Abstract
Transformation of a type I SCCmec element into Staphylococcus aureus yielded highly oxacillin-resistant transformants with a reduced growth rate. Faster-growing variants could again be selected at the cost of reduced resistance levels, demonstrating an inverse correlation between oxacillin resistance levels and growth rate.
The development of antibiotic resistance in bacteria through either the acquisition of resistance elements or mutation often occurs at the cost of reduced fitness and may result in a decreased bacterial growth rate (1). Evolution in the natural or the clinical environment usually selects for fitter variants, which compensate for the cost of resistance through the development of secondary mutations or the loss of the resistance (10).
Methicillin resistance in methicillin-resistant Staphylococcus aureus (MRSA) is mediated by the acquisition of the SCCmec element, which integrates in a site-specific manner into the staphylococcal genome (7). Besides the mecA gene, which codes for a penicillin-binding protein (PBP) with a low affinity to β-lactams, it harbors a variable set of genes unrelated to methicillin resistance and serves as an integration site for various other resistance determinants, transposons, and plasmids. Establishment of the SCCmec element in staphylococci theoretically encounters two main obstacles: one is the cost of maintenance of a relatively large additional element, of which only the mecA gene is essential for resistance; the second is the accommodation of the new PBP 2a into the staphylococcal cell wall synthesis complex. Initially, when PBP 2a enters a naïve S. aureus strain and in the absence of β-lactam pressure, PBP 2a is not beneficial and the cells select against its production (8). The ability of S. aureus to accommodate SCCmec and/or to functionally integrate PBP 2a differs from strain to strain, resulting in a wide range of resistance levels (3). Irrespective of their original oxacillin resistance levels, MRSA strains are resistant to all β-lactam antibiotics due to their ability to segregate highly resistant variants. Multiple different mutations may lead to high levels of resistance (for reviews, see references 13 and 14), but few of them have been identified (4, 9).
Our goal was to measure the in vitro cost of SCCmec on fitness by monitoring changes in growth rates. We transformed naïve, susceptible strain BB255, a derivative of the widely used strain NCTC8325 (6), by the CaCl2 method (2) with DNA originating from an MRSA strain containing a type I SCCmec element. The transformants were selected on plates containing 4 μg of cefoxitin per ml. Resistance tests were performed according to the recommendations of the National Committee for Clinical Laboratory Standards (11). The transformants were highly oxacillin resistant (MICs, 512 μg/ml). The stability of the high-level-methicillin-resistance phenotype of representative strain RA120 was tested by dilution of an RA120 culture and subsequent regeneration from single cells in nonselective medium, which demonstrated that the high-level-resistance phenotype was not an induction phenomenon resulting from selection on cefoxitin.
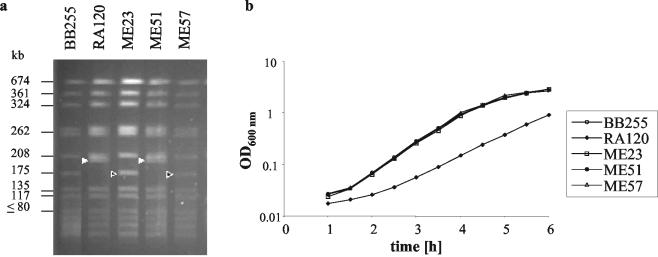
Interestingly, the transformants grew much slower than the susceptible parent, with a generation time of 40 ± 0.1 min compared to a generation time of 29 ± 0.1 min for strain BB255. Strain RA120 was cured of SCCmec by transient overexpression of ccrAB from plasmid pSR3-1, which induces precise, site-specific excision of SCCmec, as described by Ito et al. (5). The resulting susceptible strain, strain ME23, regained the doubling time and the chromosomal SmaI pulsed-field gel electrophoresis (PFGE) pattern of wild-type strain BB255 (Fig. 1), demonstrating the excision of SCCmec. This decrease in the growth rate after the introduction of SCCmec and the subsequent increase in the growth rate upon curing confirmed that SCCmec and/or the resulting high oxacillin resistance level was the cause of the decreased growth rate and, thus, the loss of fitness in vitro.
