
| mBmp6 R | ccatcacagtagttggcagcg | qRT-PCR |
| mHamp F | ttgcgataccaatgcagaaga | qRT-PCR |
| mHamp R | gatgtggctctaggctatgtt | qRT-PCR |
| mId1 F | ttggtctgtcggagcaaagcgt | qRT-PCR |
| mId1 R | cagccgttcatgtcgtagagca | qRT-PCR |
| mSmad4 F | aggtggcctgatctacacaag | qRT-PCR |
| mSmad4 R | acccgctcatagtgatatggatt | qRT-PCR |
| mRpl19 F | aggcatatgggcatagggaagag | qRT-PCR |
| mRpl19 R | ttgaccttcaggtacaggtgtg | qRT-PCR |
| hACVR1-UTR F | atcggccggcccattttcatagtgtcaagaa | Cloning |
| hACVR1-UTR R | actgtcgacgacttgaaaacagtttattt | Cloning |
| hSMAD4 BS1 F | ctagtctagagtatcttggggcaagactgcaaac | Cloning |
| hSMAD4 BS1-BS2 R | gcaggatgtcagaccctcaattctcgcagtagaagctgtgttcattccag | Cloning |
| hSMAD4 BS1-BS2 F | ctggaatgaacacagcttctactgcgagaattgagggtctgacatcctgc | Cloning |
| hSMAD4 BS2 R | atagtttaggccggccctgctgcaaggtagccaaggtc | Cloning |
| hACVR1-Mut1 F | cctgtgcttctcttctttattgttgcaggaattctttgcattcc | Mutagenesis |
| hACVR1-Mut1 R | ggaatgcaaagaattcctgcaacaataaagaagagaagcacagg | Mutagenesis |
| hACVR1-Mut2 F | gcactaggaattctttgcattccttacttgatgagttactcttaattttaaagacccaac | Mutagenesis |
| hACVR1-Mut2 R | gttgggtctttaaaattaagagtaactcatcaagtaaggaatgcaaagaattcctagtgc | Mutagenesis |
| hSMAD4 BS1-mut F | gattttttttttcttaacgtgatttgagtccaatctcagtgatg | Mutagenesis |
| hSMAD4 BS1-mut R | catcactgagattggactcaaatcacgttaagaaaaaaaaaatc | Mutagenesis |
| hSMAD4 BS2-mut F | tggctctgggttgggccagacagaacgtgactctagtttgccctctgc | Mutagenesis |
| hSMAD4 BS2-mut R | gcagagggcaaactagagtcacgttctgtctggcccaacccagagcca | Mutagenesis |
Relative miRNA or mRNA expression was determined using the ΔΔCt method (18) with RPL19 as reference for mRNA. For all qRT-PCR experiments miRNA, mRNA, or control small RNA standards were prepared by PCR amplification and used to ensure that primer sets amplified efficiently across a six 10-fold dilution series. For initial mRNA qRT-PCR experiments, HPRT and GUSB were used as reference genes in addition to RPL19, either singly or as the geometric mean of all three references, with similar results (data not shown). For all qRT-PCR experiments, no significant changes were seen in reference gene RPL19 (data not shown).
In Silico Analysis of miRNA Targets
miRNA target predictions were performed using the following internet-based programs: microRNA.org, MicroCosm, RegRNA, and TargetScan. For RegRNA analyses, minimum threshold values were set at Score ≥140 and Free_Energy ≤ −7. miRNA sequences were obtained from miRBase. Genomic sequences for the 3′UTR of putative miR-130a target genes were obtained from NCBI Aceview database (www.ncbi.nlm.nih.gov).
Plasmid Construction
Genomic DNA was extracted from the human hepatocarcinoma Hep3B cell line (ATCC) using QIAamp DNA mini kit (Qiagen) following the manufacturer's protocol. Pfu Ultra (Agilent) was used to PCR-amplify the full-length 3′UTR of ALK2 using gene-specific primers (Table 1). The 3′UTR was ligated into the pGL3-promoter plasmid (Promega) downstream of the firefly luciferase gene using FseI and SalI restriction sites. Mutation of putative miR-130a-binding sites (proximal bp 418–425; distal bp 448–454) was performed as described in the QuikChange site-directed mutagenesis protocol (Stratagene) using primers listed in Table 1.
For the SMAD4 3′UTR construct, Platinum® Pfx DNA polymerase (Invitrogen) was used to PCR-amplify 442- or 456-bp fragments containing the predicted miR-130a proximal (bp 1355–1361) and distal (bp 5167–5173) binding sites, respectively. Binding site fragments were cloned independently or in tandem into the pGl3-promoter plasmid downstream of firefly luciferase using XbaI and FseI sites (primers listed in Table 1). These constructs contain the same binding site fragments arranged in tandem as published previously (19). Mutation of putative miR-130a-binding sites was performed as described for ALK2. All constructs were confirmed by sequencing.
Transfection and Luciferase Assays
Cells were cultured in Eagle's minimal essential medium (ATCC) for Hep3B, HepG2, and HEK293 or in Dulbecco's modified Eagle's medium (Invitrogen) for Hepa 1–6 supplemented with 10% FBS (ATCC) and 2 mm l-glutamine (Invitrogen) as described previously (17). For endogenous miR-130a analysis, Hep3B, HepG2, and Hepa 1–6 cells were plated on 35-mm dishes to obtain 90–95% confluence after 18 h and harvested for miRNA. For BMP induction studies, cells were plated as above, treated with medium containing 1% FBS for 7 h, stimulated with 50 ng/ml BMP6 for 17 h, and then harvested for mRNA. For transient transfection of plasmids and miRNA mimics, 50,000–65,000 Hep3B cells were plated in 1 well of a 24-well dish. After 16 h, cells were transfected for 5 h in Opti-MEM (Invitrogen) using LipofectamineTM 2000 (Invitrogen). For 3′UTR reporter assays, Hep3B cells were transfected with 250 ng of 3′UTR firefly luciferase constructs, 25 ng of Renilla-luciferase plasmid (pRL-TK, Promega), and 2.5–10 nm of Pre-miRTM miRNA precursor mimic hsa-miR-130a-3p (miR-130a mimic) or negative control 1 or 2 (Invitrogen). Relative luciferase activity was assayed 48 h post-transfection using the Dual-Luciferase® reporter assay system (Promega) as described previously (2).
For BMP signaling and hepcidin promoter reporter assays, Hep3B cells were transfected with 250 ng of hepcidin promoter firefly luciferase reporter (Hep-luc (2)) or 250 ng of BMP-responsive firefly luciferase reporter (BRE-luc (20)), in combination with 25 ng of pRL-TK, without or with 0.5–10 nm miR-130a mimic or negative control. For ALK2 add-back experiments, cells were also co-transfected with 50 ng of pcDNA3 (Invitrogen) or pcDNA3 containing HA-tagged ALK2 cDNA without (HA-ALK2) or with the native 3′UTR (HA-ALK2–3′UTR). Twenty four hours post-transfection, cells were treated with medium containing 1% FBS for 7 h, stimulated with 5 ng/ml BMP6 (R&D Systems) for 17 h, and analyzed for relative luciferase activity as described above.
For endogenous mRNA and protein analysis, Hep3B or HEK293 cells were transfected with 2.5–10 nm miR-130a mimic or negative control as indicated, treated with BMP6 as described above, and harvested 48 h post-transfection. For ALK2 add-back experiments, cells were also co-transfected with pcDNA3, HA-ALK2, or HA-ALK2–3′UTR as above. For ALK2 mRNA half-life experiments, Hep3B cells were transfected with 10 nm miR-130a mimic or negative control. Twenty-four hours post-transfection, cells were treated with 5 μg/ml actinomycin D in DMSO (Sigma) and harvested at the indicated time points.
