Abstract
The biomedical community is increasingly migrating toward research endeavors that are dependent on large quantities of genomic and clinical data. At the same time, various regulations require that such data be shared beyond the initial collecting organization (e.g., an academic medical center). It is of critical importance to ensure that when such data are shared, as well as managed, it is done so in a manner that upholds the privacy of the corresponding individuals and the overall security of the system. In general, organizations have attempted to achieve these goals through deidentification methods that remove explicitly, and potentially, identifying features (e.g., names, dates, and geocodes). However, a growing number of studies demonstrate that deidentified data can be reidentified to named individuals using simple automated methods. As an alternative, it was shown that biomedical data could be shared, managed, and analyzed through practical cryptographic protocols without revealing the contents of any particular record. Yet, such protocols required the inclusion of multiple third parties, which may not always be feasible in the context of trust or bandwidth constraints. Thus, in this paper, we introduce a framework that removes the need for multiple third parties by collocating services to store and to process sensitive biomedical data through the integration of cryptographic hardware. Within this framework, we define a secure protocol to process genomic data and perform a series of experiments to demonstrate that such an approach can be run in an efficient manner for typical biomedical investigations.
Keywords: Cryptographic hardware, cryptography, databases, genomics, privacy, security
I. Introduction
THE integration of clinical decision support tools and high-throughput technologies into the healthcare domain is evolving the practice of medicine from a one-size-fits-all model toward a personalized system [1]. To further this goal, biomedical scientists are conducting large-scale investigations between patients’ clinical status and molecular data, such as DNA sequences, to determine how variation in the latter influences patients’ susceptibility to disease, response to treatment, and potential to succumb to adverse events (e.g., drug–drug interactions) [2], [3]. The amount of data needed to conduct such studies is often beyond the capability of a sole institution [4] and emerging regulations, such as the U.S. National Institutes of Health genome wide association study (GWAS) policy, encourage biomedical scientists to share person-specific data for reuse [5]. In order to support such regulations, various technologies have been established to facilitate data sharing, such as the database of genotypes and phenotypes (dbGaP) in the U.S. [6] and the Welcomme Trust’s biobanking program in the U.K. [7]. Notably, these systems centralize data from disparate organizations. To ensure the continued growth of such tools, it is critical to ensure the security of such systems, as well as the privacy of the records they incorporate.
Traditionally, administrators attempted to achieve privacy by “deidentifying” data through the suppression of particular attributes, such as personal names, dates, or geocodes. However, deidentification does not guarantee protection from “reidentification” [8], [9] and may obscure certain types of information that are critical to longitudinal or epidemiologic studies. As an alternative, security frameworks have been devised to manage clinical genomics data, in its most specific form, in a centralized database [10]. In such frameworks, data holders transmit encrypted versions of clinical genomics records to a third party. Subsequently, the third party administrator could integrate data coming from different sources and then execute queries on behalf of a scientist (e.g., How many records contain a diagnosis of juvenile diabetes and a particular genomic variant?) without decrypting any record in the database. To achieve this goal, the administrator at the third party evaluates a query against each stored record securely, and the results are sent to a separate third party who is in control of the decryption keys and learns only the aggregation of the result (i.e., the number of satisfying records). The result is then revealed to the researcher. One advantage of such a framework is that there is no opportunity to decrypt individual records unless both third parties collude. Thus, if a hacker breaks into one of the third party’s computer systems, he cannot expose the sensitive information.
The framework just mentioned, however, has several drawbacks which limits its general applicability. First, to facilitate evaluation of researcher’s query against the database, a computationally expensive cryptographic technique needs to be executed. Second, it may not be practical to set up multiple disparate third parties due to economic, trust, or legal concerns. Third, even when the first two problems are surmountable, there remain potential bandwidth challenges because a nontrivial amount of encrypted data must be communicated between the parties involved in the protocol.
The overarching goal of this paper is to overcome the aforementioned limitations through the application of secure hardware. We design a framework that leverages tamper-resistant hardware to enable secure clinical genomics data storage and processing at a single third party. To the best of our knowledge, this is the first work to propose such a framework for biomedical data.
The specific contributions of this paper are as follows.
We illustrate that by using the capabilities of the cryptographic hardware, different organizations can contribute their data to centralized sites, where they can be joined and queried, in a secure manner.
We show that a small program can be executed on secure hardware attached to a third party’s server that hosts the clinical genomics data. In the process, we demonstrate that the program has sufficient power to facilitate common queries used in emerging biomedical investigations, such as count queries as well as join operations.
We show that our strategy can counter various attacks. In particular, even if the system is hacked, as long as the cryptographic hardware is not compromised, no information that is stored (whether it is genomic, clinical, or demographic) will be leaked.
We provide experimental studies to demonstrate feasibility with existing hardware for real world applications.
II. Related Work
In this paper, we propose a framework to securely store, share, and query clinical genomics data using secure cryptographic hardware. Here, we provide a high-level overview of current research that is related to our approach and application.
A. Hardware Perspective
Secure cryptographic devices have been used for many tasks, including private information retrieval [11], private record linkage [12], [13], and tamper-resistant intrusion detection [14]. Recently, trusted computing platform alliance chips have been used to protect outsourced private customer data [15]. To our knowledge, none of the previous work applies secure cryptographic hardware for managing or processing clinical genomics data, which has distinct issues in terms of scale and type.
In [16], Illiev and Smith propose a system for providing obliviousness for arbitrary large computations on secure coprocessors (SCPs) for any algorithm using circuit evaluation methods in the context of secure multi-party computation (SMC). In this paper, we provide much more efficient solutions for processing genomic data in untrusted platforms.
B. Software Perspective
The encrypted data storage problem has been studied in [17]–[22]. However, the focus of these investigations has been on the various approaches that can be applied to support encrypted data within relational databases. In addition, encrypted data storage methods have been applied to personal genomic data [23]. Yet, unlike the work presented in this paper, none of the existing schemes assumes that the queries are processed under an untrusted database server attack model where the attacker is assumed to have access to the content of the disk as well as the CPU and the memory. Instead, previous work focuses on an attack model where it is assumed that the attacker has access to the storage devices but not the memories and the CPU.
Querying encrypted data under the untrusted database server attack model were first suggested in [24], where the client’s attribute domains1 are partitioned into a set of intervals. The correspondence between intervals and the original values is kept at the client site and encrypted tables with interval information are stored in the database. Efficient querying of the data is made possible by mapping original values in range and equality queries to corresponding interval values. In subsequent work, Hore et al. [25] analyzed how to create the optimal intervals for minimum disclosure risk and maximum efficiency. In [26], the potential attacks for interval-based approaches were explored and models were developed to analyze the trade-off between efficiency and disclosure risk. The major drawback of the interval-based approaches is that the data processing capability of the server is quite limited. Since the server cannot see the data in plain text format, most of the records are sent to the client for further processing, which increases both the network cost and client-side processing cost.
