Abstract
Population studies have indicated that natural resistance to flucytosine (5FC) in Candida albicans is limited to one of the five major clades, clade I. In addition, while 73% of clade I isolates are less susceptible to 5FC (MIC ≥ 0.5 μg/ml), only 2% of non-clade I isolates are less susceptible. In order to determine the genetic basis for this clade-specific resistance, we sequenced two genes involved in the metabolism of 5FC that had previously been linked to resistance (cytosine deaminase and uracil phosphoribosyltransferase), in 48 isolates representative of all clades. Our results demonstrate that a single nucleotide change from cytosine to thymine at position 301 in the uracil phosphoribosyltransferase gene (FUR1) of C. albicans is responsible for 5FC resistance. The mutant allele was found only in group I isolates. The 5FC MICs for strains without copies of the mutant allele were almost exclusively ≤0.25 μg/ml, those for strains with one copy of the mutant allele were ≥0.5 μg/ml, and those for strains with two copies of the mutant allele were ≥16 μg/ml. Thus, the two alleles were codominant. The presence of this allele is responsible for clade I-specific resistance to 5FC within the C. albicans population and thus by inference is likely to be the major underlying 5FC resistance mechanism in C. albicans. This represents the first description of the genetic mutation responsible for 5FC resistance.
Analyses of the population structure of Candida albicans have revealed five major clades (I, II, III, SA, and E), each exhibiting a degree of geographical specificity (1, 19, 20, 24). Recently, it was demonstrated that natural isolates that had been identified as resistant to flucytosine (5FC) were all members of clade I (18). Furthermore, it was demonstrated that 72% of clade I isolates exhibited reduced susceptibility to 5FC (5FC MIC ≥ 0.5 μg/ml) compared to 2% of non-clade I isolates (18). The fact that clades maintain their integrity side by side in the same geographical locale, combined with the observation that the majority of clade I isolates are less susceptible to 5FC, indicates that while recombination may occur within a clade, it may be a rare event between different clades (18, 24), an interpretation consistent with earlier studies indicating that the population structure of C. albicans is primarily clonal (4, 17).
Although it has been demonstrated that natural resistance to 5FC is confined to isolates in clade I, the genetic basis of 5FC resistance has not been identified. 5FC enters a cell through the action of cytosine permease (15). Inside the cell, 5FC is converted to 5-fluorouracil (5FU) by cytosine deaminase, encoded by the gene FCY1 (7, 16). 5FU is then converted to 5-fluorouridine monophosphate by uracil phosphoribosyltransferase (UPRTase), encoded by the gene FUR1 (7, 16). 5-Fluorouridine monophosphate is then converted to 5-fluorouridine triphosphate, which, when incorporated into RNA in place of UTP, disrupts protein synthesis (7, 16). In addition, 5FU, when converted to 5-fluorodeoxyuridine monophosphate, disrupts DNA synthesis by inhibiting thymidylate synthetase (26).
Although resistance to 5FC conceivably could develop by a number of mechanisms in C. albicans, it does so primarily by decreased activity of either cytosine deaminase or UPRTase (13, 16, 25). However, the mutations in the respective genes FCY1 and FUR1 responsible for resistance have not been identified. Here, we have analyzed the FCY1 and FUR1 gene sequences of isolates from the five clades of C. albicans in order to identify mutations responsible for decreased susceptibility and resistance to 5FC. The results reveal that both decreased susceptibility and increased resistance correlate in the majority of cases with a single change from cytosine to thymine at position 301 of the gene FUR1, which encodes phosphoribosyltransferase. This change results in an amino acid change from arginine to cysteine at position 101 in the Fur1 protein. Strains homozygous (C/C) at position 301 were susceptible to 5FC, strains heterozygous (C/T) at this position were less susceptible, and strains homozygous (T/T) at this position were resistant to 5FC. The mutation was restricted to clade I strains.
MATERIALS AND METHODS
Strains used, DNA fingerprinting, and 5FC MIC determination.
The 48 strains used in this study (Fig. 1) represent a subset of the 243 isolates analyzed previously by Pujol et al. (18). The larger collection from which these strains were derived were all DNA fingerprinted with the complex DNA probe Ca3, which separated them into five major clades (1, 19, 20). The positions of the 48 isolates used in this study within the five major clades are shown in the dendrogram of the larger collection (Fig. 1). The 5FC MICs for all of these strains had previously been determined by the reference broth microdilution method as described in the National Committee for Clinical Laboratory Standards document M27-A (12).
