Abstract
Real-time quantitative PCR was used to measure expression levels of genes encoding efflux pumps, ERG11 and two control genes, ACT1 and PMA1, in a collection of 14 fluconazole-susceptible Candida albicans isolates. For each gene, average expression levels and variations within the population were determined. These values were then used as reference points to make predictions about the molecular basis of resistance in 38 clinical isolates (the majority of which were resistant to fluconazole) obtained from 18 patients treated with posaconazole for refractory oropharyngeal candidiasis. For each of the 38 isolates, the expression levels of genes encoding efflux pumps, ERG11 and the control genes, were measured as above. Comparison of the two data sets revealed that expression of ACT1 and PMA1 did not vary significantly between the two sets of isolates. In contrast, MDR1, ERG11, CDR1, and CDR2 were overexpressed in 3, 4, 14, and 35, respectively, of the isolates from patients treated with azoles. In addition to these changes, the patient isolates all had at least one and often multiple missense mutations in ERG11. Select ERG11 alleles were expressed in Saccharomyces cerevisiae; all of the alleles tested conferred reduced susceptibility to fluconazole. Despite both the increases in pump expression and the ERG11 mutations, only one of the patient isolates exhibited a large decrease in posaconazole susceptibility.
Nearly 90% of individuals who are infected with human immunodeficiency virus or have AIDS experience at least one episode of oropharyngeal candidiasis during the course of their disease (7). Azoles have been the drugs of choice for the treatment of oropharyngeal candidiasis. Azoles inhibit the enzyme lanosterol 14α-demethylase (encoded by ERG11), which catalyzes an important step in the synthesis of ergosterol. The resulting depletion of ergosterol from the membrane, in combination with the accumulation of methylated sterols, is proposed to adversely affect membrane integrity as well as the function of some membrane-associated proteins (11, 18).
To combat the development of resistance to azoles in yeasts, in particular to fluconazole, and to expand the spectrum of susceptible pathogens, new azoles have been developed. One such agent is posaconazole, a broad-spectrum triazole in phase III trials. Posaconazole is more active than fluconazole against Candida spp. and, unlike fluconazole, is active against Aspergillus spp. (1, 14). In addition, posaconazole appears to be less affected by mutations in the gene encoding the azole target site than either voriconazole or fluconazole (D. Sanglard, F. Ischer, and J. Bille, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-221, 2002). Furthermore, although posaconazole is a substrate for the ATP-dependent efflux pumps encoded by CDR1 and CDR2, posaconazole is not a substrate for the major facilitator pumps encoded by MDR1 and FLU1 (Sanglard et al., 42nd ICAAC).
Azoles are fungistatic against yeasts, and patients often require long-term therapy; this has promoted the development of resistance. The past decade has seen numerous publications describing the molecular mechanisms of azole resistance in yeasts (most recently reviewed in references 2 and 17). The two most prevalent mechanisms of resistance are mutations in the ERG11 gene, resulting in reduced drug binding to the target enzyme and decreased intracellular drug accumulation. The latter is most often caused by increased expression of efflux pump genes, the best characterized of which are MDR1, which encodes a major facilitator pump, and CDR1 and CDR2, which encode distinct but related ATP-dependent pumps. To identify changes in gene expression, researchers have focused on sequential isolates from a single patient. In such collections, changes in gene expression are readily identified by comparing expression levels in the resistant isolates with those measured in an isogenic, azole-susceptible baseline strain. However, such analyses become problematic when a susceptible baseline isolate is not available.
To circumvent this problem, we used quantitative real-time PCR (RT-PCR) to determine the normal range of expression levels of genes known to confer azole resistance in a collection of azole-susceptible isolates. These ranges were then used as reference points to make predictions about the molecular basis of resistance in isolates from patients receiving posaconazole therapy for treatment of refractory oropharyngeal candidiasis.
(Part of this work was presented at the 103rd Annual Meeting of the American Society for Microbiology, Washington, D.C., May 2003.)
MATERIALS AND METHODS
Fungal strains.
The identity of the clinical isolates was confirmed with the Vitek Identification System with the Yeast Biochemical Card (bioMérieux Inc., Hazelwood, Mo.).
Antifungal agents and susceptibility testing.
Posaconazole was prepared at the Schering-Plough Research Institute (SPRI, Kenilworth, N.J.) as a micronized powder. Itraconazole and amphotericin B powders were obtained from Janssen Pharmaceutica Inc. (Beerse, Belgium) and Sigma Chemical Co. (St. Louis, Mo.), respectively. Voriconazole and fluconazole were obtained from Pfizer Inc. (New York, N.Y.). All drugs except voriconazole (which was dissolved in water) were dissolved in dimethyl sulfoxide, and serial dilutions were made in RPMI 1640 medium (BioWhittaker, Walkersville, Md.). MICs were determined by the procedures of the National Committee for Clinical Laboratory Standards (NCCLS) (10). When assaying the effects of expressing ERG11 alleles in Saccharomyces cerevisiae, MICs were determined in yeast nitrogen base (Qbiogene, Carlsbad, Calif.) broth supplemented with 2% raffinose and 2% galactose.
DNA typing techniques for strain identification.
Repetitive-element PCR was performed with a DiversiLab Candida kit and run on a Caliper 1000 analyzer as described by the manufacturer (Bacterial BarCodes, Houston, Tex.). The resultant data were analyzed with DiversiLab System software.
Measurement of transcript levels.
Strains were grown overnight at 35°C in YPD liquid medium (Qbiogene). Overnight cultures were diluted into fresh medium, adjusted to an optical density at 530 nm of 0.1, and grown for 3 h. Total RNA was extracted with the RNeasy minikit (Qiagen Inc., Valencia, Calif.). RNA quality (i.e., the presence of discreet 18S and 28S rRNA peaks) and quantity were measured with an Agilent Technologies (Palo Alto, Calif.) RNA 6000 Nano Assay kit run on an Agilent Technologies 2100 bioanalyzer. Multiplex RT-PCRs were run in duplicate with an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.). The 6-carboxyfluorescein-labeled test probe sets were designed in-house with Primer Express software (Applied Biosystems) and purchased from Applied Biosystems (Table 1). The 6-carboxyrhodamine 6G-labeled 18S TaqMan rRNA control reagents kit for the detection of endogenous 18S rRNA was also purchased from Applied Biosystems.