In [27], Carbunar and Sion introduce a mechanism for executing general binary join operations in an outsourced relational database framework based on a bloom filter-based technique. Our study differs by having a broader framework, which handles various types of queries, including joins and counts.
An additional difference between our study and previous studies is that in our proposed framework, we assume that the users who submit their queries do not receive any information other than the result of the query. For example, if a researcher submits an aggregation query, the researcher learns only the number of records that satisfy the query. In the previous approaches, it is assumed that the client can decrypt and further process the records which would be considered a violation in our framework.
In [28], Aggarwal et al. suggested allowing the client to partition its data across two (and more generally, any number of) logically independent database systems that are prohibited from communicating with each other. The data are first partitioned in a fashion that ensures the exposure of the contents of any particular database does not lead to a violation of privacy. The client then executes queries by transmitting appropriate subqueries to each database, and then pieces together the results at the client. A major challenge in realizing this framework in the real world is that the data owners must assume that the third parties do not collude to reveal the private data. If an attacker has access to multiple machines, the data will be compromised. By contrast, in our framework, all biomedical records are stored in an untrusted central server without requiring any such assumptions.
C. Cryptographic Techniques
In our previous research, we introduced a cryptographic framework that can be used for secure clinical genomics data storage [10]. In this framework, the clinical genomics data records are first encrypted by the data owners (e.g., hospitals) and stored in a centralized repository. Using a specific type of public key encryption, the queries of biomedical researchers (e.g., count queries) are executed by the repository managers, without decrypting the clinical genomics data.2 Instead, only the aggregated result is decrypted.
This framework provides a cryptographically secure way of executing the queries. However, this protocol required the integration of two third parties to achieve such computations, which, as we alluded to in Section I, is not always practical. Additionally, as we elaborate on in Section VI, excessive processing cost of specialized encryption techniques can make this mechanism impractical for very large datasets. In this paper, we provide a much more efficient solution by leveraging the capabilities of secure cryptographic hardware.
III. Background
In this section, we briefly summarize the topics of encrypted data storage and cryptographic hardware.
A. Encrypted Data Storage in Relational Databases
A variety of defensive mechanisms have been designed to protect against disclosures due to hacking and system compromise in the context of centralized databanks. One popular measure that is applied to reduce the effect of various attacks, and to limit disclosure risks, is data encryption. Once the data is encrypted, and the encryption keys are kept secret, attackers that hack into a system are unable to interpret the encrypted sensitive data. Due to popular demand, many commercial database management system products in the market have built-in encryption support. For example, Microsoft SQL Server 2008 provides a mechanism called transparent data encryption (TDE), which provides protection for the entire database at rest without influencing existing applications [29].
In the TDE model, the encryption and decryption operations are performed in memory, so the cryptographic keys needed for such operations must be kept in memory as well. Unless special hardware is used, all of these keys are kept in the main memory while they are in use. The implicit threat model assumed by such database products is that the server is trusted and only the disk is vulnerable to compromise. In the event that the physical storage devices are stolen, this method would effectively prevent data misuse. On the other hand, this model is not strong enough to prevent data disclosure if the attacker has access to the main memory of the server. An attacker observing memory can easily capture the master keys that encrypt and decrypt the sensitive information stored on the disk. And once the keys are revealed, all sensitive data could be compromised.
To prevent the aforementioned attack, SQL Server provides an extensible key management (EKM) module [30]. EKM enables parts of the cryptographic keys to be managed by an external cryptographic hardware device. This external hardware is leveraged to maintain the secrecy of the master keys. Unfortunately, naïve execution of such cryptographic hardware could leave stored data vulnerable to compromise. For example, if the encryption keys for a database table are leaked to memory, they could be used to recover the contents of the table.
In the framework presented in this paper, we also store the data in an encrypted format. However, one major difference between the current framework and that in [10] is that we ensure the sensitive clinical genomics data will be decrypted only within the secure cryptographic hardware. This will prevent the leakage of sensitive data.
B. SCPs
The secure cryptographic hardware we use is already commercially available in the form of SCPs. An example of an SCP is the IBM 4764 cryptographic coprocessor [31]. This is a single-board computer consisting of a CPU, memory, and special-purpose cryptographic hardware contained in a tamper-resistant shell. The hardware is certified to level 4 under FIPS PUB 140-1 [31] When installed on a server, it is capable of performing local computations that are completely hidden from the server. If tampering is detected, then the SCP clears the internal memory. Despite such enhanced security, current SCPs are limited in memory size and computation power. For example, the SCP used in this study has only 64 MB and a 266 MHz PowerPC processor. Yet, GWAS is often executed over large quantities of data. For instance, a typical GWAS study computes the correlation between 600 000 (or more) single nucleotide polymorphisms (SNPs) and a phenotype for 10 000 (or more) individuals, which is approximately 1.5 GB of data. It is not possible to fit all of the data into the SCP’s memory at the same time. In the next section, we will explain how, given its limitations, such hardware can be leveraged to craft a secure solution for clinical genomics data processing by a single third party.
IV. Description of the Proposed Framework
For convenience, we assume that the set of data owners corresponds to a set of hospitals. In our framework, each hospital can verify that the server to which they plan to transfer the clinical genomics data contains an approved SCP, and provide the SCP with the data using the SCP’s public key. For efficiency purposes, the data will be encrypted using a symmetric key encryption scheme and the key used for the encryption will itself be encrypted with the public key of the SCP.3 The trusted coprocessor can now execute join operations and run various aggregate queries over encrypted clinical genomics data. Since only the SCP can recover the key needed for decrypting the data, no individual with access to the server can interpret the stored data.
Clearly, encrypted storage can prevent the disclosure of the data even if the server is hacked. Any attempt by the server to take control of, or tamper with, the coprocessor, either through software or physically, will clear the coprocessor. In doing so, the keys will be destroyed, which eliminates the ability to decrypt any information stored or transmitted.
To guarantee that the application running on the SCP is non-malicious software and was loaded by a trusted operating system (OS), we use the remote attestation mechanism provided by the SCP [33].
The following section provides further details related to our framework.
A. Architecture
The overview of the proposed architecture is illustrated in Fig. 1. Our framework incorporates a third party called the data storage server (DS). The encrypted records of the hospitals are stored in this server.