Immunoblot
Cells or livers were lysed in RIPA Buffer (Boston BioProducts) supplemented with Halt Phosphatase Inhibitor (Thermo Scientific) and Protease Inhibitor mixture (Roche Applied Science). Lysates were prepared by centrifugation at 17,000 × g for 10 min, and protein concentration was determined using Coomassie (Bradford) protein assay kit (Thermo Scientific). Twenty five μg of whole cell lysate was separated by reducing NuPAGE® NOVEX® 4–12% bis-tris gel (Invitrogen) and transferred to nitrocellulose (Invitrogen). The following antibodies were using for immunoblots: phospho-SMAD1/SMAD5/SMAD8 (p-SMAD1/5/8) (Cell Signaling 9511); SMAD1 (Cell Signaling 9743); SMAD4 (Santa Cruz Biotechnology sc-7966); HA probe (Y-11) (Santa Cruz Biotechnology sc-805); and pan-actin (Millipore MAB1501R). p-SMAD1/5/8 and SMAD1 antibodies were incubated in MBTN Buffer (1× TBST (50 mm Tris (pH 7.4), 150 mm NaCl, 0.05% Tween 20), 10% nonfat dry milk, 1% bovine serum albumin, and 1% IgePal CA-630). All other antibodies were incubated in TBST supplemented with 5% nonfat dry milk. Chemiluminescence quantitation was performed using ImageJ 1.46r (21), with all data normalized to actin. Antibodies were validated in prior publications (2) or using siRNA knockdown experiments (data not shown).
Statistics
Microarray data were analyzed by analysis of variance and a two-tailed Student's t test with or without the Bonferroni correction for multiple comparisons. For all other data, statistical significance was determined by a two-tailed Student's t test for comparisons between two groups or by analysis of variance for comparisons between more than two groups with Newman Keuls post hoc test for pairwise multiple comparisons. p < 0.05 was considered significant.
RESULTS
miR-130a Is Up-regulated by Iron Deficiency in Vivo and Predicted to Target Multiple mRNAs in the BMP-SMAD Pathway
To identify novel miRNA regulators of systemic iron homeostasis, we performed an miRNA microarray analysis on livers from 12-week-old male and female 129S6/SvEvTac mice fed a low iron diet or a standard diet for 5 weeks (8). Hematologic and iron parameters of these mice were previously characterized (8). Significant differences in miRNA levels between low iron and control diet mouse livers were not detected using statistical analysis accounting for multiple comparisons (data not shown). Most likely, this study was underpowered to detect such differences with only three samples per group and a large within-group variation. We therefore re-analyzed the microarray data using a Student's t test without correction for multiple comparisons as an hypothesis generating method to identify potential candidate miRNAs that might be regulated by dietary iron changes, with the objective to validate potential candidates using qRT-PCR in the original cohort and a separate low iron diet cohort. Potential candidates were prioritized for validation based on the level of change relative to control and the number of predicted targets related to iron homeostasis by four internet-based target-prediction programs (see below).
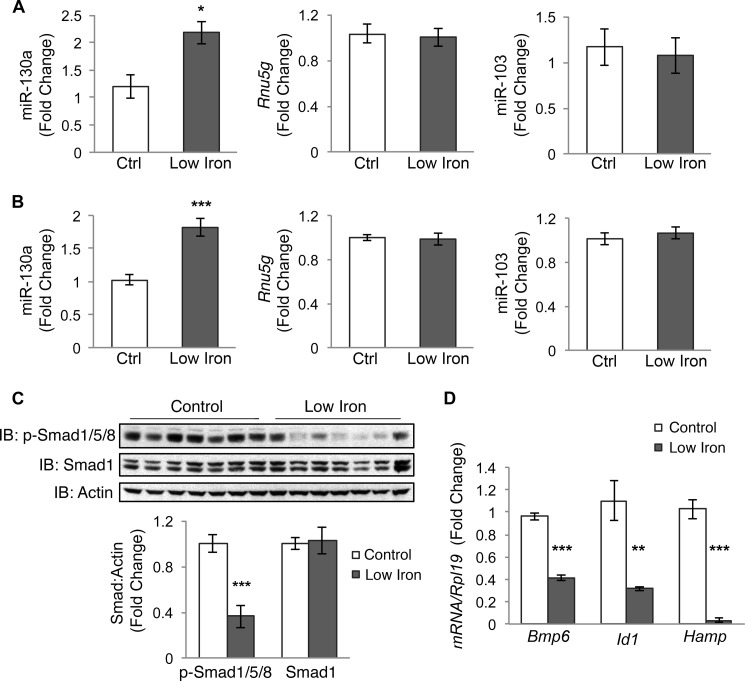
One candidate miRNA suggested to be increased in the liver by a low iron diet in the microarray was miR-130a (data not shown). Using the full original cohort of five to six mice per group (8), qRT-PCR analysis demonstrated that liver miR-130a was significantly up-regulated by ∼2-fold in the low iron diet group compared with controls, whereas we observed no change in another miRNA (miR-103) that showed no variability in our microarray or the housekeeping small RNA Rnu5g (Fig. 1A).
FIGURE 1.
Low iron diet increases liver miR-130a in mice. A, liver miR-130a, Rnu5g, and miR-103 were measured by qRT-PCR in 12-week-old male and female 129S6/SvEvTac mice after receiving a control diet (Ctrl) or a low iron diet (Low Iron) for 5 weeks (n = 5–6 per group). B–D, liver tissue from 9-week-old female C57BL/6 mice (n = 8 per group) treated with a control (Ctrl) or a low iron diet (Low Iron) for 5 weeks were analyzed for miR-130a, Rnu5g, and miR-130a by qRT-PCR (B), p-Smad1/5/8 and total Smad1 relative to actin protein expression by immunoblot (IB) and chemiluminescence quantification (C), and Bmp6, Id1, and Hamp relative to Rpl19 mRNA expression by qRT-PCR (D). Results are reported as mean ± S.E. of the fold change relative to the control group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To further verify that miR-130a is up-regulated by iron deficiency, we measured liver miR-130a levels by qRT-PCR in a different cohort of 9-week-old female C57BL/6 mice treated with a low iron or control diet for 5 weeks (n = 8 per group). Low iron diet mice exhibited evidence of iron deficiency with significantly decreased hemoglobin, mean cell hemoglobin, hemoglobin content of reticulocytes, and liver iron content and a strong trend (p = 0.05) toward decreased serum transferrin saturation compared with the control group (Table 2). Similar to the original cohort, liver miR-130a was significantly increased in the low iron diet group compared with controls, whereas miR-103 and Rnu5g showed no change (Fig. 1B). Thus, iron deficiency increased liver expression of miR-130a in two separate cohorts of mice, encompassing both genders and two different mouse strains.
TABLE 2.
Hematologic parameters and serum and tissue iron analysis of control and low iron diet mice
Red blood cell parameters, including hemoglobin (Hgb), hematocrit (Hct), mean cell volume (MCV), mean cell hemoglobin (MCH), red cell distribution width (RDW), reticulocyte count (Retic), and reticulocyte mean cell hemoglobin (Chr), as well as serum iron, transferrin saturation, and total liver iron content were measured in 9-week-old female C57BL/6 mice fed a control or low iron diet for 5 weeks. Data are presented as means ± S.E. A two-tailed Student's t test was performed to determine statistical significance.
| Control (8) | Low iron (8) | |
|---|---|---|
| Hgb (g/dl) | 14.4 ± 0.1 | 13.5 ± 0.3a |
| Hct (%) | 50.1 ± 1.0 | 51.3 ± 0.7 |
| MCV (fl) | 53.0 ± 0.7 | 51.7 ± 0.4 |
| MCH (pg) | 15.2 ± 0.1 | 13.6 ± 0.3b |
| RDW (%) | 11.9 ± 0.2 | 12.6 ± 0.3c |
| Retic (%) | 2.8 ± 0.1 | 3.2 ± 0.2a |
| Chr (pg) | 15.6 ± 0.1 | 14.2 ± 0.1d |
| Serum iron (μg/dl) | 163.3 ± 9.6 | 163.3 ± 8.7 |
| Transferrin saturation (%) | 50.9 ± 4.4 | 39.5 ± 3.0c |
| Liver iron content (μg/g) | 85.1 ± 4.1 | 38.9 ± 5.6d |
a p < 0.05.
b p < 0.01.
c p = 0.05.
d p < 0.001.