FIG. 1.
SmaI restriction patterns of the strains used and their corresponding growth curves. The SmaI fragments carrying SCCmec are indicated by filled triangles, and the corresponding fragments of the cured strain are indicated by open triangles. BB255, naïve, susceptible recipient; RA120, BB255 transformant containing SCCmec; ME23, RA120 cured of SCCmec; ME51, rapidly growing strain derived from RA120; ME57, ME51 cured of SCCmec.
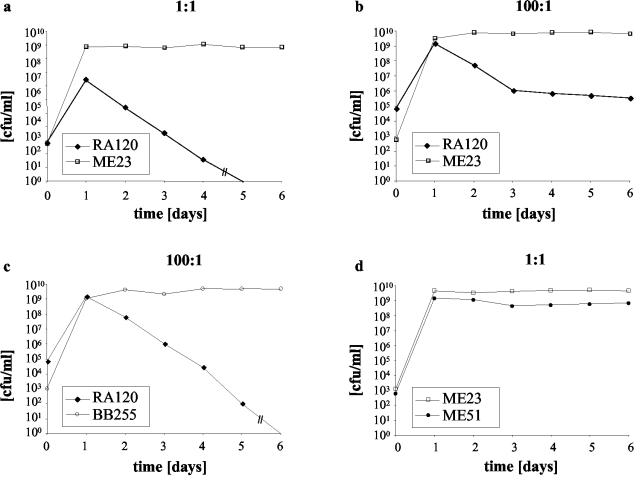
Mixed growth competition assays were performed between strain RA120 and susceptible strain ME23 by inoculating 104 CFU of each strain into 10 ml of Luria-Bertani broth in the absence of antibiotic pressure. Where indicated, the ratio of RA120 to its competitor was raised to 100:1 by increasing the RA120 inoculum to 106 CFU. Every 24 h the mixed culture was diluted by a factor of 104 with fresh broth, and the number of CFU of the susceptible strain per milliliter versus that of the resistant strain was determined by plating aliquots on nonselective plates and on plates containing 1 μg of oxacillin per ml and calculating the difference in the number of CFU. At an initial ratio of RA120 to ME23 of 1:1, the RA120 population was lost at a rate of 2 log10 CFU per day (Fig. 2a). Increasing the ratio of RA120 to ME23 to 100:1 allowed the faster-growing variants, represented by strain ME51, with a doubling time of 29 ± 0.1 min, to emerge (Fig. 2b) and to compete successfully with ME23 (Fig. 2d). The doubling time of this faster-growing variant, ME51, remained constant when it was retested 10 days later, and the strain maintained the same restriction pattern as RA120 (Fig. 1). Subculturing of RA120 alone under the same conditions did not yield faster-growing variants after the same number of days, indicating that faster-growing mutants were selected only in the presence of a competitor.
FIG. 2.
Growth competition. Survival of oxacillin-resistant strains versus that of oxacillin-susceptible strains in mixed culture after daily subculturing. (a) Strain RA120 with its cured derivative ME23 inoculated at a ratio of 1:1; (b) strain RA120 with ME23 inoculated at a ratio 100:1; (c) strain RA120 with BB255 inoculated at a ratio of 100:1; (d) strain ME51 with ME23 inoculated at a ratio 1:1.
When ME51 was cured of SCCmec, the resulting strain, strain ME57, was indistinguishable by its growth rate and PFGE pattern (Fig. 1) from BB255 and ME23. The results of competition experiments with RA120 and ME57 were identical to those of competition experiments with RA120 and ME23 (data not shown). However, when RA120 was grown in competition with naïve strain BB255 (Fig. 2c), it was more rapidly eliminated than it was in competition experiments with ME23 or ME57. This suggests that ME23 and ME57 were not as competitive against RA120 as BB255 was; this is possibly due to the acquisition of a chromosomal mutation, as has previously been postulated to occur in highly resistant MRSA strains (14). However, the introduction of an accidental mutation upon transformation cannot be ruled out.