Fig. 1.
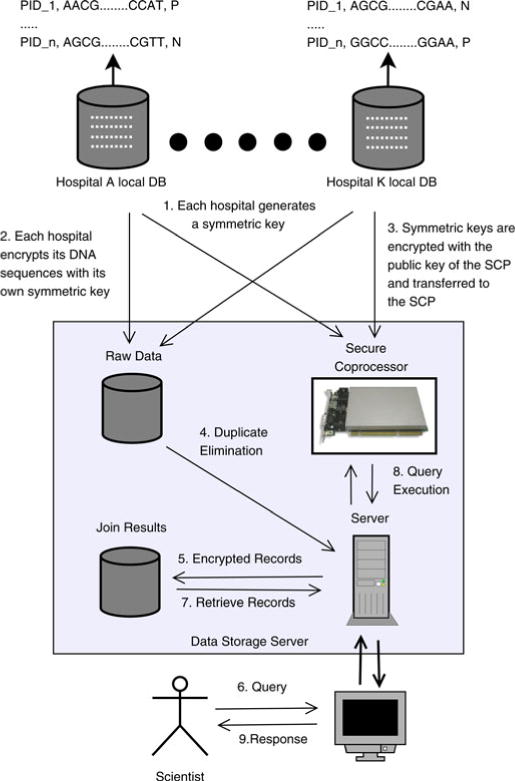
Proposed framework for management of biomedical data in third party cryptographic hardware.
We assume that the DS is untrusted.4 All of the interactions among the storage devices, the server, and the SCP are monitored by an adversary. To perform the data operations without disclosing any sensitive information, all of the data processing operations are performed within the SCP. To achieve such processing, each hospital first generates a symmetric encryption key and encrypts its records. This key is later transferred to the SCP located in the DS. The SCP provides a secure Ethernet channel through which the hospitals can communicate and transfer the key.
The information flow in this framework consists of two phases: data integration phase and query processing phase. In the data integration phase, the DS receives the patient records from the hospitals and eliminates the duplicates by executing the join operations on the SCP. In the query processing phase, the DS receives the queries of the scientists and executes these queries on the encrypted patient records with the help of the SCP.
For the data integration phase, we use the sovereign join algorithms introduced in [12]. Sovereign joins are applied to prevent adversaries from making inferences through the observation of the interactions between an SCP and the server to which it is attached. Agrawal et al. [12] formulated this problem and propose alternative techniques to prevent information leakage through patterns in I/O while maximizing the performance. We provide the implementation details of this protocol for our framework in Appendix A.
For the query processing phase, we propose a secure protocol that prevents adversaries from inferring information through interactions between the server and the SCP. We describe the details of this protocol in Section IV-C.
We describe a step-by-step walkthrough of the framework (see Fig. 1 for the corresponding protocol) in the following.
Step 1: Each hospital generates a symmetric key Ki to encrypt the data of their patients. Without loss of generality, we assume that the underlying data consist of structured clinical terms, DNA sequences, and demographics.
Step 2: The records of hospital Hi are encrypted with symmetric key Ki and transferred to DS.
Step 3: Each hospital Hi encrypts its private key Ki with the public key of the SCP and transfers the key to the SCP over the Ethernet channel of the SCP. An attacker at the server side cannot interpret the encrypted clinical genomics data because the keys are secured in tamper-resistant hardware.
Step 4: The server retrieves the records from the raw data disk for duplicate elimination.
Step 5: The server communicates with the SCP and performs the join operations on the encrypted records and eliminates duplicate records.
Step 6: The encrypted join results are written to a disk.
Step 7: Suppose that a researcher wants to learn how many records in the database contain a particular combination of SNPs when a patient is diagnosed with a certain disease. This can be represented in a logical query, such that , where j1, …, jk is an arbitrary subsequence of the data attributes. The server first receives the query.
Step 8: Next, the encrypted records are fetched from the storage device for query processing.
Step 9: The query is forwarded to the SCP along with the encrypted attributes. The SCP processes the query and returns the response to the server.
Step 10: Finally, the response is transferred to the researcher.
In the following section, we provide details about how biomedical data, with a focus on genomic sequences, are stored and queried in our framework.
B. Data Representation
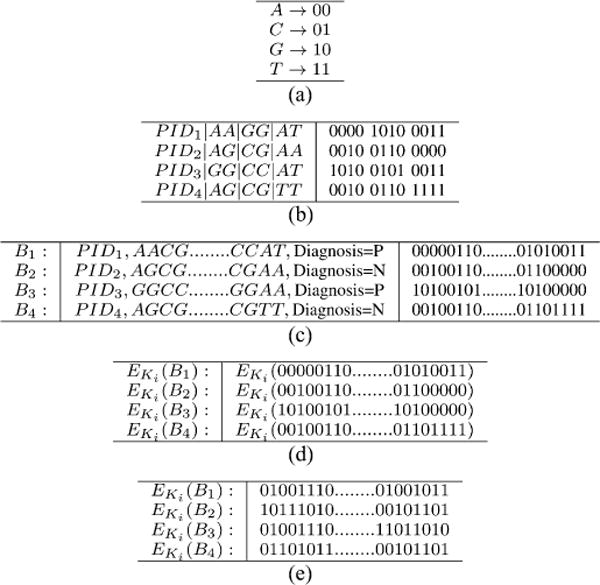
Genomic sequence data are constructed from the four letter alphabet of nucleotides {A, C, G, T}, each of which can be represented as a pair of bits as shown in Table I(a). In the context of GWAS, we adopt the model used in standard genome management software [34], where each SNP is modeled as a pair of nucleotides. Using this representation, each genomic sequence can be modeled as a series of binary values.5 Table I(b) presents four samples with three SNPs each, in the four letter alphabet along with the patient ID (PID), diagnosis information, and the corresponding binary representations [based on Table I(a)]. PIDs are used to compare the records taken from different medical institutions and eliminate duplicates.
TABLE 1.
Encryption of Clinical Genomics Data (a) Mapping From a Nucleotide Alphabet to a Two-Bit Binary Value. (b) Binary Representations of SNP Sequence Samples. (c) Original Data (Each Block Has 64 Letters). (d) Encrypting the Data. (e) The Encrypted Data
|
As mentioned earlier, we use a symmetric key encryption algorithm to encrypt the biomedical records [see Table I(c)]. Table I(d) depicts the application of encryption function and Table I(e) the resulting encrypted data.