Iron deficiency has previously been associated with decreased liver BMP6-SMAD signaling and hepcidin expression (8, 22–24), which are thought to be an important compensatory mechanism to increase iron availability. Consistent with prior studies (22–24), both low iron diet mouse cohorts exhibited significantly decreased liver Bmp6 mRNA, p-Smad1/5/8 protein, and Id1 mRNA (a target transcript of BMP-SMAD signaling (3)) levels compared with controls, suggestive of reduced BMP6-SMAD signaling (Fig. 1, C and D) (8). Low iron diet mice also exhibited significantly decreased liver hepcidin (Hamp) mRNA levels (Fig. 1D) (8). In silico analysis using four miRNA target prediction programs revealed that miR-130a was predicted to target the 3′UTR of many components of the BMP-SMAD signaling pathway involved in hepcidin regulation (Table 3), raising the possibility that up-regulation of miR-130a in the liver could have a functional role to inhibit BMP-SMAD signaling and hepcidin expression in the context of iron deficiency.
TABLE 3.
Predicted miR-130a targets
Genes involved in iron-mediated BMP-SMAD signaling and hepcidin regulation were assayed for putative miR-130a-binding sites using four target-prediction programs as follows: *, microRNA.org; &, RegRNA; $, MicroCosm; #, TargetScan. − indicates no miR-130a target predictions.
| Target genes | Human | Mouse |
|---|---|---|
| ALK2 (ACVR1) | *&$# | *&$# |
| SMAD5 | *&# | *& |
| SMAD4 | *&# | # |
| ACTRIIA (ACVR2A) | *& | *& |
| SMAD7 | * | * |
| BMP6 | *& | − |
| SMAD1 | & | − |
| TFR2 | $ | − |
| HJV (HFE2) | − | *&# |
| ALK3 (BMPR1A) | − | & |
| HFE | − | # |
| HAMP | − | − |
| SMAD6 | − | − |
| SMAD8 (SMAD9) | − | − |
| TMPRSS6 | − | − |
miR-130a Inhibits BMP-SMAD Signaling
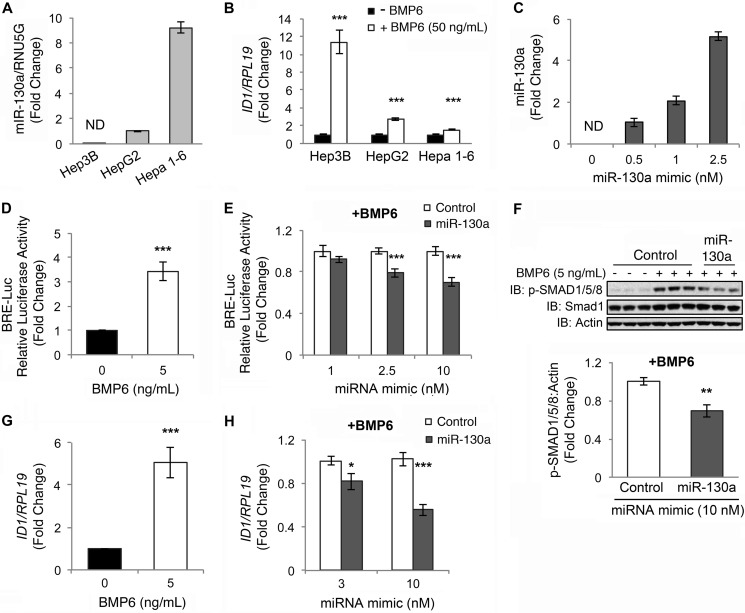
To test whether miR-130a can inhibit BMP-SMAD signaling in liver cells, we quantitated endogenous miR-130a levels by qRT-PCR in three different liver-derived cell lines, and we tested the responsiveness of these cells to BMP6 stimulation as measured by qRT-PCR of ID1 mRNA expression. Interestingly, we saw an inverse association between endogenous miR130a levels and BMP6 responsiveness in the liver-derived cell lines we tested (Fig. 2, A and B). Next, we examined whether transfection with miR-130a mimic (a synthetic double-stranded RNA construct that mimics the function of endogenous miR-130a) affected BMP-SMAD signaling in hepatoma-derived Hep3B cells. Although basal endogenous miR-130a levels were below the level of detection in this cell line, transfection with miR-130a mimic caused a dose-dependent increase in miR-130a levels as measured by qRT-PCR (Fig. 2C). As shown previously (10), BMP6 significantly increased the activity of a BMP-responsive luciferase reporter (BRE-Luc) (20), as measured by Dual-Luciferase assay (Fig. 2D). BMP6 also increased endogenous p-SMAD1/5/8 protein expression as measured by immunoblot (Fig. 2F, 1st to 6th lanes) and ID1 mRNA expression as measured by qRT-PCR (Fig. 2G). miR-130a mimic had no effect on basal BRE-Luc activity (data not shown) but significantly inhibited BMP6 activation of BRE-Luc activity compared with miRNA mimic negative control (Fig. 2E). miR-130a mimic also significantly inhibited BMP6 stimulation of endogenous p-SMAD1/5/8 protein expression (Fig. 2F, 4th to 9th lanes and bottom panel) and ID1 mRNA expression compared with negative control (Fig. 2H).
FIGURE 2.
miR-130a inhibits BMP6-SMAD signaling in hepatoma-derived cells. A, endogenous miR-130a levels relative to the small RNA RNU5G were quantified by qRT-PCR in hepatoma-derived Hep3B, HepG2, and Hepa 1–6 cells. Data are reported as the mean fold change ± S.E. relative to HepG2 cells; ND indicates not detectable. B, Hep3B, HepG2, and Hepa 1–6 cells were treated for 17 h without (black bars) or with 50 ng/ml BMP6 (white bars), followed by measurement of ID1 relative to RPL19 mRNA by qRT-PCR. Results are reported as mean ± S.E. of the fold change from untreated cells. C, Hep3B cells were transfected with increasing concentrations of miR-130a mimic and treated with 5 ng/ml BMP6 for 17 h, followed by measurement of miR-130a levels by qRT-PCR. Results are reported as the mean fold change ± S.E. relative to 0.5 nm miR-130a treated cells. D and G, Hep3B cells were transfected with BRE-Luc and control Renilla luciferase vector (D) or mock-transfected (G) and were treated without (black bar) or with 5 ng/ml BMP6 (white bar) for 17 h, followed by measurement of relative luciferase activity (D) or ID1 relative to RPL19 mRNA by qRT-PCR (G). Results are reported as mean ± S.E. of the fold change from untreated cells. E, Hep3B cells were transfected with BRE-Luc and a control Renilla luciferase vector in combination with increasing concentrations of miR-130a (gray bars) or negative control mimic (white bars) and were treated with 5 ng/ml BMP6 for 17 h, followed by measurement of relative luciferase activity. Results are reported as the mean fold change ± S.E. relative to the negative control at each concentration. F and H, Hep3B cells were transfected with miR-130a (gray bars) or negative control mimic (white bars) and were treated without or with 5 ng/ml BMP6 for 17 h, followed by measurement of phosphorylated SMAD1/5/8 (p-SMAD1/5/8) and total SMAD1 relative to actin by immunoblot (IB) and chemiluminescence quantification (F) or ID1 relative to RPL19 mRNA by qRT-PCR (H). Results are reported as the mean fold change ± S.E. relative to control at each concentration. A, results are shown for two experiments, each performed in triplicate. C, results are shown from one of two experiments performed in triplicate. For all other panels, n = 3–4 experiments, each performed in duplicate (for some immunoblots) or triplicate (for all other experiments); *, p < 0.05; **, p < 0.01; ***, p < 0.001. A representative immunoblot is shown.
