Background: Shikimate is essential for protein biosynthesis. Quinate and its derivatives are protective secondary metabolites.
Results: Members of the same gene family encode enzymes with either shikimate or quinate dehydrogenase activity.
Conclusion: The molecular genetic basis of plant quinate metabolism has been unraveled in vitro.
Significance: Identifying quinate metabolic enzymes will allow testing its ecological function and may enable biotechnological applications.
Keywords: Enzyme Kinetics, Gene Expression, Plant Biochemistry, Plant Defense, Secondary Metabolism, Primary Metabolism, Quinate Biosynthesis, Shikimate Pathway
Abstract
The shikimate pathway leads to the biosynthesis of aromatic amino acids essential for protein biosynthesis and the production of a wide array of plant secondary metabolites. Among them, quinate is an astringent feeding deterrent that can be formed in a single step reaction from 3-dehydroquinate catalyzed by quinate dehydrogenase (QDH). 3-Dehydroquinate is also the substrate for shikimate biosynthesis through the sequential actions of dehydroquinate dehydratase (DQD) and shikimate dehydrogenase (SDH) contained in a single protein in plants. The reaction mechanism of QDH resembles that of SDH. The poplar genome encodes five DQD/SDH-like genes (Poptr1 to Poptr5), which have diverged into two distinct groups based on sequence analysis and protein structure prediction. In vitro biochemical assays proved that Poptr1 and -5 are true DQD/SDHs, whereas Poptr2 and -3 instead have QDH activity with only residual DQD/SDH activity. Poplar DQD/SDHs have distinct expression profiles suggesting separate roles in protein and lignin biosynthesis. Also, the QDH genes are differentially expressed. In summary, quinate (secondary metabolism) and shikimate (primary metabolism) metabolic activities are encoded by distinct members of the same gene family, each having different physiological functions.
Introduction
The shikimate pathway initiates the synthesis of the three aromatic amino acids phenylalanine (Phe), tyrosine (Tyr), and tryptophan (Trp) (1–3). This pathway is present in bacteria, fungi, apicomplexan parasites, and plants (4–6). The absence of the shikimate pathway in animals makes this pathway a primary target for herbicides (7), live vaccines (8), antibiotics, and anti-infectious drugs (9). In plants, this pathway is important both for protein biosynthesis and for secondary metabolism. The end products and many intermediates of the pathway serve as precursors for plant hormones (salicylic acid and auxins) and for a large variety and quantity of secondary metabolites (e.g. alkaloids, benzenoids, and phenylpropanoids, including lignin), which fulfill diverse roles in plant development and chemical ecology (10–13).
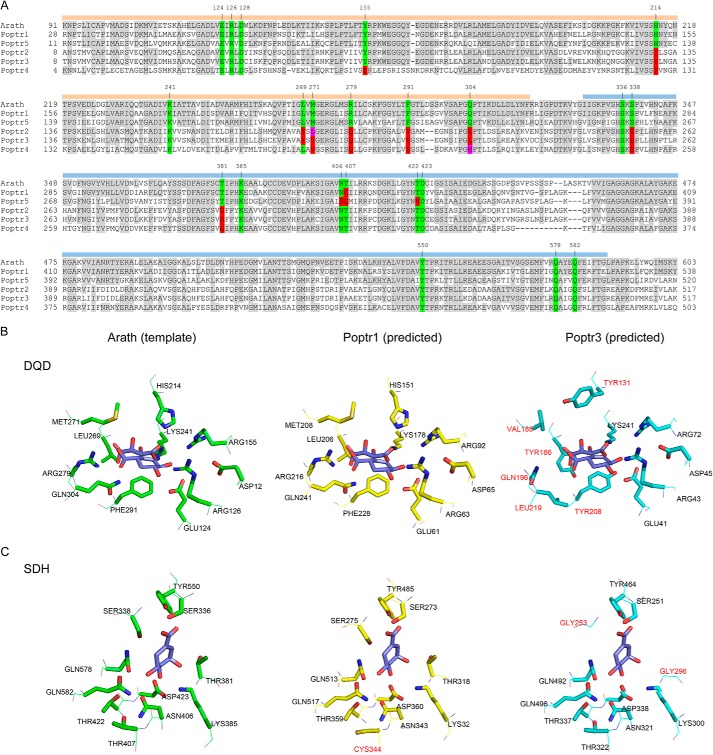
The shikimate pathway starts with the condensation of phosphoenolpyruvate and erythrose 4-phosphate and ends with the production of chorismate, the last common precursor of Trp, Tyr, and Phe (11). In plants, the whole pathway appears to be regulated primarily at the transcriptional level. It is responsive to developmental signals, light, as well as abiotic and biotic stresses (11). Most shikimate pathway enzymes are encoded by distinct genes in plants, but the conversion of 3-dehydroquinate to shikimate via 3-dehydroshikimate is catalyzed by the bifunctional enzyme dehydroquinate dehydratase/shikimate dehydrogenase (DQD3/SDH; EC 4.2.1.10 and E.C. 1.1.1.25; see Fig. 1). The DQD domain constitutes the N-terminal half of the protein and the SDH domain the C-terminal half (Fig. 2).
FIGURE 1.
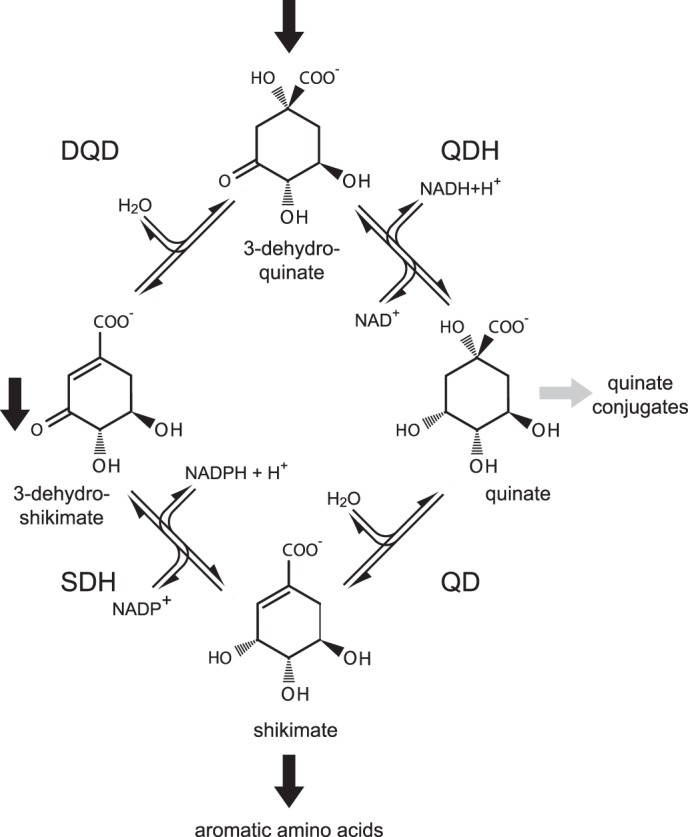
Reactions comprising the shikimate/quinate cycle. The shikimate pathway (indicated by block arrows) proceeds from 3-dehydroquinate via 3-dehydroshikimate to shikimate; the bi-functional enzyme DQD/SDH catalyzes these two reactions. Quinate can be synthesized from either 3-dehydroquinate or from shikimate; these two reactions are catalyzed by QDH and QD, respectively.
FIGURE 2.
Alignment of DQD/SDHs from P. trichocarpa (Poptr1 to Poptr5) and A. thaliana (A) and active site structure predictions of Poptr1 and Poptr3 in comparison with the known DQD/SDH structure from Arabidopsis (Protein Data Bank code 2O7Q) (B and C). A, shown is the alignment as used by Phyre for structure prediction. Note that the N-terminal signal peptide was not included in the Arabidopsis structure determination by Singh and Christendat (53) and was thus omitted from the alignment. Amino acids identical to the Arabidopsis template are shaded in gray. Active site residues of the Arabidopsis protein are highlighted in green. Identical amino acids at the respective position in the poplar sequences are also highlighted in green, and different amino acids are contrasted in red or magenta. Numbers above the sequence indicate the Arabidopsis amino acid position. The two functional domains (DQD and SDH) are highlighted by orange and blue bars, respectively. B, structure models were predicted with Phyre; DQD active sites (B) and SDH active sites (C) were visualized as stick models with PyMOL. Amino acids in the Poptr1 or Poptr3 predictions differing from the Arabidopsis template structure are highlighted in red.
DQD/SDH genes have been functionally characterized in some species, including Arabidopsis thaliana (Arabidopsis; Ref. 14), Solanum lycopersicum (tomato; Ref. 15), and Nicotiana tabacum (tobacco; Refs. 16, 17). The DQD/SDH crystal structure from Arabidopsis allowed identification of key amino acid residues involved in catalytic activities of DQD/SDH (14). Although the Arabidopsis genome encodes only a single DQD/SDH gene, in Populus trichocarpa, five putative DQD/SDH genes (here referred to as Poptr1 to Poptr5) have been identified based on sequence similarity alone (18).
