SUMMARY
TRPM7 is a ubiquitous ion channel and kinase, a unique ‘chanzyme’, required for proper early embryonic development. It conducts Zn2+, Mg2+, Ca2+ as well as monovalent cations, and contains a functional serine/threonine kinase at its carboxyl terminus. Here, we show that in normal tissues and cell lines, the kinase is proteolytically cleaved from the channel domain in a cell type-specific manner. These TRPM7 Cleaved Kinase fragments (M7CKs) translocate to the nucleus and bind multiple components of chromatin remodeling complexes, including Polycomb group proteins. In the nucleus, the kinase phosphorylates specific serines/threonines of histones. M7CK-dependent phosphorylation of H3Ser10 at promoters of TRPM7-dependent genes correlates with their activity. We also demonstrate that cytosolic free [Zn2+] is TRPM7-dependent and regulates M7CK binding to transcription factors containing zinc-finger domains. These findings suggest that TRPM7-mediated modulation of intracellular Zn2+ concentration couples ion channel signaling to epigenetic chromatin covalent modifications that affect gene expression patterns.
TRPM7 is a ubiquitously expressed cationic ion channel and serine/threonine kinase (Nadler et al., 2001; Runnels et al., 2001; Yamaguchi et al., 2001). Global disruption of Trpm7 in mice results in embryonic lethality before embryonic day 7 (E7) (Jin et al., 2008). Tissue-specific reduction of TRPM7 gene expression between E7 and E12 of development results in abnormalities of multiple organs (Jin et al., 2012). TRPM7-deficient thymocytes exhibit dysregulated synthesis of many growth factors that are necessary for the differentiation and maintenance of thymic epithelial cells (Jin et al., 2008). Loss of TRPM7 at an intermediate developmental time point alters the myocardial transcriptional profile in adulthood, impairing ventricular function (Sah et al., 2013). TRPM7 overexpression in human embryonic kidney cells alters the transcriptional profile of hundreds genes (Lee et al., 2011). These data indicate that TRPM7 is intimately associated with developmental, tissue-specific regulation of gene activity. We set out to determine how TRPM7’s channel or kinase function has such broad, tissue-specific, and age-dependent effects on gene activity.
Initially, TRPM7 was thought to mediate cellular Mg2+ homeostasis based on growth arrest in TRPM7-deficient DT-40 B cells: growth arrest could be restored by culturing these cells in media supplemented with high Mg2+ (Nadler et al., 2001; Schmitz et al., 2005; Schmitz et al., 2003). In other cell types, however, loss of TRPM7 expression resulted in defects in cell growth that could not be rescued by excess Mg2+ (Hanano et al., 2004) or that even promoted proliferation (Inoue and Xiong, 2009). In addition, TRPM7 was not essential for proliferation and maintenance of Mg2+ levels in thymocytes derived from mice with a tissue-targeted TrpM7 deletion (Jin et al., 2008). In some cases TRPM7 appears to modulate Ca2+ signaling (Du et al., 2010; Hanano et al., 2004; Middelbeek et al., 2012). Thus, the consequences of TRPM7’s channel function are not settled; it may vary by tissue type and/or by its relative localization and gating in the cell’s plasma or intracellular membranes.
TRPM7’s most unique aspect is its C-terminal active serine/threonine kinase. The functional significance of this coupling of channel and kinase is not clear. Aside from the hypothesis that channel-dependent changes in cytosolic Mg2+ may control the activity of the kinase (Schmitz et al., 2003), no experimental data suggests that kinase activity is channel-dependent. In turn, kinase activity is not essential for channel gating (Clark et al., 2006; Matsushita et al., 2005), although it may modulate channel activity (Demeuse et al., 2006; Desai et al., 2012; Schmitz et al., 2003). Although TRPM7 kinase activity may be controlled through the autophosphorylation of a serine/threonine rich region located N-terminal to the catalytic domain (Clark et al., 2008b), external signals regulating the kinase activity are not known. In vitro substrates for the TRPM7 kinase, annexin A1 and myosin IIA heavy chain (Clark et al., 2008a; Dorovkov and Ryazanov, 2004), are not necessarily substrates in vivo. More recently, the TRPM7 kinase was proposed to regulate phosphorylation of eukaryotic Elongation Factor 2 (eEF2) via activation of the eEF2-kinase (Perraud et al., 2011).
Here we show that in many cell lines and normal tissues, TRPM7’s kinase is released from the channel domain in a cell type-specific fashion. TRPM7 Cleaved Kinases (M7CKs) translocate to the nucleus and bind multiple components of chromatin remodeling complexes, including Polycomb group proteins. In the nucleus, TRPM7’s kinase modifies specific histone phosphorylation, particularly of the promoters of TRPM7-dependent genes. We show that cytosolic free zinc levels are TRPM7-dependent and regulate M7CK binding to transcription factors containing zinc-binding domains. We hypothesize that TRPM7-mediated increases in cytoplasmic zinc couples channel function to chromatin covalent modification and thus affects cellular differentiation and embryonic development.
RESULTS
The TRPM7 kinase is proteolytically cleaved from the channel domain in vivo
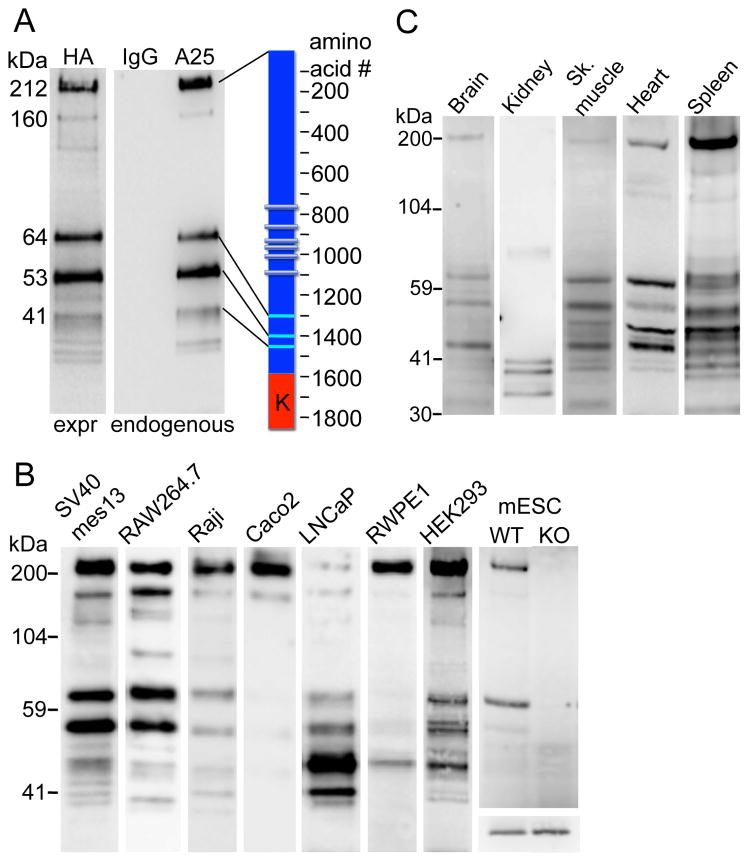
Although ubiquitously expressed, TRPM7 protein is of low abundance. In order to detect endogenous protein expression, we isolated the protein from large numbers of cells using mouse antibody for immunoprecipitation (IP) followed by western blot (WB) with rabbit antibodies to avoid antibody cross-reaction. In the course of these experiments, we noticed that in addition to a 210 kDa band corresponding to full-length TRPM7, several lower molecular weight bands were consistently detected (Figure 1A). This finding suggested that the endogenous TRPM7 molecule is proteolytically cleaved at specific sites. We refer to these fragments as TRPM7-Cleaved Kinases (M7CKs). As both antibodies recognize the TRPM7 C-terminus, the major bands corresponded to TRPM7 fragments that were cleaved between the transmembrane and kinase domains. These fragments were not the result of proteolysis during IP because their relative abundance did not vary with time over 2–17 h (Figure S1A). It is unlikely that these bands belong to proteins unrelated to TRPM7 since they were sequentially recognized with 2 different TRPM7 antibodies used for IP and WB. Yet, to rule out this possibility, we stably expressed C-terminally HA-tagged TRPM7 and used the well-characterized anti-HA-tag antibody (αHA) for IP and WB. Figure 1A shows that the ectopically expressed TRPM7-HA yielded major fragments identical to those for endogenous TRPM7, as well as a few minor lower molecular weight fragments that may have resulted from elevated protein expression. Since TRPM7-HA was ectopically expressed, this result obviates the possibility that C-terminal TRPM7 fragments were products of alternative RNA splicing. The fragments could have been generated from M7CK translation from multiple internal ribosome entry sites (IRES) present on its C-terminus, or specific post-translational proteolytic cleavage. However, IRES-dependent translation is unlikely - among more then 100 eukaryotic cellular mRNAs proposed to contain IRES, few have putative IRES sequences within exons (Thompson, 2012; see also http://iresite.org). Moreover, there were no confirmed multiple IRES within one mRNA. Thus, it is most likely that a proteolytic mechanism underlies the C-terminal fragment formation, as we previously demonstrated for caspase-mediated TRPM7 cleavage (Desai et al., 2012). Thus, a substantial fraction of TRPM7 is cleaved in specific positions of the TRPM7 C-terminus, liberating the protein kinase from the channel domain.