SMAD4 Is Not an miR-130a Target in Liver Cells
Next, we investigated the mechanism by which miR-130a inhibits BMP6-SMAD signaling in Hep3B cells. SMAD4 is a common mediator to both BMP and TGF-β signaling that was predicted to be a miR-130a target by our in silico analysis (Table 3). Interestingly, miR-130a was recently reported to bind two sites on the SMAD4 3′UTR to inhibit SMAD4 protein expression and TGF-β signaling in granulocytic cells (19). We therefore tested whether SMAD4 was an miR-130a target in our system.
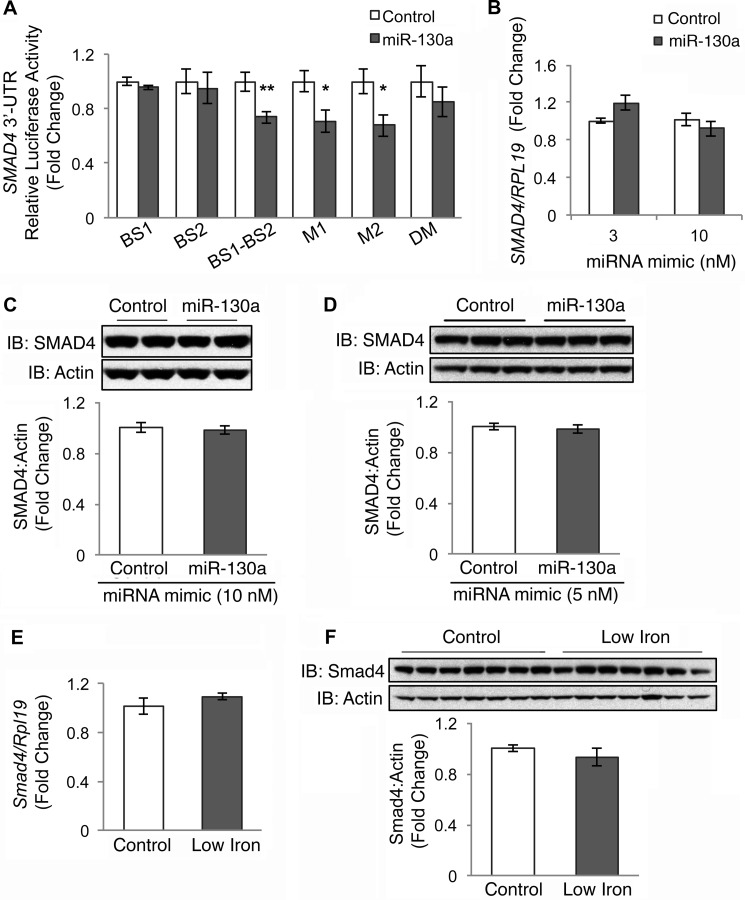
First, we tested whether miR-130a mimic transfection inhibited the activity of a firefly luciferase construct fused to the 3′UTR of SMAD4 using a Dual-Luciferase reporter assay as reported previously (19). Of note, the previous study used an artificial SMAD4 3′UTR construct in which the two predicted miR-130a-binding sites that are located almost 4 kb apart in the native 3′UTR were cloned side-by-side (19). Because of technical difficulties, we were unable to clone the predicted full-length ∼6.5-kb SMAD4 3′UTR. However, we did generate a side-by-side SMAD4 3′UTR luciferase reporter construct (BS1-BS2) similar to what was previously described (19), and we found that the miR-130a mimic inhibited 3′UTR activity of this BS1-BS2 construct compared with negative control (Fig. 3A). Although the ability of the miR-130a mimic to inhibit 3′UTR activity of this construct was abrogated to some extent when both predicted binding sites were mutated (DM), we did not see an effect when either the proximal (M1) or distal (M2) sites were mutated individually (Fig. 3A). Moreover, we did not see an effect of the miR-130a mimic on the ∼450-bp fragment SMAD4 3′UTR reporter constructs composed of either the proximal (BS1) or distal (BS2) predicted binding sites cloned in isolation (Fig. 3A).
FIGURE 3.
miR-130a mimic does not target the 3′UTR of SMAD4. A, SMAD4 3′UTR relative luciferase activity was measured by Dual-Luciferase assay in Hep3B cells transfected with 10 nm miR-130a (gray bars) or negative control mimic (white bars), in combination with control Renilla luciferase reporter and an engineered SMAD4 3′UTR firefly luciferase construct containing the proximal miR-130a binding site (BS1), the distal binding site (BS2), the two predicted miR-130a-binding sites cloned side-by-side (BS1-BS2), or a BS1-BS2 construct with mutations in the proximal (M1), distal (M2), or both predicted binding sites (DM). B–D, Hep3B (B and C) or HEK293 (D) cells were transfected with the indicated concentration of miR-130a (gray bars) or negative control mimic (white bars) and were treated for 17 h with 5 ng/ml BMP6, followed by measurement of SMAD4 relative to RPL19 mRNA by qRT-PCR (B) or SMAD4 relative to actin protein by immunoblot and chemiluminescence quantitation (C and D). A–D, results are reported as the mean fold change ± S.E. relative to negative control; n = 3–5 experiments, each performed in duplicate (C) or triplicate (A, B, and D). Representative immunoblots are shown. E and F, liver tissue from mice treated with a control (Control) or a low iron diet (Low Iron) as described in Fig. 1, B–D, were analyzed for Smad4 relative to Rpl19 mRNA (E) and Smad4 relative to actin protein expression by immunoblot and chemiluminescence quantification (F). Results are reported as the mean fold change ± S.E. relative to the control group. For all panels, significance is indicated by *, p < 0.05; **, p < 0.01.
We then tested the effects of miR-130a mimic transfection on endogenous SMAD4 mRNA and protein levels in Hep3B cells as measured by qRT-PCR and immunoblot. In contrast to previous reports in other cell systems, the miR-130a mimic had no significant effect on endogenous SMAD4 mRNA or protein levels in Hep3B cells compared with negative control (Fig. 3, B and C). We also could not reproduce the previous reported findings (19) that miR-130a inhibits SMAD4 protein levels in HEK293 cells (Fig. 3D).
Finally, we tested whether increased miR-130a in low iron diet mouse livers (Fig. 1B) was associated with reduced liver Smad4 mRNA or protein levels as measured by qRT-PCR and immunoblot. Neither Smad4 mRNA nor protein levels were significantly changed in low iron diet compared with control mice (Fig. 3, E and F).
miR-130a Mimic Targets Two Specific Binding Sites on the ALK2 3′UTR to Decrease ALK2 mRNA Stability
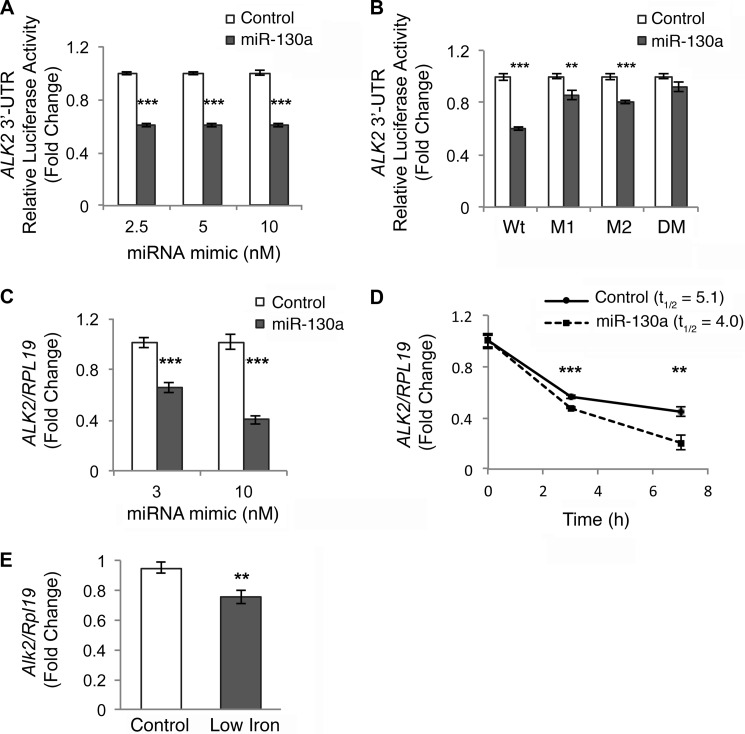
Because we could not confirm SMAD4 as a miR-130a target, at least in our biologic system, we next focused on ALK2 as the only miR-130a target predicted by all four programs in both humans and mice in our in silico analysis (Table 3). Hep3B cells were transfected with a reporter construct containing the full-length 3′UTR of human ALK2 subcloned downstream of firefly luciferase, a control Renilla luciferase vector, and increasing amounts of an miR-130a mimic or negative control, followed by measurement of relative luciferase activity. The miR-130a mimic significantly inhibited ALK2 3′UTR activity by ∼40% compared with the negative control (Fig. 4A).