From a practical perspective, encrypting each letter separately requires padding each encrypted block, which unnecessarily wastes memory. To reduce memory consumption, we construct blocks of biomedical data, which are encrypted one block at a time. We use the advanced encryption standard (AES) block cipher algorithm [35] to perform encryption, which supports a block size of 128 bits.
C. Secure Count Queries
One of the more common tasks that genome-based scientists need to perform is to determine how many samples satisfy certain characteristics. For example, scientists are interested in learning if there exist associations between SNPs and a range of clinical phenotypes [36], [37]. To enable a researcher to discover such associations, a secure framework needs to report the frequency of genomic and clinical feature occurrences. Such frequencies can be discovered by first counting the number of records the pattern occurs in and then normalizing this quantity by the total number of records in the database. Unfortunately, count queries were not designed to be executed over encrypted data in an untrusted server. Thus, we developed a secure count protocol which calculates the frequency of scientist-specified patterns without compromising the data.
The overview of the protocol is visually depicted in Fig. 2. The protocol is executed between the server and the SCP to evaluate the queries. The server has a dataset of n records with m attributes and the goal is to process a query q with k predicates defined over the columns of the dataset. Each attribute of the records is encrypted with the symmetric key (K) of the hospital using AES in counter (CTR) mode of encryption [38]. Before the execution of the query processing phase, the encryption key K is transferred to the SCP over a secure Ethernet channel. Now, the goal is to transfer these records to the SCP and determine the number that satisfy the query.
Fig. 2.
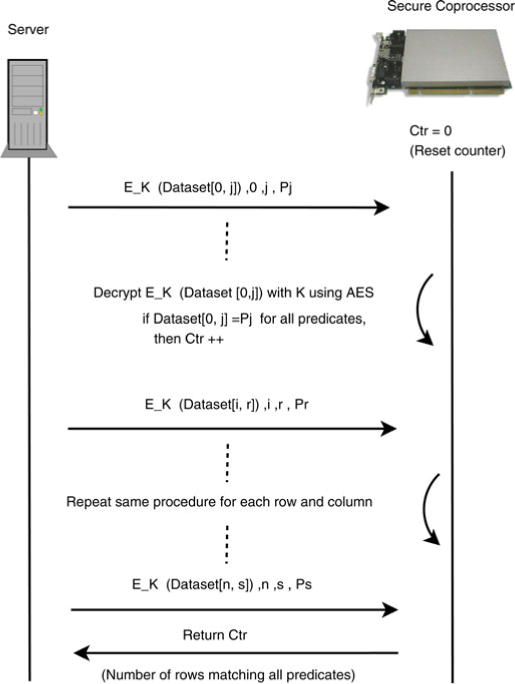
Overview of the secure count protocol.
As mentioned earlier, SCPs have limited memory. Thus, rather than sending all attributes of the records to the SCP for decryption, the server sends data from only the attributes of the predicate values defined in the query. Suppose that the first predicate, Pj is defined on the jth column of the record. As shown in Fig. 2, the server sends EK (Dataset[0, j]) to the SCP along with the row ID, column ID, and the predicate Pj. Once these values are received, the SCP decrypts EK (Dataset[0, j]) with K. After the decryption, the SCP matches the predicate values with the corresponding column’s values. The SCP keeps a counter value (or set of values) to store the number of records satisfying all predicates while processing the records. For a particular record, if all predicates are matched successfully, then the counter is incremented. After the final iteration, the counter value, which corresponds to the response of the query, is returned to the server. By applying clever buffering schemes (e.g., [12]), such an approach could handle any amount of data.
Depending on the security requirements of the application, the query results returned to a researcher can be encrypted as follows. Prior to the protocol, a secure socket layer (SSL) channel can be established between the researcher and the SCP.6 Using this channel, the query results can be transferred to the researcher without revealing the content to the server or any other observers.
V. Security Analysis
Since the operations inside the SCP are executed securely and all the sensitive biomedical data that is read or written to the server by the coprocessor are encrypted, the information leakage is possible only due to the access patterns of the algorithms running inside the SCP. In the following, we prove that for the two algorithms specified in this paper, an adversary cannot learn anything by observing the access patterns. This is achieved by proving that the access patterns of the algorithms that run on the SCP do not change based on the input [39].
Definition 1
The access pattern for algorithm A on input x (i.e., A(x)) is the sequence of accesses performed by the SCP. An algorithm A is said to run securely on the SCP, if for any two equal length inputs x, x′ of the client, the access patterns A(x) and A(x′) are computationally indistinguishable for any observer, except the SCP.
We can now prove that the count query given in Fig. 2 is secure according to Definition 1.
Theorem 1
The count protocol discussed in Fig. 2 is secure according to Definition 1.
Proof
The count protocol always reads each row of the input and outputs just one encrypted count value. Therefore, for any two equal length inputs x, x′, the access patterns of the count algorithm are exactly the same. Furthermore, due to the security of the encryption scheme, any polynomial-time adversary cannot distinguish between the different encrypted output count values. Therefore, the count protocol satisfies the Definition 1.
A similar proof for the join algorithms is provided in [12].
Traditional oblivious RAM protocols [39] provide general purpose solutions to hide the access pattern of any algorithm. In contrast, we provide a solution that is tailored for genomic data processing that is much more efficient than such generic solutions. General purpose oblivious RAM solutions require O(log2(n)) overhead for each record access, where n is the total number of records. In the proposed protocol, for count queries, for each record access, the overhead is just O(1).
VI. Experiments and Results
We developed a prototype implementation of the framework introduced in Fig. 2 to measure the performance of the data integration and query processing phases. The framework was implemented using the C programming language on a real server with an SCP. The server was an IBM x3500 server with 32 GB of main memory and an 8-core Intel Xeon CPU E5420 @ 2.50 GHz processors. The server ran a 32-bit SLES 10.2 (Linux kernel 2.6.16.60) OS. The SCP was an IBM 4764 PCI-X cryptographic coprocessor (see Section III-B). For the cryptographic operations, we used AES in CTR mode as the encryption algorithm, with a key size of 128 bits.
We organize the experimental analysis into three parts to assess various aspects of the proposed framework. First, we present the performance for the data integration phase. Second, we investigate the performance of processing the count queries. Third, we compare the performance of framework to a prior protocol based solely on cryptographic operations. In our experiments, we use synthetic binary datasets, ranging in size from 1000 to 30 000 records, and queries that involve between 10 and 50 binary attributes.
We note that the hardware specification of the server has no direct impact on the benchmarks performed because all cryptographic operations and query evaluation steps are performed on the SCP. The server is only used to submit the queries to the SCP and retrieve the output. We further point out that the performance of the experiments does not depend on whether we use a real or a synthetic dataset because the SNP values do not impact the performance of the proposed framework.
