Abstract
Type 1 diabetes (T1D) and celiac disease (CD) are autoimmune diseases with clinical and pathogenic overlap. The mean prevalence of CD in patients with T1D is about 8%. Classic intestinal symptoms of CD may not be present in T1D leading to the recommendation for active case finding in this higher risk group. Screening is done with sensitive and specific serologies including tissue transglutaminase (tTG) IgA and deaminated gliadin peptide (DGP) IgA and IgG. Positive serologies are confirmed by the presence of villous atrophy and increased intraepithelial lymphocytes on duodenal biopsy. A strict gluten free diet is recommended, although this can pose challenges for T1D patients who already have dietary restrictions. In aggregate, it appears as if the gluten free diet may help T1D management. T1D and CD have overlapping genetic and environmental risk factors. Among these, non-HLA genetic factors and the gut microbiome are among recent developments that will be discussed in this review.
Keywords: type 1 diabetes, celiac disease, HLA genetics, microbiome
Introduction
Celiac disease (CD) and type 1 diabetes (T1D) are immune-mediated diseases that share common susceptibility factors notably HLA genetics. Both have increasing incidences worldwide suggesting that, in addition to genetic factors, environmental factors also play important roles in disease pathogenesis. Indeed, emerging evidence suggests that factors such as the gut microbiome and infectious agents among others modulate innate and adaptive immunity to increase risk of both CD and T1D. CD is a polygenic systemic immune-mediated enteropathy triggered by dietary gluten characterized by a specific serum antibody response. T1D is characterized by antibody-mediated destruction of beta cells of pancreatic islets so that blood glucose levels can no longer be maintained in a physiologic range without exogenous insulin. Autoimmune conditions such as T1D occur in association with CD and screening for CD in T1D is currently advocated. Importantly, our understanding of one disease informs knowledge and management of the other and vice versa. Here, we will review overlapping clinical features and diagnostic considerations as well as highlight current knowledge of common pathogenic features such as genetics, environmental risk factors and the gut microbiome.
Clinical overlap of T1D and CD
Incidence and Prevalence
T1D and CD have variable incidence and prevalence worldwide [1,2], though overall incidence of these diseases individually is increasing [1,3]. Due to a common genetic background and interplay with environmental and immunological factors, patients with T1D are at high risk of developing other autoimmune disorders. CD is one of the most common autoimmune disorders occurring in T1D, with estimates varying between 3 and 16% with a mean prevalence of 8% [4-6] (Table 1). Although the predominance of CD in Caucasians is well studied, little is known about T1D and CD in the African American population, though there may be a protective effect in this population due to unknown factors [7].
Table 1.
Prevalence of CD in Patients with T1D
| Study (year) | T1DM Cases | Country | CD Patients, n (%) |
|---|---|---|---|
| Boudraa et al. (1996) [85] | 116 children | Algeria | 19 (16.4) |
| Peretti et al. (2004) [86] | 284 children | France | 11 (3.4%)* |
| Mahmud et al. (2005) [87] | 158 children | USA | 11 (7.0) |
| Frolich-Reiterer et al. (2008) [88] | 11,083 children/adolescents | Germany/Austria | 1238 (11.1)* |
| Salardi et al. (2008) [89] | 331 children | Italy | 22 (6.6) |
| Uibo et al. (2010) [90] | 271 children/adolescents | Estonia | 9 (3.3) |
| Fallahi et al. (2010) [91] | 96 children | Iran | 6 (6.3) |
| Djuric et al. (2010) [92] | 121 children/adolescents | Serbia | 9 (7.4) |
| Bhadada et al. (2011) [93] | 189 children/young adults | India | 21 (11.1) |
| Hanukoglu et al. (2003) [94] | 109 children | Israel | 9 (8.3) |
| Sari et al. (2010) [95] | 48 children | Turkey | 3 (6.3) |
Diagnosis made by serologic antibody testing only
(Modified from Volta et al [6])
Clinical Presentation of CD in T1D
Symptoms and signs of CD may become evident at any age through adulthood. In CD (with or without concomitant T1D), classic symptoms include diarrhea, bloating, weight loss, and growth failure (in children) (reviewed in [8,9]). CD may also present with non-classical symptoms or asymptomatically. Non-classical intestinal and extra-intestinal symptoms include constipation, heartburn, neuropathy and ataxia among others. Clinical signs of CD including iron-deficiency anemia or low bone density with or without concomitant symptoms are increasingly common presentations. Individuals with sub-clinical disease, known as silent CD, are seropositive patients with no gastrointestinal or extra-intestinal manifestations [10].
In the setting of T1D, the majority of patients do not present with classic CD signs or symptoms [11-13], though they may have mild gastrointestinal symptoms compared to diabetes patients without CD [14]. Whether symptomatic or silent, CD in children with T1D may be accompanied by growth failure and delayed puberty [10,11,17]. In 75-90% of children, T1D diagnosis usually precedes CD diagnosis with risk of CD being highest in those <4 years of age [15]. CD diagnosis is usually screen-detected at a median age of 8 years [16].
Diagnosing CD in T1D
The increased prevalence of CD in T1D along with lack of symptoms has prompted the recommendation to screen T1D patients for CD at diagnosis then annually for the first 4 years and once every 2 years for the following 6 years [18,19]. Screening for CD relies on highly sensitive and specific serology tests. While antibodies directed against native gliadin (AGA) have been in use for several decades, there is wide variability in diagnostic accuracy and they are no longer recommended for screening [20]. Currently, the most sensitive and specific serologic tests include tissue transglutaminase (tTG) IgA, endomysial (EMA) IgA, and deaminated gliadin peptide (DGP) IgA and IgG antibodies. Table 2 summarizes test characteristics for these CD serologies in general. In T1D, there is emerging evidence that spontaneous normalization of CD serologies is possible while still on gluten prompting the suggestion that if tTG is < 3 times the upper limit of normal in T1D patients, repeat testing and biopsy should be pursued to definitively diagnose CD before initiating the gluten free diet (GFD) [21].