FIGURE 4.
miR-130a mimic decreases the half-life of ALK2 mRNA by targeting two predicted binding sites in the 3′UTR. A and B, ALK2 3′UTR relative luciferase activity was measured by Dual-Luciferase assay in Hep3B cells transfected with the indicated concentrations (A) or 10 nm (B) of miR-130a (gray bars) or negative control mimic (white bars), in combination with control Renilla luciferase reporter and full-length wild type (A and B, Wt) or mutant ALK2 3′UTR firefly luciferase reporters containing mutations in the proximal (M1), distal (M2), or both (DM) predicted binding sites. Results are reported as the mean fold change ± S.E. relative to the negative control at each concentration; n = 3–9 experiments, each performed in triplicate. C, Hep3B cells were transfected with the indicated concentration of miR-130a (gray bars) or negative control mimic (white bars) and treated for 17 h with 5 ng/ml BMP6, followed by measurement of ALK2 relative to RPL19 mRNA by qRT-PCR. Results are reported as the mean fold change ± S.E. compared with control; n = 3 experiments each performed in triplicate. D, Hep3B cells were transfected with 10 nm miR-130a or negative control mimic. Twenty four hours after transfection, cells were treated with actinomycin D for 0–7 h, followed by measurement of ALK2 relative to RPL19 mRNA by qRT-PCR. Results are reported as mean ± S.E. of fold change compared with the 0 time point for each condition. Half-life (t½) calculations are based on three independent experiments, each performed in triplicate. One representative experiment is shown. E, liver tissue from mice treated with a control (Control) or a low iron diet (Low Iron) as described in Fig. 1, B–D, were analyzed for Alk2 relative to Rpl19 mRNA. Results are reported as the mean fold change ± S.E. relative to the control group. **, p < 0.01, and ***, p < 0.001.
miR-130a is predicted to bind two sites in the ALK2 3′UTR. To confirm that miR-130a specifically interacts with these predicted binding sites, we mutated these sites, either individually or together, and tested the effects of the miR-130a mimic versus negative control on 3′UTR activity by Dual-Luciferase assay. miR-130a down-regulation of ALK2 3′UTR activity was partially abrogated when either the proximal (M1) or distal (M2) predicted binding sites were mutated and was fully abolished when both binding sites (DM) were mutated (Fig. 4B). These data suggest that miR-130a interacts with both predicted binding sites on the ALK2 3′UTR.
miRNAs typically repress gene expression by increasing mRNA degradation and/or inhibiting translation efficiency (11). The former mechanism decreases both mRNA and protein expression, whereas the latter decreases protein expression without affecting mRNA expression. We therefore tested the effects of the miR-130a mimic transfection on the endogenous expression of ALK2 mRNA in Hep3B cells as measured by qRT-PCR. The miR-130a mimic significantly inhibited endogenous ALK2 relative to RPL19 mRNA levels compared with negative control (Fig. 4C). Similar effects were seen in HEK293 cells (data not shown). We were unable to test the effects of miR-130a on endogenous ALK2 protein levels because endogenous ALK2, like other BMP receptors, has a low copy number and is not detectable by immunoblot using currently available antibodies. To confirm that miR-130a mimic increased ALK2 mRNA degradation, we measured ALK2 mRNA half-life using the transcriptional inhibitor actinomycin D. As shown in Fig. 4D, miR-130a mimic significantly decreased ALK2 mRNA half-life from 5.1 ± 0.3 to 4.0 ± 0.1 h (p = 0.04).
Finally, we tested whether increased liver miR-130a in mice on a low iron diet (Fig. 1B) was associated with decreased Alk2 mRNA levels as measured by qRT-PCR. Low iron diet mice exhibited significantly reduced liver Alk2 mRNA compared with controls (Fig. 4E).
miR-130a Inhibits BMP6 Induction of Hepcidin by Targeting ALK2
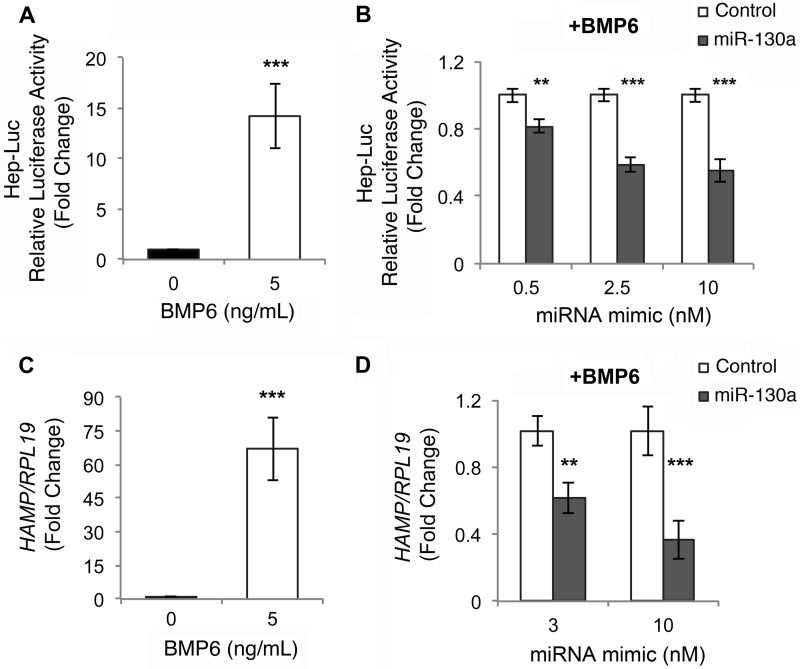
BMP6-SMAD signaling is essential for transcriptional activation of the main iron regulatory hormone hepcidin (1), and ALK2 has an important role in mediating BMP6-SMAD signaling and hepcidin expression in vivo (9). We therefore tested whether miR-130a has a functional role in regulating hepcidin transcription by examining the effects of miR-130a mimic versus negative control on BMP6 induction of hepcidin promoter luciferase activity (Hep-Luc, 2) and endogenous hepcidin (HAMP) mRNA levels in Hep3B cells. As described previously (16), BMP6 robustly stimulated Hep-Luc activity as measured by Dual-Luciferase assay (Fig. 5A) and endogenous HAMP mRNA as measured by qRT-PCR (Fig. 5C). miR-130a mimic had no effect on basal Hep-Luc activity (data not shown), but significantly inhibited BMP6 induction of Hep-Luc activity compared with negative control (Fig. 5B). miR-130a mimic also significantly inhibited BMP6 stimulation of endogenous HAMP mRNA levels as measured by qRT-PCR (Fig. 5D).
