Background: The YvcK protein is essential for Bacillus subtilis growth on gluconeogenic conditions; its overproduction rescues an mreB mutant.
Results: PrkC phosphorylates YvcK; this phosphorylation is not required for growth on gluconeogenic conditions but is necessary for mreB rescue.
Conclusion: YvcK phosphorylation is specifically involved in B. subtilis morphogenesis.
Significance: This phosphorylation-based regulatory mechanism could be widespread in bacteria.
Keywords: Antibiotic Resistance, Bacillus, Cell Wall, Gluconeogenesis, Morphogenesis, Protein Phosphorylation
Abstract
The YvcK protein has been shown to be necessary for growth under gluconeogenic conditions in Bacillus subtilis. Amazingly, its overproduction rescues growth and morphology defects of the actin-like protein MreB deletion mutant by restoration of PBP1 localization. In this work, we observed that YvcK was phosphorylated at Thr-304 by the protein kinase PrkC and that phosphorylated YvcK was dephosphorylated by the cognate phosphatase PrpC. We show that neither substitution of this threonine with a constitutively phosphorylated mimicking glutamic acid residue or a phosphorylation-dead mimicking alanine residue nor deletion of prkC or prpC altered the ability of B. subtilis to grow under gluconeogenic conditions. However, we observed that a prpC mutant and a yvcK mutant were more sensitive to bacitracin compared with the WT strain. In addition, the bacitracin sensitivity of strains in which YvcK Thr-304 was replaced with either an alanine or a glutamic acid residue was also affected. We also analyzed rescue of the mreB mutant strain by overproduction of YvcK in which the phosphorylation site was substituted. We show that YvcK T304A overproduction did not rescue the mreB mutant aberrant morphology due to PBP1 mislocalization. The same observation was made in an mreB prkC double mutant overproducing YvcK. Altogether, these data show that YvcK may have two distinct functions: 1) in carbon source utilization independent of its phosphorylation level and 2) in cell wall biosynthesis and morphogenesis through its phosphorylation state.
Introduction
Phosphorylation at serine/threonine residues by protein kinases regulates a wide range of biological functions in bacteria such as biosynthesis and metabolism of major cell wall-associated components and cell division (1–3). Bacillus subtilis encodes four Ser/Thr kinases whose function still remains largely unknown (4). PrkC, the best characterized Ser/Thr kinase, seems to be involved in signaling bacteria to exit dormancy in response to peptidoglycan fragments (5). It possesses extracellular domains composed of PASTA motifs (6) that are predicted to interact with cell wall peptidoglycans. In addition, PrkC phosphorylates various proteins such as the small ribosome-associated GTPase CpgA, the translation factors EF-Tu and EF-G, the component of the stressosome YezB, and several metabolic enzymes (4, 5, 7, 8). Thus, PrkC probably acts as a membrane kinase receptor. In B. subtilis, the prkC gene is located downstream of the prpC gene, encoding a phosphoserine/threonine phosphatase. The two proteins play opposite roles during stationary phase physiology, and PrpC was shown to dephosphorylate the phosphorylated substrates of PrkC (9–11).
Homologs of PrkC are present in a broad range of bacteria. In particular, PknB, a PrkC homolog in Mycobacterium tuberculosis, is an essential kinase involved in the regulation of cell shape and cell division (12). Interestingly, PknB phosphorylates Rv1422, a protein present in many bacteria, at the single yet non-conserved Thr-325 residue in vivo and in vitro (12, 13). Rv1422 belongs to the uncharacterized protein family related to CofD, a transferase involved in coenzyme F420 biosynthesis in archaea with high G + C content Gram-positive bacteria (14). Its role and that of its homologs in other bacteria remain unknown because these bacteria do not possess coenzyme F420. However, these proteins seem to have important functions. For example, in Staphylococcus aureus, the Rv1422 homolog is essential (15). In Listeria monocytogenes, a mutation in lmo2473, the gene encoding the Rv1422 homolog, causes pyroptosis, followed by bacterial lysis in the macrophage cytosol of the host (16). The role of Rv1422 phosphorylation remains enigmatic in M. tuberculosis, and none of its homologs has been described to be phosphorylated so far.
Interestingly, the function of the Rv1422 homolog YvcK in B. subtilis is better understood. YvcK is necessary for growth on substrates of the pentose phosphate pathway and Krebs cycle intermediates, molecules that all drive gluconeogenesis for the synthesis of cell wall precursors (17). Moreover, deconvolution fluorescence microscopy revealed that it is localized as a helix-like pattern in the cell, similar to MreB, a key actin-like cytoskeletal component in rod-shaped bacteria (18). The helical localization of MreB, which was observed by deconvolution fluorescence microscopy and reported in several publications, has recently been challenged (19). Namely, the use of high resolution fluorescence microscopy suggests that, instead of helical filamentous structures, MreB forms patches that move circumferentially around the cell and that are dependent on the cell wall synthesis machinery (20, 21). Importantly, YvcK is required for the correct localization of PBP1, a major penicillin-binding protein implicated in cell wall synthesis and cell division, and hence for maintaining the normal shape under gluconeogenic growth conditions (18). Amazingly, overproduction of YvcK rescues the morphology defects of cells lacking MreB independently of the carbon source present in the growth medium. The mreB mutant exhibits a characteristic bulging phenotype because PBP1 is mislocalized (22, 23). Overproduction of YvcK in the absence of MreB restores proper PBP1 localization and hence a normal cell shape.
In this study, we investigated whether YvcK is phosphorylated by PrkC, similar to Rv1422, which is phosphorylated by PknB. We show that PrkC phosphorylated YvcK at a single phosphorylation site (Thr-304). This phosphorylation did not affect growth under gluconeogenic conditions. Interestingly, both the bacitracin sensitivity and rescue of an mreB mutant through the proper localization of PBP1 were affected by phosphorylation of YvcK Thr-304. Our data suggest two distinct functions of YvcK in the cell.