Table 2.
CD Serology Tests
| Test | Sensitivity | Specificity | Positive Likelihood Ratio | Negative Likelihood Ratio | Comments |
|---|---|---|---|---|---|
| Tissue transglutaminase (tTG) IgA | 0.89 (95% CI, 0.82-0.94) | 0.98 (95% CI, 0.95-0.99) | 37.7 | 0.11 | Recommended test for screening |
| Deaminated gliadin peptide (DGP) IgA and IgG | 0.75 (95% CI, 0.65-0.83) | 0.94 (95% CI, 0.89-0.98) | 13.3 | 0.27 | Recommended test for screening and monitoring adherence especially if IgA deficient |
| Endomysial (EmA) IgA | 0.90 (95% CI, 0.80-0.95) | 0.99 (95% CI, 0.98-1.00) | 171 | 0.11 | High specificity; used as a confirmatory test |
| anti-gliadin IgA | 0.46-0.87 | 0.7-0.98 | 2.59-41.9 | 0.14-0.55 | Not recommended for clinical practice |
| anti-gliadin IgG | 0.25-0.93 | 0.80-0.99 | 4.38-18.6 | 0.08-0.76 | Not recommended for clinical practice |
CI, confidence interval
(Adapted from van der Windt et al [96])
Small intestinal biopsy remains the gold standard for the diagnosis of CD with at least 5 biopsies obtained from the duodenum producing the highest diagnostic yield [22]. The hallmark of CD is the presence of an abnormal duodenum characterized by reduced height of villi giving a flattened appearance, villous atrophy, crypt hyperplasia, and increased intraepithelial lymphocytes [23]. In children, recent guidelines established specific criteria by which clinicians may consider forgoing intestinal biopsy: signs or symptoms of CD, tTG greater than 10 times the upper limit of normal, and positive EMA [19]. Since T1D patients may not present with typical signs and symptoms, intestinal biopsy should likely be pursued in the majority of cases. In addition, endoscopists should obtain biopsies from the duodenal bulb and second portion of the duodenum when performing upper endoscopy for other indications in T1D patients to assess for histological changes of CD.
Treatment of CD in T1D
The recommendations for treatment of T1D with CD are the same as for all patients with CD. A strict GFD should be initiated in those with serological and histological evidence of CD. Patients should meet with a dietician for GFD teaching and should initiate a daily multivitamin given lack of certain vitamins and minerals in the GFD [24]. Most symptomatic patients will improve in 2-4 weeks on the GFD, although a portion of patients have persistent symptoms after 3-6 months referred to as non-responsive CD [25]. Serologies should be checked after 3-4 months and then yearly when normalized.
In patients with both T1D and CD, dietary compliance can be challenging. Indeed, adherence to the GFD among CD patients with diabetes ranges from 25-78% [16]. Moreover, quality of life may be lower in patients with both diagnoses. A recent study assessed quality of life in patients diagnosed with both T1D and CD and found lower scores than in matched patients with T1D alone, especially in respect to social functioning and general health perception [26]. Treatment of CD may influence the course of T1D. T1D patients with undiagnosed CD show increased risk of diabetic retinopathy [27] and nephropathy [28], although this has not been found in all studies. One study found that T1D with untreated CD had lower body mass index (BMI) and lower HbA1c scores compared to those without CD [29], although other studies have not found differences in BMI or glycemic control between T1D with and without CD [30]. Poorly controlled CD and subsequent weight loss may improve glucose control while adherence to a GFD in CD might improve nutritional absorption and thereby increase insulin requirements. However, diminished absorption of nutrients in untreated CD may increase the risk of hypoglycemia in patients with diabetes [29]. Overall, it appears that following a GFD may be beneficial (or at least not harmful) in T1D with CD; however, there is substantial heterogeneity in the literature.
New insights into pathogenic overlap of T1D and CD
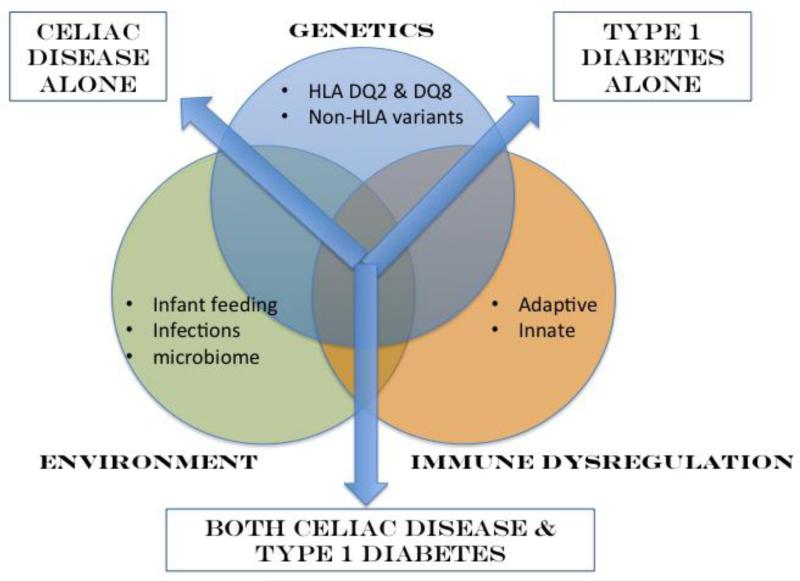
CD and T1D share a number of common risk factors including genetics, environment and immune dysregulation (Figure 1). New developments in several of these areas will be discussed below.