Many intermediates of the shikimate pathway can be branch points leading to secondary metabolic processes (19). Among these, quinate can be formed in a single step reaction from either shikimate or 3-dehydroquinate, i.e. the product or substrate of DQD/SDH (Fig. 1). Quinate is widely distributed across the plant kingdom and may accumulate to high levels (up to 10% of leave dry weight) in some plants (19). Chlorogenate, an ester of caffeate and quinate, also accumulates to high levels in some plant species such as coffee and poplars (20). It acts as a feeding deterrent because of its astringency and antimicrobial activity (21). Chlorogenic acid may also serve as an intermediate and storage reserve of lignin biosynthesis (22, 23). Chlorogenic acid is gaining interest in pharmaceutical research due to its potential role as a weight loss supplement and its association with a lower risk of type II diabetes (24) and cardiovascular diseases (25). Also, both quinate and shikimate can be used as chiral starting materials in the synthesis of antiviral drugs, including oseltamivir (Tamiflu, Ref. 26).
In bacteria, quinate can be utilized as a carbon source and channeled back to the shikimate pathway by shikimate/quinate dehydrogenase (YdiB; EC 1.1.1.282) having high affinity toward both shikimate and quinate (27). In contrast, the metabolism of quinate in plants remains largely enigmatic. The interconversion of shikimate and quinate has been demonstrated biochemically, and a role of quinate as a shikimate pathway reserve compound was proposed (28–33). Enzymes involved are either quinate dehydratase (QD; also referred to as quinate hydrolyase; no EC number assigned) or quinate dehydrogenase (QDH; EC 1.1.1.24) (11). QDHs have been (partially) purified and characterized in several angiosperms and gymnosperms (30, 34–41), but QD has been partially characterized only in pea (42). However, genes encoding these two enzymes have not been identified to date. The reaction mechanisms and the substrates of QDH and QD resemble that of SDH and DQD, respectively. SDH and QDH catalyze NADH- or NADPH-dependent reductions, and DQD and QD catalyze reversible water elimination reactions (Fig. 1).
Given the similarities between the reactions catalyzed, and because enzymes with both SDH and QDH activity have been described (27, 37), we hypothesized that QDH (and/or QD) activities may be encoded by genes similar to DQD/SDH. Here, we demonstrate that the five putative poplar DQD/SDHs represent two functionally distinct groups, one of which preferentially uses quinate (QDH) as a substrate in vitro with only residual SDH activity, although while the other has only DQD/SDH activity. Expression variation among these homologues suggests distinct functions of isoforms in both plant development and adaptive responses to stresses.
EXPERIMENTAL PROCEDURES
DQD/SDH Protein Sequence Analyses and Structure Prediction
Multiple sequence alignment of amino acid sequences (for accession numbers see Table 1) was performed using “ClustalW” implemented in “BioEdit” (43). This alignment was split into two fragments to allow separate examination of DQD and SDH domains. Pairwise sequence similarity scores for both SDH and DQD domains were calculated in BioEdit. The two alignments were used to generate Neighbor Joining trees with “Phylip” Version 3.69 (44).
TABLE 1.
DQD/SDH family members included in amino acid sequence analysis and PCR primers used
For each poplar gene, a forward (F) and reverse (R) primer was used to amplify the complete open reading frame (start and stop codons are underlined).
| Name | Species | Sequence identifier (database) | Primers sequence (5′ to 3′) |
|---|---|---|---|
| EsccoAroE | E. coli | YP_006126142.1 (GenBankTM) | |
| EsccoAroD | E. coli | YP_006124567.1 (GenBankTM) | |
| EsccoYdiB | E. coli | YP_006124566.1 (GenBankTM) | |
| Arath | A. thaliana | NP_187286.1 (GenBankTM) | |
| Solly | S. lycopersicum | NP_001234051.1 (GenBankTM) | |
| Nicta1 | N. tabacum | AAS90325.1 (GenBankTM) | |
| Nicta2 | N. tabacum | AAS90324.2 (GenBankTM) | |
| Poptr1 | P. trichocarpa | Potri.010G019000.2 (Phytozome Version 3.0) | F, GGCTTAAUATGGATTCTGCAAGCAACGTCC |
| R, GGTTTAAUCTAGTACTTTGACATGATCTTCTGAAAGAGTTC | |||
| Poptr2 | P. trichocarpa | Potri.013G029900.2 (Phytozome Version 3.0) | F, GGCTTAAUATGGGGCGTGCTGGGATC |
| R, GGTTTAAUTCAGAATTTGGCTAGAACAATCTCCC | |||
| Poptr3 | P. trichocarpa | Potri.005G043400.1 (Phytozome Version 3.0) | F, GGCTTAAUATGGGGAGTGTTGGAGTCCTGAC |
| R, GGTTTAAUTCAGAATTTGGCTAAAACAATCTCCC | |||
| Poptr4 | P. trichocarpa | Potri.014G135500.3 (Phytozome Version 3.0) | F, GGCTTAAUATGGCATTCAAGAACAACCTCTTAG |
| R, GGTTTAAUTCAAAATTGCTCCAAGACAAGC | |||
| Poptr5 | P. trichocarpa | Potri.013G029800.1 (Phytozome Version 3.0) | F, GGCTTAAUATGGATCTCCAAAGCGCTG |
| R, GGTTTAAUTTATGTATTCCTTGCTAACACATCTCTAAT |
Three-dimensional protein structures of the five putative poplar DQD/SDHs were predicted using “Phyre” (45). Model coordinates were generated using Arabidopsis DQD/SDH (Protein Data Bank ID: c2o7qA) (14) coupled with either 3-dehydroshikimate and tartrate or shikimate as a template. “PyMOL” (PyMOL Molecular Graphics System, Version 1.5. Schrödinger, LLC.) was used for visualization.
Molecular Cloning of DQD/SDH cDNAs
P. trichocarpa Nisqually-1 plant tissues (leaf, bark, xylem, and phloem) were collected from trees grown in a field at the University of Victoria. Total RNA was isolated using the cetyltrimethylammonium bromide method as described (46). cDNA synthesis was performed from 0.5 μg of total RNA using 5 μm oligo(dT) primer and 1 unit of reverse transcriptase (Invitrogen). The five putative poplar DQD/SDHs were amplified from pooled cDNA with “USER” (uracil-specific excision reagent)-modified gene-specific primers (Table 1). Proofreading “Pfu Turbo Cx Hotstart” DNA polymerase (Agilent) was used in PCR amplification according to the manufacturer's protocol (30 cycles, annealing at 56 °C for 30 s, extension at 72 °C for 120 s, and denaturation at 94 °C for 30 s). PCR amplicons were incubated with USER enzyme mixture (New England Biolabs) and cloned into pQE30-USER-His6 overexpression vectors separately following the standard protocol (47). All constructs were verified by Sanger DNA sequencing (sequencing facility at the University of Victoria). A pQE30 construct carrying the coding region of a poplar glycosyltransferase family protein (kindly provided by Dr. C. P. Constabel, University of Victoria) was used as a negative control.
Purification of Recombinant Proteins
Overexpression constructs carrying the five putative poplar DQD/SDH coding sequences were transformed into Escherichia coli (strain M15) through electroporation. 5 ml of LB broth containing selective antibiotics (100 mg/liter ampicillin and 25 mg/liter kanamycin) was inoculated with a bacterial colony and grown at 37 °C overnight with vigorous shaking. A 50-ml culture was inoculated with the pre-culture and grown at 37 °C until A600 = 0.6, and isopropyl thio-β-d-1-galactoside was added to a final concentration of 0.06 mm. Protein production was carried out at 19 °C with shaking for 16 h. Cells were harvested by centrifugation at 4000 × g for 20 min, and the cell pellet was stored at −80 °C for later use.
His6-tagged recombinant protein was extracted and affinity-purified under native conditions using nickel-NTA-agarose (Qiagen). Frozen cell pellets were resuspended in 4 ml of lysis buffer (50 mm NaH2PO3, 300 mm NaCl, and 10 mm imidazole, adjusted to pH 8 with NaOH, 1 mg/ml lysozyme) and incubated on ice for 1 h. The mixture was further lysed using a sonicator followed by centrifugation at 10,000 × g for 30 min at 4 °C. The supernatant was incubated with 1 ml of 50% nickel-NTA-agarose suspension on ice for 1 h with moderate shaking. The lysate-NTA slurry was loaded onto a polypropylene column and washed twice in wash buffer (50 mm NaH2PO3, 300 mm NaCl, NaOH to pH 8) and three times with wash buffer with 20 mm imidazole before elution (with 50 mm NaH2PO3, 300 mm NaCl, and 250 mm imidazole, NaOH to pH 8). Protein concentration was determined by Bradford assay (48) using BSA as a standard.
Denaturing SDS-PAGE was performed to assess the purity of recombinant proteins. Boiled protein samples (3 μg) were separated on 12% polyacrylamide gel and stained with GelCode Blue Stain Reagent (Thermo Scientific). Using a replicate gel, proteins were transferred to PVDF membrane (Millipore) by electroblotting. His6-tagged proteins were detected by chemiluminescence using the “SuperSignal® West HisProbeTM” kit (Thermo Scientific).
