Background: Transcriptional co-activators YAP/TAZ are pivotal effectors of the Hippo pathway and their dysfunction promote epithelial-to-mesenchymal transition (EMT) and malignant transformation.
Results: PTPN14 interacts with Kibra and activates LATS1 (upstream negative regulator of YAP).
Conclusion: PTPN14 and Kibra activate LATS1 and negatively regulate the YAP oncogenic function.
Significance: Study of the YAP regulatory mechanism is crucial for understanding its role in the physiological and pathological processes.
Keywords: Cancer, Cell Migration, Epithelial-Mesenchymal Transition (EMT), Protein Phosphatase, Protein-Protein Interaction
Abstract
The Hippo signaling pathway regulates cellular proliferation and survival, thus exerting profound effects on normal cell fate and tumorigenesis. Pivotal effectors of this pathway are YAP/TAZ, transcriptional co-activators whose dysfunction contributes to epithelial-to-mesenchymal transition and malignant transformation. Therefore, it is of great importance to decipher the mechanisms underlying the regulations of YAP/TAZ at various levels. Here we report that non-receptor tyrosine phosphatase 14 (PTPN14) interacts with the Kibra protein. The interaction between PTPN14 and Kibra is through the PPXY domain of PTPN14 and WW domain of Kibra. PTPN14 and Kibra can induce the LATS1 activation independently and cooperatively. Interestingly, activation of LATS1 by PTPN14 is dependent on the C terminus of PTPN14 and independent of the upstream mammalian STE20-like kinase (MST) proteins. Furthermore, we demonstrate that PTPN14 increases the LAST1 protein stability. Last, overexpression of Kibra rescues the increased cell migration and aberrant three-dimensional morphogenesis induced by knockdown of PTPN14, and this rescue is mediated through the activation of the upstream LATS1 kinase and subsequent cytoplasmic sequestration of YAP. In summary, our results indicate a potential regulatory role of PTPN14 in the Hippo pathway and demonstrate another layer of regulation in the YAP oncogenic function.
Introduction
In recent years, many have found that the evolutionarily conserved kinase cascade, known as the Hippo pathway, plays an important role in a wide range of cellular processes (1–5). Originally discovered through genetic screens in Drosophila melanogaster (6, 7), Hippo signaling today goes far beyond those original connections to organ size determination and growth control regulation. Indeed, it has implications in many other areas such as cell cycle progression, apoptosis, stem cell renewal, and differentiation as well as mechanotransduction (8, 9).
The Hippo signaling cascade leads to the phosphorylation and eventual cytoplasmic sequestration of its two effector molecules, YAP and TAZ, thus preventing their co-transcriptional activities. Indeed, dysregulation of the Hippo pathway has been reported to play an important role in the acquisition of certain diseases, including cancer. For example, defective upstream inhibition will allow YAP/TAZ to translocate into the nucleus, where they can bind to a number of transcription factors, most notably the TEA domain (TEAD) family of transcription factors, and activate various genes to drive cancer progression through proliferative and/or antiapoptotic effects (10, 11). In mammals, the main proteins that are known to directly phosphorylate YAP/TAZ are LATS1/2 (12–14).
We and others have recently identified the non-receptor tyrosine phosphatase, PTPN14,3 as a negative regulator of YAP activity through their direct interaction (15–18). We thought it was of interest to delineate other functions that PTPN14 may have in the Hippo pathway. As a well regulated pathway, multiple upstream effectors influence the Hippo signaling; among these is the WW domain-containing protein, Kibra (19, 20). Kibra has been shown to exert its regulation by forming a complex with two other upstream components, Merlin and Expanded (21–23). In Drosophila, Kibra was found to be able to interact with Pez, the PTPN14 homolog (24). In light of the recent findings, we decided to examine whether mammalian PTPN14 interacts with Kibra and whether they play any cooperative role together in regulating the YAP/TAZ oncogenic functions. We demonstrate here PTPN14 activates LATS1 independently or cooperatively with Kibra. Our data reveal a new negative regulatory mechanism of PTPN14 in the Hippo pathway.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
MCF10A cell culture was performed as described previously (25, 26). HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 50 units/ml penicillin/streptomycin and incubated at 37 °C with 5% CO2.
For knockdown experiments, shRNA hairpins targeting human PTPN14 were obtained from the RNAi Consortium (The Broad Institute, Boston, MA). The target sequences are listed (in the 5′-3′ direction) as follows: shPTPN14-1, GCGGTAATATACAGGTGGAAT; shPTPN14-2, CGCTCAGTACAAGTTTGTCTA; Control-shRNA, CAACAAGATGAAGAGCACCAA. Lentivirus packaging, transient transfection of 293T cells, MCF10A cell transduction, and drug selection were performed following standard protocols and were described previously (26). SMARTpool ON-TARGETplus MST1 and MST2 siRNAs targeting human MST1 and MST2 were purchased from GE Healthcare Dharmacon Inc.
Plasmid Constructs
The human HA-tagged LATS1, V5-tagged PTPN14, and their mutant expression constructs were described previously (12, 15). The human FLAG-tagged LATS1 was a gift from Dr. Xiaolong Yang at the Queen's University, Kingston, Ontario, Canada. The human Kibra ORF was cloned into pcDNA3 by EcoRI-BamHI digestion, and Kibra mutant constructs were established by PCR-based mutagenesis and confirmed by DNA sequencing.
Antibodies and Molecular Biology Analyses
Kibra, LATS1, LATS1 (phospho-Ser909), LATS1 (phospho-Thr1079), MST1, MST2, and YAP (phospho-Ser127) antibodies were purchased from Cell Signaling Technology (Beverly, MA), YAP and PTPN14 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), β-actin antibody was from Upstate Biotech Millipore (Lake Placid, NY), V5 and HA were from Life Technologies, and FLAG (M2) antibodies were from Sigma. For protein extraction, cells were washed with phosphate-buffered saline and collected with immunoprecipitation buffer: 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 20% glycerol, 0.5% NP-40, plus 1× protease inhibitor mixture (cOmpleteTM EDTA-free, Roche Diagnostics). Cell lysate was cleared by centrifugation at 14,000 rpm for 20 min at 4 °C. Lysate was loaded onto 4–15% MINI-PROTEAN TGX gel (Bio-Rad) with 4× SDS sample buffer. For immunoblot, proteins were transferred onto Immobilon-P membrane (Millipore, Billerica, MA), detected by various antibodies, and visualized with ECL Plus Western blotting detection reagents (GE Healthcare).