FIGURE 5.
miR-130a mimic inhibits BMP6 induction of hepcidin expression in Hep3B cells. A and C, Hep3B cells were transfected with Hep-Luc and control Renilla luciferase vector (A) or mock-transfected (C) and were treated without (black bar) or with 5 ng/ml BMP6 (white bar) for 17 h, followed by measurement of relative luciferase activity (A) or HAMP relative to RPL19 mRNA by qRT-PCR (C). Results are reported as the mean fold change ± S.E. relative to untreated cells. B, Hep3B cells were transfected with Hep-Luc and control Renilla luciferase vector, in combination with increasing concentrations of miR-130a (gray bars) or negative control mimic (white bars) and were treated with 5 ng/ml BMP6 for 17 h, followed by measurement of relative luciferase activity. Results are reported as the mean fold change ± S.E. relative to negative control at each concentration. D, Hep3B cells were transfected with 3 or 10 nm miR-130a (gray bars) or negative control mimic (white bars) and were treated with 5 ng/ml BMP6 for 17 h, followed by measurement of HAMP relative to RPL19 mRNA by qRT-PCR. Results are reported as the mean fold change ± S.E. relative to the negative control at each concentration. For all panels, n = 3 experiments each performed in triplicate. **, p < 0.01, and ***, p < 0.001.
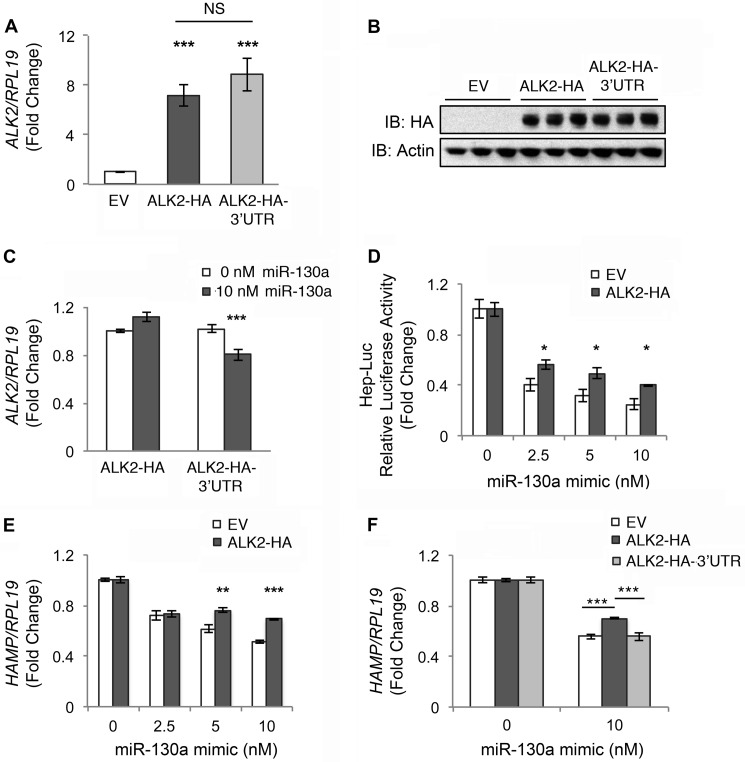
To determine whether down-regulation of ALK2 was the mechanism by which the miR-130a mimic inhibited BMP6 induction of hepcidin, we tested whether transfection of exogenous HA-tagged ALK2 cDNA without (ALK2-HA) or with the native 3′UTR (ALK2-HA-3′UTR) could reverse miR-130a effects on BMP6-stimulated hepcidin expression in Hep3B cells. Equivalent ALK2-HA expression for both constructs in transfected cells was confirmed by qRT-PCR (Fig. 6A) and immunoblot with HA antibody (Fig. 6B). Only expression of the ALK2-HA-3′UTR construct was inhibited by miR-130a mimic transfection, whereas ALK2-HA expression was not affected (Fig. 6C). ALK2-HA expression partially rescued BMP6 induction of Hep-Luc activity (Fig. 6D) as well as endogenous ID1 (data not shown) and hepcidin mRNA expression (Fig. 6E) in cells treated with miR-130a mimic. In contrast, ALK2-HA-3′UTR did not rescue BMP6 induction of hepcidin expression in cells treated with the miR-130a mimic (Fig. 6F).
FIGURE 6.
Transfection with ALK2 lacking the native 3′UTR partially reverses the inhibitory effect of miR-130a mimic on BMP6-stimulated hepcidin expression in Hep3B cells. Hep3B cells were transfected with empty vector (EV, white bars), ALK2 cDNA containing an HA tag without (ALK2-HA, dark gray bars) or with the native 3′UTR (ALK2-HA-3′UTR, light gray bars) alone (A–C, E, and F) or in combination with Hep-Luc and Renilla luciferase vectors (D), and the indicated concentrations of miR-130a mimic (C–F). Transfected cells were treated with 5 ng/ml BMP6 for 17 h, followed by measurement of ALK2 relative to RPL19 mRNA by qRT-PCR (A and C), ALK2-HA and actin protein expression by immunoblot (IB) (B), relative luciferase activity (D), and HAMP relative to RPL19 by qRT-PCR (E and F). Results are reported as the mean fold change ± S.E. relative to empty vector transfected cells (A) or to 0 nm miR-130a mimic for each construct (C–F). B, one of two experiments is shown performed in triplicate. For all other panels, n = 3–5 experiments each performed in triplicate. C, ***, p < 0.001 relative to 0 nm miR-130a mimic. For all other panels, *, p < 0.05; **, p < 0.01; and ***, p < 0.001 relative to empty vector at a given miR-130a mimic concentration or for other pairwise comparisons as indicated. NS, not significant.
DISCUSSION
Hepcidin is the master regulator of overall body iron balance, and the BMP-SMAD pathway is a central regulator of hepcidin expression in the liver (1). The mechanisms by which iron loading and iron deficiency regulate liver BMP-SMAD signaling to control hepcidin expression are incompletely understood. Here, we identified miR-130a as a novel regulator of BMP-SMAD signaling and hepcidin expression in liver cells in response to iron deficiency.
We demonstrated that miR-130a expression in the liver is up-regulated by a low iron diet in two mouse cohorts. Several links have previously been described between iron and miRNA biogenesis. miRNAs are transcribed as primary transcripts (pri-miRNA), which are cleaved by Drosha and DiGeorge critical region-8 (DGCR8) to yield stem-loop precursor miRNAs (pre-miRNAs), which are then processed by Dicer to yield functional single-stranded 20–22 ribonucleotide mature miRNAs (11). Interestingly, decreased cytosolic iron levels were recently shown to increase processing of many pre-miRNAs to mature miRNAs by increasing the association of the putative cytosolic iron chaperone poly(C)-binding protein 2 with pre-miRNAs and Dicer, although miR-130a was not specifically identified in this study (15). Additionally, heme has been associated with processing of pri-miRNAs into pre-miRNAs via the heme-binding protein DGCR8 (14). Hypoxia, which is intimately coordinated with iron homeostasis (25), was demonstrated to increase mature miR-130a but not pri- or pre-miR-130a levels in hippocampal neuronal cells (26). Future studies will be needed to determine whether these or other mechanisms are responsible for liver miR-130a up-regulation by a low iron diet.
Using multiple lines of evidence, we identified the BMP type I receptor ALK2 as a novel target of miR-130a. Mutagenesis studies confirmed that miR-130a specifically binds two sites in the ALK2 3′UTR. Consistent with a known role for miRNAs to destabilize mRNAs (11), we demonstrated that miR-130a decreases endogenous ALK2 mRNA half-life. Importantly, we showed that miR-130a has a functional effect to inhibit BMP6-SMAD signaling and BMP6-dependent hepcidin transcription in Hep3B cells.
Interestingly, miR-130a did not affect basal BRE-Luc and Hep-Luc activity in Hep3B cells. Of note, basal BMP signaling in Hep3B cells is mediated by endogenous BMP2 and BMP4 ligands and not by BMP6 (10). Although BMP6 uses the type I receptor ALK2 to signal, BMP2 and BMP4 ligands typically do not use ALK2 but rather use ALK3 and ALK6 (10). Thus, because BMP6 and therefore ALK2 do not contribute to basal BMP signaling in Hep3B cells, this can explain the lack of effect of miR-130a on basal BRE-Luc and Hep-Luc activity in this system. Another potential explanation could be that miR-130a target(s) are not limiting under basal conditions but become limiting under the condition of BMP6 stimulation.