TABLE 1.
Sequences of RT-PCR primer-probe sets
| Gene | Probe and primers | Sequence |
|---|---|---|
| ACT1 | Probe | TTGACCTTGAGATACCCAATTGAACACGGTA |
| Forward primer | TTGGTGATGAAGCCCAATCC | |
| Reverse primer | CATATCGTCCCAGTTGGAAACA | |
| PMA1 | Probe | AGATGTCCACGAAAACTACCAAAACACCGTT |
| Forward primer | TTGAAGATGACCACCCAATCC | |
| Reverse primer | GAAACCTCTGGAAGCAAATTCG | |
| ERG11 | Probe | TGCCTGACCCTGATTATAGTTCAATGGTGG |
| Forward primer | AACTACTTTTGTTTATAATTTAAGATGGACTATTGA | |
| Reverse primer | AATGATTTCTGCTGGTTCAGTAGGT | |
| MDR1 | Probe | TCGCAAGGCTAAAAGATTGAGAGCCATCA |
| Forward primer | TTACCTGAAACTTTTGGCAAAACA | |
| Reverse primer | ACTTGTGATTCTGTCGTTACCG | |
| CDR1 | Probe | TAACCCATATGTCAGAAGTGCCCGGG |
| Forward primer | TTTAGCCAGAACTTTCACTCATGATT | |
| Reverse primer | TATTTATTTCTTCATGTTCATATGGATTGA | |
| CDR2 | Probe | TCCCGGGTTTTGGATTTTCATGTACAGA |
| Forward primer | GGTATTGGCTGGTCCTAATGTGA | |
| Reverse primer | GCTTGAATCAAATAAGTGAATGGATTAC |
Expression levels as measured by RT-PCR are expressed in CT units; this is the first cycle in which the signal above a preset threshold (known as the threshold cycle) is detected. Consequently, the CT value is inversely proportional to the amount of transcript detected (i.e., the CT value for an abundant transcript will be numerically lower than the value measured for a less abundant transcript). Furthermore, a decrease in the CT value from x to x − 1 corresponds to a twofold increase in the amount of transcript detected. To control for variations in the amount of input RNA, multiplex reactions were run with the 18S rRNA probe as an internal control. Relative gene expression levels (ΔCT) were calculated as ΔCT[test gene] = CT[test gene] − CT[18S]. All probe sets were tested, alone and in a multiplex mode, with serial dilutions of cDNA. The signal from all probe sets was proportional to the amount of input cDNA over a 1,000-fold concentration range, irrespective of the presence of the 18S rRNA probe.
PCR amplification, sequencing, and cloning of ERG11 alleles.
ERG11 was PCR amplified in overlapping 600-bp segments from total genomic DNA. Both strands were sequenced by MWG-Biotech Inc, (High Point, N.C.) and compared to the sequence published in GenBank (accession number X13296). Select ERG11 alleles were PCR amplified from chromosomal DNA with PFU Turbo (Stratagene, La Jolla, Calif.) with the following oligonucleotides to generate fragments with flanking SalI and BamHI restriction sites, 5′-ACGCGTCGACAATATGGCTATTGTTGAAACTGTC-3′ and 5′-GCGGATCCTTAAAACATACAAGTTTCTCTTTT-3′. To engineer the mutation encoding the A61V substitution, the 5′ and 3′ regions of ERG11 were separately PCR amplified with the above oligonucleotides in combination with mutagenic oligonucleotides (5′-TTGGTTTGGTTCTGTAGCTTCATATGGT-3′ and 5′-ACCATATGAAGCTACAGAACCAAACCAA-3′). The resulting PCR fragments were fused together by PCR with the same oligonucleotides used for cloning ERG11. The amplified ERG11 genes were cloned into the multicopy plasmid YEp51 under control of the GAL10 promoter, sequenced to confirm accurate amplification, and transformed into S. cerevisiae strain YKKB-13 (plasmid YEp51 and S. cerevisiae strain YKKB-13 were kindly supplied by D. Sanglard, University of Vaudois, Vaudois, Switzerland) (16).
RESULTS
Clinical isolates and azole susceptibilities.
Fourteen C. albicans clinical isolates with fluconazole MICs of ≤0.5 μg/ml were randomly chosen from the SPRI culture collection (Table 2). All 14 isolates were susceptible to posaconazole, itraconazole, voriconazole, and amphotericin B. This collection of isolates is referred to below as the SPRI azole-susceptible population.
TABLE 2.