Cell Migration
Transwell cell migration assay was performed as described previously (15).
Three-dimensional Morphogenesis
MCF10A three-dimensional morphogenesis assay was performed as described previously (25, 26). Briefly, 4 × 103 MCF10A cells co-infected with Kibra and sh-Control or sh-PTPN14 lentivirus were plated into three-dimensional culture plates and cultured in growth factor-reduced reconstituted basement membrane (Matrigel; BD Biosciences). Growth medium was changed every 4 days. Images were taken at day 8. Assays were done in three independent experiments.
Immunofluorescence Microscopy
MCF10A cells were cultured on coverslips to appropriate density. Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 for 15 min. After blocking in 3% BSA for 30 min, slides were incubated with the YAP antibody diluted in 1% BSA for 1 h. After washing with PBS, slides were incubated with Alexa Fluor 488-conjugated secondary antibodies (1:1000 dilution) for 1 h.
Statistical Analysis
Statistical analysis of data were performed using the SPSS statistics software package (SPSS). All results are expressed as mean ± S.D. *, p < 0.05; **, p < 0.001; ***, p < 0.0001.
RESULTS
PTPN14 Interacts with Kibra
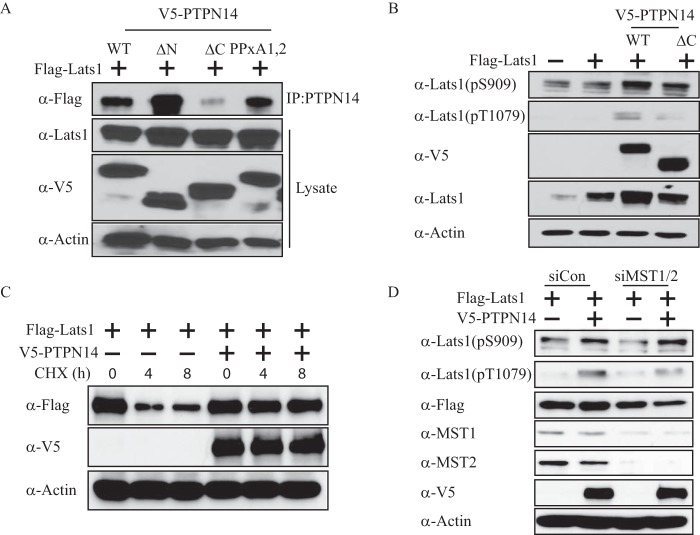
To test whether PTPN14 interacts with Kibra in mammalian cells and to identify the domain(s) critical for the interaction, we made several mutant constructs of each protein. Wild-type PTPN14 contains an N-terminal FERM (4.1, ezrin, radixin, moesin) domain, a C-terminal protein-tyrosine phosphatases (PTP) domain, and two PPXY motifs (Fig. 1A). Mutants were then created by either deleting the N or C terminus or introducing mutations at both PPXY motifs. Kibra also has a few conserved structural elements, e.g. the two WW domains that are also present in YAP and several other Hippo pathway components and have been shown to interact with the PPXY motifs. Likewise, mutations were also introduced into the Kibra protein, with either one or both WW domains deleted.
FIGURE 1.
PTPN14 interacts with Kibra. A, schematic representations of PTPN14 and Kibra, along with mutant constructs that were generated. B, PTPN14 interacts with Kibra through its C terminus and PPXY motifs. Wild-type Kibra was transfected into HEK293T cells, along with wild-type, C terminus-deleted, N terminus-deleted, or PPXY motif mutant V5-PTPN14. PTPN14 was detected by immunoblot after Kibra immunoprecipitation (IP:Kibra). C, Kibra interacts with PTPN14 through WW domains. Wild-type PTPN14 was co-transfected with wild-type Kibra, or Kibra mutants deleted of WW1, WW2, or both WW domains. Kibra was detected by immunoblot after PTPN14 immunoprecipitation (IP:PTPN14).
Using these constructs, we did co-transfection of Kibra and PTPN14 and performed co-immunoprecipitation to evaluate the interaction between these two proteins. Wild-type PTPN14 and Kibra interacted readily (Fig. 1, B and C). Furthermore, either the ΔC or the PPXY mutation of PTPN14 was able to interrupt this interaction (Fig. 1B), suggesting that both the PTP domain and the PPXY motifs of PTPN14 are involved in the interaction with Kibra. As for the domains on Kibra, no change in binding ability was seen when either WW1 or WW2 was deleted. However, interaction with PTPN14 was completely lost when both WW domains were deleted (Fig. 1C). These results indicated that PTPN14 interacts with Kibra through its C-terminal and PPXY domains, whereas Kibra interacts with PTPN14 through at least one WW domain.