Our data suggest that inhibition of ALK2 is at least part of the mechanism by which miR-130a inhibits BMP6 stimulation of hepcidin because transfection with exogenous ALK2 cDNA lacking the native 3′UTR partially rescued the effect of the miR-130a mimic on BMP6-dependent hepcidin expression. The inability of ALK2 transfection to completely reverse the effects of the miR-130a mimic could be because ALK2 levels were not fully restored by transfection. Alternatively, these data could suggest that miR-130a may target additional components of the BMP-SMAD pathway (see Table 3).
miR-130a was previously reported to bind to two predicted sites in the SMAD4 3′UTR to inhibit SMAD4 protein (but not mRNA) expression in HEK293 and other cell lines (19). In contrast, we did not see a decrease in SMAD4 protein in Hep3B cells transfected with miR-130a mimic. One explanation for the disparity could be the different biologic systems used, although we could not reproduce the previously published results in HEK293 cells. One limitation of the previous study was the use of an artificial SMAD4 3′UTR construct in which the two predicted miR-130a-binding sites that are located almost 4 kb apart in the native 3′UTR were cloned side-by-side (19). This contrasts with our data for ALK2, which used the full-length native 3′UTR. Of note, although we found that the miR-130a mimic inhibited 3′UTR activity of this SMAD4 side-by-side reporter construct, and the effects of the miR-130a mimic were reduced to some extent when both predicted binding sites were mutated, we did not see an effect from individual mutation of either the proximal or distal sites. Moreover, the miR-130a mimic had no effect on the ∼450-bp fragment SMAD4 3′UTR reporter constructs composed of either the proximal or distal predicted binding sites cloned in isolation. Thus, the apparent inhibition of SMAD4 3′UTR activity by miR-130a mimic may be an artifact of the side-by-side construct. Taken together, our data do not confirm that SMAD4 is a target of miR-130a, at least in our biologic systems.
Consistent with our in vitro studies, we demonstrated that increased liver miR-130a levels in low iron diet mice were associated with decreased liver Alk2 mRNA levels. In contrast, neither Smad4 mRNA nor protein levels were decreased in low iron diet mice, consistent with one previous report (24). The low iron diet mice also exhibited decreased liver BMP-SMAD signaling (with reduced p-Smad1/5/8 protein and Id1 mRNA) and decreased Hamp mRNA levels, as described previously in other animal models of iron deficiency (8, 22–24). Proposed mechanisms for these findings have included reduced liver Bmp6 mRNA expression (24) and increased expression of matriptase-2 protein (23), which cleaves the BMP co-receptor hemojuvelin (28). Indeed, we also found reduced liver Bmp6 mRNA levels in our model. Our data suggest that down-regulation of ALK2 by miR-130a may also contribute to decreased BMP-SMAD signaling and hepcidin expression in the context of iron deficiency. Although the change in each BMP-SMAD pathway component may be modest on its own, the combined effects of inhibiting multiple upstream components of the BMP-SMAD signaling cascade would be expected to have a robust effect on downstream signaling and hepcidin expression (Fig. 7). We hypothesize that the functional significance of this would be to maximize ferroportin cell surface expression and improve iron availability. Future studies will be needed to confirm a functional role for miR-130a in regulating liver BMP-SMAD signaling and hepcidin expression under iron-deficient conditions in vivo.
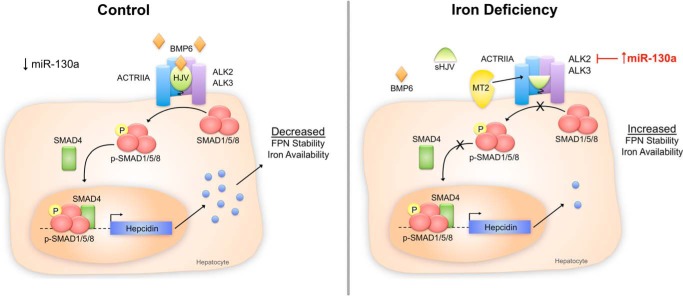
FIGURE 7.
Schematic diagram depicting the proposed role of miR-130a in BMP-SMAD signaling and hepcidin regulation under iron-deficient conditions. Under control conditions (left panel), BMP6 binds to the BMP type I receptor ALK2 or ALK3, the BMP type II receptor ACTRIIA, and the co-receptor HJV to induce phosphorylation of SMAD1/5/8 proteins, which complex with common mediator SMAD4 to increase transcription of hepcidin mRNA in hepatocytes. Hepcidin is secreted into the circulation to inhibit ferroportin (FPN) cell surface expression and decrease iron entry into the bloodstream. Under iron-deficient conditions (right panel), liver miR-130a expression is increased and targets the 3′UTR of ALK2 to inhibit expression. This acts in conjunction with a reduction in BMP6 mRNA expression and an increase in matriptase 2 (MTP2) protein expression (which is proposed to cleave HJV) to decrease signaling through the BMP-SMAD pathway, decrease hepcidin expression, increase ferroportin expression, and thereby increase iron availability.
We have previously demonstrated that BMP-SMAD pathway activators can ameliorate iron overload in an Hfe−/− mouse model of hemochromatosis (22) and that BMP-SMAD pathway inhibitors can improve iron availability and anemia in animal models of anemia of chronic disease (29). Our data raise the possibility that targeting miR-130a could represent a novel therapeutic strategy to regulate BMP-SMAD pathway signaling and hepcidin in iron homeostasis disorders. The therapeutic potential of miRNA modulators is exemplified by the fact that a miR-122 inhibitor is already in human clinical trials for treatment of hepatitis C (30). However, it is important to note that miR-130a expression is not specific to the liver (31), and miR-130a has many other reported targets (32–34). Therefore, miR-130a modulators may have other biologic effects. Interestingly, liver miR-130a was recently shown to be altered by hepatitis C (32, 33), and both miR-130a and the BMP/TGF-β superfamily have been suggested to play a role in viral replication (27, 32, 35). Whether the functional role of miR-130a as a BMP-SMAD pathway inhibitor has relevance to its role in hepatitis C is an interesting area for future investigation.
In summary, we have shown that a low iron diet up-regulates liver miR-130a in mice and that miR-130a targets ALK2 to inhibit BMP-SMAD signaling and hepcidin expression in liver cells. We propose that this pathway could contribute to the down-regulation of multiple BMP-SMAD pathway components to synergistically suppress hepcidin synthesis and thereby maximize iron availability in response to iron deficiency.
Acknowledgments
We thank Drs. Antonello Pietrangelo and Elena Corradini from University Hospital of Modena, Italy, for providing the liver tissue samples from mice on a low iron diet used for the miRNA microarray analysis. We thank Drs. Paul B. Yu from Brigham and Women's Hospital, Boston, MA, and Kohei Miyazono from University of Tokyo, Japan, for providing the ALK2 cDNA construct.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1-DK087727 (to J. L. B.) and Training Grant T32 DK007540 (to K. Z. B. and A. B. C.). This work was also supported by the William F. Milton Fund from Harvard University and a Howard Goodman fellowship award from the Massachusetts General Hospital (to J. L. B.). J. L. B. has ownership interest in the start-up company Ferrumax Pharmaceuticals, which has licensed technology from the Massachusetts General Hospital based on work cited here and in prior publications.