EXPERIMENTAL PROCEDURES
Plasmid and Strain Constructions
Standard procedures for molecular cloning and cell transformation of B. subtilis and Escherichia coli were used. All strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Primers used in this study are available upon request.
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant structures or genotypes | Ref./source |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| 3725 | trpC2, mreB::neo3427 | Ref. 22 |
| PB702 | trpC2, ΔprpC | Ref. 9 |
| BKE15760 | trpC2, prpC::erm | Genetic Stock Center |
| PB705 | trpC2, ΔprkC | Ref. 9 |
| YK706 | trpC2, amyE:: (Pxyl gfp-ponA spc) | Ref. 23 |
| SG63 | trpC2, yvcK:: (lacZ, cat, rrnBt1t2 λt0) | Ref. 17 |
| SG116 | trpC2, yvcK:: (lacZ, cat, rrnBt1t2λt0) amyE:: (Pxyl yvcK-gfp, spc) | Ref. 18 |
| SG204 | trpC2, mreB::neo3427 amyE:: (Pxyl gfp-ponA spc) | Ref. 18 |
| SG217 | trpC2, mreB::neo3427, pDG148-YvcK amyE:: (Pxyl gfp-ponA spc) | Ref. 18 |
| SG252 | trpC2, yvcK:: (lacZ, cat, rrnBt1t2λt0) amyE:: (Pxyl yvcK T304A-gfp, spc) | This study (pEFK10 → SG63) |
| SG262 | trpC2, mreB::neo3427, pDG148-YvcK T304A amyE:: (Pxyl gfp-ponA spc) | This study (pEFK12 → SG204) |
| SG264 | trpC2, mreB::neo3427, ΔprkC, amyE:: (Pxyl gfp-ponA spc) | This study (SG204 → PB705) |
| SG265 | trpC2, mreB::neo3427, ΔprkC, pDG148-YvcK amyE:: (Pxyl gfp-ponA spc) | This study (pBGM54 → SG264) |
| SG281 | trpC2, mreB::neo3427, pDG148-YvcK T304E amyE:: (Pxyl gfp-ponA spc) | This study (pEFK13 → SG204) |
| SG283 | trpC2, yvcK:: (lacZ, cat, rrnBt1t2λt0) amyE:: (Pxyl yvcK T304E-gfp spc) | This study (pEFK11 → SG63) |
| SG376 | trpC2, ycgO:: (Phyperspank-prkC erm) | T. Doan, unpublished |
| SG413 | trpC2, mreB::neo3427, pDG148-YvcK amyE:: (Pxyl gfp-ponA spc) prpC::erm | This study (BKE15760 → SG217) |
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristics (Ref./source) |
|---|---|
| pBGM54 | pDG148-YvcK (17) |
| pEFK6 | pSG1154-YvcK-GFP (18) |
| pEFK8 | pGEX-KT-YvcK (this study) |
| pEFK9 | pGEX-KT-YvcK T304A (this study) |
| pEFK10 | pSG1154-YvcK T304A-GFP (this study) |
| pEFK11 | pSG1154-YvcK T304E-GFP (this study) |
| pEFK12 | pDG148-YvcK T304A (this study) |
| pEFK13 | pDG148-YvcK T304E (this study) |
| pEFK14 | pQE30-YvcK (this study) |
For the generation of fusion proteins for the adenylate cyclase-based two-hybrid assay, the catalytic domain of the prkC gene was amplified by PCR using specific primers and inserted between the KpnI and HindIII sites in the pT18 plasmid. The yvcK gene and the catalytic domains of the prkC gene were also amplified using specific primers and inserted between the PstI and BamHI sites in the pT25 plasmid. For YvcK overproduction in E. coli, the yvcK gene was amplified by PCR using B. subtilis 168 genomic DNA as a template and a primer pair containing either PstI and BamHI or BamHI and EcoRI restriction sites. The amplified products were digested either with PstI and BamHI and then ligated to the pQE30 vector or with BamHI and EcoRI and then ligated to the pGEX-KT vector. Point mutations to substitute Thr-304 were introduced into the yvcK gene by site-directed mutagenesis by PCR amplification of plasmids pEFK8 (pGEX-KT-YvcK), pEFK6 (18), and pBGM54 (17) to give plasmids pEFK9 to pEFK12 (Table 2). For rescue of the yvcK mutant strain by yvcK mutant alleles, the SG63 strain (17) was transformed with plasmid pEFK10 or pEFK11. The mutant allele fused to the gfp gene was inserted by a double crossover event at the amyE locus and selected for spectinomycin resistance. The yvcK-gfp gene fusion was expressed from the Pxyl promoter in the presence of 0.25% xylose. Under these conditions, the mutant lacking YvcK displays both growth and cell shape defects (18). For rescue of the mreB mutant strain by overproduction of yvcK mutant alleles, the SG204 strain (18) was transformed with plasmid pEFK12 or pEFK13, in which yvcK is under the control of the Pspac promoter. In the resulting strains, yvcK alleles were expressed in the presence of 100 μm isopropyl β-d-thiogalactopyranoside (IPTG),2 whereas the gfp-ponA gene fusion was expressed from the Pxyl promoter in the presence of 0.5% xylose. All resulting constructs were verified by DNA sequencing.
Bacterial Two-hybrid Assay
The bacterial two-hybrid assay was performed as described (24, 25). The N termini of the PrkC catalytic domain (PrkCc) and YvcK were fused to the T18 or T25 catalytic domain of adenylate cyclase using plasmids pT18 and pT25 (26). Cotransformed strains of E. coli BTH101 expressing T18-PrkCc or T25 and T25-YvcK or T18 were plated on LB agar and incubated at 30 °C for 48 h. One milliliter of LB medium supplemented with 100 μg/ml ampicillin, 50 μg/ml chloramphenicol, and 0.5 mm IPTG was inoculated and incubated overnight at 30 °C. Ten microliters of the overnight culture were spotted on the LB medium plates containing appropriate antibiotics, 0.5 mm IPTG, and 40 μg/ml X-Gal. The plates were incubated overnight at 30 °C.