LATS1 Interacts with Both Kibra and PTPN14
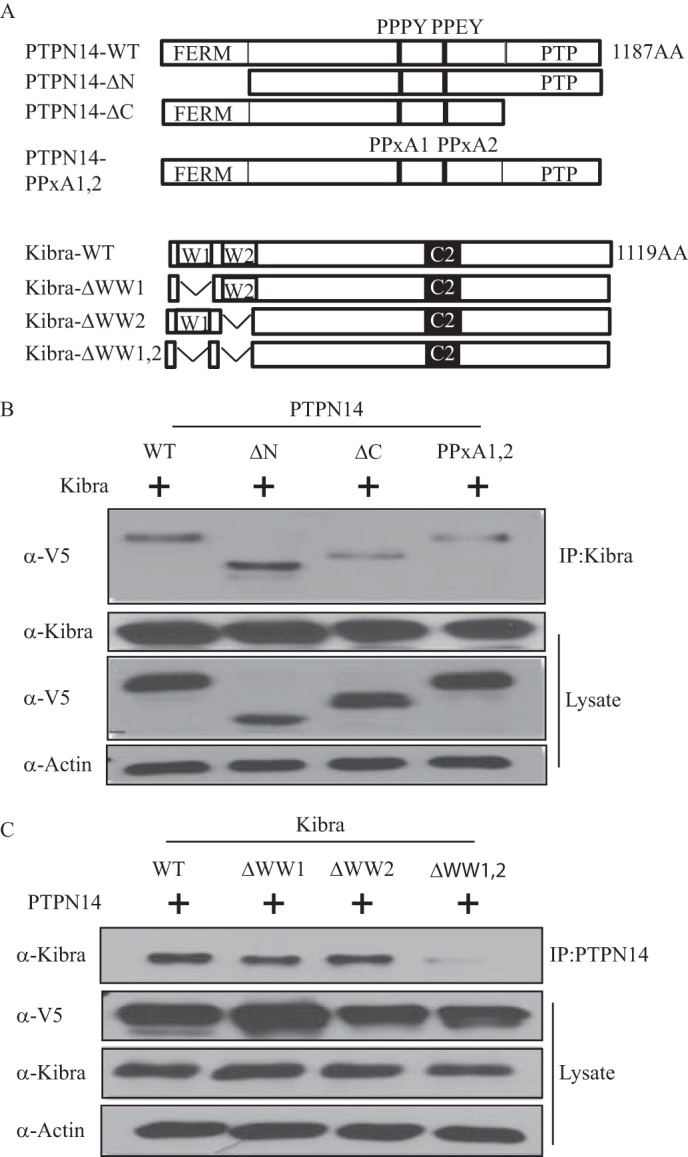
It has been reported that Kibra associates with and activates LATS1/2 by stimulating their phosphorylation on the hydrophobic motif (20). The interaction between Kibra and PTPN14 promoted us to test whether these three proteins could form a protein-protein interaction complex. As a result, when combinations of LATS1, PTPN14, and Kibra were co-transfected into HEK293T cells, PTPN14 was also pulled down after immunoprecipitation with the LATS1 antibody (Fig. 2A). The presence of Kibra along with LATS1 and PTPN14 did not affect the strength of interaction, indicating no competition between them. Similarly, Kibra was also detected with the immunoprecipitation of LATS1 (Fig. 2B). Again, no competition was observed as the level of pulled down Kibra protein was not affected by the presence or absence of PTPN14. To further test whether PTPN14, Kibra, and LATS1 form a complex, we performed immunoprecipitation using the Kibra or PTPN14 antibody after co-transfection of Kibra, PTPN14, and LATS1. As a result, we detected PTPN14 and LATS1 in the Kibra precipitates and likewise Kibra and LATS1 in the PTPN14 precipitate (Fig. 2C). To confirm these results at the endogenous level, we performed immunoprecipitation using the anti-PTPN14 antibody or the control IgG in the CAL120 breast cancer cells. It was found that Kibra and LATS1 proteins were readily detected in the PTPN14 precipitate (Fig. 2D). Taken together, we have proven that PTPN14 interacts with Kibra and LATS1 both exogenously and endogenously.
FIGURE 2.
LATS1 interacts with both Kibra and PTPN14. A, PTPN14 interacts with LATS1 independently of Kibra. HEK293T cells were transfected with vector control, LATS1 alone, or LATS1 plus Kibra, PTPN14, or both. LATS1 was immunoprecipitated (IP:LATS1), and the co-immunoprecipitated PTPN14 was detected by immunoblot using anti-V5 antibody. Kibra expression was examined using anti-Kibra antibody, and LATS1 expression was examined using anti-HA antibody. B, Kibra interacts with LATS1 independently of PTPN14. HEK293T cells were transfected with vector control, LATS1 plus PTPN14, Kibra, or both. LATS1 was immunoprecipitated, and the co-immunoprecipitated Kibra was detected by immunoblot using anti-Kibra antibody. C, Kibra, PTPN14, and LATS1 form a complex. HEK293T cells were transfected with Kibra, LATS1, and PTPN14. Kibra or PTPN14 was immunoprecipitated, and the co-immunoprecipitated LATS1 and PTPN14 were detected in Kibra precipitates or LATS1 and Kibra were detected in PTPN14 precipitates. D, PTPN14 interacts with Kibra and LATS1 endogenously. In CAL120 breast cancer cells, endogenous PTPN14 was immunoprecipitated, and the endogenous co-immunoprecipitated LATS1 and Kibra were detected by immunoblot.