Spectrophotometric Measurement of Enzyme Activities
Dehydrogenase activities of the recombinant proteins with both SDH shikimate and QDH quinate were determined photometrically by measuring NADPH or NADH production at 340 nm (17). A standard reaction mixture contained 0.01 to 0.1 mg/ml purified His6-tagged enzyme (concentrations were adjusted based on observed activity), 200 μm NADP+ or 250 μm NAD+, 75 mm Trizma base-HCl (pH 8.5), and varying concentrations of shikimate or quinate (from a minimum of 5 μm to a maximum of 5 mm). Reactions were carried out in a total volume of 1 ml and were initiated by adding shikimate or quinate. Absorbance changes at 340 nm relative to a reference lacking enzyme was measured at room temperature and recorded every 10 s for the 1st min and every minute for the following 4 min. Both shikimate and quinate dehydrogenase activities as a function of pH were determined using different buffer systems as follows: acetate buffer (pH 4–5); phosphate buffer (pH 6–7.5); Tris-HCl buffer (pH 8–8.5); and carbonate buffer (pH 9–11) (49). Other experimental conditions were as described above.
Kinetic properties (Vmax and Km values) of each enzyme for both shikimate and quinate with saturating concentrations of either NADP+ or NAD+ were determined at optimal pH (pH 8.5) using at least 12 concentrations of substrate (ranging from 0.5 μm to 5 mm). Changes in absorbance were converted to enzyme activities based on the extinction coefficient of NADPH (6.2 × 103 liter mol−1cm−1). Three replicates (independent protein purifications) were carried out for each reaction, and means were calculated. Dehydrogenase activity with 3-dehydroquinate was determined at 12 substrate concentrations ranging from 2 μm to 2 mm by measuring the consumption of NADH at 340 nm. Kinetic data of each enzyme were fitted to the Michaelis-Menten model using plotting and curve-fitting tools implemented in MATLAB to estimate maximal velocity values (Vmax) and Michaelis-Menten constants (Km). Confidence of curve fitting (i.e. 95% confidence bounds, R2 values and root mean square error) was also assessed with MATLAB. Enzyme substrate specificities were calculated by dividing Vmax by Km.
Dehydratase activity with 3-dehydroquinate (10 mm) was determined by directly measuring 3-dehydroshikimate production in the absence of a cofactor at 250 nm, which allows distinction of 3-dehydroshikimate from the other cyclitols analyzed. Enzyme activity was determined based on a standard curve of 10 3-dehydroshikimate concentrations.
Measurement of Dehydratase and Dehydrogenase Activities Using HPLC
A systematic experimental design was developed to distinguish the four potential functions (DQD, SDH, QD, and QDH). Recombinant proteins were incubated in reaction buffer at pH 8.5 with different combinations of substrates and cofactors (both at saturating concentrations). 30 min of incubation time at room temperature was allowed before reaction mixtures were filtered (0.22 μm), loaded onto an Aminex HPX-87H organic acid analysis column (300 × 7.8 mm inner diameter; 9 μm particle diameter) (50, 51), and separated on an Ultimate 3000 HPLC system equipped with a diode array detector (Dionex). Products were separated using an isocratic elution system (10 mm H2SO4) over a 40-min period with a flow rate of 0.4 ml/min. Negative control assays were prepared with boil-inactivated enzymes. Substrate and product peaks were identified through comparison with authentic standards obtained from Sigma. Peak identities were verified by their distinct retention time and UV absorption spectra.
Expression Profiling and Co-expression Analyses
Publicly available poplar Affymetrix microarray data were retrieved from various databases (ArrayExpress and NCBI GEO on July 4 and 5, 2012). The collection consists of 43 diverse experimental series for a total of 748 array hybridizations covering 17 poplar species. Microarray raw expression data were normalized using the “MAS5” algorithm (scale factor value set to 500) implemented in “R” (52). Normalized expression data were then divided into three groups (development, treatment, and transgenic) based on their sample annotations. The log2-transformed expression values were mean centered across experiments for the “development” dataset, but log2 ratios compared with the respective controls were calculated for the “treatment” and “transgenics” groups. Expression data were extracted for the five genes of interest to generate expression heat map using “HeatMapper” (52). Normalized data were also used for co-expression analysis using the “ExpressionAngler” algorithm (52). Pairwise Pearson correlation coefficients between a gene of interest and all other genes were calculated across each dataset, and the top 25 genes (r >0.5) were retained.
RESULTS
Sequence Diversity in the DQD/SDH Superfamily
The poplar genome contains five genes annotated as DQD/SDHs (Poptr1 to Poptr5) (18). Sequence similarity comparison of these proteins with characterized plant enzymes from Arabidopsis, tobacco, and tomato revealed that Poptr1 and Poptr5 share higher sequence similarity with characterized DQD/SDHs than Poptr2, Poptr3, and Poptr4 (Table 2). Poptr1 and Poptr5 share on average 66% sequence similarity with proteins from Arabidopsis, tomato, and tobacco in the SDH portion of the protein and 60% in the DQD portion. In contrast, Poptr2, Poptr3, and Poptr4 share on average only 52 and 38% similarity in the SDH and DQD portions, respectively, with characterized DQD/SDHs. The latter thus define a DQD/SDH-like group that also includes a divergent isoform from tobacco (Nicta2, Ref. 17). In comparison with bacterial SDH (AroE) and SDH/QDH (YdiB), all five poplar proteins have comparable low similarities, but plant SDHs were generally more similar to bacterial YdiB (25–30% similarity) than to AroE (21–28%).
TABLE 2.
Protein sequence similarity of DQD/SDH family members
The top right triangle shows similarities in the C-terminal SDH half of the proteins; the lower left triangle gives similarities in the N-terminal DQD half. Neighbor Joining trees were produced using the same alignments with Phylip3.69 and placed on the left sides of their corresponding tables. Sequences included are DQD/SDHs from A. thaliana (Arath), S. lycopersicum (Solly), N. tabacum (Nicta1–2), P. trichocarpa (Poptr1–5), E. coli AroD (encoding DQD only), AroE and YdiB (encoding SDH-like activity only).
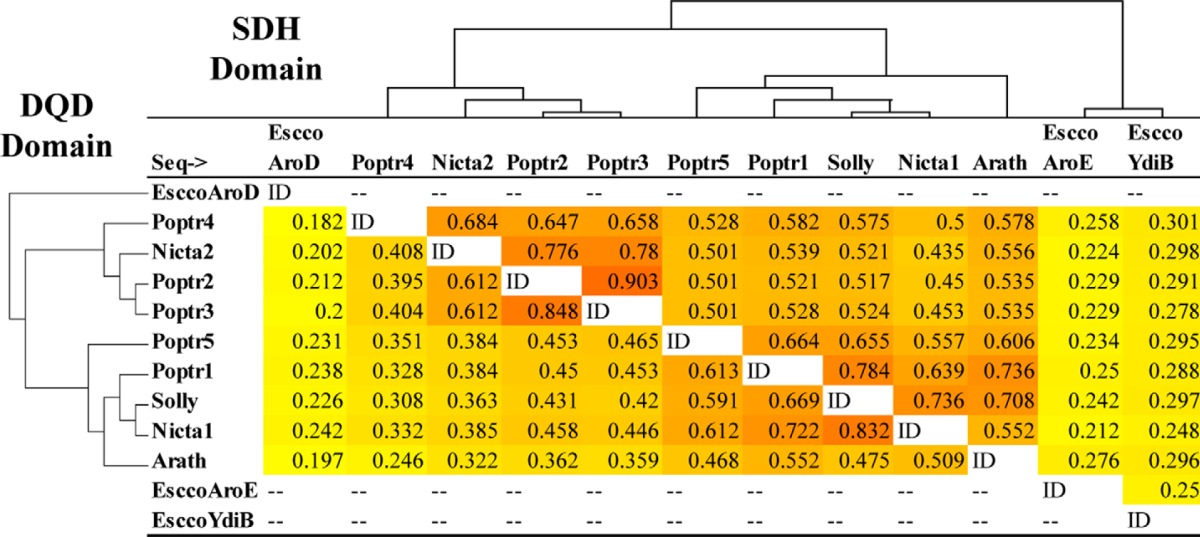
Homology-based Structure Prediction of Poplar DQD/SDHs
Structures of the five putative poplar DQD/SDHs were predicted using the homology-based modeling tool Phyre (45) based on structural data of the Arabidopsis DQD/SDH (14, 53). Reliable predictions for all five sequences were obtained (100% confidence across more than 90% of each sequence).
The N-terminal DQD domain encompasses the first 316 amino acids of the Arabidopsis protein. Singh and Christendat (14) previously showed that Lys-241 and His-214 are the major catalytic groups within the active site of the Arabidopsis DQD domain; Lys-241 forms a Schiff base intermediate with the carbonyl group of dehydroquinate during catalysis, although His-214 helps modulate the formation and breakdown of this Schiff base intermediate (14). An additional eight residues are involved in properly positioning the substrate (14). The corresponding homologous amino acids in Poptr1 and Poptr5 are highly conserved (Fig. 2A). In consequence, this results in a virtually identical inferred active site structure (Fig. 2B). In contrast, major differences were observed between the known Arabidopsis DQD domain on the one hand and the inferred structures of Poptr2, Poptr3, and Poptr4 on the other hand. Compared with each other, Poptr2, Poptr3, and Poptr4 are highly similar at most of the key residues (Fig. 2A), and therefore only the inferred active site of Poptr3 is shown (Fig. 2B). Although a lysine corresponding to Lys-241 in Arabidopsis is still present, His-214 is replaced by a Tyr in all three DQD-like domains. In addition, five of the additional eight residues previously shown to be involved in substrate binding (14) are different in all three DQD-like proteins (Fig. 2A). This creates an overall more spacious substrate-binding pocket in the predicted protein structure.