FIG.1.
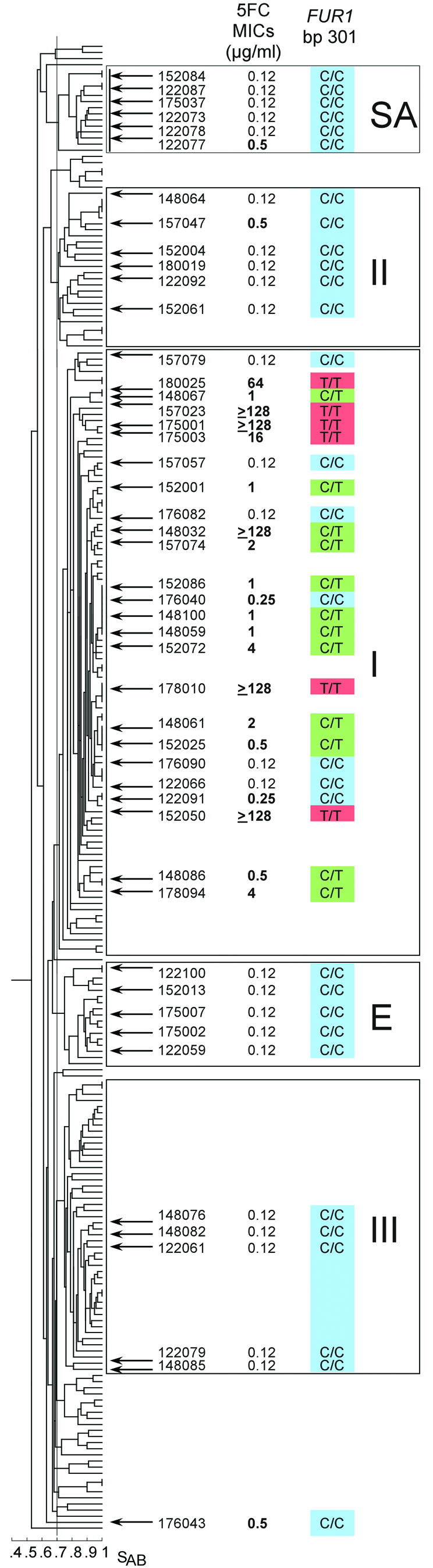
The C301T mutation is restricted to group I, as demonstrated in a cluster analysis of the initial collection of 243 C. albicans isolates. The 48 isolates selected for this study are indicated; the 5FC MICs for these isolates and their FUR1 genotypes (at position 301) are also shown. 5FC MICs that are ≥0.25 μg/ml are indicated in boldface type. FUR1 genotypes are color coded: C/C, blue; T/C, green; T/T, red.
Amplification and sequencing of FCY1 and FUR1 genes.
The complete open reading frames (ORFs) of the C. albicans genes for cytosine deaminase (FCY1) and UPRTase (FUR1) were amplified by the PCR. Primers were designed for FCY1 using GenBank accession no. U55194. The forward primer FCY1F1 (5′-ATTCTTCTTGCTTTCAACAGTCAC) was located 74 bp before the ATG codon. The 5′ end of the reverse primer FCY1R1 (5′-TGATAGTCTTCAAATGCCTGATTA) was located 281 bp after the stop codon. Those for FUR1 were designed using Stanford orf6.3823. The 5′ end of the forward primer FUR1F1 (5′-CGCAACCTGATTTTGTCCATA) was located 145 bp before the ATG codon. The 5′ end of the reverse primer FUR1R1 (5′-ATCGGAAGAATATCATGAAAATCC) was located 155 bp after the stop codon. PCRs were performed in 25-μl volumes containing 1 U of Taq DNA polymerase (New England Biolabs, Beverly, Mass.), 2.5 μl of 10× ThermoPol buffer (New England Biolabs), 0.2 mM deoxynucleoside triphosphates (Roche, Indianapolis, Ind.), 1 ng of C. albicans template DNA, and a 0.2 μM concentration (each) of either FCY1F1 and FCY1R1 or FUR1F1 and FUR1R1. DNA was extracted by the method of Scherer and Stevens (22). The PCR conditions were as follows: 7 min at 94°C; 30 cycles of 1 min at 94°C, 1 min of annealing (at 50°C for FCY1 or 52.5°C for FUR1), and 1 min at 74°C; and 10 min at 74°C. The PCR product was purified using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. The amplified ORFs were sequenced in both directions with an ABI 373A auto sequence system (Perkin-Elmer/Applied Biosystems, Foster City, Calif.), using the PCR cycle-sequencing protocol and fluorescent dye terminator dideoxynucleotides (Perkin-Elmer/Applied Biosystems). For the FCY1 ORF the sequencing primers used were FCY1SF (5′-TTTGTTTATTGCATTTATTACGAT, beginning 30 bp before the ATG codon) and FCY1R1. For the FUR1 ORF, the primers used were FUR1SF (5′-CCCTTCTTTAATTTTAATTCTAAC beginning 87 bp before the ATG codon) and FUR1R1.