MIC and RT-PCR data for SPRI azole-susceptible C. albicans isolates
| Isolate | MICa (μg/mL)
|
ΔCT
|
Substitution(s) in Erg11p | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| POS | ITZ | FLZ | VOR | ACT1 | PMA1 | ERG11 | MDR1 | CDR1 | CDR2 | ||
| C34 | 0.016 | 0.06 | 0.25 | <0.004 | 8.1 | 7.1 | 10.3 | 11.1 | 11.3 | 13.5 | None |
| C40 | 0.016 | 0.06 | 0.25 | <0.004 | 7.2 | 6.2 | 10.6 | 11.9 | 9.6 | 14.1 | D116E, K128Tb |
| C43 | <0.03 | 0.06 | 0.25 | 0.03 | 7.4 | 6.3 | 10.3 | 15.1 | 10.2 | 14.4 | NDc |
| C60 | 0.016 | 0.06 | <0.12 | <0.004 | 7.6 | 6.3 | 8.6 | 10.9 | 11.8 | 15.9 | None |
| C72 | 0.03 | 0.06 | 0.06 | <0.008 | 7.9 | 6.7 | 8.7 | 12.0 | 10.4 | 12.5 | D116E |
| C84 | 0.008 | 0.06 | <0.12 | <0.004 | 7.3 | 6.4 | 9.3 | 14.4 | 10.2 | 13.8 | D116E |
| C128 | 0.008 | 0.03 | <0.12 | <0.004 | 7.5 | 6.1 | 11.1 | 14.4 | 10.5 | 14.6 | D116E, K128T,b D496 |
| C132 | 0.016 | 0.06 | 0.25 | <0.004 | 7.5 | 5.8 | 11.3 | 13.2 | 10.0 | 14.5 | D116E, K128Tb |
| C294 | 0.008 | 0.016 | 0.25 | <0.008 | 8.9 | 8.3 | 10.9 | 12.8 | 12.3 | 16.0 | None |
| C392 | <0.004 | 0.03 | 0.5 | <0.008 | 7.9 | 6.7 | 11.1 | 14.0 | 11.2 | 14.6 | None |
| C393 | <0.004 | 0.016 | 0.25 | <0.008 | 7.8 | 6.7 | 11.3 | 13.0 | 10.7 | 14.7 | ND |
| C394 | <0.004 | 0.016 | 0.25 | <0.008 | 7.9 | 6.3 | 10.9 | 16.6 | 10.6 | 13.9 | ND |
| C395 | <0.004 | 0.016 | 0.25 | <0.008 | 7.9 | 6.8 | 11.5 | 12.4 | 10.7 | 15.6 | V437I |
| C548 | <0.004 | <0.008 | <0.125 | <0.008 | 7.6 | 6.3 | 10.0 | 14.2 | 12.3 | 14.4 | V437I |
| Avg ΔCT | 7.8 | 6.6 | 10.4 | 13.6 | 10.8 | 14.5 | |||||
| SD | 0.4 | 0.6 | 0.9 | 2 | 0.8 | 0.9 | |||||
| 3-SD range | 6.6-9.0 | 4.8-8.4 | 7.7-13.1 | 7.6-19.6 | 8.4-13.2 | 11.8-17.2 | |||||
POS, posaconazole; ITZ, itraconazole; FLZ, fluconazole; VOR, voriconazole.
Mutation in only one copy of ERG11.
ND, not done.
A second collection comprised 38 C. albicans clinical isolates cultured from 18 patients who were receiving posaconazole for treatment of refractory oropharyngeal candidiasis (Table 3). We focused on isolates that exhibited significant reductions in susceptibility to fluconazole, voriconazole, itraconazole, and posaconazole. None of the isolates showed any changes in susceptibility to amphotericin B. Relatedness among isolates from individual patients was tested by repetitive-element PCR and also inferred from the conservation of both silent and missense mutations in ERG11. The majority of the patients appeared to be colonized by a single isolate (data not shown). The baseline isolates from the 18 patients were classified according to NCCLS criteria as follows: 10 were fluconazole resistant (MIC, ≥64 μg/ml); six were fluconazole susceptible, dose dependent (S-DD) (MIC, 16 to 32 μg/ml); and two were fluconazole sensitive (MIC, ≤8 μg/ml). Of the 38 isolates analyzed, 24 were fluconazole resistant, 10 were fluconazole S-DD, and 4 were fluconazole sensitive. Of the 24 fluconazole-resistant isolates, 17 had a voriconazole MIC of >1 μg/ml, one had a posaconazole MIC of >1 μg/ml, and eight were itraconazole resistant by NCCLS criteria (MIC of >1 μg/ml).
TABLE 3.
MIC and RT-PCR data for C. albicans isolates from candidiasis patients
| Patient no. | Isolate no. | MICa (μg/ml)
|
ΔCTb
|
Substitution(s) in Erg11p | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POS | ITZ | FLZ | VOR | ACT1 | PMA1 | ERG11 | MDR1 | CDR1 | CDR2 | |||
| 1 | C482 | 0.25 | 0.5 | 16 | 0.5 | 7.9 | 7.5 | 10.4 | 13.7 | 9.2 | 9.4 | A107T |
| C486 | 0.25 | 1 | 16 | 0.25 | 8.0 | 8.1 | 9.8 | 12.3 | 11.2 | 10.1 | A107T | |
| 2 | C491 | 0.12 | 0.25 | 2 | 0.03 | 7.4 | 7.2 | 8.9 | 12.9 | 10.1 | 10.0 | D116E |
| C504 | 0.25 | 0.25 | 16 | 0.25 | 7.3 | 6.2 | 7.9 | 15.5 | 8.2 | 7.7 | F449S | |
| 3 | C498 | 0.008 | 0.06 | 1 | 0.008 | 7.4 | 5.4 | 9.6 | 12.6 | 8.2 | 10.8 | F145L, E266D, V488I |
| C502 | 0.008 | 0.06 | 1 | 0.008 | 7.2 | 5.9 | 7.4 | 11.7 | 10.6 | 10.8 | F145L, E266D, V488I | |
| C513 | 0.25 | 0.25 | 16 | 0.12 | 6.7 | 6.4 | 7.5 | 10.6 | 7.6 | 5.8 | F145L, E266D, V488I | |
| 4 | C527 | 0.12 | 0.5 | 256 | 4 | 7.5 | 6.6 | 9.1 | 12.6 | 8.9 | 8.6 | Y132H, G450E |