Figure 1. TRPM7 cleavage fragments identified in multiple cell lines and tissues.
A. TRPM7 protein cleavage fragments in mouse mesangial SV40 mes13 cells. Cells were extracted with TBS/1% NP40. Endogenous TRPM7 was immunoprecipitated (IP’d) from lysates with TRPM7 C-terminal mouse monoclonal antibody (αA25) or normal mouse IgG and probed on WB with anti-C-terminal rabbit antibody (αC47). C-terminally HA-tagged TRPM7 (expressed=expr) was IP’d with anti-HA-agarose (αHA) from SV40 mes13 cells stably expressing recombinant protein and probed on WB with αHA-peroxidase conjugate. Scale (left) indicates the molecular weight of major bands calculated from their electrophoretic mobility relative to standard molecular weight markers. Cartoon (right) shows the approximate position of cleavage sites; K indicates kinase domain.
B. TRPM7 cleavage pattern in 8 distinct cell lines. Mouse mesangial (SV40 mes13), macrophage (RAW 264.7), mESC, human B-lymphocyte (Raji), Caco-2 (colon epithelial), prostate (metastatic LNCaP and non-metastatic RWPE1), and embryonic kidney (HEK-293) cells were extracted and IP’d as described in A. Extracts demonstrate the relative amounts of cleaved TRPM7 isolated from each tissue. Information about the relative content of the full length TRPM7 and the cleaved fragments is contained in each individual lane, which are intentionally not normalized to control protein. No positive bands were found from the same tissue extracts IP’d with normal mouse IgG (not shown). mESCs were generated as described in Experimental Procedures from WT or TrpM7−/− (KO) blastocysts. The lower panel in the mESC column shows equal actin content in both mESC lysates. Samples run on different gels are combined in the figure and aligned against identical molecular weight markers.
C. TRPM7 cleavage pattern in different mouse tissues. Freshly isolated mouse organs were extracted and IP’d as described in A. Extracts demonstrate the relative amounts of cleaved TRPM7 isolated from in each tissue. No positive bands were found from the same tissue extracts IP’d with normal mouse IgG (not shown).
Different cell types display specific TRPM7 cleavage patterns seen as differences in relative expression of major fragments and generation of unique fragments (Figure 1B). This suggests there are cell type-specific mechanisms defining the cleavage site and potentially distinct functions for each cleaved fragment. This hypothesis is consistent with the observation that the TRPM7 cleavage pattern changes during differentiation of mouse embryonic stem cells (mESC) into specific progenitors (Figure S1). Upon differentiation into embryoid bodies, distinct C-terminal cleaved fragments transiently appeared on days 8–12 of differentiation (Figure S1B). Neuronal progenitors derived from mESC lack the 65 kDa C-terminal fragment detected in mESC and display only full-length TRPM7 protein (Figure S1C). Animal tissues also display a diversity of TRPM7 cleavage patterns (Figure 1C) more complex than those observed in cell lines, presumably because each tissue is composed of many cell types.
Since a substantial proportion of TRPM7 protein was found as C-terminal fragments in many different cell types (e.g. 50–70% in SV40 mes13 cells and close to 100% in mouse kidney), we expected to find equivalent amounts of the truncated channel. Thus, we immunoprecipitated TRPM7 with antibody recognizing the N-terminal epitope (NFP, amino acids 532–620) and western blotted with a C-terminal antibody (αCFP, a. a. 1277–1380) recognizing an epitope situated upstream of the C-terminal cleavage sites. This antibody combination reliably recognized transiently expressed recombinant full-length or truncated TRPM7 as well as endogenous full-length protein, but failed to detect endogenous truncated channels (Figure S1D). The lack of channel domain-containing protein suggests that the truncated channel was eliminated after C-terminal cleavage. This is consistent with results obtained using genetically modified mESC that produce a TRPM7 RNA with the stop codon placed in front of kinase domain (halting translation after amino acid 1537 (Ryazanova et al., 2010)). Patch-clamp measurements of these cells failed to detect the characteristic TRPM7 current (Ryazanova et al., 2010). Our antibody combination readily detected the full-length channel in WT mESC and the caspase-cleaved endogenous channel (Desai et al., 2012), but not the truncated channel in Δ-kinase mESC (Figure S1E). These data suggest that C-terminally truncated TRPM7 protein containing the channel domain is normally eliminated from proliferating cells.
M7CK binds multiple components of chromatin remodeling complexes
In previous experiments (Krapivinsky et al., 2006), we identified proteins that interact with TRPM7 using yeast two-hybrid screening. Evolutionarily conserved protein fragments of the human TRPM7 N- and C-terminal cytoplasmic domains were chosen as baits. Screening of a human brain library with a bait containing a portion of the TRPM7 C terminus (amino acids 1364–1865 of NP_060142.3) revealed the Ring1 and YY1 binding protein, RYBP (NP_036366.3) as a potential TRPM7 interactor. Since RYBP is a transcription factor strictly confined to the nucleus (Garcia et al., 1999) and the TRPM7 channel is a plasma membrane and vesicular protein, we assumed that it was a false positive. However, when we performed tandem affinity purification of the same TRPM7 fragment stably expressed in SV40 mes13 cells, we identified a number of nuclear proteins (see Table S1). Given that portions of the TRPM7 C-terminus are naturally cleaved from the channel and located in the nucleus (shown in Figure 3), we set out to investigate these potential interactions.
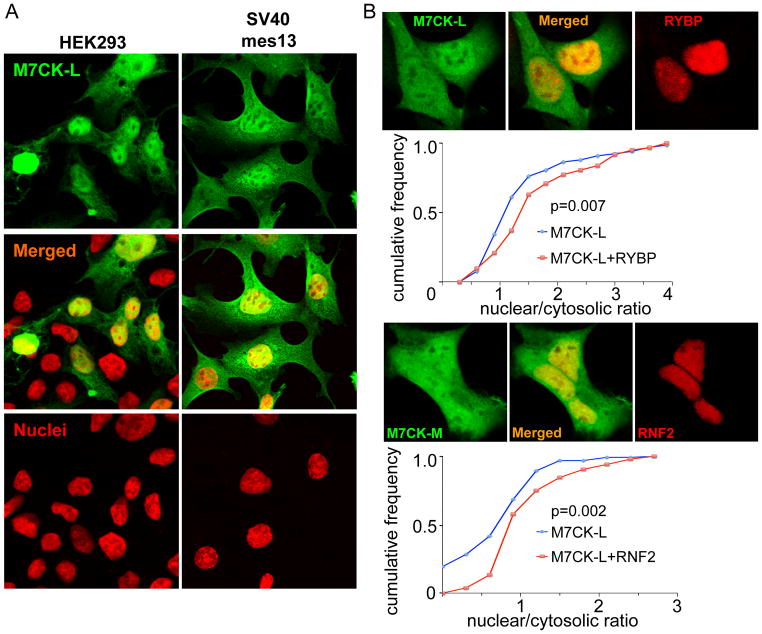
Figure 3. Nuclear location of M7CK.
A. TRPM7 cleavage fragments (M7CK) in the nucleus and cytosol. HA-tagged TRPM7 C-terminal fragment (a. a. 1299–1864; green) expressed transiently (HEK) or stably (SV40 mes13) immunostained with αHA. Nuclei stained with propidium iodide (red).
B. Bound nuclear proteins enable M7CK nuclear accumulation. HEK cells were co-transfected with the indicated HA-tagged M7CK and FLAG-tagged protein or empty vector. Relative nuclear and cytosolic M7CK concentration was measured as average fluorescence intensity in confocal images of formaldehyde-fixed cells and immunostained with epitope-tagged antibodies; 60–70 cells were measured for each condition. Cumulative frequency plots demonstrate that co-expression with the bound nuclear protein increases in the fraction of the cells with higher nuclear M7CK concentration. p, Kolmogorov-Smirnov probability.