RESULTS
Categories of susceptibility.
The breakpoints set to define clinical resistance to 5FC in C. albicans (12, 21) were the following: susceptibility, ≤4 μg/ml; intermediate resistance, 8 to 16 μg/ml; resistance, ≥32 μg/ml. From our previously published data (18), it is apparent that the 5FC MICs for susceptible clade I strains tended to be ≥0.5 μg/ml, whereas those for the majority of non-clade I strains were <0.5 μg/ml. Thus, for the purposes of this study, we defined the three following 5FC MIC categories: susceptibility, MIC ≤ 0.25 μg/ml; decreased susceptibility, MIC = 0.5 to 4 μg/ml; and resistance, MIC ≥ 8 μg/ml.
FCY1 gene sequences in clade I.
The sequences of the FCY1 gene were obtained from 15 clade I isolates, chosen to represent the complete range of susceptibility to 5FC. The isolates included five for which the MIC was 0.12 μg/ml (susceptibility), five for which the MIC was 1 μg/ml (decreased susceptibility), and five for which the MIC was ≥128 μg/ml (resistance) (Table 1).
TABLE 1.
Polymorphisms in the FCY1 and FUR1 genes of 15 group I isolatesa
| Isolate | 5FC MIC (μg/ml) | Base or polymorphism in FCY1 at position:
|
Base or polymorphism in FUR1 at position 301 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 31 | 33 | 102 | 107 | 210 | 219 | 423 | |||
| Reference sequence | A | C | C | G | G | G | G | C | |
| 122066 | 0.12 | R | Y | Y | R | R | R | R | C |
| 157057 | 0.12 | A | C | C | G | G | G | G | C |
| 157079 | 0.12 | A | C | C | G | G | G | G | C |
| 176082 | 0.12 | R | Y | Y | R | R | R | R | C |
| 176090 | 0.12 | A | C | C | G | G | G | G | C |
| 148059 | 1.0 | R | Y | Y | R | R | R | R | Y |
| 148067 | 1.0 | G | T | T | A | A | A | A | Y |
| 148100 | 1.0 | G | T | T | A | A | A | A | Y |
| 152001 | 1.0 | R | Y | Y | R | R | R | R | Y |
| 152086 | 1.0 | R | Y | Y | R | R | R | R | Y |
| 148032 | 128 | R | Y | Y | R | R | R | R | Y |
| 152050 | 128 | A | C | C | G | G | G | G | T |
| 157023 | 128 | G | T | T | A | A | A | A | T |
| 175001 | 128 | G | T | T | A | A | A | A | T |
| 178010 | 128 | G | T | T | A | A | A | A | T |
The reference sequences given are FCY1 (GenBank accession no. U55194) and FUR1 (Stanford orf6.3823). Positions are given as the number of the nucleotide in the coding sequence of the gene concerned. As C. albicans is diploid, the following codes were used to represent the bases at a given site in heterozygotes: R, A and G; Y, C and T. Polymorphisms resulting in an amino acid change are noted in boldface type. At FCY1 position 31, the polymorphism resulted in a change from isoleucine to valine; at FCY1 position 107, the polymorphism resulted in a change from glycine to aspartic acid; and at FUR1 position 301, the polymorphism resulted in a change from arginine to cysteine.