Western Blotting
After growth, bacteria were centrifuged for 10 min at 8000 rpm at 4 °C. Cell pellets were resuspended in 0.1 volume of lysis buffer containing 10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1% Nonidet P-40, 1 mm PMSF, 25 units/ml Benzonase, and 10 mg/ml lysozyme. Extracts were incubated for 10 min on ice and then heated at 100 °C for 10 min. Samples were run on a 12.5% SDS-polyacrylamide gel and transferred to nitrocellulose membrane by electroblotting. The membrane was blocked with PBS solution containing 5% (w/v) milk powder for 1 h at room temperature with shaking and then incubated with anti-YvcK antibodies (1:2500 dilution) and fluorescent Alexa Fluor 647-labeled goat anti-rabbit IgG secondary antibody (1:5000 dilution; Molecular Probes) as described previously (18). After three washes, the membrane was scanned at 700 nm using the Odyssey infrared imaging system (LI-COR Biosciences).
Protein Purifications
Plasmids overproducing His6-tagged or GST-tagged YvcK (WT or mutant) were used to transform E. coli DH5α. Purification of His6-tagged or GST-tagged recombinant proteins was performed with nickel-nitrilotriacetic acid agarose resin (Qiagen) or glutathione-Sepharose 4B (GE Healthcare), respectively, as described previously (27, 28). Recombinant soluble PrkCc and the PrpC phosphatase were expressed and purified as described (10).
Determination of YvcK Phosphorylation Site
Purified YvcK (2 μg) was incubated for 15 min at 37 °C with 1 μg of PrkCc in buffer containing 10 mm HEPES (pH 8.0), 5 mm MgCl2, 3 μg of myelin basic protein (which has been shown to stimulate PrkC kinase activity (10, 29)), and 1 mm ATP with [γ-33]ATP (1 μCi). Where indicated, 1 μg of PrpC was added after 7.5 min of incubation. The phosphorylation reaction was quenched by the addition of 5-fold concentrated loading buffer to the reaction mixtures before SDS-PAGE analysis. Gels were then dried, and phosphorylated proteins were visualized by autoradiography. Characterization of the phosphorylated amino acid residues was done as described previously (8). For determination of phosphorylation site(s), YvcK was phosphorylated in vitro by PrkCc as described above with 5 mm ATP and subjected to subsequent nanoLC-electrospray ionization-MS/MS analysis after tryptic and chymotryptic digestion as described previously (3).
Antibiotic Sensitivity Test Using Disk Diffusion Assays
Strains were grown on LB medium to an A600 of 0.8. A 150-μl aliquot of each culture was used to inoculate an LB plate, which was dried for 20 min. Filter paper disks containing 200 μg of bacitracin were then placed on the plates, which were incubated overnight at 37 °C (16 h) before observation. The overall diameter of the inhibition zones was measured along two orthogonal lines. Zones of inhibition are reported as the average diameter minus the 8.4-mm diameter of the filter paper disk.
Image Acquisition and Processing
Images and Z-stacks of 21 images were captured at a step distance range of 0.15 μm as described previously (18). Image restoration was obtained by deconvolution using Huygens Essential software (Scientific Volume Imaging). Three-dimensional visualization was performed with the Imaris software package (Bitplane).
RESULTS AND DISCUSSION
PrkC Phosphorylates YvcK Protein at Thr-304
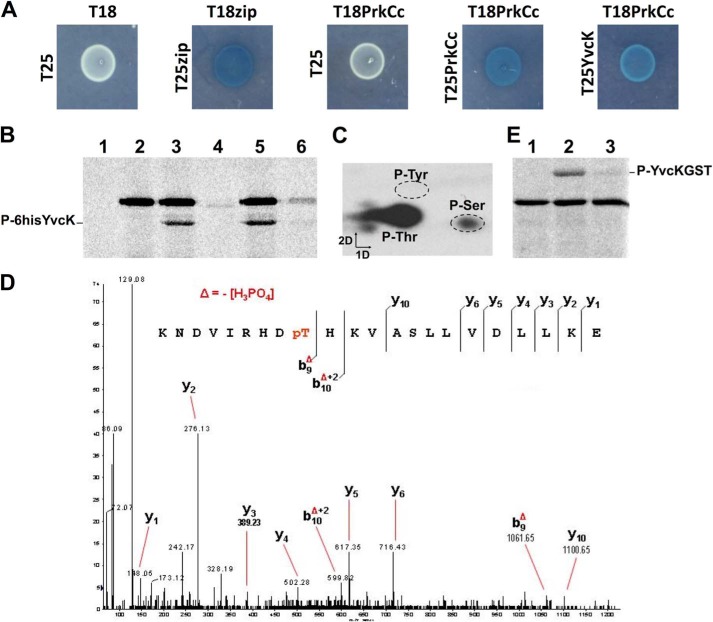
To investigate putative phosphorylation of YvcK by PrkC, we first performed bacterial two-hybrid experiments. We detected a specific interaction between PrkCc and YvcK, suggesting that YvcK could be phosphorylated by PrkC (Fig. 1A). To further investigate putative phosphorylation of YvcK by PrkC, we performed in vitro kinase assays. B. subtilis proteins YvcK and PrkCc were purified from E. coli and incubated together in the presence of [γ-33P]ATP. The reaction was analyzed by SDS-PAGE, and the phosphorylation pattern was visualized by autoradiography (Fig. 1B). The presence of two radiolabeled bands indicated that PrkCc was able to autophosphorylate, as reported previously (10, 29), and to efficiently phosphorylate YvcK. To ensure that YvcK phosphorylation was not catalyzed by a contaminating protein present in the PrkC preparation, YvcK was also incubated in the presence of purified inactive PrkCc, in which the catalytic Lys residue was replaced with Ala. Under these conditions, no PrkC autophosphorylation and phosphorylation of YvcK were detected. In addition, when YvcK previously phosphorylated by PrkCc was further incubated with the phosphatase PrpC, no phosphorylation was detected, indicating that PrpC is able to dephosphorylate the phosphorylated YvcK protein. We then determined the nature of the phosphorylated amino acid residues of YvcK. Only Thr and, to a lesser extent, Ser residues were phosphorylated (Fig. 1C). A mass spectrometry approach was used to identify which residues were phosphorylated. Analysis of tryptic and chymotryptic digests allowed 90% sequence coverage. Importantly, a single phosphorylation site corresponding to Thr-304 of YvcK was detected (Fig. 1D). Conclusive identification of Thr-304 as the single phosphorylation site was achieved by substituting Thr-304 for an unphosphorylatable Ala residue. For this, YvcK T304A was fused to the glutathione S-transferase moiety, expressed in E. coli, purified, and incubated in the presence of PrkCc and ATP, and its phosphorylation was analyzed as described above. Only a very weak signal was detected by radiolabeling with [γ-33P]ATP (Fig. 1E). NanoLC-electrospray ionization-MS/MS analysis failed to identify any additional phosphate group. These results show that PrkC phosphorylates YvcK at a single Thr residue at position 304 and that the weak radiolabeled signal detected was probably unspecific for the phosphorylated Ser residue observed in Fig. 1C.