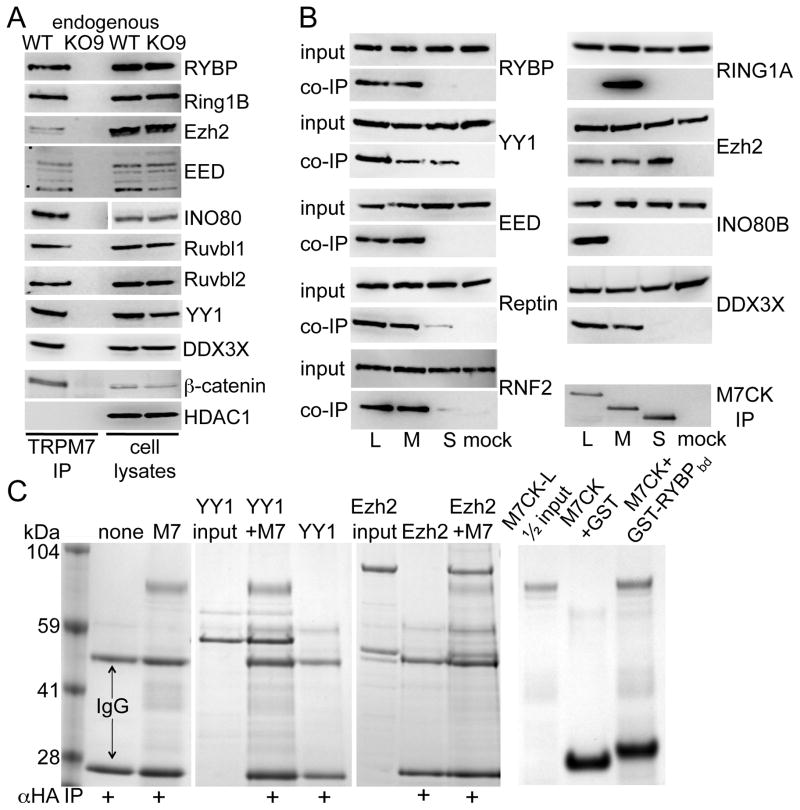
In order to verify endogenous M7CK-interacting proteins, we generated mESC lacking TrpM7 (TrpM7−/−, see Experimental Procedures, Figure 1B and Figure S2) and used these cells as negative controls in co-immunoprecipitation experiments. TRPM7 immunoprecipitation (IP) from mouse embryonic stem cells confirmed binding of endogenous TRPM7 with the nuclear proteins RYBP, Ruvbl1/pontin, Ruvbl2/reptin, DDX3X, DDB1 and DBC1 (Figure 2A and Figure S3), all constituents of chromatin remodeling complexes (see Table S2 for annotation of these proteins). RYBP is a component of Polycomb Repressor Complex 1 (PRC1) and also interacts with the transcription factor, YY1 (Garcia et al., 1999; Tavares et al., 2012). YY1 is part of the PRC2 histone methylation complex (Satijn et al., 2001) and the INO80 chromatin-remodeling complex (Cai et al., 2007; Wu et al., 2007). The RNA helicase DDX3X, and the DNA helicases Pontin and Reptin, are also components of the INO80 complex (Huen et al., 2010; Vella et al., 2012), of histone deacetylase (Gallant, 2007) and β-catenin (Bauer et al., 2000; Kim et al., 2005; Sierra et al., 2006) complexes. DDB1 is the core component of the Cul4 ubiquitin ligase complex participating in DNA damage-activated chromatin modification (Higa et al., 2006; Iovine et al., 2011). Finally, DBC1 is an inhibitor of the histone deacetylases Sirt1 (Zhao et al., 2008) and HDAC3 (Chini et al., 2010). Figure 2A and Figure S3B shows that endogenous TRPM7 specifically binds core PRC1 protein RNF2, core PRC2 proteins Ezh2 and EED1, INO80 DNA helicase, YY1, β-catenin and DDB1, but not HDAC1.
Figure 2. TRPM7 binds transcription factors and subunits of chromatin remodeling complexes.
A. Endogenous TRPM7 was IP’d from combined cytosolic+nuclear extracts of WT3 or KO9 mESC with αA25 mouse or αC47 rabbit antibody and the IP probed on WB with antibody to the indicated proteins. INO80 cell lysate lanes were run on a different gel, as indicated by gap. B. Subunits of Polycomb and INO80 chromatin remodeling complexes bind ectopically expressed M7CKs. FLAG-tagged transcription factors were transiently co-expressed in 293T cells with HA-tagged M7CK-L, -M or -S. Cytosolic+nuclear extract was IP’d with αHA-agarose and co-IP’d proteins were probed on WB with αFLAG-peroxidase. C. Affinity-purified long form of M7CK (M7CK-L) binds purified transcription factors (Coomassie). Left 3 panels: HA-tagged M7CK-L (M7) was combined with purified YY1 or Ezh2 protein complexes and IP’d with αHA-agarose. Right panel: purified GST or GST-RYBP fragment (M7CK-binding domain, a. a. 22–47, GST-RYBPbd) fusion proteins were combined with purified M7CK-L and GST pulled down with glutathione agarose.
To verify that M7CK fragments of TRPM7 bind all these molecules, we made constructs expressing portions of the TRPM7 C-terminus. Since we do not know the precise cleavage sites for endogenous TRPM7, we made a series of constructs that would express fragments of the C-terminus with molecular masses close to those of major M7CKs, as calculated from their electrophoretic mobilities. Proteins containing mouse TRPM7 a. a. 1299–1863, 1397–1863 and 1445–1863 (all 3 contain the full kinase domain) most precisely co-migrate with endogenous cleaved TRPM7 kinase (Figure S3A). We refer to these proteins M7CK-Long, -Medium and –Short, respectively. We tagged these proteins on their C-termini with an HA epitope tag and tested their ability to bind TRPM7-interacting proteins. Figure 2B shows that proteins that bound endogenous TRPM7 are specifically co-immunoprecipitated with M7CKs when co-expressed in 293T cells; (see also Figure S3B–E demonstrating M7CK to Cul4, DBC1 and Sirt1). This result indicates that the binding site for the nuclear proteins is located on the M7CK portion of TRPM7’s C-terminus. Note that there are differences in the binding of nuclear proteins to the cleaved kinases; Ezh2 and YY1 bind all 3 cleaved kinases, while RYBP, EED, RNF2 and DDX3X do not bind M7CK-S. RING1A binds only M7CK-M and INO80 binds only M7CK-L. This pattern of selective binding may rationalize the existence of multiple M7CKs in different cell types (Figure 1B).
To test for direct M7CK binding to interacting molecules, we affinity-purified HA-tagged M7CK-L produced in E. coli (Figure S3F). Purified M7CK was combined with purified recombinant YY1 or the Ezh2-EED-Suz12 complex and pulled-down with αHA agarose. As shown on Coomassie-stained gels (Figure 2C), both YY1 and the Ezh2 complex specifically bound M7CK. The TRPM7-binding domain of RYBP (a. a. 22–47, zinc-finger domain) expressed as a GST fusion protein and purified from E. coli directly bound purified M7CK-L (Figure 2C). In summary, TRPM7’s cleaved kinase fragments directly binds several components of the chromatin remodeling complexes, PRC1 and PRC2, and INO80.
Nuclear location of M7CK
Strikingly, abundant ectopically expressed TRPM7 C-terminal fragments were found in cell nuclei (Figure 3). Analysis of the TRPM7 amino acid sequences (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) predicted a putative nuclear localization signal on the kinase C-terminus (mTRPM7 amino acids 1780–1807) that may provide the basis for importin-mediated M7CK nuclear localization. Additionally, nuclear proteins bound to M7CK could facilitate its translocation from cytosol to nucleus or retain M7CK in the nucleus: both mechanisms would increase nuclear M7CK. To determine whether M7CK-binding nuclear proteins affect M7CK nuclear abundance, cells were co-transfected with HA-tagged M7CK- and FLAG-tagged nuclear binding proteins (RYBP or RNF2), or empty vector as control. After cell fixation and immunostaining, relative nuclear and cytosolic M7CK was quantified in confocal images. As shown in Figure 3B, M7CK co-expression with nuclear-localized interactors substantially increased the relative nuclear concentration of M7CKs.
Regulation of M7CK binding
The preceding experiment suggests that M7CKs can be targeted to a particular nuclear protein by cleavage at a specific C-terminal site (Figure 2B). One obvious possibility is that TRPM7’s channel activity regulates M7CK’s cleavage from the channel domain. We compared the cleavage pattern of expressed WT TRPM7 and a non-conducting pore mutant that acts as a dominant negative [inhibits the conductance of the native channel (Krapivinsky et al., 2006)]. Figure S4 shows that the cleavage pattern and extent of cleavage were identical. These data suggest that the TRPM7 C-terminal cleavage did not depend on a functional, ion-conducting pore. This implies that ions entering via the channel pore are not required to activate, for example, a localized protease that cleaves TRPM7’s channel domain from its C-terminal containing kinase domain.