Figure 1. Genetics, environment and immune dysregulation are primary risk factors identified for both celiac disease (CD) and type 1 diabetes (T1D).
While HLA DQ2 and 8 are well-known risk factors predisposing to both CD and T1D, recently performed genome wide association studies (GWAS) have identified a number of non-HLA genetic factors that are shared by CD and T1D including a number of immune genes. Environmental risk factors shared by CD and T1D include timing of cereal introduction, length of breast-feeding as well as viral infections. Recent work is also elucidating the role of the gut microbiome in both conditions. Finally, immune dysregulation, both innate and adaptive, are central to CD and T1D pathogenesis. While common risk factors predispose to CD and T1D individually, it is not yet known how these risk factors trigger both diseases in a subset of individuals. Therefore, at the present time, screening for CD in T1D and vice versa is advocated in order to identify those manifesting both conditions.
Genetics
Both T1D and CD are inherited complex diseases with strong genetic components notably HLA. Recent genome wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) associated with autoimmune conditions including T1D and CD. Overlap of genetic variants between T1D and CD (including HLA and non-HLA) underscores common pathogenic mechanisms and likely explains increased prevalence of concomitant disease.
HLA, also known as the major histocompatibility complex (MHC), is involved in antigen presentation to T cells. The HLA region is the most polymorphic observed in the human genome with thousands of unique allele sequences. The MHC class II DQ peptides are associated with both CD and T1D, while MHC class I loci have also been shown to affect T1D susceptibility but not CD [31,32]. HLA-DQ2 (also known as the haplotype DR3-DQ2) is found in about 90% of CD and 55% of T1D patients, while HLA-DQ8 is found in about 10% of CD and 70% of T1D patients [33]. In clinical support of the HLA associations, approximately 33% of HLA-DQ2 homozygous individuals with T1D express tTG autoantibodies, compared to less than 2% of the T1D patients who lacked DQ2 and DQ8 genotypes [34].
To date, about 40 loci have been associated with T1D (reviewed in [32], while there are 39 loci associated with CD (reviewed in [35]). There are several non-HLA loci that overlap between T1D and CD [35,36](Table 3). For several of these regions, there are multiple genes in the region and the causal gene has yet to be identified. The associations between CD and T1D discovered in GWAS show variable effect sizes and directions of effects. Moreover, there are likely gene-environment interactions that modulate risk, though these are yet to be determined. However, it is intriguing that several of these loci harbor genes related to immunity including IL18RAP, CTLA4, CCR5, IL2, IL21, TAGAP, PRKCQ. Future studies will help elucidate how these particular loci may increase risk of both T1D and CD.
Table 3.
Overlapping GWAS regions between T1D and CD
Overlapping environmental risk factors
Studies in both T1D and CD have found similar types of environmental risk factors such as infant feeding practices, breast-feeding and exposure to viral infections (reviewed in [37] and [38] for T1D and CD, respectively). An additional environmental risk factor is the gut microbiome to be discussed in the following section. Regarding infant feeding practices, it has been found that in genetically susceptible infants for both T1D [39,40] and CD [41], early (before 3 months) or late (after 7 months) introduction of cereal is associated with autoimmune seropositivity. Breast-feeding, especially when continued as cereal is introduced in the case of CD, appears to be protective for both diseases, although studies are mixed in their results [42,43]. Finally, infections, especially viruses, appear to increase risk of T1D and CD. In T1D, there is data implicating enteroviruses as a specific risk factor [44,45]. In CD, childhood rotavirus infection has been implicated as a risk factor [46,47]. While studies have not been entirely consistent, it does appear that several common risk factors increase risk of developing T1D or CD autoimmunity including introduction of cereals, lack of breast-feeding and viral infections. To what extent these factors predispose to development of both T1D and CD has yet to be established.
Microbiome in T1D and CD
The ecosystem of gut microbiota, called the microbiome, is among the environmental factors hypothesized to affect the development of T1D and CD. The gut microbiome is composed of bacteria that inhabit the human gastrointestinal tract, and it forms complex interactions with the host. It influences nutrition, prevents colonization by pathogens, and interacts with host immune regulation and response [48]. There are several proposed mechanisms to explain this interaction and the increased prevalence of autoimmune diseases like T1D and CD. The “hygiene hypothesis” attributes the rise in autoimmune diseases to the reduction in infections in developed nations [49]. The “fertile field hypothesis” suggests that immune reaction to gut microbes increases the exposure of self-antigens to inflammation and precipitates formation of self-reactive T cells [50]. The “old friends hypothesis” explains the rise in immune dysregulatory diseases by implicating the decreased exposure of innate immune cells to familiar, nonpathogenic organisms [51]. The “perfect storm hypothesis” states that the combined dysfunction of altered gut microbiota, porous gut mucosal barriers, and abnormal immune response all lead to development of an autoimmune phenotype [52]. Testing these hypotheses has been difficult in the past, due to limited methods for studying the enteric environment.
However, recent advances in 16S rRNA sequencing and metagenomic testing have helped make gut microbiome studies more feasible and are providing insights into the host-microbiota interaction [53].