The SDH domain spans amino acids 328–588 of the Arabidopsis protein. Lys-385 and Asp-423 were reported to likely play an important role in catalysis (14), and the homologous positions in all five poplar genes include the same amino acids (Fig. 2A). For Poptr1 and Poptr5, the remaining active site residues are also highly conserved compared with Arabidopsis (Fig. 2, A and C). In contrast, for the SDH-like enzymes, two putative active site residues were found to be divergent. Ser-338, involved in interacting with the C1 carboxylate group of the substrate in Arabidopsis (14), is replaced by Gly in the corresponding homologous positions of Poptr2, Poptr3, and Poptr4 (Fig. 2A). Based on the predicted structure, this leads to the appearance of an extra pocket in the inferred active site (Fig. 2C). This would allow more bulky substrates to fit. Quinate, with its additional hydroxyl group at the C1 position compared with shikimate, would be a good fit. In addition, instead of Thr-381 in Arabidopsis, a Gly residue is found in the homologous position of all three SDH-like poplar enzymes (Fig. 2A). This Thr is replaced by Ser, also with a smaller side chain, in YdiB from E. coli (14). Given that YdiB converts both shikimate and quinate (27), the replacement of Thr-381 with Gly in Poptr2, Poptr3, and Poptr4 also appears consistent with our hypothesis that quinate may be an alternative substrate.
Enzymatic Characterization of DQD/SDH Family Members
Full-length cDNA fragments encoding the five poplar enzymes were PCR-amplified, cloned, and expressed in E. coli as His6-tagged proteins allowing native protein purification using affinity chromatography. For Poptr4, no soluble recombinant protein was yielded despite further optimization attempts. For each of the other four enzymes, recombinant protein purification yielded a major band of the expected molecular weight, and only this band was His tag-positive in protein blot analyses (Fig. 3).
FIGURE 3.
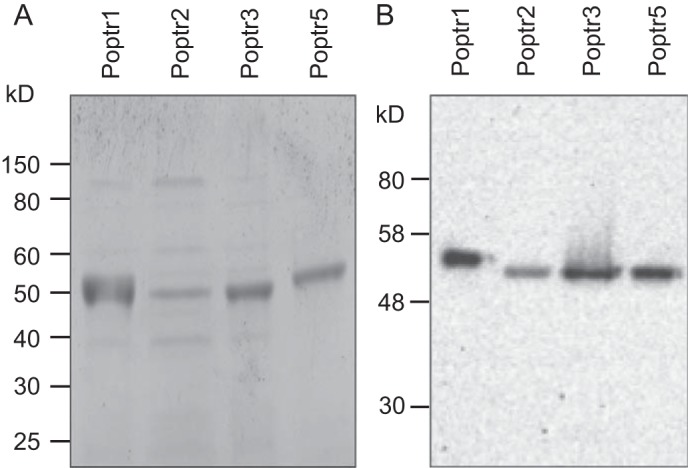
Protein gel and Western blot analyses of purified proteins. Recombinant putative DHQD/SDH from poplar (Poptr1 to Poptr5) were purified from E. coli through Ni-NTA affinity chromatography and separated on a polyacrylamide gel and stained with Coomassie Blue (A). A replicate gel was used for Western blotting and detection of His-tagged proteins using a HisProbe fused to horseradish peroxidase (B).
Dehydrogenase activities of the recombinant proteins with shikimate and quinate were first assessed photometrically by measuring NADH or NADPH production. Under saturating conditions, Poptr1 and -5 displayed strong activities with shikimate but no detectable activity with quinate even at elevated enzyme concentrations (0.1 μg/ml), thus confirming SDH activity (Fig. 4). In terms of co-factor requirement, the highest enzyme activities were observed with NADP+ for both enzymes, and activities dropped by 96% when replacing NADP+ with NAD+ for Poptr1. For Poptr5, no activity with shikimate was detectable in the presence of NAD+ even with elevated enzyme concentrations (Fig. 4). In contrast, Poptr2 and Poptr3 could act on both quinate and shikimate but displayed a much higher activity with quinate compared with shikimate at saturating conditions (Fig. 4). Poptr2 and Poptr3 preferred NAD+ because activities with both shikimate and quinate were close to the detection limit when NADP+ was used as a cofactor (Fig. 4). To validate that no endogenous E. coli SDH or QDH was co-purified, an unrelated protein of the same size (a glycosyltransferase family protein from poplar) was purified in parallel, which resulted in no SDH or QDH activity (data not shown).
FIGURE 4.
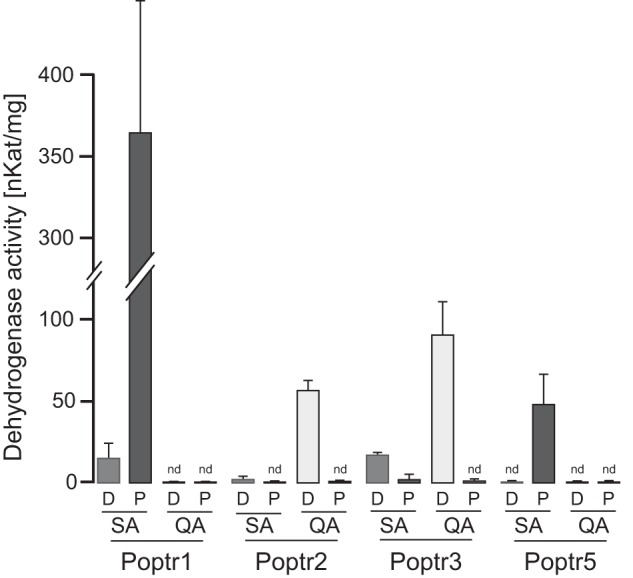
Enzyme activities of Poptr1, Poptr2, Poptr3, and Poptr5 at saturating substrate concentrations (5 mm). Either shikimate (SA) or quinate (QA) was used in conjunction with NAD+ (D) or NADP+ (P). Cofactor formation (NADH or NADPH) was measured spectrophotometrically at 340 nm and converted to dehydrogenase activities. Both SDH and QDH activities were determined in triplicate; error bars denote standard deviations. Assays yielding less than 0.02 ΔA340/min are indicated as not detectable (nd).
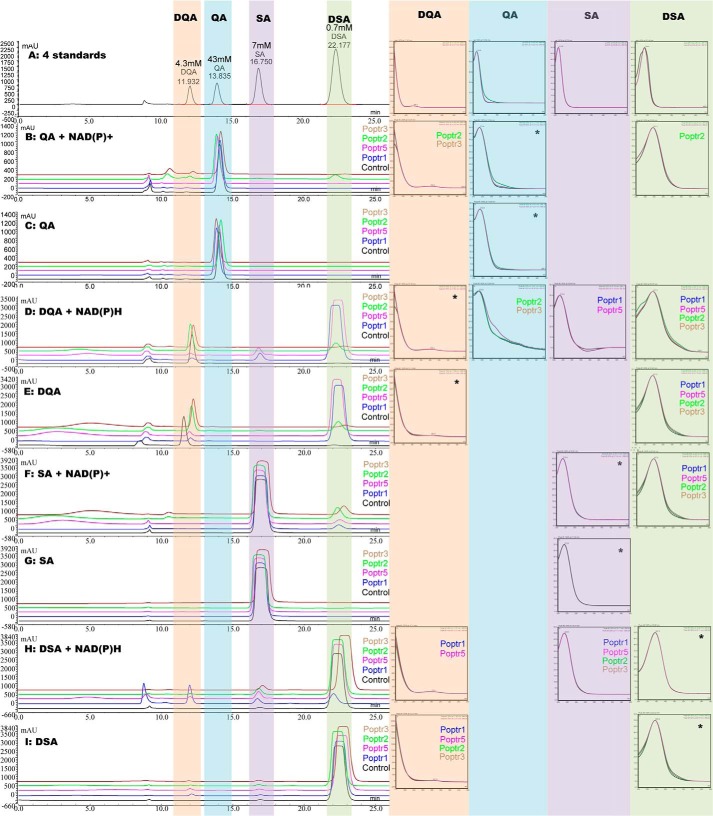
Measuring cofactor consumption alone may not be sufficient to assign QDH or SDH activities unambiguously. For example, when using quinate as a substrate, co-factor consumption can be due to QDH activity, although theoretically it can also be due to combined QD and SDH activities. Vice versa, apparent SDH activity could also be due to combined QD and QDH activities (Fig. 1). To distinguish between these possibilities, recombinant enzymes were incubated with different substrate and cofactor combinations, and reaction products were assessed by HPLC (Fig. 5). This confirmed the DQD/SDH bifunctionality of Poptr1 and -5; both were able to catalyze the production of shikimate from 3-dehydroquinate via 3-dehydroshikimate and vice versa (Fig. 5, D–E, F, H, and I). Neither enzyme was able to use or produce quinate (i.e. no apparent QD or QDH activity, Fig. 5, B–D and G). This confirmed the specificity of Poptr1 and -5 toward shikimate biosynthesis via 3-dehydroshikimate only. HPLC results also confirmed that Poptr2 and -3 were able to catalyze quinate to 3-dehydroquinate conversion using NAD+ as a cofactor (Fig. 5B), confirming QDH activity. Some DQD and SDH activities were detectable under saturating condition (Fig. 5, D–F). Surprisingly, quinate formation was barely detectable when incubating Poptr2 and Poptr3 with 3-dehydroquinate and NADH (Fig. 5D). We suspected this to be due to the very low absorption coefficient of quinate (note that 60 times more quinate was loaded in the standard mix than 3-dehydroshikimate) (Fig. 5A). Determining dehydrogenase activity with 3-dehydroquinate quantitatively by measuring NADH consumption yielded a total of 94 and 116 μm s−1 mg−1 for Poptr2 and Poptr3, respectively. This may be attributed to QDH and/or combined DQD/SDH activity. DQD activity was 0.2 μm s−1 mg−1 (Poptr2) and 0.4 μm s−1 mg−1 (Poptr3) when measuring 3-dehydroshikimate production directly. SDH activity with 3-dehydroshikimate and NADH was 0.6 and 1.1 μm s−1 mg−1 for Poptr2 and Poptr3, respectively. Thus, DQD and SDH contributed less than 1% of the total activity, leaving more than 99% of the observed dehydrogenase activity attributed to QDH activity. Serving as a control for the forward activity assays, Poptr1 had a DQD activity of 89 μm s−1 mg−1 and an SDH activity of 250 μm s−1 mg−1.