Another simple hypothesis is that TRPM7’s kinase ‘tags’ itself via autophosphorylation for nuclear targeting. We thus tested whether M7CK’s kinase activity is required for nuclear protein binding. Point mutations in TRPM7’s kinase ATP-binding site (K1646A) eliminated kinase activity as detected by TRPM7 autophosphorylation (kinase-dead; (Matsushita et al., 2005), Figure S3G). These mutations had ambiguous effects on the binding of target molecules. The kinase-dead mutations did not affect Ezh2 binding, but attenuated YY1 and RYBP binding and enhanced RNF2 binding (Figure S3H). Thus, regulation of TRPM7 kinase activity is a potential mechanism for targeting M7CK to different nuclear complexes.
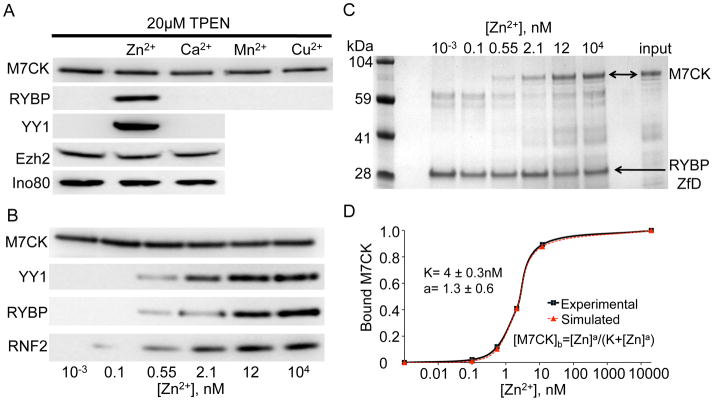
TRPM7 is permeant to divalent metal ions with the highest permeability for Zn2+ (Zn2+ ≫ Mg2+ > Mn2+ ≥ Ca2+) (Monteilh-Zoller et al., 2003) and thus may regulate cytoplasmic concentrations of these ions. Figure 4A shows that 20 μM TPEN, a heavy metal chelator, completely blocks RYBP binding to M7CK. TPEN block is relieved by Zn2+ but not by Ca2+, Mn2+ or Cu2+. Additional experiments showed that M7CK binding to proteins containing a zinc-finger (RYBP, YY1 and RNF2) is Zn2+-dependent; binding to proteins not containing a zinc-finger motif (Ezh2 and INO80) did not require Zn2+ (Figure 4A, B). The critical role of the zinc-finger structure in binding of specific proteins to M7CK is supported by the finding that the 26-amino acid RYBP sequence comprising the zinc-finger domain is sufficient for M7CK binding (Figure 3C). Half-maximal binding of RYBP, YY1 and RNF2, as well as the purified zinc-finger domain of RYBP to [Zn2+], was ~4 nM (Figure 4C), a concentration that is ~ 5–10-fold higher than resting cytosolic [Zn2+] (Krezel and Maret, 2006; Li and Maret, 2009; Taki et al., 2004; Vinkenborg et al., 2009). These levels would easily be reached by opening of TRPM7 channels, since the driving force for zinc is inward over the physiological range of voltages (EZn ≈ +80 mV). Thus, we set out to determine if TRPM7 Zn2+ conduction may regulate Zn2+-dependent M7CK targeting to specific transcription factors.
Figure 4. Zinc dependence of M7CK binding to zinc-finger domain (ZfD)-containing proteins.
A. HA-tagged M7CK was co-expressed with FLAG-tagged interacting proteins in 293T cells. The cell lysate was split into equal aliquots and supplemented with 20 μM TPEN and 50 μM of the indicated ions. M7CK was IP’d with αHA and the co-IP’d proteins probed with αFLAG.
B. FLAG-tagged ZfD-containing proteins were co-expressed in 293T cells with HA-tagged M7CK. [Zn2+] was adjusted by variation of [EGTA] and [ZnSO4]. M7CK-HA was IP’d with HA-agarose; ZfD protein binding was detected in the αFLAG WB.
C [Zn2+] dependence of M7CK binding to GST-RYBP ZfD. Purified GST fusion with RYBP fragment (a. a. 22–47 comprising the RYBP-ZfD) was combined with purified M7CK-L in the presence of varying [Zn2+] and pulled down by glutathione agarose (Coomassie-stained gel).
D. Quantification of M7CK binding shown in C. Values of integrated pixels of the bound M7CK image were normalized to maximum binding at 10 μM [Zn2+]. Fitted equation and parameters shown.
TRPM7 affects cytosolic Zn2+ concentrations
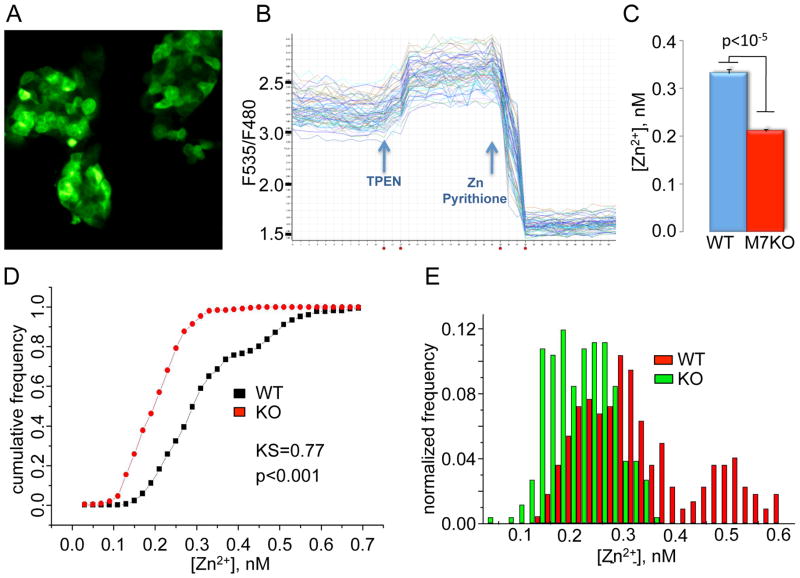
To determine whether TRPM7 activity affects cytosolic [Zn2+], we compared [Zn2+] in WT and TrpM7−/− mouse embryonic stem cells, using a stably expressed protein-encoded zinc sensor, eCALWY-4 (Vinkenborg et al., 2009). As shown in Figure 5, average cytosolic [Zn2+] is significantly lower in TrpM7−/− than in WT cells (consistent with our previous findings, cytosolic [Mg2+] was normal in TrpM7−/− cells; Figure S5). Importantly, knockout of TrpM7 resulted in the loss of the sub-population of cells with substantially higher [Zn2+] (Figure 5E). This suggests that TRPM7 channel activity was sufficient in these cells to elevate cytosolic zinc to levels that enabled targeting of M7CK to chromatin remodeling complexes.
Figure 5. Cytosolic [Zn2+] measured in WT and TrpM7−/− mESC.
A. Fluorescent image of mESC stably expressing the Zn2+ indicator, eCALWY-4.
B. eCALWY-4 fluorescence recordings from multiple cells in a single sample.
C. Calculated cytosolic [Zn2+] shown as mean ± SEM. (D) Kolmogorov-Smirnov statistics. (E) Histogram of [Zn2+] in cell populations for WT3 and TrpM7−/− (KO9) mESC 260 cells of each type were measured in 4 independent experiments.
M7CK phosphorylate histones
Since M7CKs are functional kinases (Ryazanova et al., 2004), we tested whether M7CK binding partners are substrates for M7CKs. However, extensive experiments failed to show direct phosphorylation of the verified interactors RYBP, YY1, EZH2, RING1A, Ruvbl2 and Cul4b by the kinase when the two proteins were bound (data not shown).
Many chromatin-remodeling complexes covalently modify core histones by lysine methylation, acetylation, ubiquitination or sumoylation (Badeaux and Shi, 2013; Strahl and Allis, 2000). These modifications are believed to alter chromatin structure by affecting the recruitment of non-histone proteins that modulate genomic DNA accessibility for replication, repair and for ‘tuning’ gene expression. Histones are also phosphorylated by several protein kinases; phosphorylation of specific histone residues has been associated with transcriptional regulation, apoptosis, cell cycle progression, DNA repair, chromosome condensation, and developmental gene regulation (Baek, 2011; Sawicka and Seiser, 2012) and may constitute a histone code that underlies epigenetic inheritance (Cheung et al., 2000a). The observation that M7CK is a functional protein kinase bound to several chromatin-remodeling complexes and located in the nucleus motivated us to explore whether M7CK modifies the phosphorylation state of established histone substrates for signaling kinases (Baek, 2011).