Mouse studies have provided great insights into the role of the microbiome in T1D as well as CD. In the non-obese diabetic (NOD) mouse and the Biobreeding diabetes-prone (BB-DP) mouse models T1D, germ free mice still develop T1D independent of bacterial exposure [54]. Injected bacterial antigens and exposure to particular non-pathogenic bacterial species provide a protective effect on NOD and BB-DP mice, reducing the rate of T1D suggesting that underexposure to particular bacteria eliminates an important protective element in genetically susceptible hosts [54-59]. In contrast, introduction of pathogenic strains to NOD mice increases markers of intestinal permeability, accelerates development of insulitis, and ultimately increases the rate of development of T1D [60]. Furthermore, a recent study showed that NOD mice fed a GFD had significantly different fecal microbiota than NOD mice on a gluten containing diet. The GFD was associated with a reduced incidence of hyperglycemia in NOD mice. The protective effect of the GFD, and its associated microbiome, reversed when gluten was reintroduced into the NOD mouse's diet [61].
Studies in humans with T1D show differences in the composition of the gut microbiota compared to healthy controls [62-65]. A Finnish cohort showed an abundance of the phylum Bacteroidetes in patients with T1D, as well a significantly greater prevalence of Firmicutes in healthy controls [63]. This imbalance in Bacteroidetes and Firmicutes has been observed in another study [65]. Similarly, low abundance of lactate-producing and butyrate-producing species of bacteria, a paucity of Bifidobacterium, and increased abundance of Bacteroides is also associated with beta cell autoimmunity in children [64].
Animal models have also been used to help determine the microbiome's role in CD. Similar to studies in models of T1D, introduction of nonpathogenic, commensal microbes modifies intestinal inflammatory signaling and response to gliadin [66-68]. In another similarity with T1D, exposure to pathogenic bacteria increases intestinal permeability, leading to greater translocation of gliadin fragments into the lamina propria [69]. These studies show that intestinal flora is capable of altering mucosal immune cell activity in a model of CD. Further investigation showed supplemental Lactobacillus casei not only modifies inflammatory signaling, but also induced complete recovery of villous blunting and delayed weight loss in a mouse model of gliadin-induced villous damage [68]. These results provide further evidence that the intestinal microbiome has complex effects on the host's immune function and disease states.
Similar to T1D, there are significant differences in the microbiome between patients with active CD and healthy controls that are hypothesized to influence the development and manifestations of the disease [70-77]. Many species of bacteria within the human gut are capable of utilizing gluten [78]. Patients with active CD have been found to have more abundant populations of Proteobacteria, Enterobacteriaceae, and Staphylococcaceae, while also having fewer Firmicutes and Streptococcaceae [70]. Additionally, more pathogenic bacteria with virulence features are found in the gut of patients with active CD [79]. These studies suggest that an imbalance of intestinal microbes is correlated with active CD but do not provide information on causality. It is also not clear that adherence to a GFD changes the gut microbiome back to a normal composition because studies yielded variable results [70,72,79].
The exposure of the gut to bacterial elements clearly influences innate and adaptive immunity [57,58,80-82]. A key study provided clear evidence that the gut microbiome influences autoimmune diabetes through effects on the innate immune system specifically the adaptor molecule MyD88 [82]. These investigators found that protection from T1D was dependent on commensal bacteria low in Firmicutes and Bacteroidetes and rich in Lactobacillus. Another way in which enteric microbes might modulate host immunity is through altering the balance of Th1 and T helper 17 (Th17) cells (reviewed in [83]). Th17 cells can be induced by exposure to the commensal enteric microbes, segmented filamentous bacterium and Lactobacillus johnsonii [57,58,80,81]. L. johnsonii exposure in BB-DP mice induces expression of claudin, an intestinal tight junction protein, and also enhances differentiation of Th17 cells [57,58]. The duodenal mucosa in CD also produces more IL-17A from Th17 cells than the mucosa of patients without CD. As well, these Th17 cells specifically react to gliadin [84]. Enhanced Th17 activity leads to upregulated mucosal markers of inflammation, and reduced susceptibility to pathogenic bacteria [80]. Future studies are needed to understand how the microbiome is different in individuals who develop multiple autoimmune conditions and if the microbiome could be modulated for treatment or prevention.
Conclusion
T1D and CD are autoimmune diseases with considerable clinical and pathogenic overlap. While HLA is a clear common risk factor, additional genetic and environmental factors likely play important roles in disease initiation. Given increased prevalence of having both conditions and frequent lack of classic intestinal symptoms, CD screening is currently recommended. Sensitive and specific serology tests are available, and when positive, should be confirmed by upper endoscopic biopsies showing villous changes and increased intraepithelial lymphocytes. A GFD may be challenging for some patients who already have dietary restrictions; however, it appears as if this diet may provide overall benefit (or at least no harm) in T1D patients though the data in this area is variable. Emerging evidence underscores the pathogenic overlap between these two conditions. While HLA susceptibility has been known as a common risk factor, GWAS have identified a number of non-HLA genetic risk variants. Environmental factors including introduction of solids, breast-feeding, viral infections and notably the gut microbiome appear to contribute to risk of developing both diseases individually. Additional studies are needed to elucidate how these factors predispose some individuals to getting both conditions.