A. Join Operation
We implemented the sovereign join algorithm described in Appendix A to evaluate the performance of the data integration phase. In Fig. 3, the execution time of the join operation is depicted for a buffer size ranging from 1 to 3000 records7. The results indicate that buffering the records in the SCP yields significant performance gains. Specifically, for 1000 records, a buffer size of 1000 records provides five times improvement compared to a protocol in which no buffering is used.
Fig. 3.
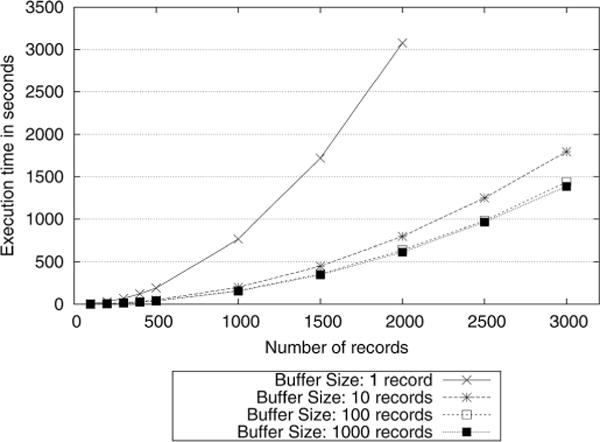
Execution time of the join operation for various buffer sizes.
The experiments show that the total execution time spent for the join operations is relatively high. However, it should be noted that the data integration phase is a one time step executed before the query execution phase. In the real applications, the system administrators could allow this step to run overnight or even a week to finish. Once the data integration phase is completed, the researchers can execute their queries in a much faster speed as we discuss the query processing performance experiments in the next section.
B. Selection Operation
In this section we evaluate the performance of executing secure count queries on the biomedical data. Fig. 4 shows the query processing time for the count queries on various datasets with different query sizes. The x-axis shows the number of predicates defined in the queries and the y-axis represents the total execution time of the protocol in seconds. We observe that the execution time increases linearly in query size. Moreover, the rate of change between the execution time and the query size increases as the number of records in the dataset grows. These results indicate that processing 10 000 records could be handled in under 1 min in our framework. Even for 30 000 records, the protocol could process a query with ten predicates in approximately 2 min.
Fig. 4.
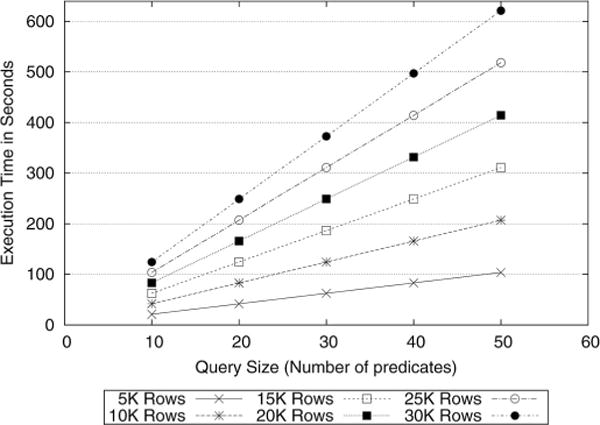
Execution time for count queries on various datasets with different query sizes (SCP-based protocol).
C. Improvements Over Prior Approaches
The solution offered in this paper is more generic in comparison to a prior approach based solely on cryptography [10]. From an analytical perspective, the multiple third party framework described in [10] leverages an encryption method that can only support count queries. In contrast, our proposed solution can directly support any online algorithm (i.e., algorithms that require one pass over the data) or multipass algorithms where the intermediate results could be kept in the memory of the SCP. More complex algorithms could be supported by modifying them to be secure in the oblivious RAM model.
From an empirical perspective, we demonstrate the runtime efficiency of our protocol with respect to the secure count algorithm [10]. For the purposes of comparison, we compute , where tserver and tSCP correspond to the amount of time required to complete a workload of count queries against a dataset on a commodity server and SCP, respectively. We refer to this computation as the improvement ratio. For the improvement ratio computations, we used the server execution times tserver reported in [10].
As Fig. 5 indicates, for a query size of ten predicates, our protocol runs about 80 times faster. We observe a decline in the improvement ratio as the number of predicates increases. Nevertheless, the difference is more than an order of magnitude for 40 predicates. One interesting point to recognize is that the improvement ratio is independent of the dataset size, which implies that our approach scales to large datasets.
Fig. 5.
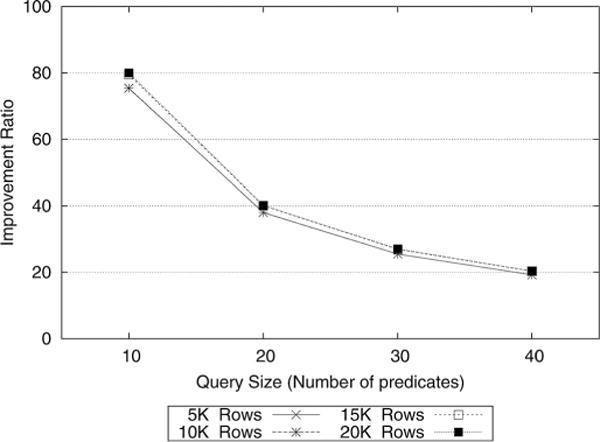
Improvement ratio compared to the multiple third party protocol in [10].
Though the current experiments were run on a different machine than the one utilized in [10], there are several reasons why this does not influence the implications of our findings. First, the queries for the proposed protocol were performed on tamper-proof hardware, which has very limited computational power and memory in comparison to the typical commodity server adopted in [10]. Second, a typical server is susceptible to many software and hardware attacks because the execution takes place in an unprotected platform. Thus, our results indicate that running the queries on an SCP is faster and more secure than the environment in [10].
VII. Discussion
In this paper, we showed that SCPs can be applied to efficiently share, store, and query structured biomedical data, such as genomic sequences. Using a real implementation, we empirically demonstrated that our framework is almost two orders of magnitude more efficient than alternative solutions based on public key cryptography.
Beyond efficiency gains, another advantage of our framework is that the SCP can facilitate the generation of secure audit logs. Consider, when a query request is sent to the cryptographic device, a new log entry could be created by the device. This entry would include the query information and a timestamp. The most recently generated log ID would always be stored within the device. Once created, the log entry would be output to the server. On the server, these logs could be stored in append-only mode and force written to write-only disk. The creation of secure logs could be utilized to satisfy some of the more stringent recommendations for the secure management of patient information as specified in regulations such as the HIPAA security rule [40].