The FCY1 gene in these 15 isolates was found to have seven polymorphic nucleotide sites dispersed throughout the gene (at bp 31, 33, 102, 210, 219, and 423) (Table 1). The substitutions described two alleles. One of these alleles was identical to that described by the sequence under GenBank accession no. U55194. It contained A, C, C, G, G, G, and G at the seven respective sites. Four isolates (152050, 157057, 157079, and 176090) were homozygous for this allele. The second allele was differentiated from the first by transitions at the seven positions (G, T, T, A, A, A, and A, respectively). The polymorphisms at positions 31 and 107 were nonsynonymous and led to amino acid changes from isoleucine to valine and glycine to aspartic acid, respectively. In both cases, this substitution was conservative, i.e., the substituted amino acid conserved either the functional group or secondary structure. Five strains were homozygous for this allele (148067, 148100, 157023, 175001, and 178010). The remaining six strains (122066, 176082, 148059, 152001, 152086, and 148032) were heterozygous, containing both alleles. There was no obvious association between the MIC and either of the alleles (Table 1). Two copies of one allele were found in both susceptible and resistant strains, while two copies of the other allele were found in both resistant strains and those with decreased susceptibility. Heterozygotes were found in all three MIC groups (Table 1).
FUR1 gene sequence in clade I.
The FUR1 gene sequence was obtained from the same 15 clade I strains analyzed for the FCY1 gene sequence. The FUR1 gene was found to have only a single polymorphic site (bp 301) among the 15 strains. Two alleles were identified, one identical to the reference sequence containing cytosine (C) at position 301 (Stanford orf6.3823) and the other containing thymine (T) at position 301 (Fig. 1). The substitution of thymine for cytosine resulted in a nonconservative amino acid change from arginine to cysteine at position 101 in the Fur1 protein. All five isolates with a 5FC MIC of 0.12 μg/ml were homozygous for the allele containing cytosine at position 301. Four of the five isolates with a 5FC MIC of 128 μg/ml were homozygous for the allele containing the thymine substitution. All five strains with decreased susceptibility and the remaining resistant strain were heterozygous (C/T) at position 301. This result strongly suggests that 5FC resistance in C. albicans is primarily due to a mutation at position 301 (C→T) in group I isolates. For the remainder of this study, which involves an analysis of the FUR1 gene in non-clade I isolates, the alleles containing cytosine or thymine at position 301 will be referred to as FURS and FURR, respectively.
FUR1 gene sequence of non-group I isolates.
To determine whether the FURR allele was specific to group I and whether it was solely found in association with 5FC resistance, the FUR1 gene was sequenced in a collection of 23 non-group I strains that included five selected randomly from each of groups II, III, E, and SA, as well as three showing the highest 5FC MIC (0.5 μg/ml) among the non-group I population analyzed by Pujol et al. (18), strain 122077 from clade SA, strain 157047 from clade II, and strain 176043, which did not group (Fig. 1).
Among these 23 strains, a further seven polymorphic sites were found in the FUR1 gene (data not shown). Only one coded for an amino acid substitution, which was of a conservative nature. This allele was found in only one strain. None of the 23 non-clade I strains presented the mutation at position 301 associated with decreased susceptibility to 5FC in group I isolates (i.e., cytosine to thymine) (Fig. 1), suggesting that the latter mutation is specific to group I strains.
FUR1 genotype as a function of susceptibility in clade I.
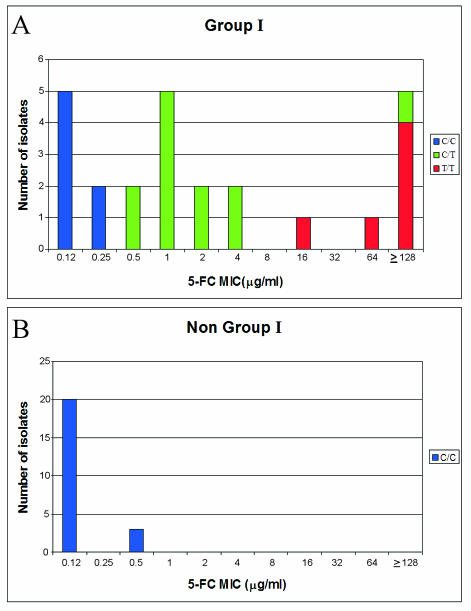
To obtain a more accurate picture of the relationship between the FUR1 alleles FURS and FURR and 5FC susceptibility, an additional 10 group I strains were analyzed, bringing to 25 the total number of group I isolates analyzed. This provided the following strain distribution of 5FC MICs according to susceptibility: five for which the MIC was 0.12 μg/ml; two for which the MIC was 0.25 μg/ml; two for which the MIC was 0.50 μg/ml; five for which the MIC was 1.0 μg/ml; two for which the MIC was 2.0 μg/ml; two for which the MIC was 4.0 μg/ml; one for which the MIC was 16 μg/ml; one for which the MIC was 64 μg/ml; and five for which the MIC was ≥ 128 μg/ml (Fig. 1). In Fig. 2A, a histogram is presented of clade I C/C (blue), C/T (green), and T/T (red) strains as a function of 5FC MIC. Except for one C/T strain for which the MIC was ≥128 μg/ml, there is a clear separation of genotypes as a function of MIC. For all C/C strains, MICs ranged from 0.12 to 0.25 μg/ml, those for all but one C/T strain ranged from 0.5 to 4, and those for all T/T strains ranged from 16 to ≥128 μg/ml. In Fig. 2B, a histogram is presented of the 23 non-clade I isolates. All non-clade I isolates were C/C. For 20 (87%) of the non-clade I isolates, the MIC was 0.12 μg/ml, while for 3 (13%) it was 0.50 μg/ml.