Compliance with Ethics Guidelines
Footnotes
Conflict of Interest
Aaron Cohn, M. Anthony Sofia, and Sonia S. Kupfer declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1•.Dabelea D. The accelerating epidemic of childhood diabetes. Lancet. 2009;373:1999–2000. doi: 10.1016/S0140-6736(09)60874-6. [An excellent review on the increasing incidence of type 1 diabetes.] [DOI] [PubMed] [Google Scholar]
- 2.Fasano A, Catassi C. Clinical practice. Celiac disease. N Engl J Med. 2012;367:2419–2426. doi: 10.1056/NEJMcp1113994. [DOI] [PubMed] [Google Scholar]
- 3•.Kang JY, Kang AH, Green A, Gwee KA, Ho KY. Systematic review: worldwide variation in the frequency of coeliac disease and changes over time. Aliment Pharmacol Ther. 2013;38:226–245. doi: 10.1111/apt.12373. [An up-to-date review on celiac disease and worldwide variation.] [DOI] [PubMed] [Google Scholar]
- 4.Dube C, Rostom A, Sy R, Cranney A, Saloojee N, et al. The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology. 2005;128:S57–67. doi: 10.1053/j.gastro.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Gillett PM, Gillett HR, Israel DM, Metzger DL, Stewart L, et al. High prevalence of celiac disease in patients with type 1 diabetes detected by antibodies to endomysium and tissue transglutaminase. Can J Gastroenterol. 2001;15:297–301. doi: 10.1155/2001/640796. [DOI] [PubMed] [Google Scholar]
- 6•.Volta U, Tovoli F, Caio G. Clinical and immunological features of celiac disease in patients with Type 1 diabetes mellitus. Expert Rev Gastroenterol Hepatol. 2011;5:479–487. doi: 10.1586/egh.11.38. [A comprehensive clinical review on celiac disease in patients with type 1 diabetes.] [DOI] [PubMed] [Google Scholar]
- 7.Kaistha A, Castells S. Celiac disease in African American children with type 1 diabetes mellitus in inner city Brooklyn. Pediatr Endocrinol Rev 5 Suppl. 2008;4:994–998. [PubMed] [Google Scholar]
- 8.Green PH. The many faces of celiac disease: clinical presentation of celiac disease in the adult population. Gastroenterology. 2005;128:S74–78. doi: 10.1053/j.gastro.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Reilly NR, Fasano A, Green PH. Presentation of celiac disease. Gastrointest Endosc Clin N Am. 2012;22:613–621. doi: 10.1016/j.giec.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Freemark M, Levitsky LL. Screening for celiac disease in children with type 1 diabetes: two views of the controversy. Diabetes Care. 2003;26:1932–1939. doi: 10.2337/diacare.26.6.1932. [DOI] [PubMed] [Google Scholar]
- 11.Holmes GK. Coeliac disease and Type 1 diabetes mellitus - the case for screening. Diabet Med. 2001;18:169–177. doi: 10.1046/j.1464-5491.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- 12.Valerio G, Maiuri L, Troncone R, Buono P, Lombardi F, et al. Severe clinical onset of diabetes and increased prevalence of other autoimmune diseases in children with coeliac disease diagnosed before diabetes mellitus. Diabetologia. 2002;45:1719–1722. doi: 10.1007/s00125-002-0923-5. [DOI] [PubMed] [Google Scholar]
- 13.Ventura A, Neri E, Ughi C, Leopaldi A, Citta A, et al. Gluten-dependent diabetes-related and thyroid-related autoantibodies in patients with celiac disease. J Pediatr. 2000;137:263–265. doi: 10.1067/mpd.2000.107160. [DOI] [PubMed] [Google Scholar]
- 14.Narula P, Porter L, Langton J, Rao V, Davies P, et al. Gastrointestinal symptoms in children with type 1 diabetes screened for celiac disease. Pediatrics. 2009;124:e489–495. doi: 10.1542/peds.2008-2434. [DOI] [PubMed] [Google Scholar]
- 15.Cerutti F, Bruno G, Chiarelli F, Lorini R, Meschi F, et al. Younger age at onset and sex predict celiac disease in children and adolescents with type 1 diabetes: an Italian multicenter study. Diabetes Care. 2004;27:1294–1298. doi: 10.2337/diacare.27.6.1294. [DOI] [PubMed] [Google Scholar]
- 16.Camarca ME, Mozzillo E, Nugnes R, Zito E, Falco M, et al. Celiac disease in type 1 diabetes mellitus. Ital J Pediatr. 2012;38:10. doi: 10.1186/1824-7288-38-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damen GM, Boersma B, Wit JM, Heymans HS. Catch-up growth in 60 children with celiac disease. J Pediatr Gastroenterol Nutr. 1994;19:394–400. doi: 10.1097/00005176-199411000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Kordonouri O, Maguire AM, Knip M, Schober E, Lorini R, et al. ISPAD Clinical Practice Consensus Guidelines 2006-2007. Other complications and associated conditions. Pediatr Diabetes. 2007;8:171–176. doi: 10.1111/j.1399-5448.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- 19••.Husby S, Koletzko S, Korponay-Szabo IR, Mearin ML, Phillips A, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54:136–160. doi: 10.1097/MPG.0b013e31821a23d0. [Society guidelines on screening and diagnosis of celiac disease recommending screening of patients with type 1 diabetes.] [DOI] [PubMed] [Google Scholar]
- 20.Leffler DA, Schuppan D. Update on serologic testing in celiac disease. Am J Gastroenterol. 2010;105:2520–2524. doi: 10.1038/ajg.2010.276. [DOI] [PubMed] [Google Scholar]
- 21••.Waisbourd-Zinman O, Hojsak I, Rosenbach Y, Mozer-Glassberg Y, Shalitin S, et al. Spontaneous normalization of anti-tissue transglutaminase antibody levels is common in children with type 1 diabetes mellitus. Dig Dis Sci. 2012;57:1314–1320. doi: 10.1007/s10620-011-2016-0. [Recent study that found over 30% of type 1 diabetes patients normalize celiac disease serologies while on a gluten free diet.] [DOI] [PubMed] [Google Scholar]