| C528 | 0.12 | 0.5 | 256 | 4 | 7.3 | 5.9 | 9.0 | 11.7 | 7.7 | 7.7 | Y132H, G450E | |
| C530 | 0.12 | 0.25 | 256 | 0.5 | 7.0 | 6.3 | 8.5 | 12.0 | 8.8 | 9.9 | G450E, G464S | |
| C534 | 0.12 | 0.5 | 256 | 4 | 7.3 | 6.4 | 8.1 | 11.0 | 8.3 | 8.8 | Y132H, G450E | |
| C546 | 0.12 | 0.25 | 128 | 2 | 8.1 | 7.5 | 9.6 | 11.9 | 9.4 | 9.4 | Y132H, G450E | |
| C536 | 0.25 | 0.5 | 256 | 4 | 8.2 | 7.7 | 9.8 | 11.7 | 9.2 | 9.4 | Y132H, G450E | |
| C572 | 0.25 | 0.25 | >256 | 8 | 8.0 | 7.4 | 9.5 | 12.1 | 9.5 | 9.2 | Y132H, G450E | |
| 5 | C481 | 0.25 | 0.5 | 32 | 0.5 | 7.9 | 7.2 | 9.9 | 15.7 | 9.4 | 9.0 | F128T, F145L |
| C483 | 0.25 | 0.5 | 8 | 0.12 | 8.2 | 7.3 | 11.0 | 14.8 | 10.0 | 9.5 | F145L | |
| 6 | C535 | 0.5 | 2 | 256 | 8 | 7.7 | 7.1 | 9.8 | 9.0 | 9.6 | 10.9 | Y132H, G448V |
| C539 | 0.25 | 2 | 128 | 8 | 7.2 | 6.8 | 11.0 | 8.3 | 6.6 | 7.1 | Y132H, G448V | |
| 7 | C516 | 0.12 | 0.25 | 32 | 0.5 | 7.3 | 7.1 | 9.8 | 14.0 | 9.7 | 9.7 | K128T, Y132F, F145L |
| 8 | C470 | 0.5 | 0.5 | 32 | 0.25 | 7.3 | 7.0 | 9.0 | 13.5 | 8.7 | 8.5 | S405F |
| C478 | 0.5 | 0.5 | 64 | 0.5 | 7.6 | 6.9 | 8.6 | 12.6 | 9.1 | 8.5 | S405F | |
| 9 | C490 | 0.5 | 1 | 32 | 0.25 | 8.3 | 7.8 | 10.6 | 9.4 | 9.0 | 9.2 | V452A |
| C497 | 0.5 | 2 | 64 | 0.5 | 8.4 | 7.7 | 9.1 | 12.5 | 10.0 | 9.3 | K128T, V452A | |
| 10 | C526 | 0.5 | 1 | 32 | 0.5 | 7.8 | 7.1 | 9.6 | 13.6 | 9.2 | 9.2 | V509M |
| C533 | 0.5 | 0.5 | 32 | 1 | 7.9 | 7.4 | 9.8 | 13.6 | 9.3 | 9.9 | V509M | |
| 11 | C594 | 0.5 | 2 | 128 | 16 | 7.2 | 6.5 | 9.0 | 11.3 | 8.2 | 7.5 | Y132H, S405F |
| C600 | 1 | 4 | >256 | 16 | 7.7 | 7.1 | 8.5 | 12.3 | 9.4 | 8.2 | Y132H, S405F | |
| 12 | C577 | 0.06 | 0.06 | 128 | 0.5 | 6.5 | 5.0 | 9.0 | 5.3 | 8.1 | 12.9 | G464S |
| C583 | 0.25 | 0.25 | 256 | 1 | 6.8 | 5.5 | 9.4 | 5.7 | 6.9 | 10.7 | G464S | |
| C587 | 0.5 | 0.5 | 256 | 4 | 7.7 | 7.3 | 9.2 | 8.2 | 10.2 | 9.6 | G464S | |
| 13 | C480 | 0.03 | 0.12 | 128 | 8 | 8.3 | 7.4 | 9.3 | 9.2 | 12.2 | 15.1 | K128T, G464S, R467I |
| 14 | C438 | 0.5 | 0.5 | 128 | 2 | 6.6 | 6.0 | 7.9 | 13.3 | 7.1 | 7.5 | Y257H, G464S |
| C439 | 1 | 4 | >256 | 16 | 7.1 | 6.2 | 7.6 | 13.5 | 7.7 | 7.5 | Y257H, G464S, G307S | |
| C440 | 4 | 8 | >256 | >16 | 7.3 | 6.4 | 8.0 | 13.5 | 7.9 | 7.7 | Y257H, G464S, G307S, A61V | |
| 15 | C444 | 0.5 | 0.5 | 256 | 0.5 | 7.0 | 5.8 | 7.4 | 13.2 | 7.8 | 7.8 | K143R |
| 16 | C489 | 0.25 | 0.25 | 128 | 1 | 7.2 | 6.1 | 8.8 | 12.8 | 10.1 | 9.5 | Y132H, Y257H, E266D |
| 17 | C477 | 1 | 0.25 | 256 | 16 | 7.6 | 6.1 | 9.3 | 7.3 | 10.5 | 15.3 | K128T, G464S, R467I |
| 18 | C507 | 1 | 2 | 64 | 8 | 6.1 | 7.2 | 8.6 | 10.5 | 8.3 | 8.4 | Y132H, H283R, G464S |
See Table 2, footnote a.
Values that fall outside the 3-SD range measured in the azole-susceptible isolates (see text for details) are shown in boldface italic.
RT-PCR measurement of gene expression in the SPRI azole-susceptible population.
Total RNA was isolated from exponentially growing cultures and analyzed by RT-PCR. The average CT value for the 18S rRNA probe was 14.9; the standard deviation (SD) was 0.4, which corresponded to a fluctuation of ±1.3-fold. The ACT1 and PMA1 genes, encoding actin and a plasma membrane ATPase, respectively, have been reported to be constitutively expressed in C. albicans (9; D. Perlin, personal communication). The average ΔCT values for ACT1 and PMA1 were 7.8 (SD, 0.4) and 6.6 (SD, 0.6), respectively. Similarly, the SDs associated with the ΔCT values for CDR1, CDR2, and ERG11 were all <1, which corresponds to a <2-fold fluctuation in expression levels. Expression of MDR1 exhibited the most fluctuation; the SD was 2, which corresponds to a ±4-fold range of expression levels.
To determine the intrastrain variability in gene expression as well as the experimental error associated with the procedure, we repeated the analysis with RNA extracted from a single isolate (C72) grown on three separate occasions. The SDs for the three measurements ranged from a high of 0.6 for CDR2 to a low of 0.3 for MDR1; the values for PMA1, ACT1, CDR1, and ERG11 all fell within this range (data not shown).