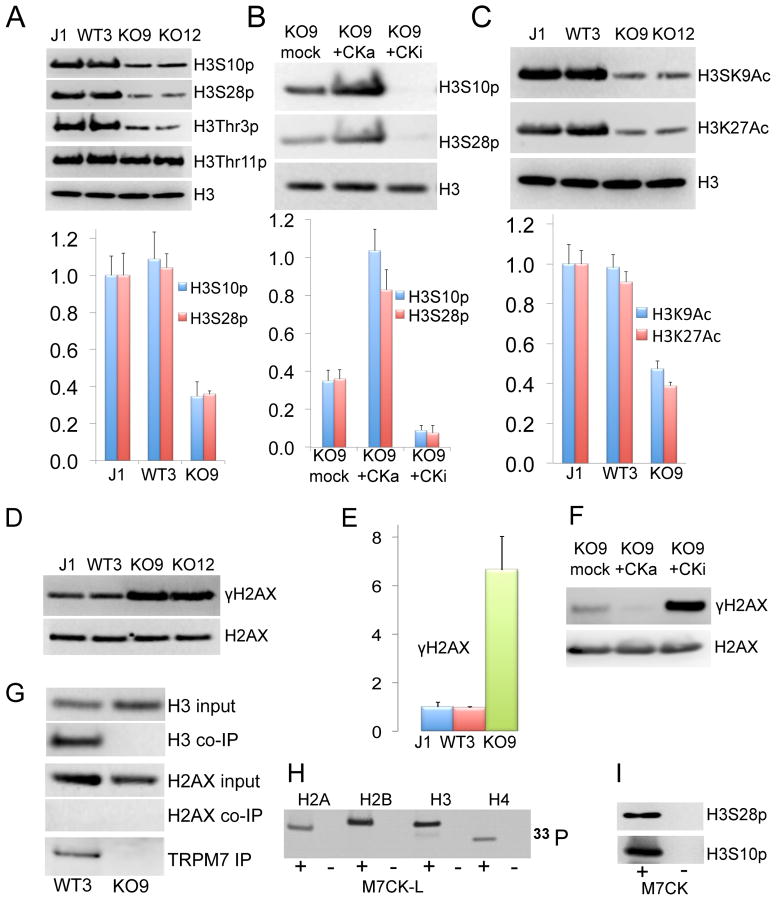
Figure 6A shows that global phosphorylation of histone H3 at serine 10 (H3S10p), serine 28 (H3S28p) and threonine 3 (H3T3p) is dramatically suppressed in TrpM7−/− mESC compared to WT cells. In contrast, phosphorylation of threonine 11 (H3T11p) was not affected and served as an intrinsic control. Differences in histone phosphorylation did not result from cell clonality because different clones with the same genotype had identical levels of histone phosphorylation (Figure 6A). H3S10p and H3S28p levels were indistinguishable in WT3 cells grown in normal media or media with an additional 10 mM MgCl2 that was required for growth of TrpM7−/− mESC (not shown).
Figure 6. TRPM7 kinase-dependent global histone phosphorylation and acetylation.
Bar graphs display histone modification data statistics (mean ± SD, normalized to J1 data) for 3 independent experiments.
A. Global H3S10, H3S28 and H3T3 phosphorylation is decreased in TrpM7−/− cells. WBs of mESC histone extracts with specific anti-phosphoserine antibodies.
B. Restoration of histone H3S10 and H3S28 phosphorylation to WT levels by stable expression of active M7CK-L in TrpM7−/− mESC (KO9+CKa). Kinase-dead M7CK mutant (KO9+CKi) expression further decreases H3S10 and H3S28 phosphorylation.
C. Decrease in global H3 histone Lys9 and Lys27 acetylation (H3K9Ac and H3K27Ac) in Trpm7−/− mESC as shown by WB with anti-acetylated lysine antibodies.
D, E. WBs and summary of global H2AX phosphorylation in mESC.
F. Attenuation of γH2AX to WT levels by stable expression of active M7CK-L in TrpM7−/− mESC. Expression of kinase-dead M7CK mutant further increases H2AX phosphorylation.
G–I. TRPM7 IP’d from mESC pulls down H3, but not H2AX (G). Purified M7CK directly phosphorylated purified histones as shown by incorporation of radioactive phosphate from [γ-33P]-ATP (H) and WB with specific phosphoserine antibody (I).
To test whether TRPM7 kinase activity underlies changes in histone phosphorylation, we generated TrpM7−/− mESC stably expressing WT M7CK-L (KO9-CKa) and its kinase-dead mutant (KO9-CKi, see Figures S2F, S3G and Experimental Procedures for description of these stable cell lines). Figure 6B shows that the expression of active kinase rescues changes in H3 histone phosphorylation, implying that the TRPM7 kinase is responsible. To our surprise, expression of the inactive M7CK mutant that was intended to serve as a negative control resulted in a further 4–6-fold decrease in phosphorylation of H3S10 and H3S27 (Figure 6B). This suggests that inactive M7CK acts as a dominant negative regulator of another kinase (likely TRPM6 kinase because of its high sequence homology to TRPM7).
H3 histone acetylation at lysines 9 and 27 activates multiple genes (Eberharter and Becker, 2002). Synergistic coupling between H3S10 and H3S28 phosphorylation and acetylation of the adjacent K9 and K27 residues has been proposed (Cheung et al., 2000b; Lau and Cheung, 2011). In agreement with this finding, TrpM7−/− cells also display a substantial decrease in H3K9 and H3K27 acetylation (Figure 6C). We also measured a substantial increase of global trimethylation of H3K9 and H3K27 in TRPM7−/− mESC. It has been surmised that H3K9me3 and H3K27me3 antibodies poorly recognize their epitopes when the adjacent residue is phosphorylated (Duan et al., 2008, Lau and Cheung, 2011). Indeed, when we pretreated cell lysates with λ phosphatase, levels of the H3K9me3 and H3K27me3 were indistinguishable in WT and TRPM7−/− mESC (not shown). The loss of TRPM7 also resulted in a 6–8-fold increase in global phosphorylation of histone H2AX at serine 139 (γH2AX) that returns to normal upon expression of active M7CK (Figure 6D). Inactive kinase expression exhibits the same pattern as the H3 histone - it enhances the effect of the TRPM7 deletion, dramatically increasing γH2AX (Figure 6F).
The TRPM7 kinase-dependent modulation of H2AX phosphorylation may be mediated by another protein kinase. Ablation of the TRPM7 kinase increased H2AX phosphorylation, while active M7CK expression caused a decrease in hyperphosphorylated γH2AX. On the other hand, global dephosphorylation of H3S10, H3T11 and H3S28, rescued by M7CK expression, is consistent with direct phosphorylation of H3 by M7CK. Since the TRPM7 C-terminal antibody specifically co-immunoprecipitates H3 histone but not H2AX histone from nuclear extracts of mESC (Figure 6G), M7CK tightly binds H3 but not H2AX. In addition, purified M7CK phosphorylated purified recombinant histones H2, H3, and H4 in vitro (shown by 33P incorporation, Figure 6H). As shown by western blot with phospho-specific antibody, purified M7CK phosphorylates purified H3 histone at S10 and S28 (Figure 6I). These results imply that M7CK directly phosphorylates selected histone residues.
M7CK-dependent histone phosphorylation correlates with gene expression
Histone phosphorylation acts as a critical intermediate step in chromosome condensation during cell division, DNA damage repair, and transcriptional regulation (Baek, 2011; Banerjee and Chakravarti, 2011; Berger, 2010). Phosphorylation of histone H2AX on Ser139 in mammalian cells is one of the early responses to double-strand DNA breaks (Xu and Price, 2011), motivating us to test whether a global increase in γH2AX reflects increased DNA damage in TrpM7−/− cells. Comparison of nuclear expression of active ATM kinase, a key element in the DNA-damage response (Bekker-Jensen and Mailand, 2010), in WT and TrpM7−/− mESC (Figure S6A) did not reveal any increase of DNA double-break foci formation, ruling out increased DNA damage in TrpM7−/− cells. The function of M7CK-dependent H2AX phosphorylation is unknown.