FIGURE 5.
HPLC elution profiles of enzyme assay products catalyzed by poplar recombinant DQD/SDHs (Poptr1 and Poptr5) and QDHs (Poptr2 and Poptr3). Purified enzymes were mixed with different combinations of substrate and cofactor as indicated. Reaction mixtures were incubated for 30 min before separation using HPLC equipped with a diode array detector. For each test, a negative control was prepared using boil-inactivated enzyme. Retention time and UV absorption spectrum were monitored and used to identify each peak in comparison with authentic standards. Standards were injected at different concentrations as indicated. Absorption spectra of standards and reaction products are shown to the right. Enzymes catalyzing the production of the respective products are noted. Spectra for the substrate used in the respective reaction are marked with a *. SA, shikimate; QA, quinate; DQA, 3-dehydroquinate; DSA, 3-dehydroshikimate.
In the absence of a cofactor, no conversion of quinate to shikimate or vice versa was detectable for either of the two enzymes (Fig. 5, C and G); thus, Poptr2 and 3 do not have detectable QD activity.
The apparent absence of QD activity for all four enzymes allowed the use of the spectrophotometric assay to distinguish SDH and QDH activities. pH optima were determined for both SDH and QDH reactions (Fig. 6). Both types of reactions had similar pH optima with all enzymes displaying high activity under basic conditions. The optimal pH for all reactions was determined to be between 8.5 and 9.5, and activities dropped quickly when pH was shifted from its optimum (Fig. 6).
FIGURE 6.
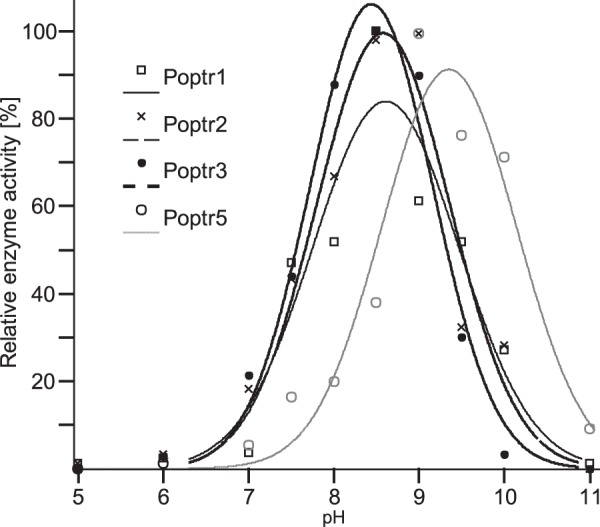
pH-dependent enzymatic activities of recombinant Poptr1, Poptr2, Poptr3, and Poptr5. Enzyme activities were determined spectrophotometrically with substrate concentrations of 100 μm. Shikimate was used as a substrate for Poptr1 and -5, and quinate was used for Poptr2 and Poptr3. Relative activities (the maximal activity of a given enzyme was set to 100%) at pH values ranging from 5 to 11 were used for curve fitting.
Michaelis-Menten kinetics of each enzyme were determined using saturating amounts of cofactor. Enzymes exhibited typical saturation kinetics, and Km and Vmax values were modeled. Among the shikimate-converting enzymes, Poptr1 had much higher Vmax (shikimate) compared with Poptr5 (Table 3). Together with a lower Km (shikimate), this results in a six times higher specificity (Vmax/Km). Both maximum velocity and affinity of Poptr1 toward shikimate dropped largely when NAD+ was used as a cofactor (Table 3). Also Poptr5 is highly specific for NADP+ as a cofactor. Even when high enzyme and shikimate concentrations were used, Poptr5 activity with NAD+ was below background (Fig. 4) preventing the determination of kinetic properties with this cofactor. Neither Poptr1 nor Poptr5 displayed any detectable activity with quinate in spectrophotometric assays. In contrast, both Poptr2 and -3 yielded similar kinetic properties in terms of Km and Vmax values for quinate (Table 3). Kinetic properties of Poptr2 and Poptr3 with shikimate were also determined in the presence of NAD+ using elevated enzyme concentrations. Both enzymes had clearly reduced maximum velocities and affinities (higher Km) toward shikimate resulting in 66 times (Poptr2) and four times (Poptr3) lower specificities for shikimate than for quinate (Table 3). Following the biosynthetic route, dehydrogenase activity of Poptr2 and Poptr3 with 3-dehydroquinate yielded Km values of 624 ± 28 and 299 ± 50 μm, and maximum velocities of 150 ± 30 and 103 ± 6 μm s−1 mg−1, respectively. Taking the very low SDH activity of Poptr2 and Poptr3 into account (above and Table 3), we attributed most of the observed dehydrogenase activity with 3-dehydroquinate to quinate synthesis (i.e. QDH activity). For both quinate-specific enzymes, activities with NADP+ as a cofactor were too low for kinetic analyses even at elevated protein concentrations. Based on these in vitro enzymatic properties, we assigned the names DQD/SDH1 and DQD/SDH2 to Poptr1 and Poptr5, and QDH1 and QDH2 to Poptr2 and Poptr3, respectively. Poptr4 was assigned the name QDH3 because of its higher sequence similarity to QDHs than to SDHs.
TABLE 3.
Kinetic properties of recombinant poplar DQD/SDH (Poptr1 and Poptr5) and QDH (Poptr2 and Poptr3)
Either shikimate or quinate was used as a substrate with at least 12 concentrations of substrate (ranging from 0.5 to 2000 μm). The tested cofactor (either NADP+ or NAD+) was kept at saturating concentrations. Vmax and Km values were estimated based on nonlinear fitting to the Michaelis-Menten model using the Curve Fitting ToolboxTM implemented in MATLAB. Vmax and Km values represent averages of three independent protein purifications. Confidence of curve fitting (i.e. 95% confidence bounds) was assessed with MATLAB. Confidence bounds are indicated as ± of the respective averages. Substrate specificities were calculates by dividing Vmax by Km.
| Substrate tested | Cofactor used | Vmax | Km | Vmax/Km | |||
|---|---|---|---|---|---|---|---|
| μm s−1 mg−1 | μm | ||||||
| Poptr1 | Shikimate | NADP+ | 323.1 | ± 11.2 | 223.1 | ± 24.4 | 1.45 |
| Shikimate | NAD+ | 17.8 | ± 3.2 | 426.9 | ± 243.8 | 0.04 | |
| Quinate | NADP+ | NDa | |||||
| Quinate | NAD+ | ND | |||||
| Poptr2 | Shikimate | NADP+ | ND | ||||
| Shikimate | NAD+ | 3.1 | ± 0.5 | 833.1 | ± 382.9 | 0.003 | |
| Quinate | NADP+ | ND | |||||
| Quinate | NAD+ | 67.0 | ± 3.5 | 334.4 | ± 47.1 | 0.20 | |
| Poptr3 | Shikimate | NADP+ | ND | ||||
| Shikimate | NAD+ | 18.0 | ± 1.4 | 391.8 | ± 92.3 | 0.05 | |
| Quinate | NADP+ | ND | |||||
| Quinate | NAD+ | 73.8 | ± 4.7 | 321.3 | ± 55.3 | 0.23 | |
| Poptr5 | Shikimate | NADP+ | 70.0 | ± 9.0 | 345.8 | ± 96.2 | 0.20 |
| Shikimate | NAD+ | ND | |||||
| Quinate | NADP+ | ND | |||||
| Quinate | NAD+ | ND | |||||
a ND means no activity detectable or too low to determine kinetic properties.