RT-PCR measurement of ACT1 and PMA1 gene expression in C. albicans isolates from oropharyngeal candidiasis patients.
The 38 C. albicans isolates exhibiting various levels of azole resistance were analyzed as above (Table 3). The average CT value for the 18S rRNA probe was 14.8 (SD, 0.4). The average ΔCT values for the control genes ACT1 and PMA1 were 7.5 (SD, 0.5) and 6.7 (SD, 0.7), respectively. These values are very similar to those measured in the 14 SPRI azole-susceptible isolates. Employing a two-tailed Student’s t test, we confirmed that the expression levels of both ACT1 and PMA1 did not differ significantly (P > 0.05) between the two populations of isolates. These data suggest that the experimental protocol (i.e., the growth conditions and extraction procedures) did not discriminate against either population.
Assignment of overexpression cutoffs for the analysis of C. albicans isolates from oropharyngeal candidiasis patients.
As shown above, the ΔCT values for test genes in the SPRI azole-susceptible population exhibited relatively minor differences in expression levels, as reflected in the SD values (the exception was MDR1). For a given test gene, the variation in ΔCT was a function of both the strain-to-strain variability in expression levels and the experimental error associated with the RT-PCR measurements. For the majority of the test genes (the exception was MDR1) in the SPRI azole-susceptible population, the variations in the ΔCT values were normally distributed. Therefore, a ΔCT value that differed from the average value by more than 3 SDs would be considered statistically different (with a confidence interval of >99%). These cutoff values (Table 2) were applied to the analysis of gene expression in the isolates from the oropharyngeal candidiasis patients; genes with a measured ΔCT value that fell outside the 3-SD range measured in SPRI azole-susceptible population (Table 3) were designated as being over- or underexpressed.
RT-PCR measurement of efflux pump and ERG11 expression levels in previously characterized C. albicans isolates.
Three sequential C. albicans isolates were previously analyzed by Sanglard and coworkers (16). The first two isolates in the series were fluconazole susceptible; Northern blot analysis confirmed that there were no significant increases in expression of ERG11, MDR1, or CDR (the probe was unable to discriminate between CDR1 and CDR2) in these isolates. The last isolate in the series was fluconazole S-DD, and Northern blot analysis revealed an eightfold increase in CDR expression. RT-PCR analysis of the same isolates confirmed these findings. In the first two isolates, the expression levels of CDR1, CDR2, MDR1, and ERG11 fell within the ranges measured in the SPRI azole-susceptible isolates (data not shown). In contrast, in the last isolate, expression of CDR2 was 16-fold higher than the average value measured in the SPRI azole-susceptible isolates. In addition, in a recent study we described an analysis of a series of C. albicans isolates exhibiting reduced susceptibility to posaconazole (3). As part of this analysis, we measured gene expression by both Northern blotting and RT-PCR (with the same probe sets employed in this analysis); the data from both techniques were in complete agreement.
Correlating changes in expression profiles and/or mutations in ERG11 with azole resistance in the oropharyngeal candidiasis isolates.
Three of the isolates from the azole-treated patients overexpressed MDR1; all three isolates were fluconazole resistant. CDR1 was upregulated in 14 patient-derived isolates, one of which was fluconazole susceptible. Similarly, of the 35 isolates overexpressing CDR2, four were fluconazole susceptible. ERG11 was overexpressed in four isolates, one of which was fluconazole susceptible. However, for all four isolates, the ΔCT values for ERG11 were only marginally greater than the cutoff values given in Table 2.
Previous work demonstrated that azole resistance in Candida spp. frequently resulted from multiple molecular mechanisms of resistance (12, 13, 17). It is clear from Table 3 that the majority of the oropharyngeal candidiasis isolates not only exhibited significant changes in efflux pump expression levels but had also acquired mutations in ERG11. In the following analysis, we focused on collections of isolates in which changes in resistance profiles appeared to result from a single molecular mechanism.
Mutations in ERG11 resulting in cross-resistance to fluconazole and voriconazole.
Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, ≥64 μg/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 μg/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (K128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). All but one of these isolates remained susceptible to posaconazole (see below). To confirm that the substitutions were responsible for the changes in susceptibility, select ERG11 alleles were cloned into a multicopy plasmid and expressed in S. cerevisiae. All of the ERG11 alleles tested conferred significant reductions in susceptibility to fluconazole, and most also conferred reduced susceptibility to voriconazole (see Table 5).
TABLE 5.
Changes in azole susceptibility resulting from expressing C. albicans ERG11 alleles in S. cerevisiae
| Original C. albicans isolate | Substitution(s) in expressed C. albicans Erg11p | MICa (μg/ml)
|
|||
|---|---|---|---|---|---|
| POS | ITZ | FLZ | VOR | ||
| None | None (vector alone) | 0.06 | 0.125 | 4 | 0.016 |
| C43 | None (wild type) | 0.06 | 0.5 | 16 | 0.125 |
| C441 | K143R | 0.25 | 1 | >256 | 1 |
| C587 | G464S | 0.25 | 1 | >256 | 1 |
| C477 | K128T, G464S, R467I | 0.125 | 0.5 | 32 | 0.25 |
| C530 | G450E, G464S | 1 | 1 | 128 | 1 |
| C600 | Y132H, S405F | 0.5 | >8 | >256 | 16 |
| C535 | Y132H, G448V | 0.25 | 1 | >256 | 0.03 |
| C572 | Y132H, G450E | 0.5 | 2 | >256 | 8 |
| C438 | Y257H, G464S | 0.5 | 1 | 128 | 4 |
| C439 | Y257H, G464S, G307S | 1 | 1 | >256 | 8 |
| C440 | Y257H, G464S, G307S, A61V | 4 | 8 | >256 | 16 |
| NAb | A61V | 0.25 | 0.5 | 16 | 0.125 |
See Table 2, footnote a.
NA, not applicable; mutation was engineered into a wild-type ERG11 gene.
