Abstract
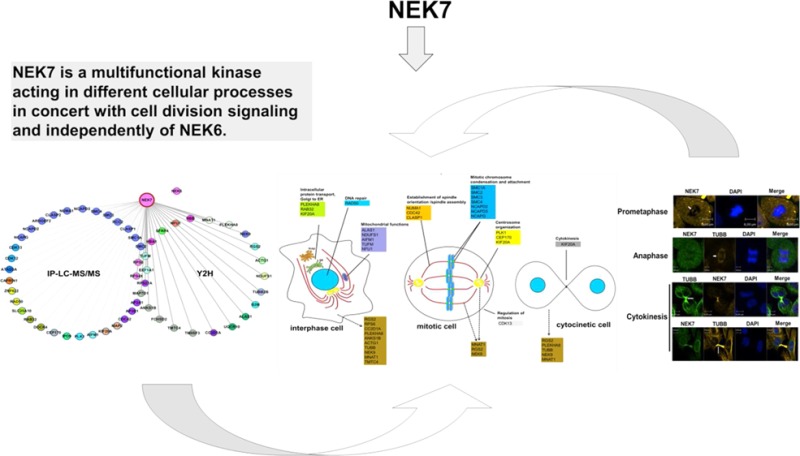
Human NEK7 is a regulator of cell division and plays an important role in growth and survival of mammalian cells. Human NEK6 and NEK7 are closely related, consisting of a conserved C-terminal catalytic domain and a nonconserved and disordered N-terminal regulatory domain, crucial to mediate the interactions with their respective proteins. Here, in order to better understand NEK7 cellular functions, we characterize the NEK7 interactome by two screening approaches: one using a yeast two-hybrid system and the other based on immunoprecipitation followed by mass spectrometry analysis. These approaches led to the identification of 61 NEK7 interactors that contribute to a variety of biological processes, including cell division. Combining additional interaction and phosphorylation assays from yeast two-hybrid screens, we validated CC2D1A, TUBB2B, MNAT1, and NEK9 proteins as potential NEK7 interactors and substrates. Notably, endogenous RGS2, TUBB, MNAT1, NEK9, and PLEKHA8 localized with NEK7 at key sites throughout the cell cycle, especially during mitosis and cytokinesis. Furthermore, we obtained evidence that the closely related kinases NEK6 and NEK7 do not share common interactors, with the exception of NEK9, and display different modes of protein interaction, depending on their N- and C-terminal regions, in distinct fashions. In summary, our work shows for the first time a comprehensive NEK7 interactome that, combined with functional in vitro and in vivo assays, suggests that NEK7 is a multifunctional kinase acting in different cellular processes in concert with cell division signaling and independently of NEK6.
Keywords: Nek7, Nek6, proteomics, N and C terminal domain, cell cycle, cancer
Introduction
The fidelity of the cell cycle is maintained, in part, by key regulatory proteins, such as kinases. Protein kinases trigger and regulate cell division events such as centrosome duplication, spindle assembly, microtubule-kinetochore attachment, as well as cytokinesis.1 The master regulators of the eukaryotic cell cycle are Cyclin-dependent kinases (CDK), members of the Aurora and Polo-like kinases (PLKs) family and the more recently discovered NimA related kinases (NEKs) family.2−4 The NEKs represent a large family of 11 serine/threonine protein kinases in mammals, named NEK1 to NEK11, that share 40–45% sequence identity with the mitotic regulator NIMA (Never in mitosis gene A) identified in the filamentous fungus Aspergillus nidulans.2,5−7 The deregulation of these proteins directly affects the cell division and has been correlated strongly with the uncontrolled cell proliferation and the appearance of tumors.8
The human protein NEK7 (NEK7) as well as the NEKs 1, 2, 6, and 9 have been importantly shown to function in mitosis contributing to the separation of centrosomes and establishment of the microtubule-based mitotic spindle.9 In fact, the depletion of NEK7 via RNAi and inactive mutants disrupted levels of γ-tubulin in interphase cells and caused an arrest in prometaphase associated with a fragile mitotic spindle,10−13 whereas its overexpression resulted in multinucleated cells and a high proportion of apoptotic cells.14 Allied to these studies, the centrosomal pericentriolar material (PCM) proteins do not accumulate at the centrosomes in NEK7-depleted cells, indicating that NEK7 is involved in the recruitment of PCM proteins, which are necessary for both centriole duplication and spindle pole formation.15 Importantly, NEK7 along with NEK6 and NEK9 constitute a mitotic signaling module in which NEK6 and NEK7 are phosphorylated and activated by NEK9, contributing to mitosis progression. Furthermore, studies have shown that the NEK7 absence leads to lethality during embryogenesis and growth retardation, indicating an indispensable role for NEK7 in development and survival16 and that the closely related protein NEK6 can not compensate for the loss of NEK7 in the organism.
Structurally, the human NEK6 and NEK7 share ∼86% identity in their C-terminal domains7,17 and only ∼20% identity in their disordered N-terminal extensions.18,19 Studies indicate that the two proteins have different roles and biological functions12 pointing to their disordered N-terminal extensions as a possible component for the differential functions of these kinases.18−21 Furthermore, NEK6 and NEK7 show differential spatiotemporal tissue distribution20 and enzymatic control,21 as well as distinct subcellular localization.13
Although all of these data suggest nonredundant roles for NEK6 and NEK7, the molecular basis for their specific roles is unknown, mainly because the protein interaction partners of NEK7 in contrast to NEK622 are so far unknown.
Therefore, we aimed to characterize the NEK7 protein interactome, shedding light on the NEK7 functions and thus providing additional information regarding the mechanisms that orchestrate its differential function and regulation from NEK6. We performed two interaction screening approaches for NEK7: a yeast two-hybrid (Y2H) system and an immunoprecipitation followed by online liquid chromatography mass spectrometry analysis (IP-LC–MS/MS). We have identified a number of novel NEK7 protein interactors belonging to a variety of biological processes not previously described to be regulated by NEK7, as well as its already reported role in cell division. Combining additional interaction and phosphorylation assays, we validated CC2D1A, TUBB2B, MNAT1, and NEK9 proteins as possible NEK7 interactors and substrates. Interestingly, RGS2, TUBB, MNAT1, NEK9, and PLEKHA8 localize with NEK7 in key sites during mitosis and cytokinesis. These results shed light on NEK7 functions in multiple molecular pathways and denote a potential involvement for its interactors in cell division. Furthermore, we obtained evidence that the closely related human kinases NEK6 and NEK7 do not share the majority of interactors and show distinct mechanisms of specific interaction, suggesting their distinct, independent, and nonredundant signaling functions in the cell. We suggest that via distinct mechanisms the N- and C-terminal domains of NEK6 and NEK7 can contribute, remarkably, to both regulation and catalysis.
Experimental Procedures
Plasmid Constructs
The coding sequence of full-length human NEK7 was amplified by PCR from a leukocyte cDNA library (Clontech) using te following primers: 5′-CGCGGATCCATGGATGAGCAATCACAAGG-3′ (forward) containing BamHI restriction site; 5′-CGGAATTCCATATGGATGAGCAATCACAAGG-3′ (forward) containing EcoRI and NdeI restriction sites; 5′-GGGGAAGCTTTTAGCTGCTTGCAGTGCATGCATG-3′ (reverse) containing HindIII restriction site; and 5′- ACGCGTCGACTTAGCTGCTTGCAGTGCAT-3′ (reverse) containing SalI restriction site. The amplified fragments corresponding to human NEK7 full-length sequence were cloned into pGEM-T Easy (Promega) (pGEMT-NEK7 construct) and were used as template to amplify the human NEK7 truncated region corresponding to its kinase domain only (NEK7Δ(1–44): the N-terminal region was deleted) with the specific primers set 5′-CGGAATTCCATATGTTTCGAATAGAAAAGAAAAT-3′ (forward) containing EcoRI and NdeI restriction sites and 5′-ACGCGTCGACTTAGCTGCTTGCAGTGCAT-3′ (reverse) containing SalI restriction site. The amplified fragments were cloned into pGEM-T Easy (Promega) (pGEMT- NEK7Δ(1–44) construct). The chimeric constructs consisting of NEK6 N-terminal-NEK7 C-terminal (N6C7) and NEK7 N-terminal-NEK6 C-terminal (N7C6) were generated by gene synthesis (GenScript Corporation, see Figure 6B for the constructs). The KpnI, EcoRI, NdeI, SalI, and ApaI restriction sites were added to the synthetic genes.
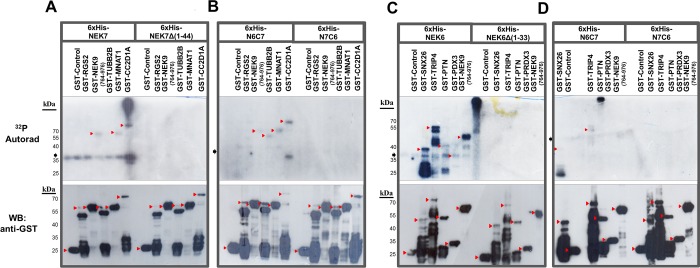
Figure 6.
Comparison of the human NEK6 and NEK7 phosphorylation profile with their kinase domains and chimeric constructs using the proteins identified by the yeast two-hybrid system. (A, B) Phosphorylation profile of NEK7full-length comparing to the NEK7Δ(1–44), N6C7, and N7C6 using NEK7 interactors RGS2, NEK9, TUBB2B, MNAT1, and CC2D1A. (C, D) Phosphorylation profile of NEK6 full-length comparing to the NEK6Δ(1–33), N6C7, and N7C6 using NEK6 interactors SNX26, TRIP4, PTN, and PRDX3, described by Meirelles et al.,22 and NEK9 (recovered in this NEK7 yeast two-hybrid screen). The phosphorylated proteins were detected by autoradiography exposition (32P Autorad) during 5 days (panels A and B) or 24 h (panels C and D). The arrowheads indicate the positions of the GST-tagged proteins or GST-control, and the arrows indicate the position of the 6×His-tagged kinases in the 32P Autorad or Western blotting (WB). The molecular weight (kDa) of the proteins is indicated. The lanes without GST-tagged proteins refer to the autophosphorylated 6×His-tagged proteins. The GST-control in normalized concentrations was not phosphorylated by any of the wild-type or mutants, indicating that the phosphorylation of substrates is specific under the assay conditions. The results are based on two independent experiments.
For the yeast two-hybrid screens we subcloned the full-length NEK7 (NEK7-pBTMK) and the chimeric constructs N6C7 (N6C7-pBTMK) and N7C6 (N7C6-pBTMK) into EcoRI/SalI restriction sites of modified yeast expression vector pBTM116K,23 in frame with the LexA DNA binding domain (Clontech). The recovered nucleotide sequences encoding the interacting proteins identified to interact with human NEK7 in yeast two-hybrid screenings were subcloned from the vector pACT2 (Clontech) into bacterial expression vector pGEX (GE Healthcare), which allows the expression of proteins in the form of a Glutathione S-transferase (GST) fusion.