FIGURE 1.
Phosphorylation of YvcK by PrkC at Thr-304. A, interaction between PrkC and YvcK by bacterial two-hybrid assay. The T18 and T25 fragments of the adenylate cyclase protein were fused to the N termini of PrkCc and YvcK. Cotransformed strains of E. coli BTH101 were spotted onto LB medium supplemented with X-Gal and IPTG and incubated overnight at 30 °C. Blue colonies indicate a positive interaction of the two moieties of the β-galactosidase enzyme and hence a positive interaction of PrkCc and YvcK. B, phosphorylation of YvcK by PrkCc. The YvcK protein was expressed either with a C-terminal His6 tag (A–C) yielding an ∼35-kDa protein, or with an N-terminal GST tag (D), yielding a 60-kDa protein. The recombinant proteins were purified and then incubated with [γ-33P]ATP and PrkCc. Myelin basic protein and [γ-33P]ATP were incubated at 37 °C for 15 min in the presence of YvcK alone (lane 1), in the presence of PrkCc alone (lane 2), in the presence of PrkCc and YvcK (lane 3), in the presence of PrkCc K40A and YvcK (lane 4), or in the presence of PrkCc and YvcK with the addition of either buffer (lane 5) or PrpC (lane 6) after 7.5 min of incubation. Samples were separated by SDS-PAGE and visualized by autoradiography. C, phosphoamino acid determination. Phosphorylated YvcK was purified and hydrolyzed, and the phosphorylated amino acid residues were separated by electrophoresis in the first dimension and by ascending chromatography in the second dimension. The positions of the unlabeled phosphoamino acid standards are circled. D, identification of Thr-304 as the phosphorylated residue in YvcK by mass spectrometry. Shown is the MS/MS spectrum of the doubly charged ion [M + 2H]2+ at m/z 656.6 of peptide-(296–317) (monoisotopic mass of 2622.42 Da). The phosphate group on Thr-304 was identified by observation of the y C-terminal daughter ion series. Starting from the C-terminal residue, all y ions lose phosphoric acid (−98 Da) after the phosphorylated Thr-304 residue. E, phosphorylation of the GST-YvcK T304A mutant. Two micrograms of purified GST (lane 1), GST-YvcK (lane 2), and GST-YvcK T304A (lane 3) were incubated with myelin basic protein, PrkCc, and [γ-33]ATP. Samples were separated by SDS-PAGE and visualized by autoradiography.
Phosphorylation of YvcK by PrkC Is Not Required for Growth under Gluconeogenic Growth Conditions
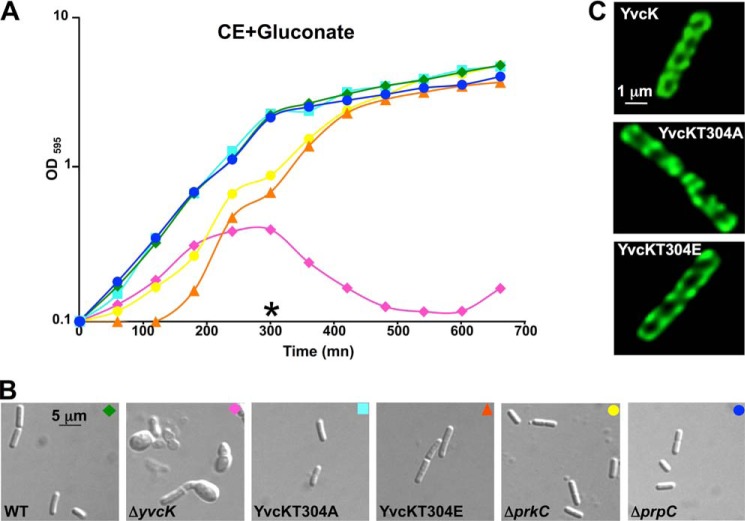
To determine the function of YvcK phosphorylation in vivo, we constructed two strains expressing either a phosphoablative (T304A) or constitutively phosphorylated mimetic (T304E) yvcK mutant allele. These substitutions are commonly used to study the physiological relevance of protein phosphorylation (30–32). The growth of both strains was analyzed on CE-gluconate minimal medium. We observed that both strains expressing YvcK in which the phosphorylation site was modified (T304A or T304E) grew normally and had a rod-shaped morphology on CE-gluconate minimal medium. In addition, similar observations were made for a prkC mutant or a prpC mutant strain (Fig. 2, A and B). These data strongly suggest that the phosphorylation of YvcK by PrkC does not affect growth under gluconeogenic conditions.