Mutations in ERG11 resulting in changes in fluconazole susceptibility.
Sixteen clinical isolates were either fluconazole S-DD or fluconazole resistant but remained susceptible to voriconazole and posaconazole (Table 3). With the exception of isolate C513 (see below), we were unable to associate a pattern of overexpression of efflux pumps or ERG11 with the changes in azole susceptibility in these isolates. However, eight of these isolates (C504, C516, C470, C478, C577, C583, C444, and C489) also had substitutions in Erg11p that were previously shown to cause a reduction in fluconazole susceptibility (12, 13, 15). The remaining seven isolates (C482, C486, C481, C490, C497, C526, and C533) also had substitutions in Erg11p; it remains to be determined if the substitutions are responsible for the change in fluconazole susceptibility.
Overexpression of CDR2 and azole resistance.
In the three isolates from patient 3, the fluconazole MIC increased from a baseline level of 1 μg/ml in isolates C498 and C502 to 16 μg/ml in C513; the MICs of posaconazole, itraconazole, and voriconazole increased similarly (Table 3). The increase in the MIC was accompanied by a 30-fold increase in expression of CDR2. Other minor variations in gene expression included a slight increase in ERG11 expression in isolates C502 and C513 and an increase in CDR1 expression in isolates C498 and C513. All three isolates had the same three missense mutations in ERG11.
Mutations in ERG11 resulting in reduced susceptibility to posaconazole.
Isolates C438, C439, and C440 were cultured from patient 14 (Table 3). The baseline isolate was resistant to fluconazole and voriconazole. Isolate C439 was resistant to itraconazole, and the final isolate exhibited reduced susceptibility to all azoles, including posaconazole. There were no significant increases in the expression levels of either ERG11 or any of the pumps in these three isolates. Sequence analysis of ERG11 revealed an ordered acquisition of mutations: isolate C438 carried two substitutions (Y257H and G464S), C439 acquired an additional substitution (G307S), and isolate C440 had acquired a fourth substitution (A61V). When the three ERG11 alleles were expressed in S. cerevisiae, they conferred the same pattern of resistance seen in the clinical C. albicans isolates (Table 4). The mutation encoding the A61V substitution was introduced into a wild type ERG11 allele and expressed in S. cerevisiae. The A61V substitution alone did not confer a significant reduction in susceptibility to posaconazole (Table 5).
TABLE 4.
Subpopulation of candidiasis patients colonized by C. albicans isolates exhibiting cross-resistance to fluconazole and voriconazolea
| Patient no. | Isolate no. | MIC (μg/ml)
|
ΔCT
|
Substitution(s) in Erg11p | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POS | ITZ | FLZ | VOR | ACT1 | PMA1 | ERG11 | MDR1 | CDR1 | CDR2 | |||
| 4 | C527 | 0.12 | 0.5 | 256 | 4 | 7.5 | 6.6 | 9.1 | 12.6 | 8.9 | 8.6 | Y132H, G450E |
| C528 | 0.12 | 0.5 | 256 | 4 | 7.3 | 5.9 | 9.0 | 11.7 | 7.7 | 7.7 | Y132H, F155L, G450E | |
| C534 | 0.12 | 0.5 | 256 | 4 | 7.3 | 6.4 | 8.1 | 11.0 | 8.3 | 8.8 | Y132H, G450E | |
| C546 | 0.12 | 0.25 | 128 | 2 | 8.1 | 7.5 | 9.6 | 11.9 | 9.4 | 9.4 | Y132H, G450E | |
| C536 | 0.25 | 0.5 | 256 | 4 | 8.2 | 7.7 | 9.8 | 11.7 | 9.2 | 9.4 | Y132H, G450E | |
| C572 | 0.25 | 0.25 | >256 | 8 | 8.0 | 7.4 | 9.5 | 12.1 | 9.5 | 9.2 | Y132H, G450E | |
| 6 | C535 | 0.5 | 2 | 256 | 8 | 7.7 | 7.1 | 9.8 | 9.0 | 9.6 | 10.9 | Y132H, G448V |
| C539 | 0.25 | 2 | 128 | 8 | 7.2 | 6.8 | 11.0 | 8.3 | 6.6 | 7.1 | Y132H, G448V | |
| 11 | C594 | 0.5 | 2 | 128 | 16 | 7.2 | 6.5 | 9.0 | 11.3 | 8.2 | 7.5 | Y132H, S405F |
| C600 | 1 | 4 | >256 | 16 | 7.7 | 7.1 | 8.5 | 12.3 | 9.4 | 8.2 | Y132H, S405F | |
| 12 | C587 | 0.5 | 0.5 | 256 | 4 | 7.7 | 7.3 | 9.2 | 8.2 | 10.2 | 9.6 | G464S |
| 13 | C480 | 0.03 | 0.12 | 128 | 8 | 8.3 | 7.4 | 9.3 | 9.2 | 12.2 | 15.1 | K128T, G464S, R467I |
| 14 | C438 | 0.5 | 0.5 | 128 | 2 | 6.6 | 6.0 | 7.9 | 13.3 | 7.1 | 7.5 | Y257H, G464S |
| C439 | 1 | 4 | >256 | 16 | 7.1 | 6.2 | 7.6 | 13.5 | 7.7 | 7.5 | Y257H, G464S, G307S | |
| C440 | 4 | 8 | >256 | >16 | 7.3 | 6.4 | 8.0 | 13.5 | 7.9 | 7.7 | Y257H, G464S, G307S, A61V | |
| 17 | C477 | 1 | 0.25 | 256 | 16 | 7.6 | 6.1 | 9.3 | 7.3 | 10.5 | 15.3 | K128T, G464S, R467I |
| 18 | C507 | 1 | 2 | 64 | 8 | 6.1 | 7.2 | 8.6 | 10.5 | 8.3 | 8.4 | Y132H, H283R, G464S |
DISCUSSION
Azole resistance in C. albicans frequently results from overexpression of genes encoding efflux pumps or from mutations in or overexpression of ERG11. Northern blot analysis has proved useful for identifying significant changes in gene expression, particularly when comparing isogenic azole-susceptible and -resistant isolates side by side on the same gel. However, such analyses are more problematic, if not impossible, if a susceptible baseline strain is unavailable. The goals of this study were twofold. First, we used RT-PCR to determine what constitutes a “normal” level of gene expression for a collection of genes that encode proteins known to confer azole resistance in azole-susceptible C. albicans isolates. The second goal was to use these population averages to identify changes in expression in a collection of C. albicans isolates exhibiting various levels of azole resistance.