To obtain higher expression levels of kinases in bacteria, we subcloned the full-length human NEK7 (6×His-NEK7 construct) into the NdeI/HindIII restriction sites; NEK7Δ(1–44) and the chimeric constructs N6C7 and N7C6 were subcloned into NdeI/SalI restriction sites in the modified vector pET28a-His-TEV (Novagen/EMD Biosciences), in fusion with a 6×His tag (constructs 6×His-NEK7Δ(1–44)), 6×His-N6C7, and 6×His-N7C6, respectively). To express NEK7 in human cells, full-length NEK7 was inserted into BamHI/SalI restriction sites in pCDNAFLAG (Invitrogen) in fusion with a FLAG tag. The sequences of all vector constructs were confirmed by restriction endonuclease analysis and DNA sequencing.
The full-length NEK6 and NEK6 kinase domain cloned into pET28a-His-TEV in fusion with a His tag (6×His-NEK6 and 6×His-NEK6Δ(1–33), respectively); full-length NEK6 cloned into pBTM116KQ in fusion with binding domain DNA LexA (pBTMK-NEK6); the interacting proteins NEK9 (NEK9), Sorting nexin 26 (SNX26), Thyroid hormone receptor interactor 4 (TRIP4), Pleiotrophin (PTN), and Peroxiredoxin 3 (PRDX3) recovered in NEK6 Y2H into pACT2 in fusion with GAL4 transcriptional activation domain (constructs: pACT-NEK9(806–979), pACT-SNX26, pACT-TRIP4, pACT-PTN and pACT-PRDX3, respectively) or subcloned into pGEX in fusion with GST (constructs: GST-SNX26, GST-PTN, GST-PDX, GST-TRIP4) were obtained as described by Meirelles et al.22
Yeast Two-Hybrid Screening (Y2H)
The pBTM116K vector was used to express the full-length human NEK7, full-length human NEK6, N6C7 and N7C6 linked to the C-terminus of LexA DNA-binding domain peptide (constructs: pBTMK-NEK7, pBTMK-NEK6, pBTMK-N6C7 and pBTMK-N7C6, respectively) or as a control (pBTM116K-control) in Saccharomyces cerevisiae strain L40 (trp1-901, his3Δ200, leu2-3, ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lac GAL4), which contains the heterologous reporter genes HIS3 and LacZ.
The yeast two-hybrid screenings were performed against the three cDNA libraries fetal brain, bone marrow, and leukocyte, cloned in pACT2 vector expressing GAL4 activation domain (AD) fusion proteins (Matchmaker System, Clontech). The autonomous activation of HIS3 gene was tested by co-transformation of yeast cells with pBTMK-NEK7 and pACT2 as a control (pACT2-control), grown in minimal medium plates without tryptophan (-W), leucine (-L), and histidine (-H) and containing 0, 5, 10, 20, 30, 50, 70, or 100 mM 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of His3p protein (Imidazoleglycerol-phosphate dehydratase).24 Although the autoactivation of HIS3 was not observed, the screenings were performed by separate co-transformation of three cDNA libraries with the pBTMK-NEK7 construct in minimal medium plates without -W, -L, and -H, containing 5 mM 3-AT. The co-transformants showing the best growth conditions had their recombinant pACT2 plasmids isolated and sequenced.
To test the growth capacity under interaction-selective conditions, the recovered clones from the yeast two-hybrid screenings for NEK7 and for NEK622 were co-transformed in the yeast strain L40 with pBTMK-NEK7, pBTMK-NEK6, pBTMK-N6C7, and pBTMK-N7C6 constructs or with the pBTM116K-control. The co-transformation products were plated in minimal medium without -W, -L, and -H, containing 3-AT gradient (1–100 mM). We considered as positive interactions the co-transformations showing a higher colony growth when compared to pBTM116K-control.
Antibodies
The following antibodies were used for either immunofluorescence staining or Western blotting purposes: goat anti-NEK7 (sc50756), goat (sc50763) or mouse (sc1004) anti-NEK9, goat anti-α-tubulin (TUBA) (sc8035) (all from Santa Cruz Biotechnology); mouse anti-NEK7 (ab68060), rabbit anti-NEK7 (ab96538), goat anti-PLEKHA8 (ab38748), rabbit anti-β-tubulin (TUBB) (ab15568), rabbit anti-RGS2 (ab36561), rabbit anti-MNAT1 (ab65125), rabbit anti-ANKS1B (ab116083), mouse anti-CC2D1A (ab68302), and mouse anti-pericentrin (PCNT) (ab28144) (all from Abcam). Mouse anti-His5 (34660, QIAGEN), mouse anti-GST (made in house), mouse anti-FLAG (F1804, Sigma-Aldrich), rabbit anti-phosphothreonine (71-8200, Invitrogen), mouse anti-PLK1 (ab17057), goat anti-SMC1 (sc-21078), and goat anti-SMC3 (sc-8198) antibodies were used for Western blotting. Secondary antibodies for immunofluorescence staining were obtained from the following sources: chicken anti-goat, anti-mouse, or anti-rabbit Alexa Fluor 488 or donkey anti-goat, anti-mouse, or anti-rabbit Alexa Fluor 546 (all from Life Technologies, Inc.); horseradish peroxidase (HRP)-conjugated anti-mouse was obtained from (Calbiochem) and HRP-linked anti-goat or anti-rabbit secondary antibodies for Western blotting (WB) purposes were obtained from Sigma-Aldrich.
Immunoprecipitation Followed by Mass Spectrometry Analysis (IP-LC–MS/MS)
To identify NEK7-associated proteins, FLAG-tagged wild-type NEK7 (FLAG-NEK7) or pCDNAFLAG as a control (FLAG-control) were used to transfect HeLa cells in exponential growth using X-tremeGENE 9 DNA Transfection Reagent (Roche).
Cells were harvested by centrifugation 24 h after transfection, and the pellets were incubated in lysis buffer (1% Triton X-100, 5 mM EDTA, 0.1 mM sodium orthovanadate, 1 μg/μL DNase and protease inhibitors cocktail diluted in PBS 1X) at 4 °C for 30 min. The lysates were sequentially cleared by centrifugation and incubated overnight with Mouse anti-FLAG M2 Affinity Gel (Sigma). The beads from samples transfected with FLAG-NEK7 or FLAG-control, in a combination of three experiments, were gently washed three times in Tris pH 8.0 buffer and were submitted for liquid chromatography-tandem MS (LC–MS/MS).
The LC–MS/MS analyses were performed on tryptic peptide samples using a nanoAcquity UPLC nanocapillary high-performance LC system (Waters Corp., Milford, MA, USA) coupled to a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion, Ithaca, NY, USA). Sample concentration and desalting were performed online using a nanoAcquity UPLC trapping column (180 μm × 20 mm, packed with 5-μm, 100-Å Symmetry C18 material; Waters) at a flow rate of 15 μL/min for 1 min. Separation was accomplished on a nanoAcquity UPLC capillary column (150 μm × 100 mm, packed with 1.7-μm, 130-Å BEH C18 material; Waters). A linear gradient of A and B buffers (buffer A: 1.5% ACN and 0.1% FA; buffer B: 98.5% ACN and 0.1% FA) from 2% to 40% buffer B over 80 min was used at a flow rate of 0.5 μL/min to elute peptides into the mass spectrometer. Columns were washed and re-equilibrated between LC–MS/MS experiments. Electrospray ionization was carried out at 1.65 kV using the NanoMate, with the Q Exactive heated transfer capillary set to 250 °C. Mass spectra were acquired in the positive-ion mode over the range m/z 400–2000 at a resolution of 70,000 (full width at half-maximum at m/z 400; ∼1 spectrum/s) and AGC target >1 × e6 (Method 1) or >3 × e6 (Method 2). Mass accuracy after internal calibration was within 1 ppm. MS/MS spectra were acquired at a rate of ∼10 MS/MS/s for the 10 most abundant, multiply charged species in each mass spectrum with a resolution of 17,500 and signal intensities >5 × e5 NL (Method1) or >2 × e5 NL (Method 2), using HCD with MS/MS collision energies set at either 24 V (Method 1) or 30 V (Method 2), nitrogen as the collision gas, and MS/MS spectra acquisition over a range of m/z values dependent on the precursor ion. Dynamic exclusion was set such that MS/MS for each precursor ion species was excluded for either 4 s (Method 1) or 20 s (Method 2) postacquisition. All spectra were recorded in profile mode for further processing and analysis.
MS and MS/MS data analysis was carried out using both Proteome Discoverer (PD) 1.4 software (Thermo Fisher Scientific) and an in-house Mascot 2.4 server (Matrix Science, London, U.K.). The MS/MS data were searched against the IPI human 3.80 amino acid sequence database for protein/peptide identification. The PD 1.4 search was set up with precursor intensity node and full tryptic peptides with a maximum of 2 missed cleavage sites with carbamidomethyl cysteine and oxidized methionine included as variable modifications. Mascot search was set up for full tryptic peptides with a maximum of 4 missed cleavage sites with carbamidomethyl cysteine and oxidized methionine included as variable modifications. The precursor mass tolerance was set to 10 ppm for Q Exactive orbitrap data, and the maximum fragment mass error was 0.8 Da. The PD1.4 searches for each Q Exactive method were uploaded into Scaffold 4.2.1 (Proteome Software, Inc.) software program while searching data via X! Tandem as described by Whelan et al.25 The quantitative data were performed with filters set at 95% minimum protein ID probability (calculated probability of correct protein identification), a minimum number of five unique peptides for one protein (five unique peptides were used to reduce the NEK7 protein interacting list to strong interacting proteins at high abundance) and a peptide threshold at 0.8% FDR (False Discovery Rate) algorithm.
In order to avoid the selection of nonspecific interactors (for example, proteins that interact with the solid-phase support, affinity reagent, or epitope tag) from affinity purification using anti-FLAG, we compared the generated list of interacting proteins from quantitative analysis with the Contaminant Repository for Affinity Purification, CRAPome (http://www.crapome.org/). Those proteins that were present in the negative controls of anti-FLAG affinity purifications under our experimental conditions were excluded from the list.
In Silico Protein–Protein Interaction (PPI) Analysis
To generate NEK7 PPI maps at proteome scale, the retrieved NEK7 interacting proteins from IP-LC–MS/MS and Y2H were integrated in interaction networks using the Integrated Interactome System (IIS) platform, developed at National Laboratory of Biosciences, Brazil (http://www.lge.ibi.unicamp.br/lnbio/IIS/).26 The enriched biological processes from the Gene Ontology database (GO, http://www.geneontology.org/) were calculated in each network using the hypergeometric distribution.26 The interaction network was visualized using Cytoscape 2.8.3 software (http://www.cytoscape.org/).27 The prediction of disordered amino acid residues in the Y2H retrieved protein sequences was calculated by PONDR VL-XT predictor (http://www.pondr.com/), and the domain composition of these sequences was obtained by Pfam (http://pfam.sanger.ac.uk/), PROSITE (http://www.expasy.ch/prosite/), or InterPro (http://www.ebi.ac.uk/interpro/) databases.