FIGURE 2.
No impact of YvcK phosphorylation on growth under gluconeogenic conditions. A, growth curves on CE-gluconate medium. Strains SG116 (yvcK-gfp), SG63 (ΔyvcK), SG252 (yvcK T304A-gfp), SG283 (yvcK T304E-gfp), PB705 (ΔprkC), and PB702 (ΔprpC) were grown overnight on LB medium. After centrifugation, cells were grown on CE-gluconate liquid medium supplemented with 0.25% xylose at 37 °C. B, cell shape observation by differential interference contrast microscopy. Cells were observed during growth on CE-gluconate liquid medium, and the images presented were recorded 5 h after inoculation, as indicated by the asterisk in the corresponding growth curves. Green diamonds, SG116; pink diamonds, SG63; cyan squares, SG252; orange triangles, SG283; yellow circles, PB705; blue circles, PB702. C, cellular localization of YvcK-GFP, YvcK T304A-GFP, and YvcK T304E-GFP. Localization was carried out in strains SG116 (YvcK-GFP), SG252 (YvcK T304A-GFP), and SG283 (YvcK T304E-GFP). YvcK-GFP localization was illustrated by the brightest image from the deconvoluted stack.
YvcK Localization Is Not Dependent on Thr-304 Phosphorylation
Phosphorylation of bacterial proteins has already been shown to affect their cellular localization (33). We therefore decided to analyze the cellular localization of the two Thr-304 mutated YvcK-GFP proteins. As shown in Fig. 2C, both constructs were organized as a helix-like structure similar to the WT protein. Furthermore, the localization of WT YvcK-GFP was also found to be similar to that in a WT strain in a prkC background (data not shown). We conclude that phosphorylation of YvcK does not affect its localization and function in growth on gluconeogenic carbon sources.
YvcK Phosphorylation Affects Bacitracin Resistance
YvcK was shown to be required for the correct localization of PBP1 under gluconeogenic growth conditions, and its overproduction in the absence of MreB restores proper PBP1 localization (18). These observations suggested that YvcK could somehow influence cell wall synthesis and assembly. In contrast, the relationship between the cell wall and PrkC is clearly established. Indeed, the kinase activity of PrkC is stimulated by peptidoglycan fragments (6). In Enterococcus faecalis, a prkC mutant was shown to exhibit an enhanced sensitivity to antibiotics that target cell wall biogenesis (34). A potential role of PrkC in antimicrobial resistance was not investigated in B. subtilis, but because the role of YvcK is still elusive and thus the effect of its PrkC-mediated phosphorylation has been difficult to determine, we decided to carry out an antibiotic screen. A prkC mutant, a prpC mutant, and a strain overproducing PrkC were tested. We observed a weak effect with bacitracin, an antibiotic that inhibits cell wall polymer biosynthesis (35). Indeed, the prkC mutant seemed to exhibit a small decrease in bacitracin sensitivity compared with the WT strain, whereas the PrkC-overproducing strain exhibited a small increase in bacitracin sensitivity (Fig. 3A). In addition, the strain lacking PrpC phosphatase had an increase in bacitracin sensitivity compared with the WT strain. This observation suggested that PrkC could phosphorylate and regulate the activity of protein(s) involved in bacitracin sensitivity. We also tested the yvcK mutant, and we observed that the absence of YvcK induced an increase in bacitracin sensitivity compared with the WT strain. The two strains expressing either the phosphoablative or constitutively phosphorylated mimetic yvcK mutant allele were then tested (Fig. 3B). The strain producing the constitutive phosphomimetic YvcK T304E mutant displayed an enhanced sensitivity for bacitracin compared with the WT strain, whereas the strain producing the phosphoablative YvcK T304A mutant exhibited a small decrease in bacitracin sensitivity compared with the WT strain (Fig. 3B). Altogether, our results suggest that YvcK could be involved, directly or indirectly, either in cell wall modification or bacitracin active release and that this YvcK function would likely be regulated in vivo by PrkC-mediated phosphorylation.
FIGURE 3.
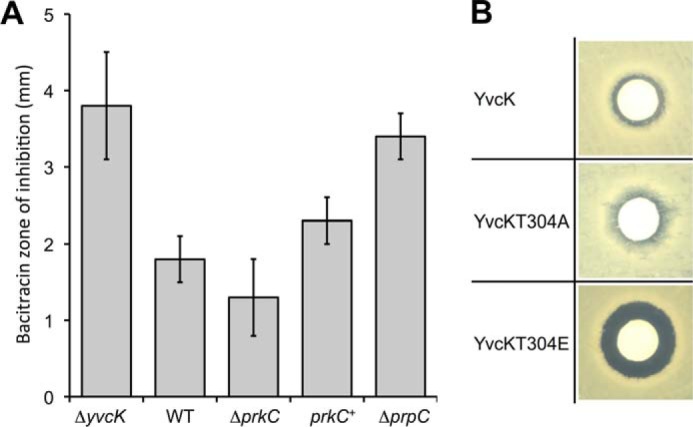
Effect of PrkC and YvcK phosphorylation on sensitivity to bacitracin. A, antibiotic sensitivity of the ΔprkC mutant strain relative to the WT strain. Strains SG63 (ΔyvcK), 168 (WT), PB705 (ΔprkC), SG376 (Phyperspank prkC), and PB702 (ΔprpC) were grown to an A600 of 0.8. A 150-μl aliquot of each culture was streaked onto LB plates, and filter paper disks containing 200 μg of bacitracin were placed on the plates, which were incubated at 37 °C. The relative sensitivity of each strain to spotted bacitracin was determined by averaging the measurements of the diameter of the zone of inhibition surrounding the patch (exclusive of the disks) after 16 h of growth at 37 °C in four independent assays. B, effect of YvcK phosphorylation on bacitracin sensitivity. The bacitracin sensitivity of strains SG116 (yvcK-gfp), SG252 (yvcK T304A-gfp), and SG283 (yvcK T304E-gfp) was tested as described above. A larger zone of inhibition represents increased sensitivity of the strains to bacitracin. Images are representative of at least six independent assays.