Though the data remain encrypted at all times within the framework (except in the tamper-resistant SCP), the results of scientists’ queries themselves can violate privacy requirements. For instance, if the answer to researcher’s query is such that there is only one record with a DNA sequence “AATCAATGAA” and a positive diagnosis of juvenile Alzheimer’s, then the researcher has uniquely pinpointed an individual’s record. In the event that uniqueness is a violation, it will be necessary to ensure that query results for a researcher do not permit the triangulation of individual’s record. This can be achieved through a process known as query restriction and its application is necessary to ensure that our framework achieves identity protection. For instance, it is possible to implement a query restriction manager that refuses to report query results if it is below some certain threshold. This topic has been studied extensively in the database security community [41]. We believe that our framework can be extended to run any compact query restriction policy manager within the SCP and intend to design such solutions in future work.
|
| |
|
Algorithm 1 Joining patient records with Sovereign Join: Server side
| |
| Input: DB1, DB2 | |
| Output: Matching records | |
| 1: | bufferCtr ← 0 |
| 2: | for rec1 in DB1 do |
| 3: | MSG ← “DB1”, recordID1, rec1 {Prepare message} |
| 4: | send_MSG(MSG) to SCP {send message} |
| 5: | get ACK1 from SCP {get acknowledgement} |
| 6: | bufferCtr ← bufferCtr + 1 {update buffer counter} |
| 7: | if bufferCtr = BUFFER_SIZE then |
| 8: | for rec2 in DB2 do |
| 9: | MSG ← “DB2”, recordID2, rec2 |
| 10: | send_MSG(MSG) to SCP |
| 11: | get ACK2 from SCP |
| 12: | outputMatchedDB1Id(ACK2) {output match result} |
| 13: | end for |
| 14: | bufferCtr ← 0 {reset buffer counter} |
| 15: | end if |
| 16: | end for |
|
| |
VIII. Conclusion
In this paper, we presented a secure framework by which person-specific biomedical data, such as genomic sequences, can be joined and queried using cryptographic hardware. In contrast to formal privacy models for biomedical data that “perturb” or “generalize” records, our methods ensure that data are shared in its most specific state. Beyond a theoretical basis, we experimentally validated that the architecture is much more efficient when compared to solutions based on expensive public key encryption protocols. We also reduced the trust requirements from multiple third parties to a single third party, which may be more viable for real world applications. We recognize this framework does not explicitly address privacy violations that can be directly extracted from the query results, but are confident that this issue can be resolved within the memory of the secure hardware and plan to address it in future work.
Fig. 6.
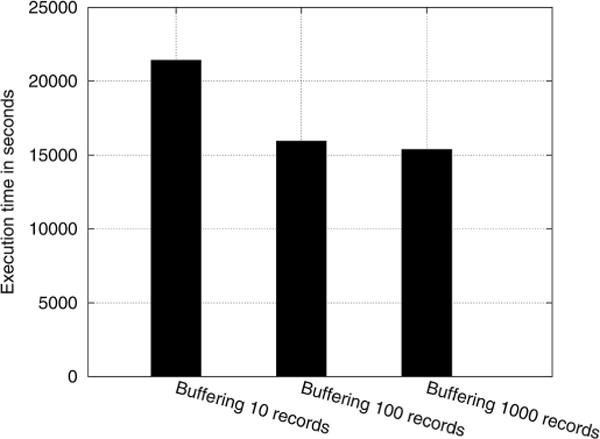
Execution time of the join operation for various buffer sizes.
Acknowledgments
This work was supported by the National Library of Medicine, National Institutes of Health under Grant R01-LM009989 and by the National Science Foundation under Grant 0845803, Grant CNS-0964350, Grant CNS-1016343, and Grant CCF-0424422.
Biographies

Murat Kantarcioglu received the B.S. degree in computer engineering from Middle East Technical University, Ankara, Turkey, and the M.S. and Ph.D degrees in computer science from Purdue University, West Lafayette, IN.
He is currently an Associate Professor in the Department of Computer Science and the Director of the UTD Data Security and Privacy Lab, the University of Texas at Dallas, Richardson. His research focuses on creating technologies that can efficiently extract useful information from any data without sacrificing privacy or security. His research has been supported by grants from the National Science Foundation (NSF), the Air Force Office of Scientific Research, the Office of Naval Research, the National Security Agency, and the National Institutes of Health.

Mustafa Canim received the B.S. degree from Bilkent University, Ankara, Turkey, and the Ph.D. degree in computer science from the University of Texas at Dallas, Richardson.
He is currently a research staff member at IBM T. J. Watson Research Center, Yorktown Heights, NY. Before joining the Ph.D. program, he was a Senior Researcher with the Database Research Group, Scientific and Technological Research Council of Turkey, a research institute in Turkey. He interned at the Database Research Group, IBM T. J. Watson Research Center in the “Solid State Sensitive Systems” project in 2008 and 2009. His research at IBM has led to several patent applications and top-level conference papers. He was also at the Engineering Tools Department, Google, as an Intern where he gained significant experience on cloud computing platforms. His research interests include the area of architecture conscious data management with a focus on privacy and performance issues and large-scale social network analysis.
Dr. Kantarcioglu is the recipient of NSF CAREER Award and Purdue CERIAS Diamond Award for academic excellence. Some of his research has been covered by the media outlets such as Boston Globe, ABC News, etc., and has received two Best Paper Awards.

Bradley Malin received the B.S. degree in biological sciences, M.S. degree in knowledge discovery and data mining, M.Phil. degree in public policy and management, and the Ph.D. degree in computer science, all from Carnegie Mellon University, Pittsburgh, PA.
He is currently an Associate Professor of Biomedical Informatics and Computer Science at Vanderbilt University, Nashville, TN, where he directs the Health Information Privacy Laboratory. His current research interests include data mining, biomedical informatics, and trustworthy computing. His research is supported by the National Science Foundation and the National Institutes of Health, with which he directs a data privacy research and consultation team for the Electronic Medical Records and Genomics Consortium.
Dr. Malin received the Presidential Early Career Award for Scientists and Engineers.
Appendix
A. Joining Patient Records
To perform join operations we implement the sovereign join algorithms introduced in [12].