With regard to the first goal, in a collection of 14 azole-susceptible isolates, five of the six test genes exhibited minor fluctuations in expression (as reflected in SD values of <1). The exception was MDR1, the SD value for which was 2, which corresponds to a ±4-fold fluctuation in expression. To estimate how much of these variations were due to experimental error versus differences between individual strains, we made replicate measurements with a single strain. Expression of MDR1 varied the least; the range of expression levels of the other test genes were similar to those measured in the SPRI azole-susceptible population. These data suggest that the fluctuations in MDR1 expression seen in our survey resulted from strain-to-strain variations rather than from inconsistencies in the experimental methodology. Lyons and White also reported that MDR1 expression varied considerably, which they ascribed to a rapid turnover of the MDR1 transcript (4). In summary, we determined that by standardizing growth conditions to avoid the previously described growth phase-dependent variations in gene expression (4), it was possible to assign an average expression level across a collection of strains.
With regard to the second goal, 38 clinical isolates from 18 patients were analyzed. The expression levels of the control genes, PMA1 and ACT1, did not vary significantly between the two populations of isolates, suggesting that the growth conditions and RNA extraction techniques were reproducible. To identify changes in expression, we designated a gene as being overexpressed if the ΔCT value measured in the patient isolate differed from the average value calculated for the susceptible isolates by more than 3 SDs. Based on these criteria, CDR2 was classified as being overexpressed in the majority of the patient isolates, including four fluconazole-susceptible isolates.
To determine if the changes in CDR2 expression were stable, select isolates were passaged for a week in liquid medium with or without fluconazole. Contrary to the findings of a previous study (6), passaging cells in the absence of fluconazole did not lead to a reversal of the changes in CDR2 expression (data not shown). Another unusual finding was that in contrast to previous reports (8, 19), CDR1 and CDR2 did not appear to be coregulated; the reason for this discrepancy is unknown. Few patient isolates exhibited overexpression of either MDR1 or ERG11; for those isolates that did, there was no clear association between azole susceptibility and changes in expression of MDR1 or ERG11. The only isolates for which there appeared to be a clear correlation between overexpression of CDR2 and a change in susceptibility were from patient 3. However, there are a number of caveats to the approach taken here. For instance, since we measured gene expression in the absence of drug, we would not have identified isolates that had acquired mutations that enable them to respond more efficiently (i.e., a more rapid and possibly larger degree of overexpression) to a drug challenge. Similarly, we cannot rule out the possibility that the isolates have acquired mutations in the efflux pump genes that enable them to transport drugs more efficiently or have changed the substrate profile of the pumps.
In addition to the changes in pump expression detailed above, all of the isolates had acquired mutations in ERG11. The ERG11 mutations in 28 of the 34 isolates that were either fluconazole resistant or fluconazole S-DD have all previously been associated with changes in azole susceptibility (5, 12, 13, 15). For the remaining six isolates, the mutations were clustered into two of the three hot-spot regions noted by Marichal and coworkers (5). We observed that a particular pattern of ERG11 mutations was associated with cross-resistance to both fluconazole and voriconazole; 16 of the 17 fluconazole- or voriconazole-resistant isolates had a substitution close to the N terminus paired with a second substitution located towards the C terminus of the protein. A similar finding was reported recently (Sanglard et al., 42nd ICAAC). We confirmed that the mutations were responsible for the change in susceptibility by expressing alleles with these mutations in S. cerevisiae; all but one of the alleles tested conferred reduced susceptibility to fluconazole and voriconazole.
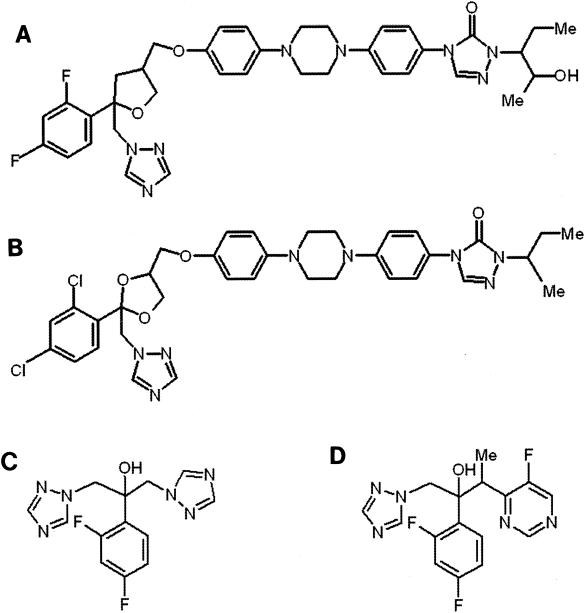
The high incidence of cross-resistance between fluconazole and voriconazole presumably results from their structural similarity (Fig. 1). Furthermore, the structural differences between the compact azoles (e.g., fluconazole and voriconazole) and those with long side chains (e.g., posaconazole and itraconazole) may explain why all but one of the 17 fluconazole- and voriconazole-resistant isolates remained susceptible to posaconazole (i.e., MIC, ≤1 μg/ml) and nine remained susceptible to itraconazole. To explain these differences, we constructed a three-dimensional model of Erg11p based on the X-ray structure of the Cyp51 orthologue from Mycobacterium tuberculosis (20). The long side chain of posaconazole and itraconazole, which is absent in fluconazole and voriconazole, is predicted to make extensive hydrophobic interactions with Erg11p and may serve to stabilize binding of these drugs to some of the mutated forms of the protein. Consistent with our model, the A61V mutation in isolate C440, which specifically impacted the binding of posaconazole and itraconazole (as reflected in the MIC change), is predicted to interfere with binding of the long side chain (20). However, it is important to note that the A61V substitution alone does not confer significant levels of resistance to posaconazole; some or all of the additional substitutions seen in isolate C440 are required to cause the observed change in susceptibility to posaconazole (Table 5).