Global increases of H3S10p and H3S28p in mitotic cells demonstrates temporal and spatial associations with chromosome condensation (Goto et al., 2002; Wei et al., 1999) and is sufficiently reliable that H3S10p is widely used as a mitotic marker. If H310p and H3S28p dephosphorylation in TrpM7−/− cells reflects a state of mitotic histone phosphorylation, it would be detected as decrease in the proportion of mitotic cells. No decrease in the percentage of mitotic cell phosphohistone immunofluorescence (Figure S6B) was observed in counts of H310p and H3S28p immunofluorescence-positive WT and TrpM7−/− cells. Thus, M7CK phosphorylates histones in interphase. Our results suggest that TRPM7 kinase has no major effect on cell cycle, consistent with its lack of effect on cell proliferation rates (Figure S2).
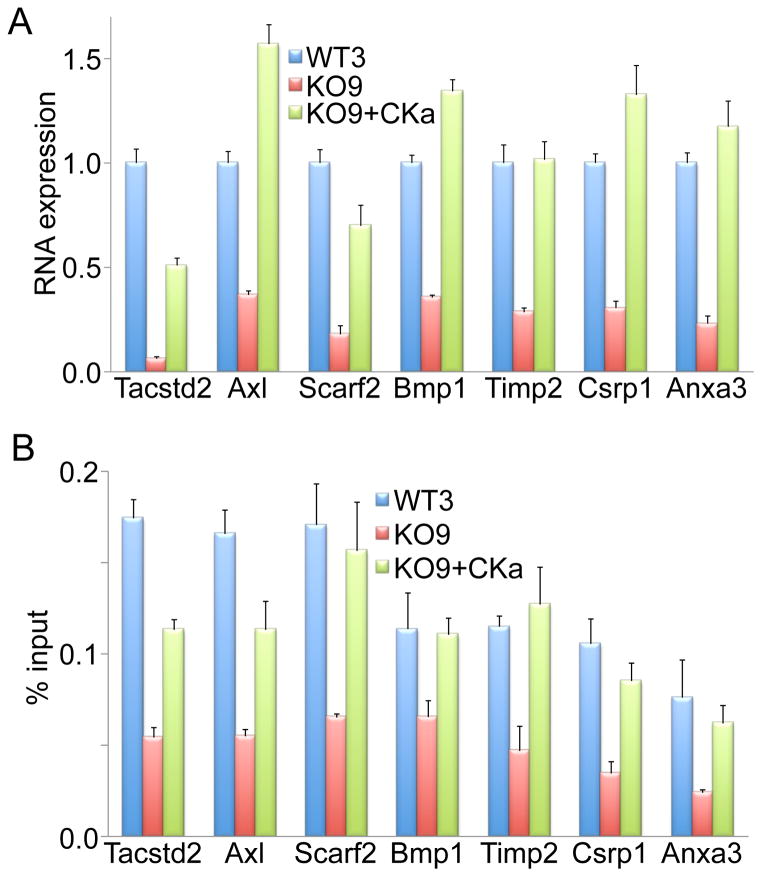
Accumulating evidence suggests that local histone phosphorylation on specific residues, particularly H3S10, affects expression of specific genes (Baek, 2011; Berger, 2010; Yang et al., 2012). We next determined if M7CK activity affected gene expression and if specific gene expression correlated with M7CK-dependent histone phosphorylation of the affected genes. We performed microarray analysis for differential gene expression in WT and TrpM7−/− mESC (Table S3). For further validation of the microarray data, we selected several genes with maximally down-regulated expression levels in TrpM7−/− cells and performed gene expression analysis with quantitative PCR (qPCR). qPCR comparison of gene expression in mESC WT, TrpM7−/− and TrpM7−/− stably expressing M7CK, confirmed the microarray data and demonstrated that gene down-regulation in TrpM7−/− cells was rescued by M7CK expression (Figure 7A). This result argues that gene regulation was TRPM7 kinase-dependent. Chromatin immunoprecipitation with antibody specifically recognizing phosphorylated H3S10 followed by qPCR with primers amplifying proximal promoter sequences of the same genes (ChIP-qPCR) showed that H3S10p content in promoters of the genes tested was substantially decreased in TrpM7−/− cells and this decrease was rescued by M7CK expression (Figure 7B). These results suggest that M7CK-mediated phosphorylation of H3S10 regulates gene expression.
Figure 7. M7CK-dependent H3S10 histone phosphorylation in gene promoters correlates with gene expression.
A. M7CK-L expression in TrpM7−/− mESC rescues activity of down-regulated genes. Expression of selected genes was quantified with RT-qPCR in WT3, TrpM7−/− (KO9) and KO9 stably expressing M7CK-L (KO9+CKa) mESC.
B. H3S10 phosphorylation in promoters of genes affected by TRPM7 deletion in mESC was attenuated in TrpM7−/− (KO9) cells and restored by M7CK expression (KO9+CKa), as determined by ChIP-qPCR.
DISCUSSION
Here we showed that a portion of TRPM7’s C-terminus containing the functional kinase is cleaved from the channel moiety and accumulates in the nucleus by binding to transcription factors composing chromatin-remodeling complexes. In the nucleus, M7CK phosphorylates histones on specific residues involved in essential cellular functions such as transcription regulation, DNA repair, and mitotic chromatin condensation. We also demonstrated that TRPM7 increases cytosolic free [Zn2+] and that binding of zinc-finger containing M7CK interactors is zinc-dependent. These findings suggest a signaling mechanism in which TRPM7 activation increases cytosolic [Zn2+], that in turn stimulates binding of M7CK to zinc-dependent transcription factors. M7CK translocation to the nucleus and subsequent phosphorylation of nuclear histones changes the chromatin covalent modification landscape affecting cell differentiation and embryonic development.
The linkage of protein kinase and channel domains in the evolution of TRPM channels suggests that these functions are coupled. We propose that this function is to link zinc entry and chromatin-modifying functions, in particular through the zinc dependence of M7CK binding to particular nuclear proteins. Since zinc concentrations that enable M7CK binding (EC50 = 4 nM) are only 5–10 fold higher then resting intracellular [Zn2+], and since nuclear [Zn2+] closely follows cytosolic [Zn2+] (Miranda et al., 2012), it is likely that TRPM7 channel function precedes initiation of M7CK binding to nuclear proteins. This is supported by our data that shows that cells with inactive TRPM7 channel have lower cytosolic [Zn2+].
M7CK formation depends on cell lineage, conditions, and time, suggesting that specific M7CKs are made in the course of development and cell differentiation. Whether this mechanism is based on cell-specific activation/expression of selective proteases, or the presence of specific proteins protecting the cleavage sites, is not known. Interestingly, after truncation, the portion of TRPM7 containing the channel domain is eliminated. One potential rationalization for this observation may be that the kinase-deleted channel stimulates apoptosis (Desai et al., 2012). That the channel is deleted after kinase cleavage suggests that the channel is required for kinase function, not vice versa.
Chromatin remodeling complexes, and particularly Polycomb complexes, are transcriptional modifiers that regulate a number of key processes in cell differentiation via covalent histone modifications (Fisher and Fisher, 2011; Surface et al., 2010). M7CKs interact with several transcription factors and nuclear proteins composing chromatin-modifying complexes. We found several potential regulatory mechanisms that may tune the specificity of M7CK binding to particular nuclear complexes. M7CK binding to different interactors depends on the cleavage site, kinase activity, and cytosolic [Zn2+]. The combination of these factors may provide specific, cell context-dependent M7CK nuclear complex formation. Although logical to assume that M7CK binding partners were potential substrates for the M7 kinase, our in vitro experiments failed to show direct M7CK phosphorylation of RYBP, YY1, EZH2, RING1A, Ruvbl2, or Cul4b when they were in complex with M7CK. We cannot exclude, however, the possibility that some other M7CK binding partners are phosphorylated. Nevertheless, M7CK activity is important for nuclear protein binding, perhaps because M7CK autophosphorylation (Clark et al., 2008b) is critical for these interactions. In summary, our findings demonstrate that nuclear protein binding to M7CK increases nuclear concentrations of M7CK where it phosphorylates histones, indicating that these proteins retain M7CKs in nucleus. These results strongly suggest that M7CK association with transcription factors may help target M7CK to a specific genomic location to provide site-specific histone phosphorylation.
Association of M7CK with subunits of PRC2 may seem at odds to the existing paradigm that PRC2-dependent H3K27 methylation and PRC2 binding to promoters represses transcriptional activity while H3S28 phosphorylation facilitates H3K27 acetylation and transcriptional activation. However, recent findings indicate that PRC2 (Jacob et al., 2011) and the Ezh2 methyltransferase subunit (Li et al., 2009) can also activate transcription (Simon and Kingston, 2013). In particular, H3K27me3 and H3S28p can coexist on the same histone molecule and H3K27me3S28 phosphorylation displaces PRC2 from the promoter (Gehani et al., 2010). In a similar scenario, M7CK binding of PRC2 may target it to a specific site where it phosphorylates the critical H3S28 and displaces PRC2 to activate transcription.