- 22.Rostom A, Murray JA, Kagnoff MF. American Gastroenterological Association (AGA) Institute technical review on the diagnosis and management of celiac disease. Gastroenterology. 2006;131:1981–2002. doi: 10.1053/j.gastro.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Chand N, Mihas AA. Celiac disease: current concepts in diagnosis and treatment. J Clin Gastroenterol. 2006;40:3–14. doi: 10.1097/01.mcg.0000190644.01661.2b. [DOI] [PubMed] [Google Scholar]
- 24.Miranda J, Lasa A, Bustamante MA, Churruca I, Simon E. Nutritional Differences Between a Gluten-free Diet and a Diet Containing Equivalent Products with Gluten. Plant Foods Hum Nutr. 2014 doi: 10.1007/s11130-014-0410-4. [DOI] [PubMed] [Google Scholar]
- 25••.Rubio-Tapia A, Hill ID, Kelly CP, Calderwood AH, Murray JA. ACG clinical guidelines: diagnosis and management of celiac disease. Am J Gastroenterol. 2013;108:656–676. doi: 10.1038/ajg.2013.79. quiz 677. [American society guidelines for screening and diagnosis of celiac disease recommending active case finding of celiac disease in individuals with type 1 diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakker SF, Pouwer F, Tushuizen ME, Hoogma RP, Mulder CJ, et al. Compromised quality of life in patients with both Type 1 diabetes mellitus and coeliac disease. Diabet Med. 2013;30:835–839. doi: 10.1111/dme.12205. [DOI] [PubMed] [Google Scholar]
- 27••.Mollazadegan K, Sanders DS, Ludvigsson J, Ludvigsson JF. Long-term coeliac disease influences risk of death in patients with type 1 diabetes. J Intern Med. 2013;274:273–280. doi: 10.1111/joim.12092. [This large Swedish study found that in patients with both celiac and type 1 diabetes with >15 years of diabetes increases risk of death 2.8 fold.] [DOI] [PubMed] [Google Scholar]
- 28.Leeds JS, Hopper AD, Hadjivassiliou M, Tesfaye S, Sanders DS. High prevalence of microvascular complications in adults with type 1 diabetes and newly diagnosed celiac disease. Diabetes Care. 2011;34:2158–2163. doi: 10.2337/dc11-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amin R, Murphy N, Edge J, Ahmed ML, Acerini CL, et al. A longitudinal study of the effects of a gluten-free diet on glycemic control and weight gain in subjects with type 1 diabetes and celiac disease. Diabetes Care. 2002;25:1117–1122. doi: 10.2337/diacare.25.7.1117. [DOI] [PubMed] [Google Scholar]
- 30.Scaramuzza AE, Mantegazza C, Bosetti A, Zuccotti GV. Type 1 diabetes and celiac disease: The effects of gluten free diet on metabolic control. World J Diabetes. 2013;4:130–134. doi: 10.4239/wjd.v4.i4.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krolewski AS, Warram JH, Rand LI, Kahn CR. Epidemiologic approach to the etiology of type I diabetes mellitus and its complications. N Engl J Med. 1987;317:1390–1398. doi: 10.1056/NEJM198711263172206. [DOI] [PubMed] [Google Scholar]
- 32•.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007732. doi: 10.1101/cshperspect.a007732. [Up-to-date review of genetic discoveries in type 1 diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermann R, Turpeinen H, Laine AP, Veijola R, Knip M, et al. HLA DR-DQ-encoded genetic determinants of childhood-onset type 1 diabetes in Finland: an analysis of 622 nuclear families. Tissue Antigens. 2003;62:162–169. doi: 10.1034/j.1399-0039.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 34.Sollid LM, Jabri B. Is celiac disease an autoimmune disorder? Curr Opin Immunol. 2005;17:595–600. doi: 10.1016/j.coi.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 35•.Kumar V, Wijmenga C, Withoff S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin Immunopathol. 2012;34:567–580. doi: 10.1007/s00281-012-0312-1. [Recent review on genetic associations in celiac and type 1 diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Knip M, Simell O. Environmental triggers of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007690. doi: 10.1101/cshperspect.a007690. [Recent review of environmental risk factors in type 1 diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kupfer SS, Jabri B. Pathophysiology of celiac disease. Gastrointest Endosc Clin N Am. 2012;22:639–660. doi: 10.1016/j.giec.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norris JM, Barriga K, Klingensmith G, Hoffman M, Eisenbarth GS, et al. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA. 2003;290:1713–1720. doi: 10.1001/jama.290.13.1713. [DOI] [PubMed] [Google Scholar]
- 40.Ziegler AG, Schmid S, Huber D, Hummel M, Bonifacio E. Early infant feeding and risk of developing type 1 diabetes-associated autoantibodies. JAMA. 2003;290:1721–1728. doi: 10.1001/jama.290.13.1721. [DOI] [PubMed] [Google Scholar]
- 41.Norris JM, Barriga K, Hoffenberg EJ, Taki I, Miao D, et al. Risk of celiac disease autoimmunity and timing of gluten introduction in the diet of infants at increased risk of disease. JAMA. 2005;293:2343–2351. doi: 10.1001/jama.293.19.2343. [DOI] [PubMed] [Google Scholar]
- 42.Virtanen SM, Knip M. Nutritional risk predictors of beta cell autoimmunity and type 1 diabetes at a young age. Am J Clin Nutr. 2003;78:1053–1067. doi: 10.1093/ajcn/78.6.1053. [DOI] [PubMed] [Google Scholar]