The join operations in our framework require interactions between the server and the SCP. The implementation of the algorithms for the server and SCP is provided in Algorithm 1 and 2, respectively. These algorithms describe how records from two hospitals can be joined using a buffering approach that minimizes the number of passes over the inner relation. The set of records of the first and second hospital is depicted as DB1 and DB2, respectively.
|
| |
|
Algorithm 2 Joining patient records with Sovereign Join: SCP side
| |
| Input: Request messages (MSG) | |
| Output: Matching results | |
| 1: | bufferCtr ← 0 |
| 2: | while TRUE do |
| 3: | wait for new message MSG in blocked state |
| 4: | if get_DB_ID(MSG) = “DB1” then |
| 5: | dataDB1 [bufferCtr] ← decrypt(get_record(MSG)) {buffer the record} |
| 6: | recordDB1 [bufferCtr] ← get_recordID(MSG) |
| 7: | bufferCtr ← bufferCtr + 1 {update buffer counter} |
| 8: | else |
| 9: | if get_DB_ID(MSG) = “DB2” then |
| 10: | dataDB2 ← decrypt(get_record(MSG)) |
| 11: | recordDB2 ← get_recordID(MSG) |
| 12: | for i = 0 to BUFFER_SIZE do |
| 13: | if distanceMetric(dataDB1[i], dataDB2) < 1 then |
| 14: | matchedDB1Id ← recordDBl [j] {send match result} |
| 15: | end if |
| 16: | matchedDB1Id ← “No match” |
| 17: | send_ACK2(matchedDB1Id) {no match found} |
| 18: | end for |
| 19: | bufferCtr ← 0 {reset buffer counter} |
| 20: | end if |
| 21: | end if |
| 22: | end while |
|
| |
Algorithm 1 summarizes the execution of the join operation on the server side. For all records in DB1, the server prepares a message request MSG, including the record ID and the encrypted record. Using a blocking function called “send_MSG” provided by the server, the server sends MSG to the SCP and waits to receive an acknowledgement from the SCP. Using a counter variable bufferCtr, the server tracks the number of records buffered in the SCP. If the number of DB1 records transferred to the SCP exceeds the buffer size, the server starts sending the encrypted records of DB2 together with their record IDs. For all records in DB2, an acknowledgement is received from the SCP including the knowledge of whether there is a match. After making a full pass over DB2 records, bufferCtr is reset to continue with buffering new records of DB1.
Algorithm 2 summarizes the substeps of the join operation executed on the SCP side. The application running on the SCP waits for new messages to be received in a blocking state. Once a new message is received, the database identifier of the message is checked using the function get_DB_ID. If the message includes a record of DB1, the record is decrypted and buffered in the internal memory of the SCP. For decryption, the application programming interface (API) provided by the SCP is used. Also, the record ID information is buffered in an array called recordDB1. The get_recordID function is used to extract the record identifier from the message. If the message contains a record from DB2, the record is decrypted first and stored in another buffer. The decrypted record is then compared with all other records of DB1 iteratively using a preferred distance metric. If there is a match, an acknowledgement message including the matching identifier information is sent to the server.
Footnotes
In a relational model, an attribute domain is defined as the set of values allowed in an attribute.
In public key encryption, two keys are used: a public key and a private key. The public key is made generally available and is invoked to encrypt messages by anyone who wishes to send a message to the person that the key belongs to. The private key is kept secret and is used to decrypt received messages.
In symmetric key encryption, both sender and receiver share the same key to encrypt/decrypt messages. Symmetric key encryption is typically much faster than public key encryption. See [32] for more details.
As discussed previously, an SCP has many features that makes it secure against potential physical tampering attacks. Therefore, it could be used in environments where the DS is untrusted. This enables applications where encrypted data can be sent directly to a DS that is controlled by researchers. In such applications, researchers can run the queries directly at the DS without ever viewing the data. In this paper, we focus on a common trusted DS that can support multiple researchers.
We note the framework can handle various representations of genomic and health data and we show SNPs without loss of generality.
IBM 4764 SCP provides functionality to establish SSL channel to communicate with remote applications.
Contributor Information
Mustafa Canim, Email: canim@utdallas.edu, Department of Computer Science, University of Texas at Dallas, Richardson, TX 75083 USA.
Murat Kantarcioglu, Email: muratk@utdallas.edu, Department of Computer Science, University of Texas at Dallas, Richardson, TX 75083 USA.
Bradley Malin, Email: b.malin@vanderbilt.edu, Department of Biomedical Informatics, Vanderbilt University, Nashville, TN 37203 USA.
References
- 1.Khoury M, Rich EC, Randhawa G, Teutsch S, Niederhuber J. Comparative effectiveness research and genomic medicine: An evolving partnership for 21st century medicine. Genet Med. 2009;11(10):707–711. doi: 10.1097/GIM.0b013e3181b99b90. [DOI] [PubMed] [Google Scholar]
- 2.Green E, Guyer M, National Human Genome Research Institute Charting a course for genomic medicine from base pairs to bedside. Nature Genet. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 3.Gurwitz D, Lunshof J, Altman R. A call for the creation of personalized medicine databases. Nature Rev Drug Discov. 2006;5(1):23–26. doi: 10.1038/nrd1931. [DOI] [PubMed] [Google Scholar]
- 4.Burton P, Hansell A, Fortier I, Manalio T, Khoury M, Little J, Elliott P. Size matters: Just how big is big?: Quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2009;38(1):263–273. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institutes of Health. Policy for sharing of data obtained in NIH supported or conducted genome-wide association studies (GWAS) 2007 Aug; NOT-OD-07-088. [Google Scholar]
- 6.Mailman M, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang Z, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry S. The NCBI dbGaP database of genotypes and phenotypes. Nature Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peakman T, Elliott P. The UK biobank sample handling and storage validation studies. Int J Epidemiol. 2008;37(Suppl 1):i2–i6. doi: 10.1093/ije/dyn019. [DOI] [PubMed] [Google Scholar]
- 8.Malin B. An evaluation of the current state of genomic data privacy protection technology and a roadmap for the future. J Amer Med Informat Assoc. 2005;12(1):28–34. doi: 10.1197/jamia.M1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benitez K, Malin B. Evaluating re-identification risks with respect to the HIPPA Privacy Rule. J Amer Med Informat Assoc. 2010;17(2):169–177. doi: 10.1136/jamia.2009.000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantarcioglu M, Jiang W, Liu Y, Malin B. A cryptographic approach to securely share and query genomic sequences. IEEE Trans Inf Technol Biomed. 2008 Sep;12(5):606–617. doi: 10.1109/TITB.2007.908465. [DOI] [PubMed] [Google Scholar]
- 11.Asonov D, Freytag J. Almost optimal private information retrieval. Proc 2nd Int Workshop Privacy Enhancing Technol. 2002:209–223. [Google Scholar]
- 12.Agrawal R, Asonov D, Kantarcioglu M, Li Y. Sovereign joins. Proc 22nd IEEE Int Conf Data Eng. 2006:26–37. [Google Scholar]
- 13.Iliev A, Smith SW. Private information storage with logarithmspace secure hardware. Proc Int Inf Security Workshops. 2004:199–214. [Google Scholar]
- 14.Zhang X, et al. Secure coprocessor-based intrusion detection. Proc ACM SIGOPS Eur Workshop. 2002 Sep;:239–242. [Google Scholar]
- 15.Mont MC, Pearson S, Bramhall P. Towards accountable management of identity and privacy: Sticky policies and enforceable tracing services. Proc 14th Int Workshop Database Expert Syst Appl. 2003:377–382. [Google Scholar]
- 16.Iliev A, Smith SW. Small, stupid, and scalable: Secure computing with faerieplay. Proc fifth ACM Workshop on Scalable Trusted Computing. 2010:41–52. [Online]. Available: http://doi.acm.org/10.1145/1867635.1867643.