We identified two modes of TRPM7-dependent histone phosphorylation. Phosphorylation of histone H3 residues was dramatically attenuated when TrpM7 was genetically silenced but increased when M7CK was restored in TrpM7−/− cells. In conjunction with experiments demonstrating tight binding of M7CK to H3 histone and in vitro H3 phosphorylation by M7CK, these data strongly suggest that M7CK directly phosphorylates H3 histone in vivo. A second mode of TRPM7-dependent histone phosphorylation was observed as a substantial increase of histone H2AX-Ser139 phosphorylation in TrpM7−/− cells that was restored to WT levels by M7CK. This indirect action suggests that ATM, ATR and DNA-PK, the protein kinases known to phosphorylate H2AX (Banerjee and Chakravarti, 2011), may be targets of M7CK.
In summary, the chanzyme TRPM7 is cleaved in a cell- and time-specific fashion. The cleaved fragment, containing an N-terminal intrinsically disordered region and C terminal kinase, binds to chromatin-remodeling complex proteins in a Zn2+-dependent fashion. In the nucleus the kinase phosphorylates histones on specific residues known to modify chromatin affecting cell differentiation and embryonic development.
Experimental Procedures
Full details are provided in the Extended Experimental Procedures. Mice were treated in accordance with guidelines approved by the Boston Children’s Hospital Animal Care and Use Committee (IUACUC).
cDNA constructs
HA- and FLAG-tagged M7CK constructs were made by cloning PCR-amplified mouse TrpM7 sequences into modified phCMV3 (Genlantis) and further subcloning of tagged cDNA into a pPyCAG-IP vector (from Ian Chambers, University of Edinburgh (Chambers et al., 2003)), or the modified bacterial expression vector pET32 (Novagen) containing an N-terminal fusion of 10xHis-GB1 sequence (from Martin Kurtev, BCH). The NTAP vector, made in a pcDNA4TO backbone (Invitrogen), contained a sequence encoding Protein G (PrG), a TEV protease cleavage site, and streptavidin-binding peptide (SBP), followed by a multiple cloning site, IRES and EGFP (Burckstummer et al., 2006). NTAP-M7CK cDNA was made by in-frame subcloning of the mouse TrpM7 sequence encoding a. a. 1364–1863 and C-terminal HA tag into the NTAP vector.
Derivation and culture of mESC
Blastocysts were isolated from TrpM7+/− mice (Jin et al., 2008) 3.5 d. postcoitus and cultured on a layer of γ-irradiated mouse embryonic fibroblasts using standard procedures (Bryja et al., 2006) in media containing an additional 10 mM MgCl2. Single ESC colonies were expanded, genotyped, and transfected with linearized cDNA using nucleofection ES solution (protocol A-23; Lonza); stably transfected cells were selected by antibiotic incubation.
Biochemical procedures
See Extended Experimental Procedures for cell extracts for tandem affinity purification, TRPM7 IP, histone Western blots, antibodies, and buffers. NTAP-M7CK was purified as described in (Burckstummer et al., 2006) in lysis buffer; purified proteins were separated on SDS PAGE and proteins digested with trypsin. MALDI-TOF MS and MS/MS peptide analyses were carried out (Krapivinsky et al., 2011) in the proteomic core facility at BCH. Purified histones (New England Biolabs) were phosphorylated in vitro with purified M7CK-L in buffer containing [γ-33P]-ATP. GST-RYBP protein was purified from expressing BL21 bacteria using Glutathione Sepharose (GE Healthcare). M7CK-L mouse TRPM7 protein containing N-terminal 10xHis-GB1 (Hammarstrom et al., 2002) and C-terminal HA-epitope tag was purified from lysates of BL21 bacteria expressing the protein on a cobalt resin (Talon, Clontech). Samples run on different gels are combined in figures and aligned against identical molecular weight markers (Figures 1, 2, 4, 6, S1, S3 and S4) as indicated by blank spaces between lanes.
Fluorescence microscopy
mES cells stably expressing eCALWY4 were background-corrected 480/535nm ratio images for cytosolic [Zn2+]. Free [Zn2+] was calculated for each cell according to the equation: (Dittmer et al., 2009) where Rmax = maximum fluorescence ratio obtained with 50 μM TPEN and Rmin = minimum fluorescence ratio obtained with 100 μM ZnCl2 in the presence of the 5 μM Zn2+ ionophore, pyrithione; KD=0.63 nM (Vinkenborg et al., 2009). For cytosolic [Mg2+], mES cells were loaded with 4 μM Mag-Fura/AM (Molecular Probes) at 37°C for 30 min in culture medium. Dye was excited at 350 nm and 380 nm and emission recorded at λ > 480 nm. Free [Mg2+] was calculated according to: [Mg2+]=Kd [(R − Rmin)/(Rmax − R)] × β, where R = ratio of fluorescence at 350 and 380 nm, Rmax=1.46 and Rmin=0.19 are the ratios for Mag-Fura free acid at 350 and 380 nm in the presence of saturating [Mg2+] and nominally 0 [Mg2+], respectively; β = 4.9, the ratio of fluorescence of Mag-Fura-2 at 380 nm in nominally free and saturating [Mg2+]. Mag-Fura KD for Mg2+ =1.5 mM (Raju et al., 1989).
Microarray Expression Profiling was performed on Illumina MouseWG-6 v2.0 Expression BeadChip arrays at the BCH microarray facility using GenomeStudio software. These data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE55578, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55578).
Chromatin immunoprecipitation (ChIP) analysis
Cells were fixed in PBS in 1% paraformaldehyde (7 min, 25 °C), quenched with 125 mM glycine, and sonicated. Mouse H3S10p (#17–685 Millipore) antibody and Protein G-Dynabeads (Invitrogen) were used for overnight chromatin immunoprecipitation. The negative ChIP control with normal mouse IgG produced signals <0.01% of the input.
Supplementary Material
Highlights.
The ubiquitous chanzyme, TRPM7, is cleaved in a cell-type specific fashion.
Cleaved kinase translocates to the nucleus and binds transcription factors.
Zinc entry via TRPM7 increases kinase binding to transcription factors.
The kinase phosphorylates H3S10, H3S28 and H3T3 to alter transcription.
Acknowledgments
We thank the Margaret Thompson and members of the Mouse Gene Manipulation Facility of the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (IDDRC; NIHP30-HD 18655) for assistance with generation of mESC and mice, Bimal Desai for helpful and inspiring discussions, Nat Blair for computational assistance, and Rajan Sah for assistance in hyperovulating mice.