PTPN14 and Kibra Activate LATS1 Cooperatively
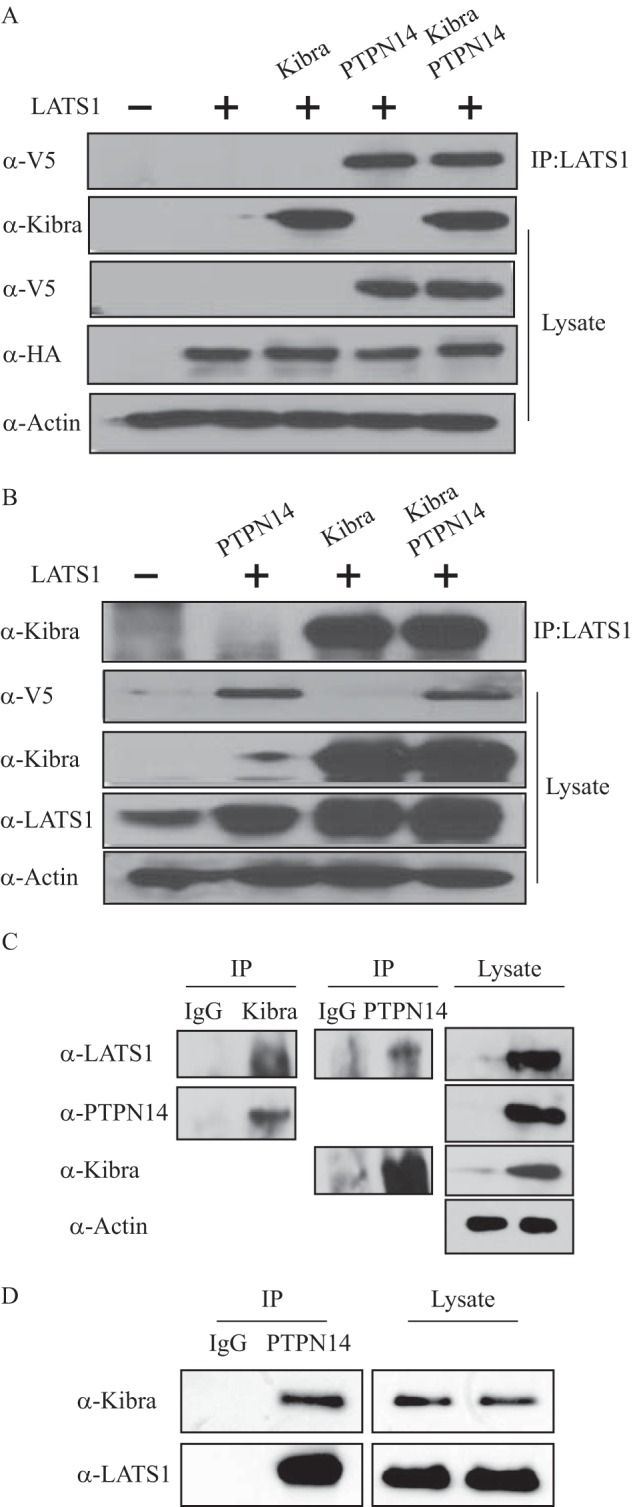
To further characterize the Kibra and LATS1 interaction, we investigated the important domains involved in their interaction. It was found that Kibra deleted of either WW domain still produced an interaction with LATS1 as strong as the wild-type Kibra. However, deletion of both Kibra WW domains largely obliterated their interactions, indicating that both WW domains are essential for the interaction between Kibra and LATS1 (Fig. 3A). LATS1/2 have been known to be serine/threonine protein kinases. Phosphorylation of an autophosphorylation site (Ser909 for LATS1 and Ser872 for LATS2) is required for their kinase activity (13). As mentioned above, Kibra associates with and activates LATS1/2 by stimulating their phosphorylation at the hydrophobic motif (20). We then first set out to examine whether PTPN14 activates LATS1 in a Kibra-dependent or -independent manner. We co-transfected the LATS1 and PTPN14 constructs with the wild-type or ΔWW-mutant Kibra into HEK293T cells. Indeed, we found that PTPN14 activated LATS1 and increased the YAP-Ser127 phosphorylation even in the presence of ΔWW-mutant Kibra (Fig. 3B), indicating that activation of LATS1 by PTPN14 and subsequent phosphorylation of YAP-Ser127 can occur in a Kibra-independent manner.
FIGURE 3.
PTPN14 and Kibra activate LATS1. A, Kibra interacts with LATS1 through its WW domains. LATS1 and Kibra mutants with deleted WW1, WW2, or both domains were co-transfected into HEK293T cells. Kibra was immunoprecipitated (IP:Kibra), and the co-immunoprecipitated LATS1 was examined by immunoblot. B, PTPN14 activates LATS1 independently of Kibra. HEK293T cells were co-transfected with control vectors, with LATS1 and PTPN14, or with LATS1 and PTPN14 plus wild-type or mutant Kibra missing two WW domains. LATS1 activation (p-Ser909 (pS909)) and YAP phosphorylation (p-Ser127 (pS127)) were detected by immunoblot. C, PTPN14 and Kibra cooperatively activate LATS1. HEK293T cells were transfected with vector control, LATS1 plus PTPN14, Kibra, or both. Activation of LATS1 (p-Ser909 (pS909)) was examined by immunoblot.
Next, to test whether the PTPN14 and Kibra interaction can also enhance LATS1 activation, LATS1 was co-expressed with either PTPN14 or Kibra or both (Fig. 3C). Intriguingly, we observed an increase in the level of phospho-LATS1 (Ser909) when either PTPN14 or Kibra was co-transfected with LATS1. Furthermore, when both PTPN14 and Kibra were present, LATS1 activation was further increased. Taken together, these results suggest that PTPN14 and Kibra can work independently, but can also cooperatively or synergistically activate LATS1.