- BMP
- bone morphogenetic protein
- miRNA
- microRNA
- HJV
- hemojuvelin
- Hep-Luc
- hepcidin promoter firefly luciferase reporter
- BRE-Luc
- BMP responsive firefly luciferase reporter
- qRT-PCR
- reverse transcription and quantitative real time PCR
- HAMP
- hepcidin mRNA
- pri-miRNA
- primary transcript miRNA
- pre-miRNA
- precursor miRNA
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Ganz T., Nemeth E. (2012) Hepcidin and iron homeostasis. Biochim. Biophys. Acta 1823, 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Babitt J. L., Huang F. W., Wrighting D. M., Xia Y., Sidis Y., Samad T. A., Campagna J. A., Chung R. T., Schneyer A. L., Woolf C. J., Andrews N. C., Lin H. Y. (2006) Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat. Genet. 38, 531–539 [DOI] [PubMed] [Google Scholar]
- 3. Andriopoulos B., Jr., Corradini E., Xia Y., Faasse S. A., Chen S., Grgurevic L., Knutson M. D., Pietrangelo A., Vukicevic S., Lin H. Y., Babitt J. L. (2009) BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 41, 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meynard D., Kautz L., Darnaud V., Canonne-Hergaux F., Coppin H., Roth M. P. (2009) Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat. Genet. 41, 478–481 [DOI] [PubMed] [Google Scholar]
- 5. Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corradini E., Meynard D., Wu Q., Chen S., Ventura P., Pietrangelo A., Babitt J. L. (2011) Serum and liver iron differently regulate the bone morphogenetic protein 6 (BMP6)-SMAD signaling pathway in mice. Hepatology 54, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kautz L., Meynard D., Monnier A., Darnaud V., Bouvet R., Wang R. H., Deng C., Vaulont S., Mosser J., Coppin H., Roth M. P. (2008) Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood 112, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 8. Corradini E., Garuti C., Montosi G., Ventura P., Andriopoulos B., Jr., Lin H. Y., Pietrangelo A., Babitt J. L. (2009) Bone morphogenetic protein signaling is impaired in an HFE knockout mouse model of hemochromatosis. Gastroenterology 137, 1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steinbicker A. U., Bartnikas T. B., Lohmeyer L. K., Leyton P., Mayeur C., Kao S. M., Pappas A. E., Peterson R. T., Bloch D. B., Yu P. B., Fleming M. D., Bloch K. D. (2011) Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood 118, 4224–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xia Y., Babitt J. L., Sidis Y., Chung R. T., Lin H. Y. (2008) Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood 111, 5195–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sayed D., Abdellatif M. (2011) MicroRNAs in development and disease. Physiol. Rev. 91, 827–887 [DOI] [PubMed] [Google Scholar]
- 12. Castoldi M., Muckenthaler M. U. (2012) Regulation of iron homeostasis by microRNAs. Cell. Mol. Life Sci. 10.1007/s00018-012-1031-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castoldi M., Vujic Spasic M., Altamura S., Elmén J., Lindow M., Kiss J., Stolte J., Sparla R., D'Alessandro L. A., Klingmüller U., Fleming R. E., Longerich T., Gröne H. J., Benes V., Kauppinen S., Hentze M. W., Muckenthaler M. U. (2011) The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Invest. 121, 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faller M., Matsunaga M., Yin S., Loo J. A., Guo F. (2007) Heme is involved in microRNA processing. Nat. Struct. Mol. Biol. 14, 23–29 [DOI] [PubMed] [Google Scholar]
- 15. Li Y., Lin L., Li Z., Ye X., Xiong K., Aryal B., Xu Z., Paroo Z., Liu Q., He C., Jin P. (2012) Iron homeostasis regulates the activity of the microRNA pathway through poly(C)-binding protein 2. Cell Metab. 15, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Babitt J. L., Huang F. W., Xia Y., Sidis Y., Andrews N. C., Lin H. Y. (2007) Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J. Clin. Invest. 117, 1933–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meynard D., Vaja V., Sun C. C., Corradini E., Chen S., López-Otín C., Grgurevic L., Hong C. C., Stirnberg M., Gütschow M., Vukicevic S., Babitt J. L., Lin H. Y. (2011) Regulation of TMPRSS6 by BMP6 and iron in human cells and mice. Blood 118, 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang A. S., Xiong S., Tsukamoto H., Enns C. A. (2004) Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood 103, 1509–1514 [DOI] [PubMed] [Google Scholar]
- 19. Häger M., Pedersen C. C., Larsen M. T., Andersen M. K., Hother C., Grønbæk K., Jarmer H., Borregaard N., Cowland J. B. (2011) MicroRNA-130a-mediated down-regulation of Smad4 contributes to reduced sensitivity to TGF-β1 stimulation in granulocytic precursors. Blood 118, 6649–6659 [DOI] [PubMed] [Google Scholar]
- 20. Korchynskyi O., ten Dijke P. (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277, 4883–4891 [DOI] [PubMed] [Google Scholar]
- 21. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corradini E., Schmidt P. J., Meynard D., Garuti C., Montosi G., Chen S., Vukicevic S., Pietrangelo A., Lin H. Y., Babitt J. L. (2010) BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice. Gastroenterology 139, 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang A. S., Anderson S. A., Wang J., Yang F., DeMaster K., Ahmed R., Nizzi C. P., Eisenstein R. S., Tsukamoto H., Enns C. A. (2011) Suppression of hepatic hepcidin expression in response to acute iron deprivation is associated with an increase of matriptase-2 protein. Blood 117, 1687–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Theurl I., Schroll A., Nairz M., Seifert M., Theurl M., Sonnweber T., Kulaksiz H., Weiss G. (2011) Pathways for the regulation of hepcidin expression in anemia of chronic disease and iron deficiency anemia in vivo. Haematologica 96, 1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mole D. R. (2010) Iron homeostasis and its interaction with prolyl hydroxylases. Antioxid. Redox Signal. 12, 445–458 [DOI] [PubMed] [Google Scholar]
- 26. Saito K., Kondo E., Matsushita M. (2011) MicroRNA 130 family regulates the hypoxia response signal through the P-body protein DDX6. Nucleic Acids Res. 39, 6086–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakamoto N., Yoshimura M., Kimura T., Toyama K., Sekine-Osajima Y., Watanabe M., Muramatsu M. (2007) Bone morphogenetic protein-7 and interferon-α synergistically suppress hepatitis C virus replicon. Biochem. Biophys. Res. Commun. 357, 467–473 [DOI] [PubMed] [Google Scholar]
- 28. Silvestri L., Pagani A., Nai A., De Domenico I., Kaplan J., Camaschella C. (2008) The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 8, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Theurl I., Schroll A., Sonnweber T., Nairz M., Theurl M., Willenbacher W., Eller K., Wolf D., Seifert M., Sun C. C., Babitt J. L., Hong C. C., Menhall T., Gearing P., Lin H. Y., Weiss G. (2011) Pharmacologic inhibition of hepcidin expression reverses anemia of chronic inflammation in rats. Blood 118, 4977–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janssen H. L., Reesink H. W., Lawitz E. J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A. J., Patick A. K., Chen A., Zhou Y., Persson R., King B. D., Kauppinen S., Levin A. A., Hodges M. R. (2013) Treatment of HCV infection by targeting MicroRNA. N. Engl. J. Med. 368, 1685–1694 [DOI] [PubMed] [Google Scholar]
- 31. Liang Y., Ridzon D., Wong L., Chen C. (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhanja Chowdhury J., Shrivastava S., Steele R., Di Bisceglie A. M., Ray R., Ray R. B. (2012) Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J. Virol. 86, 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu N., Shen C., Luo Y., Xia L., Xue F., Xia Q., Zhang J. (2012) Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem. Biophys. Res. Commun. 425, 468–472 [DOI] [PubMed] [Google Scholar]
- 34. Boll K., Reiche K., Kasack K., Mörbt N., Kretzschmar A. K., Tomm J. M., Verhaegh G., Schalken J., von Bergen M., Horn F., Hackermüller J. (2013) miR-130a, miR-203, and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 32, 277–285 [DOI] [PubMed] [Google Scholar]
- 35. Zhang X., Daucher M., Armistead D., Russell R., Kottilil S. (2013) MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS One 8, e55733. [DOI] [PMC free article] [PubMed] [Google Scholar]