PrkC Phosphorylation of Overproduced YvcK Rescues an mreB Mutant
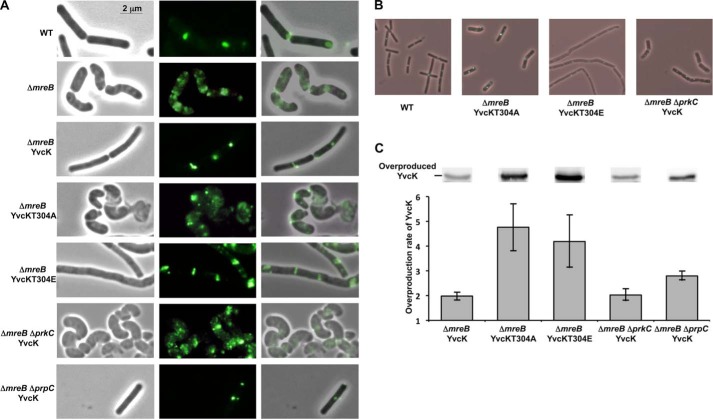
Because overproduction of YvcK rescues the morphology defects of mreB mutant cells by restoring the proper localization of PBP1 (18), we tested whether YvcK phosphorylation could affect its ability to rescue an mreB mutant. We constructed mreB mutant strains expressing a GFP-PBP1 fusion (18) and overproducing the mutant allele of either yvcK T304A or yvcK T304E. Interestingly, we observed that the mreB mutant overproducing the phosphoablative YvcK T304A protein had a bulging phenotype (22), similar to the mreB mutant without any YvcK overproduction. Cell shape was abnormal, and PBP1 was mislocalized (Fig. 4A). In contrast, the mreB mutant overproducing the constitutively phosphorylated mimetic YvcK T304E protein did not show any bulging phenotype, and cells did not swell like mreB-deficient cells. Rather, they exhibited long filaments and were clearly different in shape from WT cells. This observation indicates that, in the case of YvcK, the substitution of Thr-304 with a glutamic acid residue is not equivalent to the phosphorylation of Thr-304 at the physiological level. We speculate that the long filaments observed for the mreB mutant overproducing YvcK T304E could be due to affected cell division and/or cell elongation. Importantly, in these filaments, PBP1 seemed to be correctly localized at the septum and at the poles of the bacteria (Fig. 4A). If the PBP1 mislocalization and morphological defects were actually due to the absence of MreB and to an altered phosphorylation state of overexpressed YvcK (Fig. 4A), we hypothesized that these defects should be rescued in medium supplemented with high concentrations of Mg2+. Indeed, the addition of magnesium to the growth medium was previously shown to rescue strains affected in their cell wall integrity and to allow normal growth of mreB mutants (22). To check this, mreB mutants overproducing WT and mutant alleles of yvcK were grown in the presence of high Mg2+ concentrations and analyzed by microscopy. Our results revealed that, except for the mreB mutant cells overproducing yvcK T304E (which still looked like “spaghetti”), the cell shape and PBP1 localization were indeed rescued by high levels of Mg2+ (Fig. 4B). In addition, to check that the observed phenotypes were due to the replacement of the phosphorylation site and not to a difference in YvcK expression, we carried out Western blot analysis of YvcK expression in mutant and WT cells. We observed that the level of YvcK overproduction seemed to be higher for strains expressing the mutant proteins (Fig. 4C). We thus carried out quantitative Western blotting using an internal standard. We observed that YvcK T304A and YvcK T304E were produced in similar amounts, but overproduction induced different phenotypes (Fig. 4C). Overproduction of YvcK T304A could not rescue the absence of mreB, and the cells had a bulging phenotype. In contrast, the constitutively phosphorylated mimetic YvcK T304E mutant rescued the MreB phenotype but displayed a cell cycle defect phenotype, suggesting that an excess of phosphorylated YvcK is detrimental to the cell. These results suggest that YvcK phosphorylation is necessary for cell morphology in the absence of MreB.
FIGURE 4.
Impact of phosphorylation on the ability of YvcK to rescue an mreB mutant. A, cell shape and cellular localization of GFP-PBP1 in mreB mutant strains upon YvcK overproduction. Strains were grown on LB medium, and YvcK proteins were expressed from the inducible Pspac promoter in the presence of 100 μm IPTG, whereas the gfp-ponA gene fusion was expressed from the inducible Pxyl promoter in the presence of 0.5% xylose. The cell shape (left panels) and PBP1 localization (middle panels) were analyzed by microscopy for strains YK706 (WT), SG204 (ΔmreB), SG217 (ΔmreB, pDG148-YvcK), SG262 (ΔmreB, pDG148-YvcK T304A), SG281 (ΔmreB, pDG148-YvcK T304E), SG265 (ΔmreB ΔprkC, pDG148-YvcK), and SG413 (ΔmreB ΔprpC, pDG148-YvcK). Merged images of phase and fluorescent micrographs are shown (right panels). B, of cell shape and intracellular localization of GFP-PBP1 upon either YvcK T304A or YvcK T304E overproduction in an mreB mutant strain. Strains were grown on LB medium supplemented with 25 mm MgSO4, and YvcK proteins were expressed from the inducible promoter in the presence of 100 μm IPTG, whereas the gfp-ponA gene fusion was expressed from the inducible promoter in the presence of 0.5% xylose. Cell shape and PBP1 localization were analyzed by microscopy, and an overlay of phase and fluorescent micrographs is presented for strains YK706 (WT), SG262 (ΔmreB, pDG148-YvcK T304A), SG281 (ΔmreB, pDG148-YvcK T304E), and SG265 (ΔmreB ΔprkC, pDG148-YvcK). C, analysis of YvcK overproduction by Western blotting. Strain SG204 (ΔmreB) and overproducing strains SG217 (ΔmreB, pDG148-YvcK), SG262 (ΔmreB, pDG148-YvcK T304A), SG281 (ΔmreB, pDG148-YvcK T304E), SG265 (ΔmreB ΔprkC, pDG148-YvcK), and SG413 (ΔmreB ΔprpC, pDG148-YvcK) were grown on Difco Antibiotic Medium 3 (PAB) medium supplemented with 0.3 m sucrose and 25 mm MgSO4 to allow normal growth of mreB mutants (20) and with 100 μm IPTG until A600 = 1. After centrifugation, the pellets were resuspended in 0.1 volume of lysis buffer. For each, 8 and 16 μl of crude extract were separated by SDS-PAGE. After blotting, YvcK was detected using antibodies directed against YvcK as described previously (16). For each overproducing strain, the image shows the signal obtained for overproduced YvcK from 16 μl of crude extract. To estimate the relative quantity of YvcK in crude extract and to compare the different lanes and different blots (for each strain analyzed, 16 and 8 μl of crude extract were loaded on the gel), we used an internal standard, the YpmB protein, which was detected using specific antibodies (1:15,000 dilution). The level of YvcK overproduction is the ratio of YvcK quantities determined using ImageJ in an overproducing strain and in the non-overproducing strain SG204.