Activation of LATS1 by PTPN14 Is Dependent on the C Terminus of PTPN14 and Independent of the MST1/2 Proteins
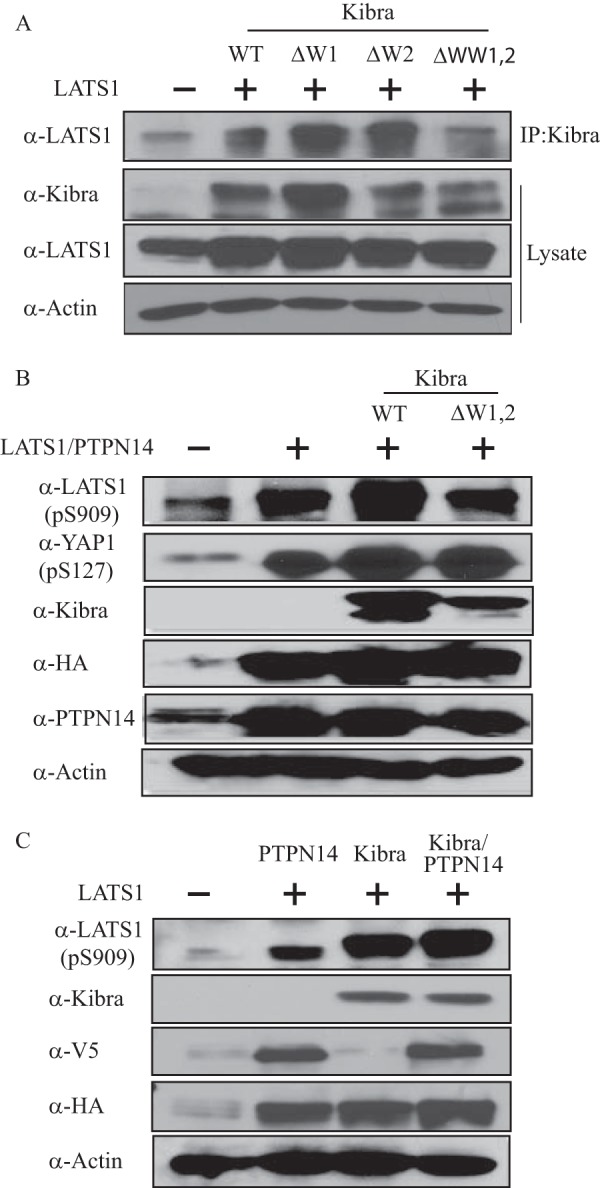
To understand the mechanism of the activation of LATS1 by PTPN14, we first investigated the critical domains involved in their interaction. It was found that deletion of the N terminus of PTPN14 or mutations of the PPXY domain had no effect on its interaction with LATS1 (Fig. 4A). However, deletion of the C terminus of PTPN14 largely obliterated their interaction (Fig. 4A). Consistently, loss of the PTPN14 C terminus could not activate LATS1 (Fig. 4B). On the other hand, we had previously shown that overexpression of PTPN14 increases the protein level of LATS1 (15). To test whether PTPN14 prevents LATS1 from degradation, we measured the LATS1 half-life in the presence or absence of overexpressed PTPN14. To do this, we first co-transfected LATS1 as well as control vector or PTPN14 into cells and treated the cells 48 h later with cycloheximide to inhibit protein synthesis. Of note, in the presence of overexpressed PTPN14, the half-life of transfected FLAG-tagged LATS1 was significantly prolonged (Fig. 4C), indicating that PTPN14 stabilizes LATS1 at the protein level.
FIGURE 4.
The C terminus of PTPN14 is important for LATS1 activation. A, PTPN14 interacts with LATS1 through its C-terminal motifs. Wild-type LATS1 was transfected into HEK293T cells, along with wild-type, C terminus-deleted, N terminus-deleted, or PPXY motif mutant V5-PTPN14. LATS1 was detected by immunoblot after PTPN14 immunoprecipitation (IP:PTPN14). B, activation of LATS1 by PTPN14 depends on the PTPN14 C terminus. Control vector or wild-type LATS1 was transfected into HEK293T cells, along with vector, wild-type, or C terminus-deleted V5-PTPN14. LATS1 (p-Ser909 (pS909)) and LATS1 (p-Thr-1079 (pT1079)) were detected by immunoblot. C, PTPN14 lengthens the LATS1 half-life. Wild-type LATS1 was transfected into HEK293T cells, along with vector or wild-type V5-PTPN14. Forty-eight hours after transfection, cells were treated with 20 μg/ml cycloheximide (CHX) at different time points as indicated. The LATS1 protein level was determined by immunoblot. D, activation of LATS1 by PTPN14 is independent of the MST proteins. Wild-type LATS1 and wild-type PTPN14 were co-transfected into HEK293T cells, along with siControl (siCon) or siMST1/2. LATS1 (p-Ser909 (pS909)) and LATS1 (p-Thr-1079 (pT1079)) were detected by immunoblot.
LATS1 protein contains two conserved key regulatory phosphorylation sites, i.e. Ser909 and Thr1079, which are essential for the LATS1 kinase activity (27). In addition, the upstream MST proteins are important for LATS1 Thr1079 phosphorylation (28, 29). To our interest, we detected LATS1 activation under the condition of MST1/2 knockdown, and siMST1/2 had no effect on the PTPN14-induced LATS1 phosphorylation (Fig. 4D). Taken together, these data indicate that the activation of LATS1 through PTPN14 is independent of the upstream MST proteins, but dependent on the C terminus of PTPN14.
Kibra Rescues the Malignant Phenotypes Induced by Knockdown of PTPN14
We and others have previously shown that PTPN14 directly interacts with YAP and negatively regulates its target genes and oncogenic functions (15–18). To test whether Kibra has any effect on the oncogenic phenotype induced by the loss-of-function of PTPN14, we used two independent shRNAs to knock down PTPN14 in human non-transformed MCF10A epithelial cells in the presence of overexpressed Kibra (Fig. 5A).
FIGURE 5.
Overexpression of Kibra rescues the malignant phenotypes induced by knockdown of PTPN14. A, effective knockdown of PTPN14 and overexpression of Kibra in MCF10A cells as revealed by immunoblot. B, overexpression of Kibra abolishes the activation of YAP targets, connective tissue growth factor (CTGF), and CYR61. Quantitative RT-PCR analysis of the YAP target genes in shPTPN14 or overexpressed Kibra/shPTPN14 MCF10A cells was performed. GAPDH was used as an internal control. Error bars equal ± S.D. (**, p < 0.001) C, overexpression of Kibra inhibits the cell migration induced by knockdown of PTPN14 in a cell migration assay. Mean number of migrated cells per field of four fields from each triplicate well of three independent experiments is shown. Error bars equal ± S.D. (***, p < 0.0001). D, overexpression of Kibra obliterates the aberrant acinar formation induced by knockdown of PTPN14 in three-dimensional culture. Representative phase contrast images from three independent experiments are shown.
We found that knockdown of PTPN14 induced significant expression of the YAP targets, such as connective tissue growth factor (CTGF) and CYR61. In addition, this induction was abolished by overexpressed Kibra (Fig. 5B). Furthermore, overexpression of Kibra significantly inhibited the cell migration induced by knockdown of PTPN14 (Fig. 5C).