Protein Expression and Purification
The proteins identified in the NEK7 yeast two-hybrid screens, β-tubulin-2B chain (TUBB2B), Coiled-coil and C2 domain-containing protein 1A (CC2D1A), and Regulator of G-protein signaling 2 (RGS2) in fusion with GST (GST-TUBB2B, GST-CC2D1A and GST-RGS2, respectively), were expressed in E. coli BL21 (DE3/pRARE) using 0.5 mM IPTG at 37 °C for 4 h. The GST-tagged proteins NEK9, comprising its regulatory region from 764 to 976 amino acid residues (GST-NEK9(764–976)) and CDK-activating kinase assembly factor MAT1 (GST-MNAT1) were expressed in E. coli BL21 (DE3) cells using 1 mM IPTG at 25 and 30 °C, respectively, all for 4 h. The full-length human NEK7 protein (6×His-NEK7) or kinase domain NEK7 (6×His-NEK7Δ(1–44)) and chimeric constructs N6C7 (6×His-N6C7) and N7C6 (6×His-N7C6) were expressed in E. coli BL21 (DE3/pRARE) or BL21 (DE3) cells, respectively. Expression was induced for 4 h using 1 mM isopropyl-β-d-thio-galactoside (IPTG) at 28 °C to 6×His-NEK7 and 6×His-N7C6 constructs and 0.5 mM and 1 mM IPTG at 25 °C to 6×His-NEK7Δ(1–44) and 6×His-N6C7 constructs, respectively. Induced cells were harvested and lysed by sonication in extraction buffer (50 mM HEPES pH 7.5, 5 mM sodium phosphate, 300 mM NaCl, 5% glycerol) supplemented with 1 mM PMSF and 625 μg/mL lysozyme. The proteins GST-SNX26, GST-TRIP4, GST-PTN, and GST-PRDX3 were expressed as described by Meirelles et al.22 The cell lysates were separated by centrifugation at 16,000 × g for 10 min at 4 °C in order to obtain the supernatant. Cleared fractions from protein expression in fusion with GST as well as GST as a control (control-GST) were purified by GST affinity chromatography medium binding to Glutathione Sepharose 4 Fast Flow followed by elution in buffer containing 50 mM Tris-HCl and 50 mM reduced glutathione pH 8.0, according to the protocol described for Batch Purification (GE Healthcare). Cleared fractions of 6×His-NEK7, 6×His-NEK7Δ(1–44), 6×His-N6C7, and 6×His-N7C6 obtained by lysis were purified by affinity liquid chromatography using a HiTrap Chelating Affinity Chromatography Column (GE Healthcare) followed by elution with a linear concentration gradient of imidazole (1 to 100 mM) in extraction buffer. All of the eluted recombinant proteins were dialyzed against kinase buffer containing 50 mM MOPS pH 7.4, 300 mM NaCl, 10 mM MgCl2, and 0.1 mM PMSF. The purified proteins 6×His-NEK6 and 6×His-NEK6Δ(1–33) were obtained as described by Meirelles et al.22 The expression of all proteins was confirmed by WB using monoclonal mouse anti-GST clone 5.3.3 (produced in house) or mouse anti-5×His primary antibodies and Horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibodies, and the bands were detected by a luminol chemiluminescence detection system (Santa Cruz Biotech) or by SDS-PAGE stained with Coomassie blue (SDS-PAGE).
Pull-Down Assay
To validate some of the candidate NEK7 interacting proteins recovered by yeast two-hybrid and IP-LC–MS/MS, we performed affinity purification (pull-down assay) using 6×His-NEK7 or 6×His-RARA as bait. 6×His-RARA was used as a control since the protein RARA (Retinoic acid receptor alpha) is not described to interact with NEK7 or its potential interactor ANKS1B and NEK9. For this purpose, human embryonic kidney (HEK) 293T cells (HEK293T) were collected by gently shaking off the flask and centrifuged. Next, HEK293T extracts produced by homogenization in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and protease inhibitor cocktail) were incubated overnight at 4 °C with 6×His-NEK7, 6×His-RARA bound to Ni-NTA agarose resin (Qiagen), or Ni-NTA agarose resin, previously equilibrated in wash buffer (50 mM Tris-HCl pH 8.0, 1% NP40, and protease inhibitor cocktail). The resins containing the complexes were washed three times in wash buffer, and bound proteins were analyzed by WB using specific antibodies, as previously described.28
In Vitro Protein Phosphorylation
To test the ability of NEK7 to catalyze 32P or Pi transfer to NEK7 itself or exogenous substrates, GST-tagged proteins as substrates or GST-control were previously equilibrated in kinase buffer and incubated at 30 °C in 100 μM ATP and 185 kBq [γ-32P]ATP (222 TBq/mmol; NEN). Then 6×His-tagged NEK6Δ(1–33), NEK7Δ(1–44), N6C7, or N7C6 proteins were added to this reaction for 30 min. The autophosphorylation was performed by 6×His-tagged proteins incubation in 100 μM ATP and 185 kBq [γ-32P]ATP (222 TBq/mmol; NEN) without being added to the GST-tagged proteins, for 30 min. Assays were carried out in a final volume of 50 μL. Electrophoresis sample buffer was added to stop the reaction, and the proteins were resolved by SDS-PAGE. 32P incorporation was detected by autoradiography. In the case of the chemiluminescence assay, GST-NEK9(764–976) was incubated with or without 200 μM ATP, in the presence or absence of 1 μg of 6×His-NEK7 in kinase buffer for 1 h in a reaction volume of 50 μL. The reaction was stopped by addition of electrophoresis sample buffer, and the proteins were resolved by SDS-PAGE. Phosphoamino acids were detected by Western blotting using polyclonal rabbit anti-phosphothreonine primary antibody diluted 1/250 in 3% bovine serum albumin (BSA) and goat anti-rabbit secondary antibodies conjugated to Horseradish Peroxidase (Sigma). The protein bands were detected by Coomassie blue staining or luminol chemiluminescence detection system (Santa Cruz Biotech) according to the instructions supplied by the manufacturer.
In Vitro Kinase Activity Assay
The in vitro kinase activity assay was performed using LANCE Ultra Kinase Activity Assay (Product N° TRF0126-D/TR0126-M, PerkinElmer) containing ULight-p70 S6K (Thr389) Peptide (phosphorylation motif LGFTYVAP). The assay was optimized with 80 nM 6×His-NEK7, 6×His-NEK7Δ(1–44), 6×His-N6C7, 6×His-N7C6 6×His-NEK6, or 6×His-NEK6Δ(1–33) purified proteins, 50 nM ULight-p70 S6K (Thr389) Peptide, and 100 μM ATP, prediluted in kinase buffer (50 mM HEPES pH 7.5, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, and 0.01% Tween-20). Ten microliters total volume of kinase reaction was added to the wells of a 384-well OptiPlate. The kinase reactions were incubated for 60 min at 23 °C and stopped by the addition of 10 mM EDTA. For the detection of the phospho-substrate, the Eu-anti-phospho-p70 S6K (Thr389) antibody diluted in Detection Buffer was added to a final concentration of 2 nM, and the reactions were then incubated for 16 h at 4 °C. A reaction without addition of ULight-p70 S6K (Thr389) Peptide was perfomed and used as a control. The signal was measured on a 2104 EnVision Multilabel Microplate Reader. Excitation wavelength was set to 320 nm, and emission was recorded at 665 nm.
Immunocytochemistry
HeLa cells were grown and evaluated for cell viability by trypan blue exclusion (Invitrogen). The cell count was performed using Countess Automated Cell Counter (Invitrogen). A total of 20–30,000 cells per well were seeded in 384-cell Carrier plates (PerkinElmer) to a final volume of 50 μL.
The cells in a confluence of 70% were fixed and permeabilized at room temperature for 20 min in a solution containing 3.7% formaldehyde, 0.2% Triton X-100 in PBS 1X supplemented with 50 mM EGTA and 30 μg/mL taxol. The cells were washed twice with PBS 1X to remove the fixing solution. After removing the fixing solution, cells were incubated for 30 min with blocking solution containing 3% BSA and 0.8% Triton X-100 diluted in PBS 1X. The fixation and permeabilization steps were made using the JANUS Modular Dispense Technology (MDT) Automated Workstation (PerkinElmer). Then, the cells were washed twice with PBS 1X, and proteins were detected by incubating the cells for 1 h at room temperature with goat anti-NEK7, goat or mouse anti-NEK9, goat anti-TUBA, mouse anti-NEK7, rabbit anti-NEK7, goat anti-PLEKHA8, rabbit anti-TUBB, rabbit anti-RGS2, rabbit anti-MNAT1, rabbit anti-ANKS1B, mouse anti-CC2D1A, or mouse anti-PCNT, diluted 1:100 in blocking solution. Primary antibodies were removed by washing three times with a solution containing PBS 1X and 0.05% Tween 20, and the proteins were revealed by incubating for 2 h at room temperature with chicken anti-goat, anti-mouse, or anti-rabbit Alexa Fluor 488 or donkey anti-goat, anti-mouse, or anti-rabbit Alexa Fluor 546 diluted 1:300 in blocking solution. The labeling was stopped by removing the secondary antibody by washing three times with a solution containing PBS 1X and 0.05% Tween 20 (PBS-T), and the nucleus was stained for 5 min with DAPI 0.5 mM diluted 1:100 in PBS-T. The cells were observed by fluorescence microscopy using Operetta High Content Imaging System (PerkinElmer). The images edition was performed using Volocity Demo 6.1.1 software.
Results
Identification of Human NEK7 Interacting Proteins by Yeast Two-Hybrid and Mass Spectrometry
To clarify NEK7 biological functions, we report here two screening approaches to identify proteins that interact with NEK7: the first was based on direct interactions using a Y2H29,30 system, and the second was based on IP-LC–MS/MS.
In order to follow the functional distribution pattern of NEK7 across tissues, the Y2H was employed using full-length wild-type NEK7-LexA fusion protein as bait against three cDNA libraries: human fetal brain, bone marrow, and leukocyte. A total of 8.5 × 103 screened co-transformants for fetal brain, 4.1 × 104 for bone marrow, and 1.66 × 104 for leukocyte yielded 88 positive clones for the HIS3 reporter gene that were amplified in E. coli and sequenced. To test if human NEK7 could, in fact, interact with the proteins recovered in the Y2H and reduce false-positive clones, the 88 retrieved interacting proteins were tested in yeast cells for their growth capacity in minimal medium plates without tryptophan, leucine, and histidine but containing 1–100 mM 3-AT gradient. The screen resulted in the identification of 25 positive interacting proteins for NEK7 (Table 1). Interestingly, we recovered the regulatory domain of NEK9 (amino acids 764–976) previously described to interact with NEK7,7 in a way validating our approach.