- 43.Akobeng AK, Ramanan AV, Buchan I, Heller RF. Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies. Arch Dis Child. 2006;91:39–43. doi: 10.1136/adc.2005.082016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. Bmj. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stene LC, Oikarinen S, Hyoty H, Barriga KJ, Norris JM, et al. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY). Diabetes. 2010;59:3174–3180. doi: 10.2337/db10-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stene LC, Honeyman MC, Hoffenberg EJ, Haas JE, Sokol RJ, et al. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. The American Journal of Gastroenterology. 2006;101:2333–2340. doi: 10.1111/j.1572-0241.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 47.Zanoni G, Navone R, Lunardi C, Tridente G, Bason C, et al. In celiac disease, a subset of autoantibodies against transglutaminase binds toll-like receptor 4 and induces activation of monocytes. PLoS medicine. 2006;3:e358. doi: 10.1371/journal.pmed.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bach JF. Six questions about the hygiene hypothesis. Cell Immunol. 2005;233:158–161. doi: 10.1016/j.cellimm.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 50.von Herrath MG, Fujinami RS, Whitton JL. Microorganisms and autoimmunity: making the barren field fertile? Nat Rev Microbiol. 2003;1:151–157. doi: 10.1038/nrmicro754. [DOI] [PubMed] [Google Scholar]
- 51.Rook GA, Adams V, Hunt J, Palmer R, Martinelli R, et al. Mycobacteria and other environmental organisms as immunomodulators for immunoregulatory disorders. Springer Semin Immunopathol. 2004;25:237–255. doi: 10.1007/s00281-003-0148-9. [DOI] [PubMed] [Google Scholar]
- 52.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57:2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Atkinson MA, Chervonsky A. Does the gut microbiota have a role in type 1 diabetes? Early evidence from humans and animal models of the disease. Diabetologia. 2012;55:2868–2877. doi: 10.1007/s00125-012-2672-4. [Recent review of the gut microbiome and development of type 1 diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McInerney MF, Pek SB, Thomas DW. Prevention of insulitis and diabetes onset by treatment with complete Freund's adjuvant in NOD mice. Diabetes. 1991;40:715–725. doi: 10.2337/diab.40.6.715. [DOI] [PubMed] [Google Scholar]
- 56.Satoh J, Shintani S, Oya K, Tanaka S, Nobunaga T, et al. Treatment with streptococcal preparation (OK-432) suppresses anti-islet autoimmunity and prevents diabetes in BB rats. Diabetes. 1988;37:1188–1194. doi: 10.2337/diab.37.9.1188. [DOI] [PubMed] [Google Scholar]
- 57.Valladares R, Sankar D, Li N, Williams E, Lai KK, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One. 2010;5:e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 59.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 60.Lee AS, Gibson DL, Zhang Y, Sham HP, Vallance BA, et al. Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia. 2010;53:741–748. doi: 10.1007/s00125-009-1626-y. [DOI] [PubMed] [Google Scholar]
- 61••.Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, et al. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS One. 2013;8:e78687. doi: 10.1371/journal.pone.0078687. [Marietta and colleagues compared the effect of feeding a gluten free diet and gluten containing diet on the microbiome and incidence of hyperglycemia in NOD mice and found a reduced rate of hyperglycemia while on the gluten free diet.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soyucen E, Gulcan A, Aktuglu-Zeybek AC, Onal H, Kiykim E, et al. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr Int. 2013 doi: 10.1111/ped.12243. [DOI] [PubMed] [Google Scholar]
- 63.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laparra JM, Olivares M, Gallina O, Sanz Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS One. 2012;7:e30744. doi: 10.1371/journal.pone.0030744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papista C, Gerakopoulos V, Kourelis A, Sounidaki M, Kontana A, et al. Gluten induces coeliac-like disease in sensitised mice involving IgA, CD71 and transglutaminase 2 interactions that are prevented by probiotics. Lab Invest. 2012;92:625–635. doi: 10.1038/labinvest.2012.13. [DOI] [PubMed] [Google Scholar]
- 68.D'Arienzo R, Stefanile R, Maurano F, Mazzarella G, Ricca E, et al. Immunomodulatory effects of Lactobacillus casei administration in a mouse model of gliadin-sensitive enteropathy. Scand J Immunol. 2011;74:335–341. doi: 10.1111/j.1365-3083.2011.02582.x. [DOI] [PubMed] [Google Scholar]
- 69.Cinova J, De Palma G, Stepankova R, Kofronova O, Kverka M, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS One. 2011;6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez E, Donat E, Ribes-Koninckx C, Fernandez-Murga ML, Sanz Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl Environ Microbiol. 2013;79:5472–5479. doi: 10.1128/AEM.00869-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanchez E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal Staphylococcus spp. and virulent features associated with coeliac disease. J Clin Pathol. 2012;65:830–834. doi: 10.1136/jclinpath-2012-200759. [DOI] [PubMed] [Google Scholar]
- 72.Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol. 2009;62:264–269. doi: 10.1136/jcp.2008.061366. [DOI] [PubMed] [Google Scholar]