The three-dimensional culture system allows epithelial cells to organize into structures resembling their in vivo architectures, making it a unique tool to study potential cancer genes and pathways in a biologically relevant context (30). Knockdown of PTPN14 has been reported to induce remarkably the aberrant three-dimensional acinar formation (17). To further test whether Kibra overexpression can rescue the aforementioned phenotype, we studied the MCF10A three-dimensional acinar formation in the presence of overexpressed Kibra. Interestingly, overexpression of Kibra completely rescued the shPTPN14-induced aberrant acinar formation (Fig. 5D). Overall, these results indicated that overexpression of Kibra rescues the malignant phenotypes induced by loss of function PTPN14.
We hypothesized that the above effects are due to the activation of LATS1/2 by Kibra, which leads to the inactivated YAP oncogenic function. To confirm this hypothesis, we measured the levels of inactivated YAP (p-Ser127) and activated LATS1 (p-Ser909) by immunoblot. As expected, we detected the decreased YAP (p-Ser127) and LATS1 (p-Ser909) in shPTPN14 cells (Fig. 6A). In contrast, overexpression of Kibra in these cells reversed the pattern (Fig. 6A). Using an immunostaining approach, we further demonstrated that Kibra also induced the YAP cytoplasmic sequestration (Fig. 6B). In summary, our data reinforce the function of Kibra as a tumor suppressor and suggest its crosstalk with PTPN14 in the Hippo signaling pathway (Fig. 6C).
FIGURE 6.
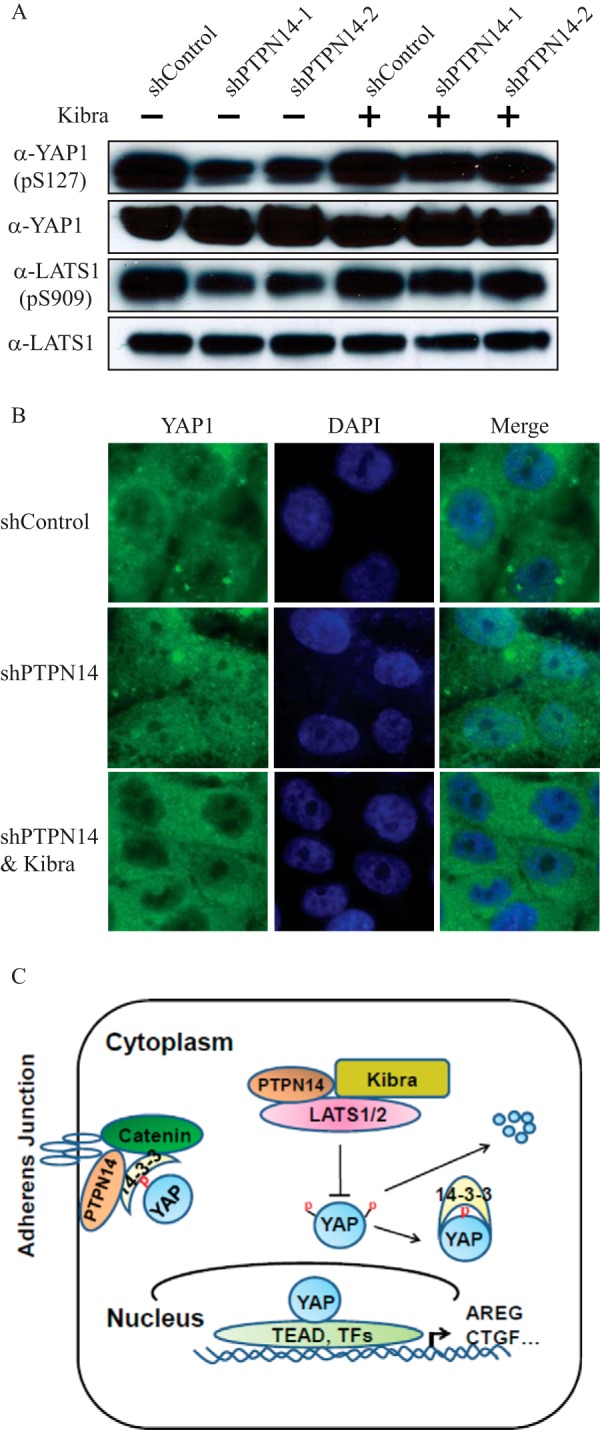
Overexpression of Kibra results in activation of LATS1 and thus inactivates YAP. A, overexpression of Kibra restored the level of activated LATS1 (p-Ser909 (pS909)) and inactivated YAP (p-Ser127 (pS127)) in shPTPN14 cells as revealed by immunoblot. B, overexpression of Kibra results in YAP nuclear exclusion. Immunofluorescence microscopy (magnification, ×63) shows YAP localization in the nucleus of shPTPN14 MCF10A cells versus its nuclear exclusion upon Kibra overexpression. C, a working model for the regulation of YAP by PTPN14. PTPN14 regulates the YAP oncogenic function through two layers: 1) PTPN14 directly binds to YAP, leading to YAP sequestration in the cytoplasm or at cell-cell junctions; and 2) PTPN14 itself or through a complex with Kibra activates LATS proteins and subsequently inactivates the YAP function.
DISCUSSION
We and others have recently demonstrated that PTPN14 negatively regulates the YAP oncogenic function through direct interaction and subsequent sequestration of YAP in the cytoplasm (15–18). Our present study reveals another potential mechanism underlying the regulatory roles of PTPN14 in the YAP oncogenic function. First, we showed that PTPN14 interacts with Kibra, through the PPXY and PTP motifs of PTPN14 as well as the WW domains of Kibra. Second, both PTPN14 and Kibra activate LATS1. Third, PTPN14 interacts with and activates LATS1 in a Kibra-independent manner, whereas both proteins can also increase the LATS1 activity cooperatively. Fourth, the activation of LATS1 by PTPN14 is independent of the upstream MST proteins, but dependent on the C terminus of PTPN14. Last, knockdown of PTPN14 induces the malignant phenotypes of increased cell migration and aberrant acinar formation in three-dimensional cultures, which can be rescued by overexpression of Kibra. Overall, our study demonstrates a novel regulatory mechanism of the Hippo pathway components.