Table 1. Human NEK7 Interacting Proteins Identified by the Yeast Two-Hybrid System Screensa.
| redundancy in libraryc |
|||||||
|---|---|---|---|---|---|---|---|
| protein interacting with Nek7b | gene | Uniprot accession | coded protein residues (retrieved/complete sequence) | FB | BM | L | growth scored |
| Beta-2-microglobulin | B2M | P61769 | 1–119/119 | 1 | 0 | 0 | 1/0 |
| Serine/threonine-protein kinase Nek9 | NEK9 | Q8TD19 | 764–976/979 | 3 | 3 | 2 | 4/0 |
| Regulator of G-protein signaling 2 | RGS2 | P41220 | 1–211/211 | 0 | 0 | 1 | 1/1 |
| CDK-activating kinase assembly factor MAT1 | MNAT1 | P51948 | 1–309/309 | 1 | 2 | 0 | 1/1 |
| Secreted frizzled-related protein 4 | SFRP4 | Q6FHJ7 | 1–346/346 | 0 | 4 | 0 | 3/0 |
| Cytochrome b-c1 complex subunit 9 | UQCR10 | Q9UDW1 | 1–63/63 | 0 | 1 | 0 | 1/0 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondria | NDUFS1 | P28331 | 451–727/727 | 0 | 2 | 0 | 1/0 |
| 5-Aminolevulinate synthase, nonspecific, mitochondrial | ALAS1 | P13196 | 94–640/640 | 0 | 1 | 0 | 1/0 |
| NFU1 iron-sulfur cluster scaffold homologue, mitochondrial | NFU1 | Q9UMS0 | 22–208/254 | 1 | 1 | 4 | 1/0 |
| Tubulin beta-2B chain | TUBB2B | Q9BVA1 | 247–430/445 | 0 | 0 | 1 | 1/1 |
| Hemoglobin subunit alpha | HBA1 | P69905 | 1–142/142 | 0 | 2 | 0 | 1/0 |
| Hemoglobin subunit beta | HBB | P68871 | 1–147/147 | 0 | 2 | 0 | 1/0 |
| Elongation factor 1-alpha 1 | EEF1A1 | P68104 | 340–432/462 | 0 | 1 | 0 | 1/0 |
| Elongation factor Tu, mitochondrial | TUFM | P49411 | 13–455/455 | 1 | 0 | 0 | 1/1 |
| Pleckstrin homology domain-containing family A member 8 isoform 2 | PLEKHA8 | Q96JA3 | 103–418/459 | 0 | 0 | 2 | 1/0 |
| Actin, cytoplasmic 2 | ACTG1 | P63261 | 187–375/375 | 0 | 0 | 3 | 1/1 |
| 40S ribosomal protein S6 | RPS6 | P62753 | 1–228/249 | 0 | 1 | 0 | 1/1 |
| Coiled-coil and C2 domain-containing protein 1A | CC2D1A | Q6P1N0 | 501–940/951 | 0 | 1 | 0 | 1/1 |
| 40S ribosomal protein S4, X isoform | RPS4X | P62701 | 5–263/263 | 0 | 2 | 0 | 1/1 |
| Ubiquitin-40S ribosomal protein S27a | RPS27A | P62979 | 1–156/156 | 0 | 2 | 0 | 1/0 |
| Ankyrin repeat and sterile alpha motif domain-containing protein 1B | ANKS1B | Q7Z6G8 | 6–426/426 | 0 | 1 | 0 | 3/0 |
| FCH and double SH3 domains protein 2 | FCHSD2 | O94868 | 627–740/740 | 1 | 0 | 0 | 1/1 |
| MAP7 domain-containing protein 1 | MAP7D1 | Q3KQU3 | 715–841/841 | 1 | 0 | 0 | 1/1 |
| Transmembrane and TPR repeat-containing protein 4 | TMTC4 | Q5T4D3 | 467–741/741 | 0 | 0 | 9 | 4/0 |
| Transmembrane 9 superfamily member 3 | TM9SF3 | Q9HD45 | 398–589/589 | 0 | 0 | 2 | 1/0 |
The proteins were selected from yeast cells under growth conditions in minimal medium without tryptophan, leucine, and histidine and containing 3-AT.
Results from BLASTX (GenBank).
Absolute number of sequences retrieved of each human library (FB: fetal brain, BM: bone marrow, L: leukocyte).
Growth test in yeast cells transfected with recovered sequences in the vector pACT2 and pBTM116K-NEK7/empty pBTM116KQ (concentration of 3AT = 5 mM).
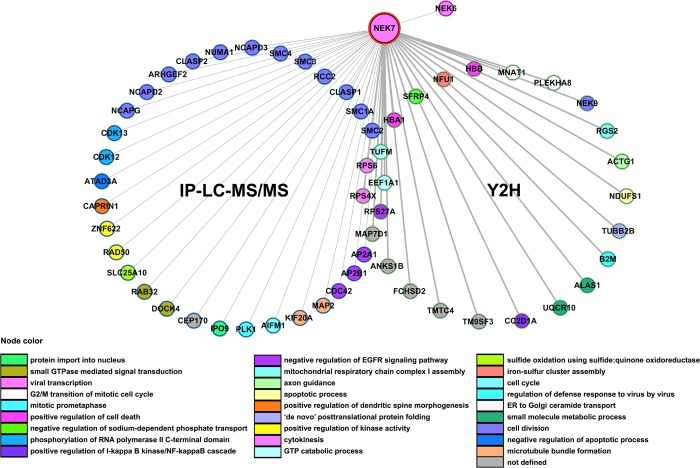
The Y2H may fail to detect bona fide interactors that are unable to associate with NEK7 in yeast or may also yield false-positives that only associate in the context of this type of assay, and likewise, not all NEK7 interacting proteins may have been detected using this approach. We therefore decided to use a second assay based on IP-LC–MS/MS. We reasoned that this complementary approach would generate a second list of candidates that could be compared and supplemented to the Y2H screens to identify other potential candidate NEK7 interactors. To better achieve the NEK7 interaction/functional profile, FLAG-NEK7 or FLAG-control were immunoprecipitated from HeLa cells in triplicate experiments and analyzed by mass spectrometry. The HeLa cervical cancer-derived cell line was used since the interference with NEK7 function induces growth inhibition, mitotic arrest, and apoptosis,12 immediately suggesting potential NEK7 functions in this cell system. While uploading IP-LC–MS/MS data in Scaffold software using a stringent filter setting (see Experimental Procedures), proteins that co-immunoprecipitated with NEK7-FLAG but not FLAG-control in the triplicate experiments and that were not deposited in the Contaminants Repository for Affinity Purification (CRAPome) or those that overlapped with Y2H results were considered potential NEK7 interactors, yielding 36 proteins (Table 2). Notably, six Y2H and IP-LC–MS/MS overlapping proteins were identified (Isoform 1 of MAP7 domain-containing protein 1 (MAP7D1), 40S ribosomal protein S4, X isoform (RPS4X), 40S ribosomal protein S6 (RPS6), Tu translation elongation factor, mitochondrial precursor (TUFM), and Similar to Elongation factor 1-alpha 1 (EEF1A1)), indicating them as potential NEK7-specific interactors (Table 2, Figure 1). To further explore the NEK7 signaling pathways and its interacting proteins, we performed Protein–Protein Interaction (PPI) functional network analysis using both Y2H and IP-LC–MS/MS proteins, totaling 61 interactors. The analysis resulted in the enrichment of diverse biological processes and cellular components based on the Gene Ontology database (Figure 1 and Table S1, Supporting Information). Furthermore, we analyzed the retrieved Y2H sequences of NEK7 interacting proteins for their secondary structure concerning the domain composition and presence of disordered amino acid residues. Consistent with the bioinformatic prediction, NEK7 protein partners were shown to be organized in a variety of domain families and disordered segments (Table S2, Supporting Information). Disordered protein regions control the degree of motion between domains and, interestingly, may enable the binding of different partners as well as be targets of post-translational modifications such as phosphorylation.31,32 Together, our results indicate that NEK7 may potentially interact with and phosphorylate distinct proteins, composed of varied domains and disordered regions, therefore pointing to an involvement of NEK7 in a broad range of signaling pathways and hence highlighting it as a multifunctional kinase.
Table 2. NEK7 Interacting Proteins Identified by IP-LC–MS/MSa.
| proteinb | gene | Uniprot accession | fold change TICc | FLAG-control total spectra | FLAG-NEK7 total spectra |
|---|---|---|---|---|---|
| Serine/threonine-protein kinase Nek7 | NEK7 | Q8TDX7 | 9.10 × 109 | 1291 | |
| cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier | SLC25A10 | Q9UBX3 | 2.60 × 108 | 68 | |
| Importin-9 | IPO9 | Q96P70 | 1.00 × 108 | 88 | |
| ATPase family AAA domain-containing protein 3A isoform 3 | ATAD3A | Q9NVI7 | 9.90 × 107 | 94 | |
| Protein RCC2 | RCC2 | Q9P258 | 4.90 × 107 | 80 | |
| Isoform 3 of Apoptosis-inducing factor 1, mitochondrial | AIFM1 | O95831 | 3.80 × 107 | 43 | |
| Ubiquitin-40S ribosomal protein S27a | RPS27A | P62979 | 3.50 × 107 | 36 | |
| Isoform 1 of AP-2 complex subunit beta | AP2B1 | P63010 | 3.30 × 107 | 22 | |
| Isoform 1 of Structural maintenance of chromosomes protein 2 | SMC2 | O95347 | 2.80 × 107 | 35 | |
| Isoform 1 of Caprin-1 | CAPRIN1 | Q14444 | 2.80 × 107 | 37 | |
| Isoform 1 of Structural maintenance of chromosomes protein 4 | SMC4 | Q9NTJ3 | 2.70 × 107 | 69 | |
| Isoform 1 of CLIP-associating protein 1 | CLASP1 | Q7Z460 | 2.50 × 107 | 89 | |
| Isoform 1 of CLIP-associating protein 2 | CLASP2 | O75122 | 1.60 × 107 | 20 | |
| Ras-related protein Rab-32 | RAB32 | Q13637 | 1.50 × 107 | 25 | |
| Isoform 1 of Centrosomal protein of 170 kDa | CEP170 | Q5SW79 | 1.20 × 107 | 33 | |
| Isoform 1 of Rho guanine nucleotide exchange factor 2 | ARHGEF2 | Q92974 | 1.20 × 107 | 24 | |
| Structural maintenance of chromosomes protein 1A | SMC1A | Q14683 | 1.20 × 107 | 18 | |
| Isoform 1 of Microtubule-associated protein 2 | MAP2 | P11137 | 1.10 × 107 | 17 | |
| Zinc finger protein 622 | ZNF622 | Q969S3 | 6600000 | 32 | |
| Isoform 1 of Cyclin-dependent kinase 13 | CDK13 | Q14004 | 6300000 | 17 | |
| Isoform 2 of Nuclear mitotic apparatus protein 1 | NUMA1 | Q14980 | 5600000 | 23 | |
| Structural maintenance of chromosomes protein 3 | SMC3 | Q9UQE7 | 5600000 | 24 | |
| Isoform 1 of Dedicator of cytokinesis protein 4 | DOCK4 | Q8N1I0 | 5400000 | 11 | |
| Isoform 1 of MAP7 domain-containing protein 1 | MAP7D1 | Q3KQU3 | 5000000 | 17 | |
| Condensin complex subunit 1 | NCAPD2 | Q15021 | 4700000 | 28 | |
| Condensin-2 complex subunit D3 | NCAPD3 | P42695 | 3300000 | 31 | |
| Serine/threonine-protein kinase PLK1 | PLK1 | P53350 | 3000000 | 13 | |
| Kinesin-like protein KIF20A | KIF20A | O95235 | 2900000 | 9 | |
| Isoform 1 of Cyclin-dependent kinase 12 | CDK12 | Q9NYV4 | 2700000 | 92 | |
| Isoform 1 of DNA repair protein RAD50 | RAD50 | Q92878 | 2100000 | 10 | |
| Isoform B of AP-2 complex subunit alpha-1 | AP2A1 | O95782 | 2000000 | 15 | |
| cDNA FLJ54776, highly similar to Cell division control protein 42 homologue | CDC42 | P60953 | 1700000 | 17 | |
| Condensin complex subunit 3 | NCAPG | Q9BPX3 | 830000 | 6 | |
| 40S ribosomal protein S4, X isoform | RPS4X | P62701 | 1,9 | 115 | 236 |
| 40S ribosomal protein S6 | RPS6 | P62753 | 1,4 | 71 | 149 |
| Tu translation elongation factor, mitochondrial precursor | TUFM | P49411 | 1,1 | 54 | 114 |
| Similar to Elongation factor 1-alpha 1 | EEF1A1 | P68104 | 0,8 | 44 | 63 |
The proteins that co-immunoprecipitated with NEK7-FLAG but not FLAG-control and that were not deposited in the CRAPome or the proteins that overlapped with Y2H are listed.