Expression Profiling of Poplar DQD/SDHs and QDHs
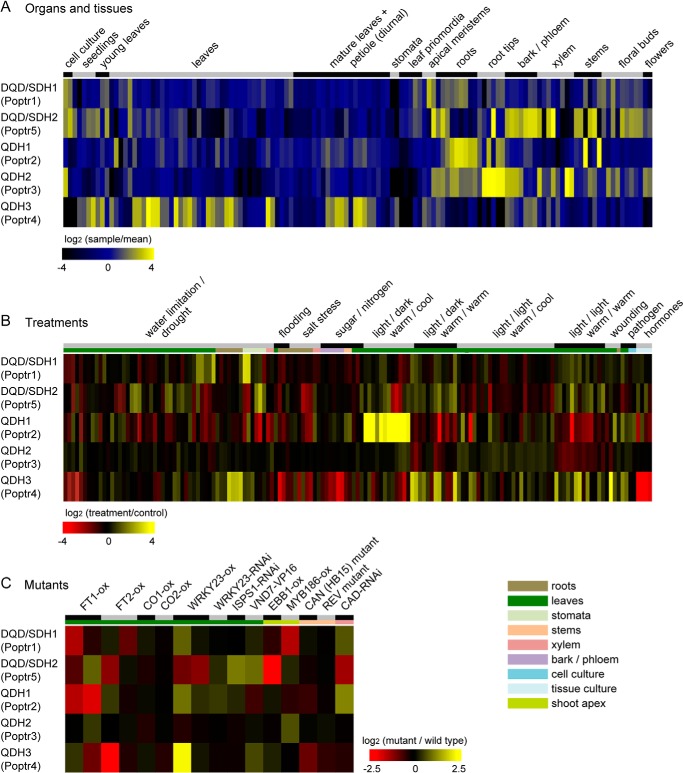
To assess expression profiles of Poplar DQD/SDHs and QDHs across different organs and their induction in response to environmental stimuli, publicly available poplar Affymetrix microarray data (748 hybridizations) were retrieved, normalized, and grouped into developmental samples, treatments (compared with the respective controls), and mutant (transgenics) versus wild type comparisons. Processed data were extracted from each group to generate expression profiles of DQD/SDHs and QDHs. DQD/SDH1 (Poptr1) was constitutively expressed (but at low levels) across a wide range of tissues and is expressed to highest levels in reproductive organs and actively growing tissues, such as shoot apices and leaf primordia (Fig. 7A). DQD/SDH1 was found to be co-expressed with three putative shikimate pathway genes as well as a phenylalanyl-tRNA synthetase and other tRNA synthetases (data not shown), which suggested a role in aromatic amino acid biosynthesis primarily for protein production. In contrast, DQD/SDH2 (Poptr5) is expressed to high levels in lignified tissues, and its expression in most other tissue types was negligible. QDH1 and QDH2 (Poptr2 and Poptr3) were found highly expressed in roots and root tips, respectively. QDH2 is also highly expressed in bark and some vascular tissues. QDH3 (Poptr4) showed a distinct expression pattern with predominant expression in leaves, seedlings, and stomata and some expression in bark and differentiating tissues (Fig. 7A). DQD/SDHs and QDHs respond differently to environmental stresses and genetic modifications. DQD/SDH1 expression was induced by water limitation especially in cotelydons and stomata (Fig. 7B). QDH1 showed high sensitivity toward light conditions and temperature changes, and its expression was induced by cold temperature at night. QDH3 showed a marked response to wounding and fungal infection, and its expression level was also increased in plants overexpressing the WRKY23 transcription factor involved in plant defense (Fig. 7C).
FIGURE 7.
Organ-specific expression of poplar DQD/SDHs and QDHs (A), their response to changing environmental factors (B), and their response to genetic perturbations (C). Publicly available large-scale microarray expression data for any Populus species were compiled, normalized, and grouped based on experiment annotations. Log-transformed expression ratios were visualized as heatmap. Organ expression data are shown relative to the mean expression of the respective gene across all experiments, treatment and mutant expression data are shown relative to the respective control experiments. Detailed descriptions of each sample and database accessions for the respective datasets can be found in supplemental Table 1.
DISCUSSION
The poplar DQD/SDH family encompasses two functionally distinct groups as follows: one specific for shikimate (DQD/SDHs) and the other specific for quinate (QDHs) in vitro. The three poplar QDHs were originally annotated as DQD/SDHs due to the fact they share significant sequence similarity with previously characterized DQD/SDHs (18). However, in-depth sequence and inferred structure comparison with functionally characterized DQD/SDHs revealed that QDHs are indeed distinct, and this was validated through in vitro enzymatic analysis. In addition, DQD/SDHs and QDHs showed different developmental expression patterns and stress responses indicative of different physiological and ecological roles of distinct isoforms. It needs to be stressed, however, that our annotations are based on biochemical in vitro data only, which will need to be validated through mutant analyses in vivo.
Amino Acid Residues Putatively Involved in Substrate Discrimination
Comparison of poplar QDH sequences with the characterized Arabidopsis SDH active site regions (14) revealed two distinct sites specific for QDHs. Ser-338, which is essential for SDH function in Arabidopsis (14), is replaced by Gly in the homologous position of all poplar QDHs and also in one of the two tobacco isoforms (annotated as NtDQD/SDH2). NtDQD/SDH2 has reduced SDH activity compared with NtDQD/SDH1 (17). The ability of NtDQD/SDH2 to use quinate as a substrate was not tested, but Ding et al. (17) suggested that the preferred substrate may be a shikimate derivative with a larger group at the C1 position, and quinate fits this description. The Ser-to-Gly conversion may generate extra space in the inferred QDH active sites that could accommodate the hydroxyl group at the C1 position of quinate. Instead of Thr at position 381 in Arabidopsis, a Gly residue is present at the homologous position of all poplar QDHs. This Thr residue is replaced by Ser in YdiB having both SDH and QDH activities (14, 27). Both Gly and Ser have smaller side groups compared with Thr and thus may leave more space in the active sites to accommodate quinate's distinct conformation. These sequence and structural considerations were in agreement with our hypothesis, and thus in vitro biochemical analyses were warranted
DQD/SDHs and QDHs in Poplar
The observed Km (shikimate) values for poplar DQD/SDHs are within the range of other functionally characterized DQD/SDHs (ranging from 130 to 860 μm) (14, 17, 37, 54). In contrast, the poplar QDH enzymes characterized here have lower Km (quinate) values (255 and 300 μm) compared with previously characterized QDHs partially purified from loblolly pine and Siberian larch, which range from 1.84 to 3.6 mm (36, 37). This higher affinity of poplar QDHs toward quinate may indicate highly specialized functions in quinate (derivative) metabolism in poplar. Both SDHs and QDHs share similar pH optima (between 8.5 and 9.5), which is consistent with the pH range observed in tobacco (9.0–9.4) (17). In terms of cofactor preference, NADP+ is generally considered to be the exclusive cofactor for SDHs since Yaniv and Gilvarg (55) described the first partial purification of SDH from E. coli and noted that “DPN [diphosphopyridine nucleotide, i.e. NAD+] could not replace TPN [triphosphopyridine nucleotide, i.e. NADP+] as electron acceptor.” The absence of detectable SDH activity with NAD+ was then confirmed for partially purified SDHs from mung bean and pea in 1961 (56, 57). These three studies also noted an absence of detectable dehydrogenase activity with quinate and other structurally related compounds. In consequence, subsequent studies reported exclusively on shikimate and NADP+ when characterizing plant SDHs (14, 15, 17, 58–60). Here, we confirm the absence of detectable SDH activity with NAD+ for DQD/SDH2 (Poptr5), but likely because of its unusually high dehydrogenase activity, we were able to detect minute SDH activity with NAD+ for DQD/SDH1 (Poptr1). However, the estimated Km (shikimate) nearly doubles, and maximum velocity is less than 5% of the SDH activity observed with shikimate when using NADP+.
In contrast to SDH, variations in cofactor preference have been reported for QDHs. For both poplar QDHs, activity was only detectable when using NAD+ as a cofactor. Likewise, QDH protein extracts from other angiosperms such as bean and maize, and those from most bacteria, use NAD+ exclusively or preferentially as a cofactor (35, 38, 41). In contrast, QDH enzyme fractions from gymnosperms prefer NADP+ (61). Differences in cofactor preference of SDH and QDH may allow separate regulation of carbon flux into either shikimate or quinate metabolism from their common precursor 3-dehydroquinate.
SDH and QDH activities are present in distinct protein fractions in maize and kiwi (41, 62), although in loblolly pine both activities are present in a single protein fraction (37). The two poplar QDHs do have residual SDH activity. Because gymnosperms appear to evolve at a lower rate than angiosperms (63), this may suggest an ongoing specialization of the QDH isoforms toward quinate in plants.
Absence of QD Activity
Quinate can be taken up by intact leucoplasts from pea roots and converted directly into shikimate via a QD activity (also referred to as quinate hydrolyase) (42, 64, 65). Because of the similarity of the QD reaction mechanism with that of DQD, we originally hypothesized that QD may be encoded by DQD-like genes in poplar. However, in vitro enzyme assays failed to support this hypothesis; QD activity was not detectable and instead only residual DQD activity was maintained in QDHs. It remains open whether the DQD activity is being lost and the N-terminal half of the QDH protein is maintained only for structural reasons, or whether another function not tested here has been gained by this domain.