PTPN14 is suggested to play roles in a wide variety of cell functions. Because we had previously identified it as a YAP-interacting protein that negatively regulates the YAP activity (15), it became of interest to explore its other interactions and abilities to influence the regulation of the Hippo pathway. PTPN14 (Pez) was first cloned in 1995 while a group was searching for PTPs that are expressed in normal breast tissue (31). It is a non-receptor tyrosine phosphatase from the FERM family of proteins. Although it had not yet been linked directly to the involvement of the Hippo pathway in cancer formation and progression, PTPN14 has long been known to be somehow involved in cell proliferation (32). It has also been linked to TGFβ and epithelial-to-mesenchymal transition in early organ development, as well as to cell motility and cell-cell adhesion through interactions with β-catenin (33, 34). A role of PTPN14 in cancer has been implicated as a number of its mutations are reported in breast and colorectal tumors (36, 37). In particular, a gene expression profiling of mouse pancreatic tumors revealed PTPN14 to be significantly down-regulated (p < 10-10.8) in the tumor invasion front and liver metastasis as compared with the primary tumor, with the finding validated by immunohistochemistry (38). We have demonstrated here that PTPN14 could interact with LATS1, the upstream negative regulator of YAP, and such interaction activates LATS1 protein and increases the inactive form of YAP (p-Ser127). Intriguingly, we also showed that PTPN14 interacts with Kibra and activates LATS1, adding another layer to the complicated Hippo pathway regulation. It has been shown by Xiao et al. (20) that LATS2 interacts with Kibra through its PPXY domain and amino acids 667–788 and the WW domains of Kibra. It is thus tempting to hypothesize that PTPN14, LATS1, and Kibra potentially form a trimeric complex. Further studies will be required to test this hypothesis.
On the other hand, our data indicate that PTPN14 promotes the LATS1 activation independently of the upstream MST proteins. The subcellular localization of LATS1 plays an important role in the regulation of its activity because membrane-localized LATS1 has an increased activity (29). We further hypothesize that PTPN14 interacts with and stabilizes LATS1 protein, as well as translocates LATS1 to the membrane to facilitate its phosphorylation and activation. Detailed analysis of this hypothesis is warranted in future studies.
Kibra (WWC1) is a WW domain-containing protein that has been found to be involved with memory performance and development of Alzheimer disease (39, 40). It has also been linked to the Hippo pathway, acting as a tumor suppressor (4). Originally found to work in conjunction with the upstream molecules Merlin and Expanded (21–23), Kibra has recently been under investigation for tumor-suppressive effects through other mechanisms. Most notably, it has been shown to be able to interact with and activate the LATS proteins to regulate YAP (19, 20).
It has been shown that the first WW domain of Drosophila Kibra is important for its binding to Pez (Drosophila PTPN14) (24). Interestingly, we found that either WW domain of mammalian Kibra is dispensable for its interaction with PTPN14, whereas deletion of both WW domains disrupted the interaction. On the other hand, we found that both the PPXY motif and the PTP domain of PTPN14 are required for its interaction with Kibra, in contrast to the Drosophila Pez in which both the PPXY and the PTP domains are dispensable for its interaction with the Drosophila Kibra (24). The amino acid 1121–1127 in the PTP domain of PTPN14 has been proposed as a substrate binding motif. It will be of interest to further test whether this motif contributes to the PTPN14 and Kibra interaction and whether Kibra is a direct substrate of PTPN14. Our results indicate that although the interaction between PTPN14 and Kibra is conserved from Drosophila to mammals, the detailed interaction structures may have diverged during the evolution. Interestingly, as our present manuscript was in revision, Wang et al. (41) also reported the interaction between mammalian Kibra and PTPN14, furthering reinforcing our findings here.
Taken together, our data suggest important regulatory mechanisms in the Hippo pathway that have yet to be thoroughly examined. There appear to be many levels of regulation throughout the pathway, and we have presented here one more mechanism. Most importantly, although the tumor-suppressive natures of PTPN14 (15, 17), Kibra (21–23), and LATS1 (4) have been previously noted separately, our results indicate that they have the ability to work together to more efficiently regulate the YAP oncogenic functions. The effect of their interaction can be better elucidated with further studies, e.g. the effect of PTPN14 overexpression can be examined in Kibra knockdown cells. It was recently shown that knockdown of Kibra in MCF10A cells can induce the features of epithelial-to-mesenchymal transition, similar to what we observed in our study of PTPN14 (19). These findings make it reasonable to propose that PTPN14 and Kibra may work together to elicit a downstream effect on their regulation of the YAP oncogenic function.
Acknowledgment
We thank Dr. Xiaolong Yang at the Queen's University, Kingston, Ontario, Canada for helpful discussion and kindly sharing the FLAG-tagged LATS1 construct.
This work was supported by an American Cancer Society Institutional Grant (ACS-IRG) to Roswell Park Cancer Institute (RPCI), a Roswell Park Alliance Grant, and National Institutes of Health Grant P30 CA016056 through the NCI (to J. Z.).
- PTPN14
- protein-tyrosine phosphatase non-receptor type 14
- MST
- mammalian STE20-like kinase
- LATS
- large tumor suppressor
- p
- phospho.