The overlapping proteins found by mass spectrometry (this table) and by yeast two-hybrid (Table 1) are underlined.
Fold change, as determined by the Scaffold program;25 TIC = total ion count.
Figure 1.
Interaction network of human NEK7 protein partners identified by IP-LC–MS/MS and Y2H system screens. Two circles represent the interactions retrieved by both methods. Y2H interactors also retrieved by IP-MS/MS are depicted in the intersection between the two circles. Thicker edges correspond to direct interactions retrieved by the Y2H screens. The proteins’ color code refers to their biological function given by the top enriched Gene Ontology biological processes (p ≤ 0.05). The proteins included in these interaction analyses were selected from the IP-LC–MS/MS by the following criteria: exclusive peptides (no peptides in replicated FLAG-control), overlapping proteins with Y2H and proteins not deposited in the Contaminant Repository for Affinity Purification (CRAPome). NEK9 (also identified by the Y2H screening) and NEK6 were already described to interact with NEK7.7,39 The protein–protein interaction network was built using the Integrated Interactome System (IIS) platform26 and visualized using the Cytoscape software.27
Human NEK7 Binds to CC2D1A, TUBB, MNAT1, NEK9, ANKS1B, SMC1, SMC3, and PLK1 and Phosphorylates CC2D1A, TUBB2B, MNAT1, and NEK9 Proteins in Vitro
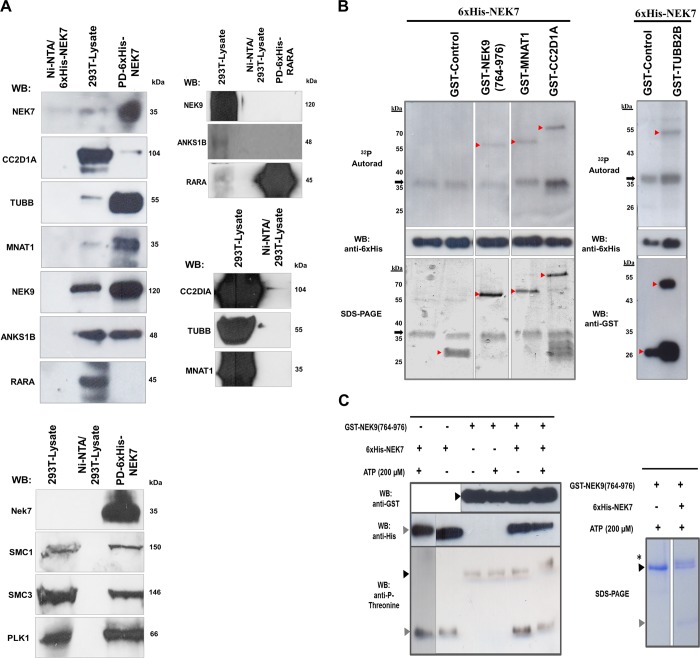
To confirm the specificity of NEK7 novel interacting proteins and to test if they could also behave as substrates, some proteins of diverse functions recovered in the Y2H system direct interaction screens were selected, and additional assays of pull-down and in vitro phosphorylation were carried out. For pull-down assays, HEK293T cell extracts were incubated with various 6×His-NEK7 or 6×His-RARA constructs immobilized on Ni-NTA agarose beads, and the bound protein complexes were subjected to Western blotting using specific antibodies. Instead of using cell lines related to the tissues used to construct the cDNA libraries of the Y2H screens, we used HEK293T cells since the NEK7 interactions may vary according to cell type and in the course of development and differentiation. The results revealed that NEK7 could interact with all proteins tested (CC2D1A, TUBB, MNAT1, NEK9, ANKS1B, SMC1, SMC3, and PLK1) but not with RARA or Ni-NTA agarose resin (Figure 2A). Moreover, 6×His-RARA did not pull-down the NEK7 interactor ANKS1B and NEK9 (Figure 2A, right panel), and all interactors showed no interaction with Ni-NTA agarose resin, thereby showing the effectiveness of the assay. Specifically, we confirmed that TUBB, MNAT1, NEK9, and ANKS1B co-precipitated very strongly with 6×His-NEK7 whereas CC2D1A co-precipitated weakly (Figure 2A).
Figure 2.
In vitro interaction and phosphorylation assay of candidate interactors of human NEK7. (A) Western blotting (WB) analysis from pull-down (PD) of recombinant NEK7 binding to HEK293T (293T) endogenous CC2D1A, TUBB (β-tubulin), MNAT1, NEK9, ANKS1B, SMC1, SMC3, and PLK1 and not to the RARA or Ni-NTA agarose resin (Ni-NTA). ANKS1B and NEK9 do not bind to 6×His-RARA (right panel), and all proteins show no interaction with Ni-NTA agarose resin (Ni-NTA), therefore demonstrating the assay specificity. Molecular weight (kDa) of the proteins is indicated. The pull-down assay results are based on two independent experiments, and phosphorylation assay results are based on three independent experiments. (B) Human Nek7 in vitro autophosphorylation and phosphorylation of proteins NEK9, MNAT1, CC2D1A, and TUBB2B, retrieved in the yeast two-hybrid screens. The arrowheads indicate the positions of the GST-tagged proteins or GST-control, whereas the arrows indicate the position of the 6×His-NEK7 detected in the autoradiography (32P Autorad), WB, or SDS-PAGE. Molecular weight (kDa) of the proteins is indicated. (C) Western blotting analyses shows the NEK7 requirement for the NEK9 majority phosphorylation. Recombinant NEK9 was incubated in the presence (+) or absence (−) of 6×His-NEK7 or 200 μM ATP. The black arrowheads indicate the positions of GST-NEK9(764–976), gray arrowheads indicate the positions of 6×His-NEK7 in the WB, and the asterisk points to the recombinant NEK9 upper mobility bands (∼51 kDa) in the SDS-PAGE (right panel).
For the proposed in vitro kinase assays, 6×His-NEK7 was incubated with kinase buffer supplemented with ATP and [γ-32P]ATP in the presence or absence of their possible substrates, NEK9, MNAT1, CC2D1A, and TUBB2B, in fusion with GST or GST-control. The 6×His-NEK7 was able to phosphorylate recombinant NEK9 at ∼50 kDa, MNAT1 at ∼62 kDa, CC2D1A at ∼76 kDa, and TUBB2B at ∼50 kDa. Furthermore, 6×His-NEK7 could autophosphorylate in the form of a monomer (∼36 kDa) (Figure 2B, 32P Autorad). No activity of 6×His-NEK7 was detected on GST-control (Figure 2B, 32P Autorad, GST-control lane), thus allowing the conclusion that the phosphorylation was specific for the tested substrates.
In addition, to further assess the NEK9 phosphorylation by NEK7, we performed an in vitro phosphorylation assay using chemiluminescence in which recombinant GST-NEK9(764–976) was incubated with or without ATP, in the presence or absence of 6×His-NEK7. Interestingly, we observed that GST-NEK9(764–976) presented minimal phosphorylation when preincubated with or without ATP or with 6×His-NEK7 without ATP. Importantly, we detected phosphorylated GST-NEK9(764–976), using the anti-phosphothreonine antibody and GST-NEK9(764–976) as a double band detected by SDS-PAGE in the presence of 6×His-NEK7 and ATP (Figure 2C, anti-P-threonine and SDS-PAGE, respectively). Therefore, these results suggest a likely requirement of NEK7 for NEK9 phosphorylation and point to a possible NEK7 positioning as upstream NEK9 regulator, in a regulatory feedback mechanism.7
Subcellular Localization of Human NEK7 and Its Interacting Proteins Throughout the Cell Cycle
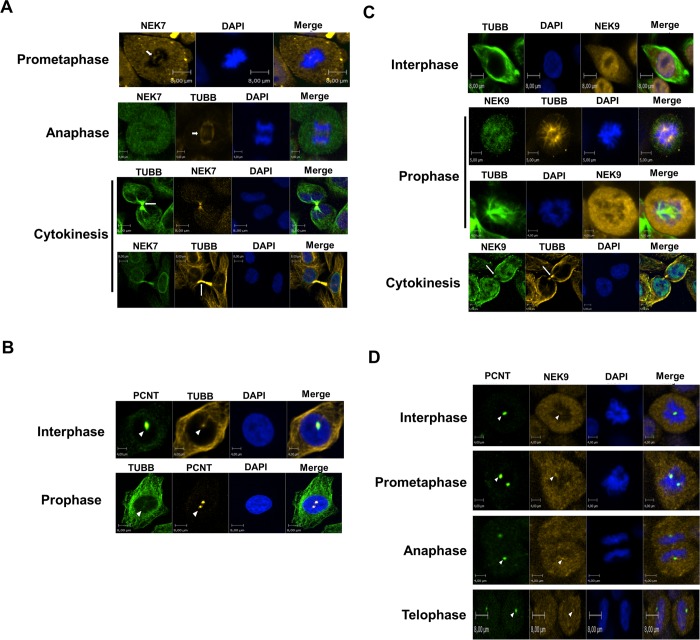
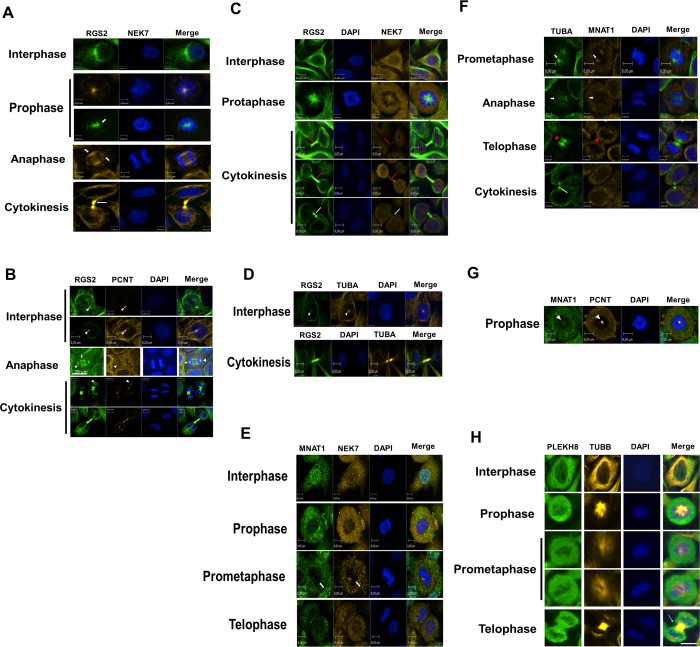
Immunocytochemistry staining of endogenous NEK7 and α-tubulin in cells after depolymerization of the microtubule demonstrated a centrosomal localization of NEK7, independent of the microtubule network and mitotic spindle integrity14,15 pointing to a NEK7 centrosomal function. To further explore this and better understand the functional correlation of NEK7 and its interacting proteins identified in the Y2H, we examined by confocal immunofluorescence microscopy the endogenous subcellular localization of these proteins with microtubule proteins and with the known centrosome component pericentrin (PCNT),33 throughout the different stages of the cell cycle.