PBP1 is the main bifunctional transglycosylase/transpeptidase in B. subtilis. It is required for peptidoglycan synthesis in cell division and elongation (36). Its localization changes during the cell cycle. PBP1 shuttles between the septum and the lateral wall. This dynamic behavior is highly regulated by a complex mechanism implicating various cell division proteins, including FtsZ, EzrA, and GpsB (36–38) and MreB (23). In the absence of MreB, we postulated that any defect in YvcK phosphorylation could hinder the correct localization of PBP1, thus leading to aberrant bulging morphology. To test this hypothesis, we constructed a strain overproducing WT YvcK in an mreB prkC double mutant and another strain overproducing WT YvcK in an mreB prpC double mutant. We determined the amounts of YvcK overproduced (Fig. 4C). We observed that, even if the level of WT YvcK overproduction was similar in all the strains, YvcK overproduction could not rescue the absence of mreB in the absence of prkC (Fig. 4A). These bacteria still had a bulging shape and an abnormal localization of PBP1 (Fig. 4A), similar to that displayed by mreB mutant cells overproducing the phosphoablative form of YvcK. In contrast, overproduction of WT YvcK in the presence of PrkC and in the absence or presence of PrpC or overproduction of YvcK T304E (consistent with a hyperphosphorylation phenotype) rescued the localization of PBP1 in the absence of MreB. However, overproduction of the constitutively phosphorylated mimetic form of YvcK somehow affected its ability to work in a coordinated manner with the other components involved in peptidoglycan synthesis, thus leading to abnormal cell elongation or cell division and increased sensitivity to bacitracin. In addition, cells overproducing WT YvcK in the absence of both PrpC and MreB did not show a hyperphosphorylation phenotype but had a normal shape, suggesting that YvcK phosphorylation is still relatively weak under these conditions. Taken together, our results suggest that PrkC-dependent phosphorylation of YvcK is necessary for proper PBP1 localization and its ability to function in peptidoglycan synthesis inside either the divisome or the elongasome, thus allowing mreB-deficient cells to achieve normal cell shape.
In conclusion, we have shown that YvcK is phosphorylated by PrkC from B. subtilis. In addition, our data are consistent with two distinct functions for YvcK. Although the function of YvcK in carbon source utilization appears to be independent of its phosphorylation state, its role in bacitracin sensitivity and its contribution to cell morphogenesis in the absence of MreB, via the correct positioning of PBP1, require phosphorylation by PrkC. Finally, we observed, by carrying out a sequence conservation analysis using STRING (version 9.0), that PrkC and YvcK are conserved together in several bacteria (data not shown). Hence, we speculate that PrkC-mediated phosphorylation has a widespread role in the regulation of cell morphogenesis in bacteria.
Acknowledgments
We thank the Common Center for Microanalysis of Proteins (Institut de Biologie et Chimie des Protéines, FR 3302) for expertise and technical assistance in mass spectrometry analysis. We also thank S. J. Séror and C. W. Price for the kind gift of strains, T. Doan for the kind gift of the unpublished SG376 strain and critical reading of the manuscript, and B. Khadaroo for editing the manuscript.
This work was supported by CNRS, the Agence Nationale de la Recherche (P-loop and PiBaKi), and Aix-Marseille Université.
- IPTG
- isopropyl β-d-thiogalactopyranoside
- PrkCc
- PrkC catalytic domain.