References
- Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell. 2011;42:274–284. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol Cell Biol. 2011;31:4858–4873. doi: 10.1128/MCB.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. Embo J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Berger SL. Cell signaling and transcriptional regulation via histone phosphorylation. Cold Spring Harb Symp Quant Biol. 2010;75:23–26. doi: 10.1101/sqb.2010.75.044. [DOI] [PubMed] [Google Scholar]
- Bryja V, Bonilla S, Arenas E. Derivation of mouse embryonic stem cells. Nat Protoc. 2006;1:2082–2087. doi: 10.1038/nprot.2006.355. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000a;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000b;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Chini CC, Escande C, Nin V, Chini EN. HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem. 2010;285:40830–40837. doi: 10.1074/jbc.M110.153270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, Figdor CG, Moolenaar WH, Jalink K, van Leeuwen FN. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. Embo J. 2006;25:290–301. doi: 10.1038/sj.emboj.7600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Middelbeek J, Lasonder E, Dulyaninova NG, Morrice NA, Ryazanov AG, Bresnick AR, Figdor CG, van Leeuwen FN. TRPM7 Regulates Myosin IIA Filament Stability and Protein Localization by Heavy Chain Phosphorylation. Journal of Molecular Biology. 2008a;378:788–801. doi: 10.1016/j.jmb.2008.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Middelbeek J, Morrice NA, Figdor CG, Lasonder E, van Leeuwen FN. Massive autophosphorylation of the Ser/Thr-rich domain controls protein kinase activity of TRPM6 and TRPM7. PLoS ONE. 2008b;3:e1876. doi: 10.1371/journal.pone.0001876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeuse P, Penner R, Fleig A. TRPM7 channel is regulated by magnesium nucleotides via its kinase domain. J Gen Physiol. 2006;127:421–434. doi: 10.1085/jgp.200509410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai BN, Krapivinsky G, Navarro B, Krapivinsky L, Carter BC, Febvay S, Delling M, Penumaka A, Ramsey IS, Manasian Y, et al. Cleavage of TRPM7 releases the kinase domain from the ion channel and regulates its participation in Fas-induced apoptosis. Dev Cell. 2012;22:1149–1162. doi: 10.1016/j.devcel.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. Genetically encoded sensors to elucidate spatial distribution of cellular zinc. J Biol Chem. 2009;284:16289–16297. doi: 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorovkov MV, Ryazanov AG. Phosphorylation of annexin I by TRPM7 channel-kinase. J Biol Chem. 2004;279:50643–50646. doi: 10.1074/jbc.C400441200. [DOI] [PubMed] [Google Scholar]
- Du J, Xie J, Zhang Z, Tsujikawa H, Fusco D, Silverman D, Liang B, Yue L. TRPM7-mediated Ca2+ signals confer fibrogenesis in human atrial fibrillation. Circ Res. 2010;106:992–1003. doi: 10.1161/CIRCRESAHA.109.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Chen H, Costa M, Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. The Journal of biological chemistry. 2008;283:33585–33590. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:1–7. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Gallant P. Control of transcription by pontin and reptin. Trends Cell Biol. 2007;17:187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. Embo J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- Hanano T, Hara Y, Shi J, Morita H, Umebayashi C, Mori E, Sumimoto H, Ito Y, Mori Y, Inoue R. Involvement of TRPM7 in cell growth as a spontaneously activated Ca2+ entry pathway in human retinoblastoma cells. J Pharmacol Sci. 2004;95:403–419. doi: 10.1254/jphs.fp0040273. [DOI] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Huen J, Kakihara Y, Ugwu F, Cheung KL, Ortega J, Houry WA. Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem Cell Biol. 2010;88:29–40. doi: 10.1139/o09-122. [DOI] [PubMed] [Google Scholar]
- Inoue K, Xiong ZG. Silencing TRPM7 promotes growth/proliferation and nitric oxide production of vascular endothelial cells via the ERK pathway. Cardiovasc Res. 2009;83:547–557. doi: 10.1093/cvr/cvp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine B, Iannella ML, Bevilacqua MA. Damage-specific DNA binding protein 1 (DDB1): a protein with a wide range of functions. Int J Biochem Cell Biol. 2011;43:1664–1667. doi: 10.1016/j.biocel.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Jacob E, Hod-Dvorai R, Ben-Mordechai OL, Boyko Y, Avni O. Dual function of polycomb group proteins in differentiated murine T helper (CD4+) cells. J Mol Signal. 2011;6:5–20. doi: 10.1186/1750-2187-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Desai BN, Navarro B, Donovan A, Andrews NC, Clapham DE. Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science. 2008;322:756–760. doi: 10.1126/science.1163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wu LJ, Jun J, Cheng X, Xu H, Andrews NC, Clapham DE. The channel kinase, TRPM7, is required for early embryonic development. Proc Natl Acad Sci U S A. 2012;109:E225–233. doi: 10.1073/pnas.1120033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Stotz SC, Manasian Y, Clapham DE. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc Natl Acad Sci U S A. 2011;108:19234–19239. doi: 10.1073/pnas.1117231108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Mochida S, Krapivinsky L, Cibulsky SM, Clapham DE. The TRPM7 ion channel functions in cholinergic synaptic vesicles and affects transmitter release. Neuron. 2006;52:485–496. doi: 10.1016/j.neuron.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Krezel A, Maret W. Zinc-buffering capacity of a eukaryotic cell at physiological pZn. J Biol Inorg Chem. 2006;11:1049–1062. doi: 10.1007/s00775-006-0150-5. [DOI] [PubMed] [Google Scholar]
- Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Hong SE, Lim HH, Kim do H, Park CS. Alteration of the transcriptional profile of human embryonic kidney cells by transient overexpression of mouse TRPM7 channels. Cell Physiol Biochem. 2011;27:313–326. doi: 10.1159/000327958. [DOI] [PubMed] [Google Scholar]
- Li X, Gonzalez ME, Toy K, Filzen T, Merajver SD, Kleer CG. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009;175:1246–1254. doi: 10.2353/ajpath.2009.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Maret W. Transient fluctuations of intracellular zinc ions in cell proliferation. Exp Cell Res. 2009;315:2463–2470. doi: 10.1016/j.yexcr.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Matsushita M, Kozak JA, Shimizu Y, McLachlin DT, Yamaguchi H, Wei FY, Tomizawa K, Matsui H, Chait BT, Cahalan MD, et al. Channel function is dissociated from the intrinsic kinase activity and autophosphorylation of TRPM7/ChaK1. J Biol Chem. 2005;280:20793–20803. doi: 10.1074/jbc.M413671200. [DOI] [PubMed] [Google Scholar]
- Middelbeek J, Kuipers AJ, Henneman L, Visser D, Eidhof I, van Horssen R, Wieringa B, Canisius SV, Zwart W, Wessels LF, et al. TRPM7 is required for breast tumor cell metastasis. Cancer Res. 2012;72:4250–4261. doi: 10.1158/0008-5472.CAN-11-3863. [DOI] [PubMed] [Google Scholar]
- Miranda JG, Weaver AL, Qin Y, Park JG, Stoddard CI, Lin MZ, Palmer AE. New alternately colored FRET sensors for simultaneous monitoring of Zn(2)(+) in multiple cellular locations. PLoS ONE. 2012;7:e49371. doi: 10.1371/journal.pone.0049371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteilh-Zoller MK, Hermosura MC, Nadler MJ, Scharenberg AM, Penner R, Fleig A. TRPM7 provides an ion channel mechanism for cellular entry of trace metal ions. J Gen Physiol. 2003;121:49–60. doi: 10.1085/jgp.20028740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler MJ, Hermosura MC, Inabe K, Perraud AL, Zhu Q, Stokes AJ, Kurosaki T, Kinet JP, Penner R, Scharenberg AM, et al. LTRPC7 is a Mg.ATP-regulated divalent cation channel required for cell viability. Nature. 2001;411:590–595. doi: 10.1038/35079092. [DOI] [PubMed] [Google Scholar]
- Perraud AL, Zhao X, Ryazanov AG, Schmitz C. The channel-kinase TRPM7 regulates phosphorylation of the translational factor eEF2 via eEF2-k. Cell Signal. 2011;23:586–593. doi: 10.1016/j.cellsig.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju B, Murphy E, Levy LA, Hall RD, London RE. A fluorescent indicator for measuring cytosolic free magnesium. Am J Physiol. 1989;256:C540–548. doi: 10.1152/ajpcell.1989.256.3.C540. [DOI] [PubMed] [Google Scholar]
- Runnels LW, Yue L, Clapham DE. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science. 2001;291:1043–1047. doi: 10.1126/science.1058519. [DOI] [PubMed] [Google Scholar]
- Ryazanova LV, Dorovkov MV, Ansari A, Ryazanov AG. Characterization of the protein kinase activity of TRPM7/ChaK1, a protein kinase fused to the transient receptor potential ion channel. J Biol Chem. 2004;279:3708–3716. doi: 10.1074/jbc.M308820200. [DOI] [PubMed] [Google Scholar]
- Ryazanova LV, Rondon LJ, Zierler S, Hu Z, Galli J, Yamaguchi TP, Mazur A, Fleig A, Ryazanov AG. TRPM7 is essential for Mg(2+) homeostasis in mammals. Nat Commun. 2010;1:109–117. doi: 10.1038/ncomms1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R, Mesirca P, Mason X, Gibson W, Bates-Withers C, Van den Boogert M, Chaudhuri D, Pu W, Mangoni ME, Clapham DE. The Timing of Myocardial Trpm7 Deletion during Cardiogenesis Variably Disrupts Adult Ventricular Function, Conduction and Repolarization. Circulation. 2013;128:101–14. doi: 10.1161/CIRCULATIONAHA.112.000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka A, Seiser C. Histone H3 phosphorylation - A versatile chromatin modification for different occasions. Biochimie. 2012;94:2193–2201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C, Dorovkov MV, Zhao X, Davenport BJ, Ryazanov AG, Perraud AL. The channel kinases TRPM6 and TRPM7 are functionally nonredundant. J Biol Chem. 2005;280:37763–37771. doi: 10.1074/jbc.M509175200. [DOI] [PubMed] [Google Scholar]
- Schmitz C, Perraud AL, Johnson CO, Inabe K, Smith MK, Penner R, Kurosaki T, Fleig A, Scharenberg AM. Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7. Cell. 2003;114:191–200. doi: 10.1016/s0092-8674(03)00556-7. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Molecular cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Taki M, Wolford JL, O’Halloran TV. Emission ratiometric imaging of intracellular zinc: design of a benzoxazole fluorescent sensor and its application in two-photon microscopy. J Am Chem Soc. 2004;126:712–713. doi: 10.1021/ja039073j. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA. 2012;3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40:3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–267. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.