We verified that NEK7 and TUBB localized at the spindle midzone in anaphase and at the cytoplasmic bridge in cytokinesis (Figure 3A). TUBB mostly localized with PCNT in interphase and prophase (Figure 3B) and with NEK9 at the cytoplasmic bridge in cytokinesis (Figure 3C), while NEK9 localized with PCNT in prophase, prometaphase, anaphase, and telophase (Figure 3D). RGS2 was notably localized at the perinuclear region in interphase (presumably the microtubule-organizing center (MTOC)), at nucleating microtubules in prophase, at the spindle midzone and asters in anaphase, and at the cytoplasmic bridge during cytokinesis (Figure 4A and B). In addition, RGS2 showed staining with PCNT during interphase, anaphase, and cytokinesis (Figure 4B). We observed evident staining of RGS2 with NEK7 in prophase (Figure 4C). During cytokinesis, a staining of RGS2 with NEK7 (Figure 4B) and with α-tubulin (TUBA) (Figure 4D) was specifically observed at the cytoplasmic bridge, probably midbody. These observations indicate a probable involvement of RGS2 and NEK7 in key events during cell cycle, especially in mitosis and cytokinesis. MNAT1 localized with NEK7 and TUBA at the spindle pole during prometaphase (Figure 4E and F) and at the spindle midzone in anaphase but decreased at the site of cleavage furrow ingression in telophase and disappeared in the abscission point in cytokinesis (Figure 4F). In addition, we observed a staining of MNAT1 with PCNT in prophase (Figure 4G). We verified that NEK7 was distributed with PLEKHA8 in a diffuse manner in the cytoplasm until prometaphase but was accumulated presumably at the contractile ring with TUBB during telophase (Figure 4H). Taken together, these data denote a potential involvement of NEK7 and its interacting proteins in molecular and functional mechanisms throughout the cell cycle, especially in mitosis and cytokinesis. Table S3, Supporting Information, summarizes the localization of NEK7 and its interacting proteins in the different subcellular components throughout the cell cycle.
Figure 3.
Subcelullar localization in HeLa cells of NEK7, NEK9, TUBB, and PCNT (Pericentrin) throughout the cell cycle. Endogenous proteins were detected with primary antibodies against indicated proteins and revealed with Alexa Fluor 488 or Alexa Fluor 546 (Molecular Probes Inc.) secondary antibodies. The images were visualized by confocal fluorescence microscopy using an Operetta High Content Imaging System (PerkinElmer). PCNT was used to stain the centrosome. The nucleus was stained by Hoechst. The images were edited using Volocity Demo version 6.1.1 software (PerkinElmer). The images represent analyses of at least 25 cells in each cell cycle phase from three independent experiments, and all cells showed the localization pattern represented in the images. Magnitude: 40X. The scale bars are indicated. Short arrows denote spindle pole, long arrows indicate cytoplasmic bridge, and arrowheads denote centrosome staining.
Figure 4.
Subcelullar localization in HeLa cells of NEK7, RGS2, MNAT1, PLEKH8, TUBA (α-tubulin), and PCNT (Pericentrin) throughout the cell cycle. Endogenous proteins were detected with primary antibodies (Santa Cruz Biotechnology or Abcam), revealed with Alexa Fluor 488 or Alexa Fluor 546 (Molecular Probes Inc.) secondary antibodies, and visualized by confocal fluorescence microscopy using an Operetta High Content Imaging System (PerkinElmer). PCNT (Pericentrin) was used to stain the centrosome and TUBA (α-tubulin) to stain the microtubules and/or cytoskeleton. The nucleus was stained by Hoechst. The images were edited using Volocity Demo version 6.1.1 software (PerkinElmer). The specific cell cycle phases and scale bars are indicated. At least 25 cells were analyzed in each cell cycle phase from three independent experiments, and all cells showed the localization pattern represented in the images. Magnitude: 60X in panels D and G; 40X in the other images. Short arrows denote spindle midzone and pole, white long arrows denote cytoplasmic bridge, red long arrows indicate possible midbody, white arrowheads denote centrosome, and red arrowheads denote contractile ring localization.
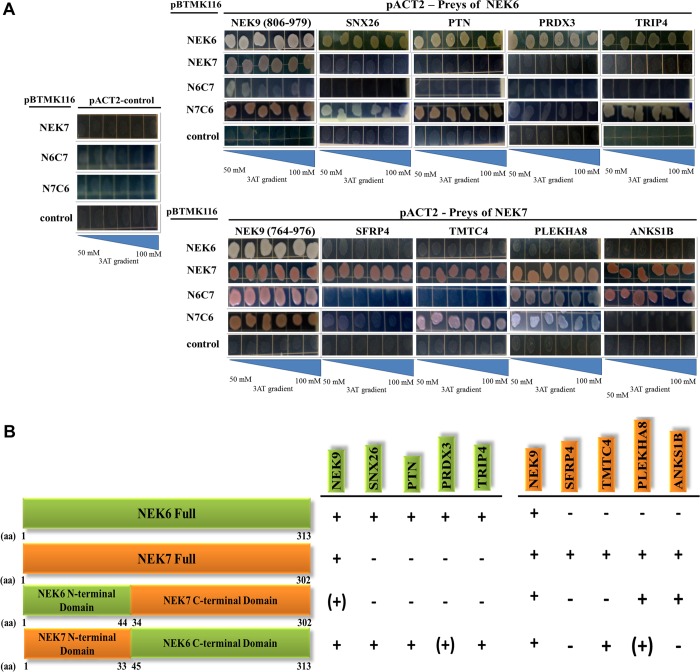
Differential Human NEK7 and NEK6 Interaction Profiles and the Role of N- and C-Terminal Domains in Protein Recognition
In the search for a better understanding of the structural underpinnings that determine the regulation and catalysis, as well as in the possible independent roles of human NEK6 and NEK7, we compared the interaction profile of five NEK6 interacting proteins (NEK9, SNX26, TRIP4, PTN, and PRDX3), described by Meirelles et al.22 with five NEK7 interacting proteins (NEK9, SFRP4, TMTC4, PLEKHA8, and ANKS1B) in a 3-AT gradient growth selection assay using the yeast two-hybrid system. Interaction profile comparison revealed that aside from NEK9, which is common to both NEK6 and NEK7, all other interactors were specific for each NEK (Figure 5A), indicating that NEK6 and NEK7 do not share common interactors, with the exception of NEK9, thereby pointing to independent and nonredundant cellular functions for NEK6 and NEK7.
Figure 5.
Comparison of the interaction profile of human NEK6 and NEK7 and the chimeric constructs N6C7 and N7C6 using proteins identified by the yeast two-hybrid system. (A) The upper panel shows the interaction profile of NEK6, NEK7, N6C7, and N7C6 with NEK6 interacting proteins SNX26 (Sorting nexin 26), TRIP4 (Thyroid hormone receptor interactor 4), PTN (Pleiotrophin isoform CRAc), PRDX3 (Peroxiredoxin 3), and NEK9, as described by Meirelles et al.22 The lower panel shows the interaction profile of NEK6, NEK7, N6C7, and N7C6 with NEK7 interacting proteins SFRP4, TMTC4, PLEKHA8, ANKS1B, and NEK9 obtained by our yeast two-hybrid screen. The tests were performed in triplicate using growth selection by 3-amino-1,2,4-triazole (3-AT) gradient in minimal medium without tryptophan, leucine, and histidine. (B) A schematic representation of relative positions of NEK6 and NEK7 domains (as described by O’ Regan et al.12 and Meirelles et al.19) and N6C7 and N7C6 chimeric constructs is shown. The results of the interaction profiles are indicated as follows: + = strong growth; (+) = reduced growth; – = no growth. aa: amino acid residues.
Previously, our group reported that the specific interactions and activity of human NEK6 and NEK7 could be regulated by their disordered N-terminal domain.19 However, the participation of the N- and C-terminal domains of NEK6 and NEK7 in regulation and catalysis is still unclear. This prompted us to test the interaction capacity of chimeric constructs consisting of NEK6 N-terminus/NEK7 C-terminus (N6C7) and NEK7 N-terminus/NEK6 C-terminus (N7C6) (Figure 5B) with common NEK9 and four specific human NEK6 and NEK7 interacting proteins, each. As shown in Figure 5A (upper panel), the N7C6 chimera was able to interact strongly with three (SNX26, PTN, and TRIP4) and weakly with one (PRDX3) of the NEK6 specific interacting proteins and, in addition, with two (TMTC4 and PLEKHA8) NEK7 specific interacting proteins (Figure 5A, lower panel). These findings demonstrate that the C-terminal catalytic domain of NEK6 may have an important role in recognition of the specific NEK6 interacting proteins. Furthermore, the presence of the NEK7 N-terminal could transplant some capacity to the chimera to interact with two specific NEK7 partners, suggesting that for NEK7 both N- and C-terminal domains have important contributions in the interacting proteins recognition.
Conversely, the N6C7 chimera did not interact with any of the four NEK6 specific interactors (Figure 5A, upper panel) and showed no interaction with SFRP4 and TMTC4 out of four NEK7 specific interactors (Figure 5A, lower panel). These results may suggest that in contrast to the NEK7 N-terminal, the NEK6 N-terminal cannot graft interacting capacity with NEK6 specific interactors to the NEK7 kinase domain. Accordingly, NEK7 may depend more on the N-terminal region, which is only ∼20% conserved with NEK6, and less on the conserved C-terminal catalytic region to promote its interactions. On the other hand, NEK6 N-terminal extension seems to be essential to promote its interactions but depends mostly on the ∼20% region that is similar to NEK7 and seems to depend also on its C-terminal, particularly on its exclusive region. Together, our results suggest that NEK6 and NEK7 display distinct interaction mechanisms in which the NEK7 N-terminus and the NEK6 C-terminus seem to play specific roles in contributing to binding specificity of each kinase.
Noteworthy, the NEK6 interacting partner NEK9 (amino acid residues 806–979)22 presented a reduced interaction with the N6C7 chimera (Figure 5A, upper panel), while NEK7 interacting partner NEK9 (amino acid residues 764–976) presented similar interaction affinity with both chimeras (Figure 5A, lower panel). These data further suggest that the NEK9 amino acid residues 764–806 comprise the most important region for ligation affinity with both NEK6 and NEK7.
N- and C-Terminal Domains of Human NEK6 and NEK7 Are Important for Phosphorylation
On the basis of our interaction analyses results, we proposed to investigate the N- and C-terminal structural role of NEK6 and NEK7 for phosphorylation. For this purpose, we compared the relative activities of 6×His-NEK6, 6×His-NEK7, and mutants constructs 6×His-NEK6Δ(1–33), 6×His-NEK7Δ(1–44), 6×His-N6C7, and 6×His-N7C6 on some NEK7 interacting proteins confirmed by pull-down and NEK6 interacting proteins described by Meirelles et al.,22 all in fusion with GST, using an in vitro phosphorylation assay. We verified that NEK7 phosphorylated its interacting proteins NEK9, MNAT1, and CC2D1A, but not RGS2 and TUBB2B (Figure 6A). Surprisingly, the N6C7 chimera was able to phosphorylate NEK7 interactors NEK9, MNAT1, CC2D1A, and importantly TUBB2B (Figure 6B) and even some of the NEK6 interactors SNX26 and TRIP4 (Figure 6D). These findings are in agreement with our observations that the chimera N6C7 can still interact with two of the specific NEK7 substrates in the Y2H (Figure 5A, lower panel), and in the case of SNX26 and TRIP4, we reasoned that the interaction may be transient or weak in vivo, but that the phosphorylation still can occur.