- 17.Agrawal R, Kiernan J, Srikant R, Xu Y. Order-preserving encryption for numeric data. ACM SIGMOD Int. Conf. Manage. Data; Paris, France. Jun 13–18; 2004. [Google Scholar]
- 18.Iyer B, Mehrotra S, Mykletun E, Tsudik G, Wu Y. A framework for efficient storage security in RDBMS. Proc 9th Int Conf Extend Database Technol. 2004:627–628. [Google Scholar]
- 19.Elovici Y, Shmueli E, Waisenberg R, Gudes E. A structure preserving database encryption scheme. Proc Workshop Secure Data Manage Connected World. 2004 Aug 30; [Online] Available: http://www.extra.research.philips.com/sdm-workshop/RonenSDM.pdf.
- 20.Canim M, Kantarcioglu M. Design and analysis of querying encrypted data in relational databases. (ser Lecture Notes in Computer Science).Proc 21st IFIP WG 11.3 Working Conf Data Appl Security. 2007;4602:177–194. [Google Scholar]
- 21.Canim M, Kantarcioglu M, Hore B, Mehrotra S. Building disclosure risk aware query optimizers for relational databases. Proc VLDB Endowment. 2010;3(1–2):13–24. [Google Scholar]
- 22.Canim M, Kantarcioglu M, Inan A. Query optimization in encrypted relational databases by vertical schema partitioning. Proc Secure Data Manage. 2009:1–16. [Google Scholar]
- 23.Adida B, Kohane I. GenePING: Secure, scalable management of personal genomic data. BMC Genom. 2006;7(1):93. doi: 10.1186/1471-2164-7-93. [Online]. Available: http://www.biomedcentral.com/1471-2164/7/93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacigumus H, Iyer BR, Li C, Mehrotra S. Proc 2002 ACM SIGMOD Int Conf Manage Data. Madison, WI: 2002. Jun 4–6, Executing SQL over encrypted data in the database-service-provider model; pp. 216–227. [Online] Available: http://doi.acm.org/10.1145/564691.564717. [Google Scholar]
- 25.Hore B, Mehrotra S, Tsudik G. A privacy-preserving index for range queries. Proc 30th Int Conf Very Large Data Bases. 2004:720–731. [Google Scholar]
- 26.Damiani E, Vimercati SDC, Jajodia S, Paraboschi S, Samarati P. Balancing confidentiality and efficiency in untrusted relational DBMSs. Proc 10th ACM Conf Comput Commun Security. 2003:93–102. [Online]. Available: http://doi.acm.org/10.1145/948109.948124.
- 27.Carbunar B, Sion R. Joining privately on outsourced data. Proc 7th Secure Data Manage Workshop. 2010:70–86. [Google Scholar]
- 28.Aggarwal G, Bawa M, Ganesan P, Garcia-Molina H, Kenthapadi K, Motwani R, Srivastava U, Thomas D, Xu Y. Two can keep a secret: A distributed architecture for secure database services. Proc 2nd Biennial Conf Innovative Data Syst Res. 2005:186–199. 0002. [Google Scholar]
- 29.Understanding transparent data encryption (TDE) 2010 [Online]. Available: http://msdn.microsoft.com/en-us/library/bb934049.aspx.
- 30.Understanding extensible key management (EKM) 2010 [Online]. Available: http://technet.microsoft.com/en-us/library/bb895340.aspx.
- 31.IBM. IBM 4764 cryptographic coprocessor. 2004 [Online] Available: http://www.ibm.com/security/cryptocards.
- 32.Schneier B. Applied Cryptography: Protocols, Algorithms and Source Code in C. New York: Wiley; 1996. [Google Scholar]
- 33.Goldman K, Valdez E. Matchbox: Secure data sharing. IEEE Internet Comput. 2004 Nov-Dec;8(6):18–24. [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, Maller J, Sklar P, de Bakker P, Daly M, Sham P. PLINK: A tool set for whole-genome association and population-based linkage analyses. Amer J Human Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Institute of Standards and Technology. Advanced encryption standard (AES) National Institute of Standards and Technology; 2001. (Tech Rep NIST Special Publication FIPS-197). [Online] Available: http://csrc.nist.gov/publications/fips/fips197/fips-197.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denny J, Ritchie M, Basford M, Pulley J, Bastarache L, Brown-Gentry K, Wang D, Masys D, Roden D, Crawford D. PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manolio T. Collaborative genome-wide association studies of diverse diseases: Programs of the NHGRI’s office of population genomics. Pharmacogenomics. 2009;10(2):235–241. doi: 10.2217/14622416.10.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipmaa H, Rogaway P, Wagner D. Ctr-mode encryption. Proc NIST, Comput Security Resource Center, First Modes Operat Workshop. 2000 [Online] Available: http://csrc.nist.gov/CryptoToolkit/modes/workshop1/papers/lipmaa-ctr.pdf.
- 39.Pinkas B, Reinman T. Oblivious ram revisited. Proc 30th Annu Conf Adv Cryptol. 2010:502–519. [Online]. Available: http://portal.acm.org/citation.cfm?id=1881412.1881447.
- 40.Dept. of Health and Human Services. Security standards for the protection of electronic protected health information. Federal Register. 2003 Feb 20;45(164) [Google Scholar]
- 41.Adam NR, Worthmann JC. Security-control methods for statistical databases: A comparative study. ACM Comput Surveys. 1989;21(4):515–556. [Google Scholar]