Distinct Physiological Roles of Poplar DQD/SDHs and QDHs
Shikimate derivatives function in both primary metabolism (e.g. protein biosynthesis) and secondary metabolism (e.g. phenylpropanoid metabolism) (6). DQD/SDH1 (Poptr1) is broadly expressed with highest levels in actively growing tissues. Together with the co-expression analysis, this suggests that DQD/SDH1 is likely involved in housekeeping primary functions, i.e. protein production. DQD/SDH2 (Poptr5) is highly expressed in vascular tissues, which suggests a more specific role in providing precursors for lignin biosynthesis. QDH1 (Poptr2) and QDH2 (Poptr3) are preferentially expressed in roots, suggesting a role of quinate and its derivatives in the rhizosphere. Chlorogenic acid, a common derivative of quinate, accumulates to high levels in the root tissue of many plant species, including sweet potato, carrot, lettuce, and burdock (66–70). It may be involved in developmental processes, such as root hair formation (66), or in defense against pests, such as root aphids in lettuce (69). In aerial parts, quinate and chlorogenic acid are formed mainly in dormant buds, young leaves, and bark of poplar trees, and its production is induced by herbivore and pathogen infestation (71–73). Although attempts to biochemically characterize QDH3 proved unsuccessful, its predominant expression in leaves and its induction in response to wounding and fungal infection suggest a defensive role in leaves through production of quinate and its derivatives such as chlorogenic acid (74). Chlorogenic acid has both anti-herbivore and anti-microbial activities (75–77), and quinate might also act as an astringent herbivore deterrent in the above-mentioned organs (78). The possible role of QDH3 in defense is further highlighted by its increased expression in poplar plant overexpressing WRKY23, as transcription factors involved in the regulation of plant defense against pathogens and abiotic stresses (79).
In conclusion, we showed that quinate and shikimate metabolic activities are encoded by distinct members of the same gene family in poplar. This now sets the groundwork for exploring the molecular evolutionary history of the gene family to decipher how genes involved in primary metabolism evolved to adopt a function in plant secondary metabolism. It also enables studying the in vivo physiological role of QDH in plant development and chemical ecology using reverse genetic approaches.
Acknowledgments
We thank Johannes Moquenco for technical assistance and Dr. C. Peter Constabel for critical reading of the manuscript. We appreciate the help from Dr. Martin Boulanger with protein structure modeling and interpretation. We are thankful to Dr. Osao Adachi for providing 3-dehydroshikimate prior to it becoming commercially available. Laboratory infrastructure was supported by a Leaders Opportunity Fund from the Canadian Foundation for Innovation (to J. E.).
This work was supported in part by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (to J. E.).

This article contains supplemental Table S1.
- DQD
- dehydroquinate dehydratase
- SDH
- shikimate dehydrogenase
- QDH
- quinate dehydrogenase
- QD
- quinate dehydratase
- NTA
- nitrilotriacetic acid.
REFERENCES
- 1. Floss H. G. (1979) The shikimate pathway. Recent Adv. Phytochem. 12, 59–89 [Google Scholar]
- 2. Herrmann K. M. (1995) The shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7, 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmid J., Amrhein N. (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39, 737–749 [Google Scholar]
- 4. Roberts F., Roberts C. W., Johnson J. J., Kyle D. E., Krell T., Coggins J. R., Coombs G. H., Milhous W. K., Tzipori S., Ferguson D. J., Chakrabarti D., McLeod R. (1998) Evidence for the shikimate pathway in apicomplexan parasites. Nature 393, 801–805 [DOI] [PubMed] [Google Scholar]
- 5. Tohge T., Watanabe M., Hoefgen R., Fernie A. R. (2013) Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maeda H., Dudareva N. (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105 [DOI] [PubMed] [Google Scholar]
- 7. Steinrücken H. C., Amrhein N. (1980) The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 94, 1207–1212 [DOI] [PubMed] [Google Scholar]
- 8. O'Callaghan D., Maskell D., Liew F. Y., Easmon C. S., Dougan G. (1988) Characterization of aromatic- and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect. Immun. 56, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X., Zhang S., Hao F., Lai X., Yu H., Huang Y., Wang H. (2005) Expression, purification and properties of shikimate dehydrogenase from Mycobacterium tuberculosis. J. Biochem. Mol. Biol. 38, 624–631 [DOI] [PubMed] [Google Scholar]
- 10. Herrmann K. M. (1995) The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 107, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herrmann K. M., Weaver L. M. (1999) The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503 [DOI] [PubMed] [Google Scholar]
- 12. Weaver L. M., Herrmann K. M. (1997) Dynamics of the shikimate pathway in plants. Trends Plant Sci. 2, 346–351 [Google Scholar]
- 13. Dewick P. M. (1995) The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 12, 579–607 [DOI] [PubMed] [Google Scholar]
- 14. Singh S. A., Christendat D. (2006) Structure of Arabidopsis dehydroquinate dehydratase shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway. Biochemistry 45, 7787–7796 [DOI] [PubMed] [Google Scholar]
- 15. Bischoff M., Schaller A., Bieri F., Kessler F., Amrhein N., Schmid J. (2001) Molecular charaterization of tomato 3-dehydroquinate dehydratase-shikimate: NADP oxidoreductase. Plant Physiol. 125, 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonner C. A., Jensen R. A. (1994) Cloning of cDNA encoding the bifunctional dehydroquinase. Shikimate dehydrogenase of aromatic- amino-acid biosynthesis in Nicotiana tabacum. Biochem. J. 302, 11–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding L., Hofius D., Hajirezaei M. R., Fernie A. R., Börnke F., Sonnewald U. (2007) Functional analysis of the essential bifunctional tobacco enzyme 3-dehydroquinate dehydratase/shikimate dehydrogenase in transgenic tobacco plants. J. Exp. Bot. 58, 2053–2067 [DOI] [PubMed] [Google Scholar]
- 18. Hamberger B., Ehlting J., Barbazuk B., Douglas C. J. (2006) Comparative genomics of the shikimate pathway in Arabidopsis, Populus trichocarpa and Oryza sativa: shikimate pathway gene family structure and identification of candidates for missing links in phenylalanine biosynthesis. Recent Adv. Phytochem. 40, 85–113 [Google Scholar]
- 19. Bentley R. (1990) The shikimate pathway–a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25, 307–384 [DOI] [PubMed] [Google Scholar]
- 20. Farah A., Monteiro M., Donangelo C. M., Lafay S. (2008) Chlorogenic acids from green coffee extracts are highly bioavailable in humans. J. Nutr. 138, 2309–2315 [DOI] [PubMed] [Google Scholar]
- 21. Lou Z., Wang H., Zhu S., Ma C., Wang Z. (2011) Antibacterial activity and mechanism of action of chlorogenic acid. J. Food Sci. 76, M398–M403 [DOI] [PubMed] [Google Scholar]
- 22. Hoffmann L., Besseau S., Geoffroy P., Ritzenthaler C., Meyer D., Lapierre C., Pollet B., Legrand M. (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16, 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maher E. A., Bate N. J., Ni W., Elkind Y., Dixon R. A., Lamb C. J. (1994) Increased disease susceptibility of transgenic tobacco plants with suppressed levels of preformed phenylpropanoid products. Proc. Natl. Acad. Sci. U.S.A. 91, 7802–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paynter N. P., Yeh H. C., Voutilainen S., Schmidt M. I., Heiss G., Folsom A. R., Brancati F. L., Kao W. H. (2006) Coffee and sweetened beverage consumption and the risk of type 2 diabetes mellitus: the atherosclerosis risk in communities study. Am. J. Epidemiol. 164, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 25. Morton L. W., Abu-Amsha Caccetta R., Puddey I. B., Croft K. D. (2000) The chemistry and biological effects of dietary phenolic compounds: relevance to cardiovascular disease. Clin. Exp. Pharmacol. Physiol. 27, 152–159 [DOI] [PubMed] [Google Scholar]
- 26. Nie L., Shi X. (2009) A novel asymmetric synthesis of oseltamivir phosphate (Tamiflu) from (−)-shikimic acid. Tetrahedron Asymmetry 20, 124–129 [Google Scholar]
- 27. Lindner H. A., Nadeau G., Matte A., Michel G., Ménard R., Cygler M. (2005) Site-directed mutagenesis of the active site region in the quinate/shikimate 5-dehydrogenase YdiB of Escherichia coli. J. Biol. Chem. 280, 7162–7169 [DOI] [PubMed] [Google Scholar]
- 28. Minamikawa T., Yoshida S., Hasegawa M. (1969) Alicyclic acid metabolism in plants 3. Fate of 14C-shikimate and 14C-quinate in mung bean plants. Plant Cell Physiol. 10, 283–289 [Google Scholar]
- 29. Yoshida S. (1969) Biosynthesis and conversion of aromatic amino acids in plants. Annu. Rev. Plant Physiol. 20, 41–62 [Google Scholar]