As opposed to NEK7, NEK6 could phosphorylate all of its specific interacting proteins (Figure 6C), while the N7C6 chimera could not phosphorylate any protein, neither NEK7 nor NEK6 specific interactors (Figure 6B and D, respectively). Furthermore, the deletion of the N-terminus from both NEK7 and NEK6 completely ablated their catalytic activity toward any of the NEK6 and NEK7 specific substrates (Figure 6A and C, respectively). In addition, the N-terminal domain of NEK6 could provide catalytic activity for NEK7 toward its specific substrates, indicating an important role for the N-terminus of NEK6 in catalysis. Together, these observations indicate that the structural preservation of the NEK6 and NEK7 N-terminal domain is absolutely necessary for an effective catalytic activity.
Role of the N-Terminal Domain of Human NEK7 and NEK6 in Phosphorylation
A previous study demonstrated that recombinant NEK6 and NEK7 phosphorylate p70 S6 kinase at Thr412 and other sites and activate it in vitro and in vivo.34 Here, we examined the relative kinase activity of full-length NEK6 and NEK7 and mutants NEK7Δ(1–44), NEK6Δ(1–33), N6C7 and N7C6 in fusion with 6×His tag, on the substrate ULight-p70S6K peptide (LGFTYVAP). The 6×His-NEK7Δ(1–44) protein exhibited a reduced catalytic activity compared with 6×His-NEK6Δ(1–33) (Figure 7), pointing once more to the importance of the N-terminal domain in the catalytic activity of NEK6 and NEK7, having a particularly higher impact on NEK7 catalytic regulation.
Figure 7.
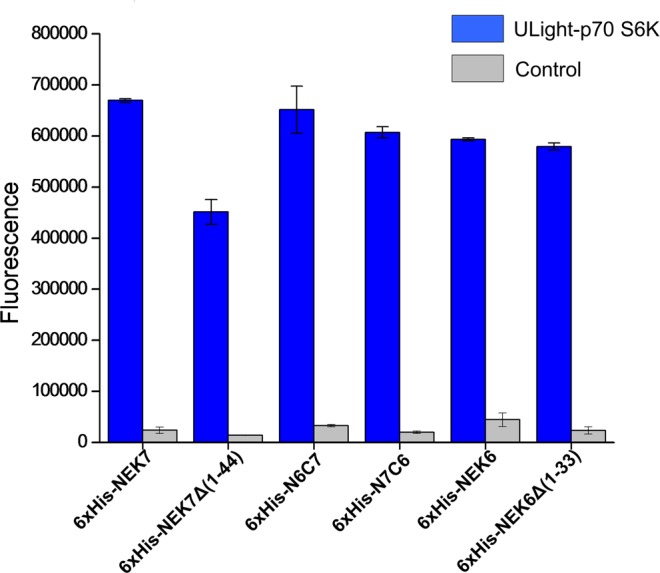
In vitro kinase activity assay of full-length NEK7 (6×His-NEK7), NEK7 kinase domain (6×His-NEK7Δ(1–44)), chimeras N6C7 (6×His-N6C7) and N7C6 (6×His-N7C6), full-length NEK6 (6×His-NEK6), and NEK6 kinase domain (NEK6Δ(1–33)). The enzymes were incubated at a concentration of 80 nM with 50 nM ULight-p70 S6K Peptide, and the phosphorylation was scored in the presence of 100 μM ATP. Kinase reactions were terminated after 60 min by the addition of EDTA. A reaction without addition of ULight-p70 S6K Peptide was used as a control (Control). Data represent the mean and standard deviation of three independent experiments. The signal was measured on a 2104 EnVision Multilabel Microplate Reader. Excitation wavelength was set to 320 nm, and fluorescence emission was recorded at 665 nm.
Discussion
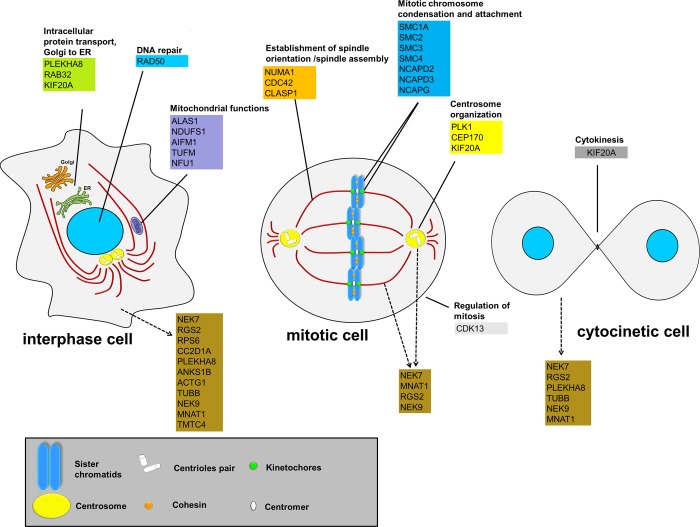
This study represents the first global functional proteomics approach to identify the proteins that interact with human NEK7. Our combined studies that employed Y2H and IP-LC–MS/MS revealed 61 potential NEK7 interacting proteins involved in different biological processes and cellular components, many of them not previously known to associate with NEK7. As shown in Figure 1 and Table S1, Supporting Information, these proteins largely fall into unsuspected processes not previously described for NEK7, such as RNA processing, DNA repair, mitochondrial regulation, and intracellular protein transport. On the other hand, we identified overlapping classes, on the basis of the cellular processes in which NEK7 is already described to be involved, such as cell division and mitotic cell cycle. In this context, we highlight important proteins belonging to different subprocesses from manual literature curation in combination with GO term analysis, such as establishment of spindle orientation (NUMA1 and CLASP1), mitotic chromosome condensation (different components of the human condensing and cohesin complexes), regulation of mitosis (CDK13), centrosome organization (PLK1 and CEP170), and cytokinesis (KIF20A) (Figure 8 and Table S1, Supporting Information), pointing to an extensive NEK7 regulatory role in the cell as well as on novel and multiple important components of the cell cycle.
Figure 8.
Schematic representation of human NEK7 interactors throughout the cell cycle. The scheme depicts the components of NEK7 and its interactors throughout the cell cycle according to biological processes (filled line) and subcellular localization (dashed arrows). The subprocesses were manually classified according to the literature in combination with GO term analysis. See detailed legend for symbols at the bottom of the figure. ER: endoplasmic reticulum.
This notion is supported by the fact that we were able to show that several of the NEK7 interacting proteins serve indeed as its in vitro substrates, including TUBB2B, MNAT1, RGS2 (not shown here), CC2D1A, and NEK9, and by the observation that several of these interacting proteins, including NEK9, TUBB, MNAT1, RGS2, and PLEKH8, localized along NEK7 in specific subcellular regions through the cell cycle (Figures 3, 4, and 8 and Table S3, Supporting Information). The major specific sites of localization include the spindle and spindle pole during mitosis, the contractile ring during telophase, and the cytoplasmic bridge (presumably midbody) during cytokinesis (Figures 3 and 4 and Table S3, Supporting Information). In this scenario, the important potential NEK7 targets such as TUBB2B, PLK1, SMC1A, and SMC3 were validated for interaction with NEK7 by in vitro pull-down assay (Figure 2A). It is relevant to emphasize that PLK1 was previously demonstrated to control the mitotic centrosome separation through phosphorylation/activation of NEK9 and NEK6/7-dependent sequential phosphorylation of kinesin KIF11.35 Furthermore, the potential NEK7 interactor and substrate, TUBB2B, is an isoform of β-tubulin class II that promotes the regulation of microtubule dynamics in human cells when phosphorylated,36,37 indicating a direct NEK7 role in regulating microtubules themselves. Together, these data support the idea that NEK7 may potentially act as a multifunctional kinase that regulates proteins categorized into distinct or shared biological processes, specifically in the cell division signaling.
The evolutionarily conserved NEK6 and NEK7 are the result of a duplication event during early chordate evolution, and their high similarity to each other could suggest highly equivalent functions.16 Nevertheless, structural and functional studies have indicated possible differential functions for these kinases.13,17,18,20,21,35 The defining feature of NEKs is a conserved kinase domain in the N-terminal region and a C-terminal region highly divergent in composition and length but that contains all the motifs that are typical of a regulatory domain such as coiled coil motifs.38 NEK6 and NEK7 break this rule having a nonconserved short N-terminal extension followed immediately by a C-terminal domain that is the kinase domain itself. Studies performed by our group suggest that the interactions of human NEK6 and NEK7 with their specific/different partners could be dependently regulated by their distinct and disordered N-terminal domains.19 Then, it is plausible that the N- and C-terminal domains of NEK6 and NEK7 likely act in concert to recognize specific interactors/substrates, via distinct mechanisms.
To investigate the role of N- and C-terminal domains of NEK6 and NEK7 in specific protein recognition and phosphorylation as well as to differentiate their functions, we developed chimeric constructs of NEK6 and NEK7 and conducted an interaction comparative experiment by Y2H. We observed that, aside from NEK9, all other proteins interacted specifically with each NEK, providing evidence that NEK6 and NEK7 could play independent and nonredundant roles in the cell. In this respect, we suggest that NEK7 could physically interact with various proteins within disparate cellular microenvironments to regulate cellular functions differently from NEK6. We also demonstrate that N7C6 chimera presented the broad binding capacity with both NEK6 and NEK7 interactors, indicating a role of NEK6 C-terminal catalytic domain and NEK7 N-terminal in protein recognition, in light of the fact that deletion of the N-terminus from NEK6 had abrogated all interactions with its interactors.22 These studies delineate NEK6 and NEK7 N-terminal domains as important regulatory structural components, corroborating previous data,18,19 and highlight a surprising role for their C-terminal catalytic domain in binding affinity and/or specificity. In addition, the evidence that NEK6 and NEK7 N- and C-terminal domains present different binding properties indicates distinct mechanisms for NEK6 and NEK7 interaction and regulation. The relatively distinct N-terminal structures of NEK6 and NEK7 flanking the kinase domains may cause the majority of the differential response.
Regarding phosphorylation, our results demonstrate a higher activity of NEK6 on its specific substrates compared to NEK7. Importantly, N6C7 chimera showed a greater phosphorylation spectrum compared to wild-type NEK7, whereas N7C6 chimera together with NEK6Δ(1–33) and NEK7Δ(1–44) mutants did not exhibit catalytic activity on different proteins substrates. These observations suggest that the NEK6 N-terminal domain is important for its catalytic function and provides gain-of-function phenotypes for NEK7 kinase domain over nonspecific substrates. These findings are in line with our observations of the reduced catalytic activity on specific peptide substrates presented by NEK7Δ(1–44). In this regard, the catalytic domain alone could be intrinsically autoinhibited, regulated in a protein interaction-specific manner or conceivably through interactions with the regulatory N-terminal domain. Indeed, NEK7 structural studies showed that the N-terminal extension of NEK7 (residues 20–33) adopts a highly unusual structure sitting on the core catalytic domain,18 which may contribute to stabilizing the active conformation of the kinase. Therefore, we hypothesized that NEK6 and NEK7 N- and C-terminal domains can contribute to both regulation and catalysis, via distinct mechanisms. Structural and functional studies using truncated constructions and site-directed mutagenesis of NEK7 and NEK6 would further dissect the region(s) of their N- and C-terminal domains, involved in determining the regulation and catalysis mechanisms. This would help us to further understand the independent signaling functions of NEK6 and NEK7 in mammalian cells.