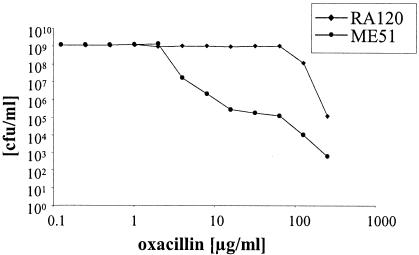
An interesting observation was that strain ME51, as well as 24 of 24 other fast-growing oxacillin-resistant single colonies analyzed from the competition of strains RA120 and ME23, all had reduced levels of oxacillin resistance compared to that of RA120. However, at this stage we do not know whether the fast-growing population comprised a single clone or whether several events resulted in faster growth. The population analysis profile revealed that RA120 had changed from a homogeneously, highly resistant MRSA strain into a heterogeneously resistant MRSA strain (Fig. 3) for which the oxacillin MIC was 64 μg/ml. Highly resistant subclones of ME51, isolated and purified from plates containing 128 or 256 mg of oxacillin per ml, again showed a reduced growth rate, with a doubling time of 39.5 ± 2.7 min. Analogous transformations into BB255 were done with other SCCmec type I or type IV elements (data not shown), and analysis of the generation time confirmed a correlation between oxacillin resistance levels and growth rate.
FIG. 3.
Population analysis profiles. The numbers of colonies that formed after 48 h in the presence of different concentrations of oxacillin after overnight cultures of strain RA120 or ME51 were plated on increasing concentrations of oxacillin are indicated.
The cost of SCCmec may be compensated for in nature. This is reflected by the rapid rate of growth of community-acquired MRSA strains upon which pressures other than antibiotics may act and which, interestingly, have generally been found to exhibit lower oxacillin resistance levels (12). We cannot exclude the possibility that the experimentally demonstrated interrelationship between oxacillin resistance levels and growth rate may also be compensated for in clinical isolates. Oxacillin resistance levels appear to have a higher impact on the growth rate than the addition of the extra DNA comprising the SCCmec element.
Acknowledgments
We thank T. Ito for plasmid pSR3-1.
This work was supported by the Swiss National Science Foundation grants NRP 49-063201 and 31-63552.00. R. P. Adhikari was supported by a grant from the Paul Ehrlich Gesellschaft, Bonn, Germany.
REFERENCES
- 1.Andersson, D. I., and R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bächi, B., and M. L. Kohler. 1983. A novel site on the chromosome of Staphylococcus aureus influencing the level of methicillin resistance: genetic mapping. FEMS Microbiol. Lett. 20:305-309. [Google Scholar]
- 3.Figueiredo, A. M., E. Ha, B. N. Kreiswirth, H. de Lencastre, G. J. Noel, L. Senterfit, and A. Tomasz. 1991. In vivo stability of heterogeneous expression classes in clinical isolates of methicillin-resistant staphylococci. J. Infect. Dis. 164:883-887. [DOI] [PubMed] [Google Scholar]
- 4.Fujimura, T., and K. Murakami. 1997. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J. Bacteriol. 179:6294-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of the three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenni, R., and B. Berger-Bächi. 1998. Teichoic acid content in different lineages of Staphylococcus aureus NCTC8325. Arch. Microbiol. 170:171-178. [DOI] [PubMed] [Google Scholar]
- 7.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama, Y., H. Z. Zhang, D. Hong, and H. F. Chambers. 2003. Jumping the barrier to beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 185:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo, N., K. Kuwahara-Arai, H. Kuroda-Murakami, E. Tateda-Suzuki, and K. Hiramatsu. 2001. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob. Agents Chemother. 45:815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagaev, I., J. Björkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 6th ed. NCCLS document M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Okuma, K., K. Iwakawa, J. D. Turnidge, W. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohrer, S., and B. Berger-Bächi. 2003. What makes resistance to methicillin heterogeneous? J. Med. Microbiol. 52:605-607. [DOI] [PubMed] [Google Scholar]
- 14.Ryffel, C., A. Strässle, F. H. Kayser, and B. Berger-Bächi. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:724-728. [DOI] [PMC free article] [PubMed] [Google Scholar]