REFERENCES
- 1. Fleurie A., Manuse S., Zhao C., Campo N., Cluzel C., Lavergne J. P., Freton C., Combet C., Guiral S., Soufi B., Macek B., Kuru E., VanNieuwenhze M. S., Brun Y. V., Di Guilmi A. M., Claverys J. P., Galinier A., Grangeasse C. (2014) Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 10, e1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Molle V., Kremer L. (2010) Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077 [DOI] [PubMed] [Google Scholar]
- 3. Fiuza M., Canova M. J., Zanella-Cléon I., Becchi M., Cozzone A. J., Mateos L. M., Kremer L., Gil J. A., Molle V. (2008) From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J. Biol. Chem. 283, 18099–18112 [DOI] [PubMed] [Google Scholar]
- 4. Pietack N., Becher D., Schmidl S. R., Saier M. H., Hecker M., Commichau F. M., Stülke J. (2010) In vitro phosphorylation of key metabolic enzymes from Bacillus subtilis: PrkC phosphorylates enzymes from different branches of basic metabolism. J. Mol. Microbiol. Biotechnol. 18, 129–140 [DOI] [PubMed] [Google Scholar]
- 5. Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yeats C., Finn R. D., Bateman A. (2002) The PASTA domain: a β-lactam-binding domain. Trends Biochem. Sci. 27, 438. [DOI] [PubMed] [Google Scholar]
- 7. Absalon C., Obuchowski M., Madec E., Delattre D., Holland I. B., Séror S. J. (2009) CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155, 932–943 [DOI] [PubMed] [Google Scholar]
- 8. Pompeo F., Freton C., Wicker-Planquart C., Grangeasse C., Jault J. M., Galinier A. (2012) Phosphorylation of CpgA protein enhances both its GTPase activity and its affinity for ribosome and is crucial for Bacillus subtilis growth and morphology. J. Biol. Chem. 287, 20830–20838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaidenko T. A., Kim T. J., Price C. W. (2002) The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J. Bacteriol. 184, 6109–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Madec E., Laszkiewicz A., Iwanicki A., Obuchowski M., Séror S. (2002) Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46, 571–586 [DOI] [PubMed] [Google Scholar]
- 11. Obuchowski M., Madec E., Delattre D., Boël G., Iwanicki A., Foulger D., Séror S. J. (2000) Characterization of PrpC from Bacillus subtilis, a member of the PPM phosphatase family. J. Bacteriol. 182, 5634–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005) The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C. M., Bemis G., Church G. M., Steen H., Husson R. N. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graupner M., Xu H., White R. H. (2002) Characterization of the 2-phospho-l-lactate transferase enzyme involved in coenzyme F420 biosynthesis in Methanococcus jannaschii. Biochemistry 41, 3754–3761 [DOI] [PubMed] [Google Scholar]
- 15. Chaudhuri R. R., Allen A. G., Owen P. J., Shalom G., Stone K., Harrison M., Burgis T. A., Lockyer M., Garcia-Lara J., Foster S. J., Pleasance S. J., Peters S. E., Maskell D. J., Charles I. G. (2009) Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sauer J. D., Witte C. E., Zemansky J., Hanson B., Lauer P., Portnoy D. A. (2010) Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7, 412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Görke B., Foulquier E., Galinier A. (2005) YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 151, 3777–3791 [DOI] [PubMed] [Google Scholar]
- 18. Foulquier E., Pompeo F., Bernadac A., Espinosa L., Galinier A. (2011) The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol. Microbiol. 80, 309–318 [DOI] [PubMed] [Google Scholar]
- 19. Chastanet A., Carballido-Lopez R. (2012) The actin-like MreB proteins in Bacillus subtilis: a new turn. Front. Biosci. 4, 1582–1606 [DOI] [PubMed] [Google Scholar]
- 20. Domínguez-Escobar J., Chastanet A., Crevenna A. H., Fromion V., Wedlich-Söldner R., Carballido-López R. (2011) Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 [DOI] [PubMed] [Google Scholar]
- 21. Garner E. C., Bernard R., Wang W., Zhuang X., Rudner D. Z., Mitchison T. (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Formstone A., Errington J. (2005) A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. 55, 1646–1657 [DOI] [PubMed] [Google Scholar]
- 23. Kawai Y., Daniel R. A., Errington J. (2009) Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71, 1131–1144 [DOI] [PubMed] [Google Scholar]
- 24. Gilmour M. W., Gunton J. E., Lawley T. D., Taylor D. E. (2003) Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49, 105–116 [DOI] [PubMed] [Google Scholar]
- 25. Battesti A., Bouveret E. (2012) The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334 [DOI] [PubMed] [Google Scholar]
- 26. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galinier A., Haiech J., Kilhoffer M. C., Jaquinod M., Stülke J., Deutscher J., Martin-Verstraete I. (1997) The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. U.S.A. 94, 8439–8444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hakes D. J., Dixon J. E. (1992) New vectors for high level expression of recombinant proteins in bacteria. Anal. Biochem. 202, 293–298 [DOI] [PubMed] [Google Scholar]
- 29. Madec E., Stensballe A., Kjellström S., Cladière L., Obuchowski M., Jensen O. N., Séror S. J. (2003) Mass spectrometry and site-directed mutagenesis identify several autophosphorylated residues required for the activity of PrkC, a Ser/Thr kinase from Bacillus subtilis. J. Mol. Biol. 330, 459–472 [DOI] [PubMed] [Google Scholar]
- 30. Molle V., Gulten G., Vilchèze C., Veyron-Churlet R., Zanella-Cléon I., Sacchettini J. C., Jacobs W. R., Jr., Kremer L. (2010) Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol. Microbiol. 78, 1591–1605 [DOI] [PubMed] [Google Scholar]
- 31. Jani C., Eoh H., Lee J. J., Hamasha K., Sahana M. B., Han J. S., Nyayapathy S., Lee J. Y., Suh J. W., Lee S. H., Rehse S. J., Crick D. C., Kang C. M. (2010) Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 10, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bird T. H., MacKrell A. (2011) A CtrA homolog affects swarming motility and encystment in Rhodospirillum centenum. Arch. Microbiol. 193, 451–459 [DOI] [PubMed] [Google Scholar]
- 33. Jers C., Pedersen M. M., Paspaliari D. K., Schütz W., Johnsson C., Soufi B., Macek B., Jensen P. R., Mijakovic I. (2010) Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol. Microbiol. 77, 287–299 [DOI] [PubMed] [Google Scholar]
- 34. Kristich C. J., Wells C. L., Dunny G. M. (2007) A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. U.S.A. 104, 3508–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Storm D. R., Strominger J. L. (1973) Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248, 3940–3945 [PubMed] [Google Scholar]
- 36. Scheffers D. J., Errington J. (2004) PBP1 is a component of the Bacillus subtilis cell division machinery. J. Bacteriol. 186, 5153–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheffers D. J., Jones L. J., Errington J. (2004) Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51, 749–764 [DOI] [PubMed] [Google Scholar]
- 38. Claessen D., Emmins R., Hamoen L. W., Daniel R. A., Errington J., Edwards D. H. (2008) Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68, 1029–1046 [DOI] [PubMed] [Google Scholar]