In summary, in the present study we have characterized a comprehensive protein interactome for human NEK7 and identified 61 proteins in diverse sets of pathways and processes; this result, combined with functional in vitro and in vivo assays, reinforces NEK7 participation in the regulation of key components of cell division and reveals novel possible NEK7 interactors and functions. We showed that the closely related human kinases NEK6 and NEK7 do not share common interactors, with the exception of NEK9, and display distinct modes of protein interaction. In this regard, given the previously reported importance of NEK6 and NEK7 N-terminal domains in protein regulation, we provided evidence that, via distinct mechanisms for NEK6 and NEK7, the N- and C-terminal domains are both important structural components for regulation and catalysis of these kinases. Our data suggest distinct, independent, and nonredundant signaling functions for NEK6 and NEK7 in the cell and provide a novel conceptual framework for future investigation of the structural and biochemical basis of a distinct NEK6 and NEK7 regulation and catalysis.
Acknowledgments
The authors thank Maria Eugênia Camargo for technical assistance. We thank Dr. Artur T. Cordeiro and Gustavo F. Mercaldi for assistance with the assays performed using the EnVision Multilabel Microplate Reader, Tereza C. L and Silva for assistance with the Operetta High Content Imaging System (PerkinElmer) and Mariana Bertini Teixeira by kindly providing the 6×His-RARA protein. This work was supported by Fundação de Amparo à Pesquisa do Estado São Paulo (FAPESP), Conselho Nacional de Pesquisa e Desenvolvimento (CNPq) and Centro Nacional de Pesquisa em Energia e Materiais (CNPEM). We also acknowledge the U.S. National Institutes of Health contract HHSN268201000031C from NHLBI and grant P41 GM104603 from NIGMS
Supporting Information Available
Tables of node attributes from NEK7 first neighbors network, in silico analysis of protein interaction with NEK7 retrieved by the yeast two-hybrid system screens, and proposed cellular localization and colocalization of NEK7 with interacting partners throughout the cell cycle. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ma H. T.; Poon R. Y. How protein kinases co-ordinate mitosis in animal cells. Biochem. J. 2011, 435, 17–31. [DOI] [PubMed] [Google Scholar]
- Fry A. M.; Meraldi P.; Nigg E. A. A centrosomal function for the human NEK2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998, 17, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz L.; Dutt P.; Haider N.; Stambolic V. NEK family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson C. A.; Haq T.; Yeoh S.; Fry A. M.; Bayliss R. The structural mechanisms that underpin mitotic kinase activation. Biochem. Soc. Trans. 2013, 41, 1037–1041. [DOI] [PubMed] [Google Scholar]
- Morris N. R. Mitotic mutants of Aspergillus nidulans. Genet. Res. 1975, 26, 237–254. [DOI] [PubMed] [Google Scholar]
- Oakley B. R.; Morris N. R. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 1983, 96, 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belham C.; Roig J.; Caldwell J. A.; Aoyama Y.; Kemp B. E.; Comb M.; Avruch J. A mitotic cascade of NIMA family kinases. Nercc1/NEK9 activates the NEK6 and NEK7 kinases. J. Biol. Chem. 2003, 278, 34897–34909. [DOI] [PubMed] [Google Scholar]
- Li J. J.; Li S. A. Mitotic kinases: the key to duplication, segregation, and cytokinesis errors, chromosomal instability, and oncogenesis. Pharmacol Ther. 2006, 111, 974–984. [DOI] [PubMed] [Google Scholar]
- Upadhya P.; Birkenmeier E. H.; Birkenmeier C. S.; Barker J. E. Mutations in a NIMA-related kinase gene, NEK1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L. M.; Mahjoub M. R. Caught NEK-ing: cilia and centrioles. J. Cell Sci. 2005, 118, 5161–5169. [DOI] [PubMed] [Google Scholar]
- Kim S.; Lee K.; Rhee K. NEK7 is a centrosomal kinase critical for microtubule nucleation. Biochem. Biophys. Res. Commun. 2007, 360, 56–62. [DOI] [PubMed] [Google Scholar]
- O’Regan L.; Blot J.; Fry A. M. Mitotic regulation by NIMA-related kinases. Cell Div. 2007, 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L.; Fry A. M. The NEK6 and NEK7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 2009, 29, 3975–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yissachar N.; Salem H.; Tennenbaum T.; Motro B. NEK7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 2006, 580, 6489–6495. [DOI] [PubMed] [Google Scholar]
- Kim S.; Kim S.; Rhee K. NEK7 is essential for centriole duplication and centrosomal accumulation of pericentriolar material proteins in interphase cells. J. Cell Sci. 2011, 124, 3760–3770. [DOI] [PubMed] [Google Scholar]
- Salem H.; Rachmin I.; Yissachar N.; Cohen S.; Amiel A.; Haffner R.; Lavi L.; Motro B. NEK7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene 2010, 29, 4046–4057. [DOI] [PubMed] [Google Scholar]
- Kandli M.; Feige E.; Chen A.; Kilfin G.; Motro B. Isolation and characterization of two evolutionarily conserved murin kinases (NEK6 and NEK7) related to the fungal mitotic regulator, NIMA. Genomics 2000, 68, 187–196. [DOI] [PubMed] [Google Scholar]
- Richards M. W.; O’Regan L.; Mas-Droux C.; Blot J. M.; Cheung J.; Hoelder S.; Fry A. M.; Bayliss R. An autoinhibitory tyrosine motif in the cell-cycle-regulated NEK7 kinase is released through binding of NEK9. Mol. Cell 2009, 36, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles G. V.; Silva J. C.; Mendonça Yde A.; Ramos C. H.; Torriani I. L.; Kobarg J. Human NEK6 is a monomeric mostly globular kinase with an unfolded short N-terminal domain. BMC Struct. Biol. 2011, 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige E.; Motro B. The related murine kinases, NEK6 and NEK7, display distinct patterns of expression. Mech. Dev. 2002, 110, 219–223. [DOI] [PubMed] [Google Scholar]
- Minoguchi S.; Minoguchi M.; Yoshimura A. Differential control of the NIMA related kinases, NEK6 and NEK7, by serum stimulation. Biochem. Biophys. Res. Commun. 2003, 301, 899–906. [DOI] [PubMed] [Google Scholar]
- Meirelles G. V.; Lanza D. C. F.; Silva J. C.; Bernachi J. S.; Leme A. F. P.; Kobarg J. Characterization of NEK6 interactome reveals an important role for its short n-terminal domain and colocalization with proteins at the centrosome. J. Proteome Res. 2010, 9, 6298–6316. [DOI] [PubMed] [Google Scholar]
- Bartel P. L.; Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995, 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Devault A.; Martinez A. M.; Fesquet D.; Labbe J. C.; Morin N.; Tassano J. P.; Nigg E. A.; Digger J. C.; Doree M. MNAT1 (‘ménage à trois’) a new RING finger protein subunit stabilizing ciclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995, 14, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S. A.; He J.; Lu M.; Souda P.; Saxton R. E.; Faull K. F.; Whitelegge J. P.; Chang H. R. Mass spectrometry (LC–MS/MS) identified proteomic biosignatures of breast cancer in proximal fluid. J. Proteome Res. 2012, 11, 5034–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazzolle M. F.; de Carvalho L. M.; Slepicka H. H.; Vidal R. O.; Pereira G. A.; Kobarg J.; Meirelles G. V. IIS - Integrated Interactome System: A Web-based platform for the annotation, analysis and visualization of protein-metabolite-gene-drug interactions by integrating a variety of data sources and tools. PLoS One 2014, 9, e100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpili M. J.; Delben T. M.; Kobarg J. Identification of proteins that interact with the central coiled-Ccoil region of the human protein kinase NEK1. Biochemistry 2003, 42, 15369–15376. [DOI] [PubMed] [Google Scholar]
- Bartel P, Chien C, Sternglanz R, Fields S.. Cellular Interactions in Development: A Practical Approach; Hartley D.A., Ed.; Oxford University Press: Oxford, 1993; pp 153–179. [Google Scholar]
- Park S.; Lim B. B.; Perez-Terzic C.; Mer G.; Terzic A. Interaction of asymmetric ABCC9-encoded nucleotide binding domains determines KATP channel SUR2A catalytic activity. J. Proteome Res. 2008, 7, 1721–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde M. L.; Hazra T. K.; Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cell. Mol. Life Sci. 2010, 67, 3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T.; Kay L. E.; Forman-Kay J. D. Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 2010, 23, 105–116. [DOI] [PubMed] [Google Scholar]
- Doxsey S. J.; Stein P.; Evans L.; Calarco P. D.; Kirschner M. Pericentrin highly conserved centrosome protein involved in microtubule organization. Cell 1994, 76, 639–650. [DOI] [PubMed] [Google Scholar]
- Belham C.; Comb M. J.; Avruch J. Identification of the NIMA family kinases NEK6/7 as regulators of the p70 ribosomal S6 kinase. Curr. Biol. 2011, 11, 1155–1167. [DOI] [PubMed] [Google Scholar]
- Rapley J.; Nicolàs M.; Groen A.; Regué L.; Bertran M. T.; Caelles C.; Avruch J.; Roig J. The NIMA-family kinase NEK6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 2008, 121, 3912–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourest-Lieuvin A.; Peris L.; Gache V.; Garcia-Saez I.; Juillan-Binard C.; Lantez V.; Job D. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol. Biol. Cell 2006, 17, 1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Xi B.; Wu J.; An H.; Zhu J.; Abassi Y.; Feinstein S. C.; Gaylord M.; Geng B.; Yan H.; Fan W.; Sui M.; Wang X.; Xu X. Natural product derivative bis(4-fluorobenzyl) trisulfide inhibits tumor growth by modification of beta-tubulin at Cys 12 and suppression of microtubule dynamics. Mol. Cancer Ther. 2009, 8, 3318–3330. [DOI] [PubMed] [Google Scholar]
- Meirelles G. V.; Perez A. M.; Souza E. E.; Basei F. L.; Papa P. F.; Hanchuk T. D. M.; Cardoso V. B.; Kobarg J. “Stop Ne(c)king around”: How systems biology can help to characterize the functions of NEK family kinases from cell cycle regulation to DNA damage response. World J. Biol. Chem. 2014, 5, 141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing R. M.; Chu P.; Elisma F.; Li H.; Taylor P.; Climie S.; McBroom-Cerajewski L.; Robinson M. D.; O’Connor L.; Li M.; Taylor R.; Dharsee M.; Ho Y.; Heilbut A.; Moore L.; Zhang S.; Ornatsky O.; Bukhman Y. V.; Ethier M.; Sheng Y.; Vasilescu J.; Abu-Farha M.; Lambert J. P.; Duewel H. S.; Stewart I. I.; Kuehl B.; Hogue K.; Colwill K.; Gladwish K.; Muskat B.; Kinach R.; Adams S. L.; Moran M. F.; Morin G. B.; Topaloglou T.; Figeys D. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.