- 30. Boudet A. M. (1980) Studies on quinic acid biosynthesis in Quercus pedunculata Ehrh. seedlings. Plant Cell Physiol. 21, 785–792 [Google Scholar]
- 31. Gamborg O. L. (1967) Aromatic metabolism in plants—IV: the interconversion of shikimic acid and quinic acid by enzymes from plant cell cultures. Phytochemistry 6, 1067–1073 [Google Scholar]
- 32. Weinstein L. H., Porter C. A., Laurencot H. J. (1961) Role of quinic acid in aromatic biosynthesis in higher plants. Contrib. Boyce Thompson Inst. 21, 201–214 [Google Scholar]
- 33. Goldschmidt O., Quimby G. R. (1964) The role of quinic acid. Tappi 47, 528–533 [Google Scholar]
- 34. Gamborg O. L. (1966) Aromatic metabolism in plants III. Quinate dehydrogenase from mung bean cell suspension culture. Biochim. Biophys. Acta 128, 483–491 [DOI] [PubMed] [Google Scholar]
- 35. Kang X., Scheibe R. (1993) Purification and characterization of the quinate: oxidoreductase from Phaseolus mungo sprouts. Phytochemistry 33, 769–773 [Google Scholar]
- 36. Ossipov V., Chernov A., Zrazhevskaya G., Shein I. (1995) Quinate:NAD(P)+-oxidoreductase from Larix sibirica: purification, characterization and function. Trees 10, 46–51 [Google Scholar]
- 37. Ossipov V., Bonner C., Ossipova S., Jensen R. (2000) Broad-specificity quinate (shikimate) dehydrogenase from Pinus taeda needles. Plant Physiol. Biochem. 38, 923–928 [Google Scholar]
- 38. Minamikawa T. (1977) Quinate:NAD oxidoreductase of germinating Phaseolus mungo seeds: partial purification and some properties. Plant Cell Physiol. 18, 743–752 [Google Scholar]
- 39. Refeno G., Ranjeva R., Boudet A. M. (1982) Modulation of quinate: NAD+ oxidoreductase activity through reversible phosphorylation in carrot cell suspensions. Planta 154, 193–198 [DOI] [PubMed] [Google Scholar]
- 40. Graziana A., Boudet A. M. (1983) in Groupe Polyphénols Journées Internationales d'Etudes et Assemblées Générales (Boudet A. M., Ranjeva R., eds) pp. 120–125 [Google Scholar]
- 41. Graziana A., Boudet A., Boudet A. M. (1980) Association of the quinate: NAD+ oxidoreductase with one dehydroquinate hydro-lyase isoenzyme in corn seedlings. Plant Cell Physiol. 21, 1163–1174 [Google Scholar]
- 42. Leuschner C., Herrmann K. M., Schultz G. (1995) The metabolism of quinate in pea roots (purification and partial characterization of a quinate hydrolyase). Plant Physiol. 108, 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall T. A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 44. Felsenstein J. (1989) PHYLIP: Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166 [Google Scholar]
- 45. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 46. Haruta M., Major I. T., Christopher M. E., Patton J. J., Constabel C. P. (2001) A Kunitz trypsin inhibitor gene family from trembling aspen (Populus tremuloides Michx.): cloning, functional expression, and induction by wounding and herbivory. Plant Mol. Biol. 46, 347–359 [DOI] [PubMed] [Google Scholar]
- 47. Geu-Flores F., Nour-Eldin H. H., Nielsen M. T., Halkier B. A. (2007) USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 49. Diaz J., Merino F. (1997) Shikimate dehydrogenase from pepper (Capsicum annuum) seedlings, purification, and properties. Physiol. Plant. 100, 147–152 [Google Scholar]
- 50. Mousdale D. M., Coggins J. R. (1985) High-performance liquid chromatography of shikimate pathway intermediates. J. Chromatogr. 329, 268–272 [Google Scholar]
- 51. Givry S., Bliard C., Duchiron F. (2007) Selective ketopentose analysis in concentrate carbohydrate syrups by HPLC. Carbohydr. Res. 342, 859–864 [DOI] [PubMed] [Google Scholar]
- 52. Toufighi K., Brady S. M., Austin R., Ly E., Provart N. J. (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 43, 153–163 [DOI] [PubMed] [Google Scholar]
- 53. Singh S. A., Christendat D. (2007) The DHQ-dehydroshikimate-SDH-shikimate-NADP(H) complex: insights into metabolite transfer in the shikimate pathway. Cryst. Growth Des. 7, 2153–2160 [Google Scholar]
- 54. Muir R. M., Ibáñez A. M., Uratsu S. L., Ingham E. S., Leslie C. A., McGranahan G. H., Batra N., Goyal S., Joseph J., Jemmis E. D., Dandekar A. M. (2011) Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 75, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yaniv H., Gilvarg C. (1955) Aromatic biosynthesis: XIV. 5-dehydroshikimic reductase. J. Biol. Chem. 213, 787–795 [PubMed] [Google Scholar]
- 56. Balinsky D., Davies D. D. (1961) Aromatic biosynthesis in higher plants. 1. preparation and properties of dehydroshikimic reductase. Biochem. J. 80, 292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nandy M., Ganguli N. C. (1961) Studies on 5-dehydroshikimic reductase from mung bean seedlings (Phaseolus aureus). Arch. Biochem. Biophys. 92, 399–408 [DOI] [PubMed] [Google Scholar]
- 58. Balinsky D., Dennis A. W., Cleland W. W. (1971) Kinetic and isotope-exchange studies on shikimate dehydrogenase from Pisum sativum. Biochemistry 10, 1947–1952 [DOI] [PubMed] [Google Scholar]
- 59. Dowsett J. R., Corbett J. R. (1971) The purification and properties of shikimate dehydrogenase. Biochem. J. 123, 23P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Davies D. D., Teixeira A., Kenworthy P. (1972) The stereospecificity of nicotinamide-adenine dinucleotide-dependent oxidoreductases from plants. Biochem. J, 127, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ossipov V., Shein I. (1986) The role of quinate-dehydrogenase in quinic acid metabolism in coniferous plants. Biochimiya 51, 230–236 [Google Scholar]
- 62. Marsh K. B., Boldingh H. L., Shilton R. S., Laing W. A. (2009) Changes in quinic acid metabolism during fruit development in three kiwifruit species. Funct. Plant Biol. 36, 463–470 [DOI] [PubMed] [Google Scholar]
- 63. Buschiazzo E., Ritland C., Bohlmann J., Ritland K. (2012) Slow but not low: genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol. Biol. 12, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Leuschner C., Schultz G. (1991) Uptake of shikimate pathway intermediates in chloroplasts. Phytochemistry 30, 2203–2207 [Google Scholar]
- 65. Schmidt C. L., Gründemann D., Groth G., Müller B., Hennig H., Schultz G. (1991) Shikimate pathway in non-photosynthetic tissues. Identification of common enzymes and partial purification of dehydroquinate hydrolyase, shikimate oxidoreductase and chorismate mutase from roots. J. Plant Physiol. 138, 51–56 [Google Scholar]
- 66. Pilet P. E. (1964) Effect of chlorogenic acid on the auxin catabolism and the auxin content of root tissues. Phytochemistry 3, 617–621 [Google Scholar]
- 67. Cole R. A. (1985) Relationship between the concentration of chlorogenic acid in carrot roots and the incidence of carrot fly larval damage. Ann. Appl. Biol. 106, 211–217 [Google Scholar]
- 68. McClure T. T. (1960) Chlorogenic acid accumulation and wound healing in sweet potato roots. Am. J. Bot. 47, 277–280 [Google Scholar]
- 69. Cole R. A. (1984) Phenolic acids associated with the resistance of lettuce cultivars to the lettuce root aphid. Ann. Appl. Biol. 105, 129–145 [Google Scholar]
- 70. Maruta Y., Kawabata J., Niki R. (1995) Antioxidative caffeoylquinic acid derivatives in the roots of burdock (Arctium lappa L.). J. Agric. Food Chem. 43, 2592–2595 [Google Scholar]
- 71. Edwards P. J., Wratten S. D. (1983) Wound induced defences in plants and their consequences for patterns of insect grazing. Oecologia 59, 88–93 [DOI] [PubMed] [Google Scholar]
- 72. Johnson G., Schaal L. A. (1952) Relation of chlorogenic acid to scab resistance in potatoes. Science 115, 627–629 [DOI] [PubMed] [Google Scholar]
- 73. Friend J., Reynolds S. B., Aveyard M. A. (1973) Phenylalanine ammonia lyase, chlorogenic acid and lignin in potato tuber tissue inoculated with Phytophthora infestans. Physiol. Plant Pathol. 3, 495–507 [Google Scholar]
- 74. Sung W. S., Lee D. G. (2010) Antifungal action of chlorogenic acid against pathogenic fungi, mediated by membrane disruption. Pure Appl. Chem. 82, 219–226 [Google Scholar]
- 75. De Sotillo D. R., Hadley M., Wolf-Hall C. (1998) Potato peel extract a nonmutagenic antioxidant with potential antimicrobial activity. J. Food Sci. 63, 907–910 [Google Scholar]
- 76. Ma C. M., Kully M., Khan J. K., Hattori M., Daneshtalab M. (2007) Synthesis of chlorogenic acid derivatives with promising antifungal activity. Bioorg. Med. Chem. 15, 6830–6833 [DOI] [PubMed] [Google Scholar]
- 77. Leiss K. A., Maltese F., Choi Y. H., Verpoorte R., Klinkhamer P. G. (2009) Identification of chlorogenic acid as a resistance factor for thrips in Chrysanthemum. Plant Physiol. 150, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rogiers S. Y., Knowles N. R. (1997) Physical and chemical changes during growth, maturation, and ripening of saskatoon (Amelanchier alnifolia) fruit. Can. J. Bot. 75, 1215–1225 [Google Scholar]
- 79. Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258 [DOI] [PubMed] [Google Scholar]