REFERENCES
- 1. Varelas X., Wrana J. L. (2012) Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 22, 88–96 [DOI] [PubMed] [Google Scholar]
- 2. Zhao B., Tumaneng K., Guan K. L. (2011) The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 13, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pan D. (2010) The Hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harvey K. F., Zhang X., Thomas D. M. (2013) The Hippo pathway and human cancer. Nat Rev. Cancer 13, 246–257 [DOI] [PubMed] [Google Scholar]
- 5. Lin J. I., Poon C. L., Harvey K. F. (2013) The Hippo size control pathway: ever expanding. Sci. Signal. 6, pe4. [DOI] [PubMed] [Google Scholar]
- 6. Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 [DOI] [PubMed] [Google Scholar]
- 7. Xu T., Wang W., Zhang S., Stewart R. A., Yu W. (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 8. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 [DOI] [PubMed] [Google Scholar]
- 9. Ramos A., Camargo F. D. (2012) The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 22, 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan S. W., Lim C. J., Loo L. S., Chong Y. F., Huang C., Hong W. (2009) TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J. Biol. Chem. 284, 14347–14358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J., Smolen G. A., Haber D. A. (2008) Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 68, 2789–2794 [DOI] [PubMed] [Google Scholar]
- 13. Hao Y., Chun A., Cheung K., Rashidi B., Yang X. (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 [DOI] [PubMed] [Google Scholar]
- 14. Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L. (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP. Genes Dev. 24, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X., Yang N., Figel S. A., Wilson K. E., Morrison C. D., Gelman I. H., Zhang J. (2013) PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 32, 1266–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang J. M., Nagatomo I., Suzuki E., Mizuno T., Kumagai T., Berezov A., Zhang H., Karlan B., Greene M. I., Wang Q. (2013) YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene 32, 2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang W., Huang J., Wang X., Yuan J., Li X., Feng L., Park J. I., Chen J. (2012) PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 26, 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Michaloglou C., Lehmann W., Martin T., Delaunay C., Hueber A., Barys L., Niu H., Billy E., Wartmann M., Ito M., Wilson C. J., Digan M. E., Bauer A., Voshol H., Christofori G., Sellers W. R., Hofmann F., Schmelzle T. (2013) The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS One 8, e61916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moleirinho S., Chang N., Sims A. H., Tilston-Lünel A. M., Angus L., Steele A., Boswell V., Barnett S. C., Ormandy C., Faratian D., Gunn-Moore F. J., Reynolds P. A. (2013) KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 32, 1821–1830 [DOI] [PubMed] [Google Scholar]
- 20. Xiao L., Chen Y., Ji M., Dong J. (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 286, 7788–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu J., Zheng Y., Dong J., Klusza S., Deng W. M., Pan D. (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baumgartner R., Poernbacher I., Buser N., Hafen E., Stocker H. (2010) The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell 18, 309–316 [DOI] [PubMed] [Google Scholar]
- 23. Genevet A., Wehr M. C., Brain R., Thompson B. J., Tapon N. (2010) Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poernbacher I., Baumgartner R., Marada S. K., Edwards K., Stocker H. (2012) Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr. Biol. 22, 389–396 [DOI] [PubMed] [Google Scholar]
- 25. Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 26. Yang N., Morrison C. D., Liu P., Miecznikowski J., Bshara W., Han S., Zhu Q., Omilian A. R., Li X., Zhang J. (2012) TAZ induces growth factor-independent proliferation through activation of EGFR ligand amphiregulin. Cell Cycle 11, 2922–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. (2006) NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7, 253–264 [DOI] [PubMed] [Google Scholar]
- 28. Chan E. H., Nousiainen M., Chalamalasetty R. B., Schäfer A., Nigg E. A., Silljé H. H. (2005) The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076–2086 [DOI] [PubMed] [Google Scholar]
- 29. Hergovich A., Schmitz D., Hemmings B. A. (2006) The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 345, 50–58 [DOI] [PubMed] [Google Scholar]
- 30. Debnath J., Brugge J. S. (2005) Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 5, 675–688 [DOI] [PubMed] [Google Scholar]
- 31. Smith A. L., Mitchell P. J., Shipley J., Gusterson B. A., Rogers M. V., Crompton M. R. (1995) Pez: a novel human cDNA encoding protein tyrosine phosphatase- and ezrin-like domains. Biochem. Biophys. Res. Commun. 209, 959–965 [DOI] [PubMed] [Google Scholar]
- 32. Wadham C., Gamble J. R., Vadas M. A., Khew-Goodall Y. (2000) Translocation of protein tyrosine phosphatase Pez/PTPD2/PTP36 to the nucleus is associated with induction of cell proliferation. J. Cell Sci. 113, 3117–3123 [DOI] [PubMed] [Google Scholar]
- 33. Wadham C., Gamble J. R., Vadas M. A., Khew-Goodall Y. (2003) The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates β-catenin. Mol. Biol. Cell 14, 2520–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wyatt L., Wadham C., Crocker L. A., Lardelli M., Khew-Goodall Y. (2007) The protein tyrosine phosphatase Pez regulates TGFβ, epithelial-mesenchymal transition, and organ development. J. Cell Biol. 178, 1223–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deleted in proof.
- 36. Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 37. Wang Z., Shen D., Parsons D. W., Bardelli A., Sager J., Szabo S., Ptak J., Silliman N., Peters B. A., van der Heijden M. S., Parmigiani G., Yan H., Wang T. L., Riggins G., Powell S. M., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., Velculescu V. E. (2004) Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304, 1164–1166 [DOI] [PubMed] [Google Scholar]
- 38. Niedergethmann M., Alves F., Neff J. K., Heidrich B., Aramin N., Li L., Pilarsky C., Grützmann R., Allgayer H., Post S., Gretz N. (2007) Gene expression profiling of liver metastases and tumour invasion in pancreatic cancer using an orthotopic SCID mouse model. Br. J. Cancer 97, 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bates T. C., Price J. F., Harris S. E., Marioni R. E., Fowkes F. G., Stewart M. C., Murray G. D., Whalley L. J., Starr J. M., Deary I. J. (2009) Association of KIBRA and memory. Neurosci. Lett. 458, 140–143 [DOI] [PubMed] [Google Scholar]
- 40. Papassotiropoulos A., Stephan D. A., Huentelman M. J., Hoerndli F. J., Craig D. W., Pearson J. V., Huynh K. D., Brunner F., Corneveaux J., Osborne D., Wollmer M. A., Aerni A., Coluccia D., Hänggi J., Mondadori C. R., Buchmann A., Reiman E. M., Caselli R. J., Henke K., de Quervain D. J. (2006) Common Kibra alleles are associated with human memory performance. Science 314, 475–478 [DOI] [PubMed] [Google Scholar]
- 41. Wang W., Li X., Huang J., Feng L., Dolinta K. G., Chen J. (2014) Defining the protein-protein interaction network of the human Hippo pathway. Mol. Cell. Proteomics 13, 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]