FIG. 1.
Structures of posaconazole (A), itraconazole (B), voriconazole (C), and fluconazole (D).
Interestingly, there was a marked difference in the distribution of silent mutations between the ERG11 alleles from the sensitive and resistant populations. For example, seven isolates from the SPRI azole-susceptible population had silent mutations in the ERG11 alleles; in six of these isolates the mutations were distributed heterogenously between the two ERG11 alleles. In contrast, 23 of the isolates cultured from patients treated with azoles had at least two and frequently more than five silent mutations in ERG11. In all 23 isolates, the silent mutations were strictly conserved between the two ERG11 alleles. We hypothesize that the absence of heterozygosity at the ERG11 locus in the patient-derived isolates is a result of the selective pressures associated with azole treatment. More specifically, following the acquisition of an ERG11 mutation that confers reduced susceptibility to azoles, there is presumably an additional growth advantage conferred on isolates that undergo a gene conversion event that results in introduction of the ERG11 mutation, and thereby the linked silent mutations, into both copies of ERG11.
In summary, it is apparent that more than one mechanism of resistance may contribute to the overall resistance phenotype in many isolates. Other recent studies have made similar observations (11, 13, 19). It remains to be determined in what temporal order these individual mechanisms arise and to what degree each individual resistance mechanism contributes to the overall resistance phenotype. Such an analysis would require systematic elimination of each resistance mechanism by gene knockouts. At present, such manipulations are not feasible in clinical C. albicans isolates. The occurrence of multiple mechanisms of resistance presumably reflects both the long periods of drug exposure and the static mode of action of azoles against yeasts. From a therapeutic point of view, posaconazole appears to offer advantages over fluconazole and voriconazole, since posaconazole appears to be less affected by either mutations in ERG11 or the overexpression of specific efflux pumps.
Acknowledgments
We thank Todd Black (SPRI) for helpful comments, David Perlin and Steven Park (PHRI, Newark, N.J.) for sharing unpublished results, Michelle Treitel (SPRI) and Scott Walker (SPRI) for editing the manuscript, and Robyn Hawkinson (SPRI) for technical help.
REFERENCES
- 1.Cacciapuoti, A., D. Loebenberg, E. Corcoran, F. Menzel, Jr., E. L. Moss, Jr., C. Norris, M. Michalski, K. Raynor, J. Halpern, C. Mendrick, B. Arnold, B. Antonacci, R. Parmegiani, T. Yarosh-Tomaine, G. H. Miller, and R. S. Hare. 2000. In vitro and in vivo activities of SCH 56592 (posaconazole), a new triazole antifungal agent, against Aspergillus and Candida. Antimicrob. Agents Chemother. 44:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canuto, M. M., and F. G. Rodero,2002. Antifungal drug resistance to azoles and polyenes. Lancet Infect. Dis. 2:550-563. [DOI] [PubMed] [Google Scholar]
- 3.Li, X., N. Brown, A. S. Chau, J. L. López-Ribot, C. A. Mendrick, D. Loebenberg, R. S. Hare, B. J. DiDomenico, and P. M. McNicholas. 2004. Changes in susceptibility to posaconazole (POS) in clinical isolates of Candida albicans. J. Antimicrob. Chemother. 53:74-80. [DOI] [PubMed] [Google Scholar]
- 4.Lyons, C. N., and T. C. White,2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. Ramaekers, F. C. Odds, and H. V. Bossche. 1999b. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 6.Marr, K. A., C. N. Lyons, T. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meunier, F. 1989. Candidiasis. Eur. J. Clin. Microbiol. Infect. Dis. 8:438-447. [DOI] [PubMed] [Google Scholar]
- 8.Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 9.Morschhauser, J., S. Michel, and J. Hacker. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257:412-420. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. Document M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Nozawa, Y., and T. Morita. 1986. Molecular mechanisms of antifungal agents associated with membrane ergosterol: dysfunction of membrane ergosterol and inhibition of ergosterol biosynthesis, p. 111-122. In K. Iwata and H. Vanden Bossche (ed.), In vitro and in vivo evaluation of antifungal agents. Elsevier Science Publishers, B. V., Amsterdam, The Netherlands.
- 12.Perea, S., Lopez- J. L. Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perea, S., J. L. Lopez-Ribot, B. L. Wickers, W. R. Kirkpatrick, R. K. McAtee, O. P. Dib, S. P. Bachmann, S. Keller, M. Martinez, and T. F. Patterson. 2001. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller, M. A., S. A. Messer, R. J. Hollis, and R. N. Jones. 2001. In vitro activities of posaconazole (SCH 56592) compared with those of itraconazole and fluconazole against 3685 clinical isolates of Candida spp. and Cryptococcus neoformans. Antimicrob. Agents Chemother. 45:2862-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanglard, D., K. Kuchler, F. Ischer, J.-L. Pagani, M. Monod, and J. Bille. 1998. Mechanisms of resistance to azole antifungal gents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 18.Vanden Bossche, H., G. Willemsens, and P. Marichal. 1987. Anti-Candida drugs-the biochemical basis for their activity. Crit. Rev. Microbiol. 15:57-72. [DOI] [PubMed] [Google Scholar]
- 19.White, T. C., S. Holleman, F. Dy, F. D. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao, L., V. Madison, A. S. Chau, D. Loebenberg, R. E. Palermo, and P. M. McNicholas. Three dimensional models of the wild type and mutated forms of the cytochrome P450 14α-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into the binding of posaconazole. Antimicrob. Agents Chemother. 48:568-574. [DOI] [PMC free article] [PubMed]