- 73.Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56:1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- 74.De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010;10:63. doi: 10.1186/1471-2180-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng J, Kalliomaki M, Heilig HG, Palva A, Lahteenoja H, et al. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. 2013;13:113. doi: 10.1186/1471-230X-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wacklin P, Kaukinen K, Tuovinen E, Collin P, Lindfors K, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. 2013;19:934–941. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 77.Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, et al. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiol. 2010;10:175. doi: 10.1186/1471-2180-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caminero A, Herran AR, Nistal E, Perez-Andres J, Vaquero L, et al. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. 2014 doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- 79.Sanchez E, Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, et al. Reduced diversity and increased virulence-gene carriage in intestinal enterobacteria of coeliac children. BMC Gastroenterol. 2008;8:50. doi: 10.1186/1471-230X-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 82.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sorini C, Falcone M. Shaping the (auto)immune response in the gut: the role of intestinal immune regulation in the prevention of type 1 diabetes. Am J Clin Exp Immunol. 2013;2:156–171. [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez S, Molina IJ, Romero P, Gonzalez R, Pena J, et al. Characterization of gliadin-specific Th17 cells from the mucosa of celiac disease patients. Am J Gastroenterol. 2011;106:528–538. doi: 10.1038/ajg.2010.465. [DOI] [PubMed] [Google Scholar]
- 85.Boudraa G, Hachelaf W, Benbouabdellah M, Belkadi M, Benmansour FZ, et al. Prevalence of coeliac disease in diabetic children and their first-degree relatives in west Algeria: screening with serological markers. Acta Paediatr Suppl. 1996;412:58–60. doi: 10.1111/j.1651-2227.1996.tb14254.x. [DOI] [PubMed] [Google Scholar]
- 86.Peretti N, Bienvenu F, Bouvet C, Fabien N, Tixier F, et al. The temporal relationship between the onset of type 1 diabetes and celiac disease: a study based on immunoglobulin a antitransglutaminase screening. Pediatrics. 2004;113:e418–422. doi: 10.1542/peds.113.5.e418. [DOI] [PubMed] [Google Scholar]
- 87.Mahmud FH, Murray JA, Kudva YC, Zinsmeister AR, Dierkhising RA, et al. Celiac disease in type 1 diabetes mellitus in a North American community: prevalence, serologic screening, and clinical features. Mayo Clin Proc. 2005;80:1429–1434. doi: 10.4065/80.11.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frohlich-Reiterer EE, Hofer S, Kaspers S, Herbst A, Kordonouri O, et al. Screening frequency for celiac disease and autoimmune thyroiditis in children and adolescents with type 1 diabetes mellitus--data from a German/Austrian multicentre survey. Pediatr Diabetes. 2008;9:546–553. doi: 10.1111/j.1399-5448.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- 89.Salardi S, Volta U, Zucchini S, Fiorini E, Maltoni G, et al. Prevalence of celiac disease in children with type 1 diabetes mellitus increased in the mid-1990 s: an 18-year longitudinal study based on anti-endomysial antibodies. J Pediatr Gastroenterol Nutr. 2008;46:612–614. doi: 10.1097/MPG.0b013e31815d697e. [DOI] [PubMed] [Google Scholar]
- 90.Uibo O, Heilman K, Rago T, Shor R, Paal M, et al. Symptomless celiac disease in type 1 diabetes: 12-year experience in Estonia. Pediatr Int. 2010;52:230–233. doi: 10.1111/j.1442-200X.2009.02955.x. [DOI] [PubMed] [Google Scholar]
- 91.Fallahi GH, Ahmadian JH, Rabbani A, Yousefnezhad AS, Rezaei N. Screening for celiac disease in diabetic children from Iran. Indian Pediatr. 2010;47:268–270. doi: 10.1007/s13312-010-0048-8. [DOI] [PubMed] [Google Scholar]
- 92.Djuric Z, Stamenkovic H, Stankovic T, Milicevic R, Brankovic L, et al. Celiac disease prevalence in children and adolescents with type 1 diabetes from Serbia. Pediatr Int. 2010;52:579–583. doi: 10.1111/j.1442-200X.2010.03085.x. [DOI] [PubMed] [Google Scholar]
- 93.Bhadada SK, Kochhar R, Bhansali A, Dutta U, Kumar PR, et al. Prevalence and clinical profile of celiac disease in type 1 diabetes mellitus in north India. J Gastroenterol Hepatol. 2011;26:378–381. doi: 10.1111/j.1440-1746.2010.06508.x. [DOI] [PubMed] [Google Scholar]
- 94.Hanukoglu A, Mizrachi A, Dalal I, Admoni O, Rakover Y, et al. Extrapancreatic autoimmune manifestations in type 1 diabetes patients and their first-degree relatives: a multicenter study. Diabetes Care. 2003;26:1235–1240. doi: 10.2337/diacare.26.4.1235. [DOI] [PubMed] [Google Scholar]
- 95.Sari S, Yesilkaya E, Egritas O, Bideci A, Cinaz P, et al. Prevalence of Celiac disease in Turkish children with type 1 diabetes mellitus and their non-diabetic first-degree relatives. Turk J Gastroenterol. 2010;21:34–38. doi: 10.4318/tjg.2010.0045. [DOI] [PubMed] [Google Scholar]
- 96••.van der Windt DA, Jellema P, Mulder CJ, Kneepkens CM, van der Horst HE. Diagnostic testing for celiac disease among patients with abdominal symptoms: a systematic review. JAMA. 2010;303:1738–1746. doi: 10.1001/jama.2010.549. [Excellent review of diagnostic testing strategies for celiac disease.] [DOI] [PubMed] [Google Scholar]
- 97.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. The New England journal of medicine. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]