FIG. 2.
Distribution of the polymorphisms present at position 301 of the FUR1 gene in relation to 5FC MIC. The 25 clade I isolates (A) and the 23 non-clade I isolates (B) are represented separately. The different genotypes are color coded.
Comparison of Fur1 from other eukaryotes.
Alignment and comparison of the protein sequences of described and predicted Fur1 proteins from the fungi C. albicans, Saccharomyces cerevisiae (GenBank accession no. NP 011996), Neurospora crassa (NP 594785), and Schizosaccharomyces pombe (O13867), as well as the parasite Toxoplasma gondii (Q26998) were conducted using ClustalW (available at http://clustalw.genome.ad.jp/). This comparison revealed the region around the Arg101 residue to be highly conserved (Fig. 1). The Arg101 residue itself was conserved in all five proteins.
DISCUSSION
The clinical use of 5FC is limited, in part, by the perceived high frequency of primary resistance and the rate at which secondary resistance develops during treatment. Recent large scale studies, however, have revealed primary resistance of C. albicans to be in the range of 3.0 to 6.5% (2, 14). It has recently been shown that intrinsic resistance to 5FC is associated with only one of the major clades of C. albicans (18). Here we describe a single nucleotide polymorphism in the FUR1 gene of C. albicans that is present in only group 1 and is responsible for resistance to 5FC in this clade. We believe this to be the first identification of a single mutation underlying natural 5FC resistance in C. albicans.
The epidemiological results presented here suggest that the substitution of thymine for cytosine at nucleotide position 301, resulting in a change from arginine to cysteine at amino acid position 101, is likely to be the most important mechanism of 5FC resistance found in C. albicans populations. From the data presented here for clade I, the presence of one allele with the thymine substitution (FURR) confers a 5FC MIC of ≥0.5 μg/ml, whereas the presence of two copies of this allele results in MICs that are ≥16 μg/ml. Our results demonstrate that the two alleles, FURS and FURR, are codominant. The range of 5FC MICs present in isolates with one or two FURR copies is likely explained by the presence of other 5FC resistance mechanisms in the same strain. The role of the FURR allele in 5FC resistance will be further elucidated by mutational analysis. The presence of additional 5FC resistance mechanisms is dramatically illustrated by strain 148032, which is heterozygous for FUR1 but for which the MIC of 5FC is ≥128 μg/ml. However, it appears that the alterations in the 5FC MIC produced by other mechanisms are superimposed on the basic 5FC MIC conferred by the thymine substitution at nucleotide position 301 in FUR1.
It had previously been hypothesized that decreased susceptibility to 5FC could be due to heterozygosity for a single resistant allele, and that resistance could then be conferred by homozygosis to two resistant alleles (3). This hypothesis has been verified. Over 70% of clade I isolates, which are distributed worldwide (1, 19, 20, 24), exhibit reduced susceptibility (MIC ≥ 0.5 μg/ml) or resistance (MIC ≥ 8 μg/ml) to 5FC, while only 2% of all non-clade I isolates exhibit these characteristics (18). We have demonstrated here that all tested clade I isolates for which the MICs were ≥0.5 μg/ml are either heterozygous or homozygous for FURR. Since the majority of clade I isolates are less susceptible (0.5 ≤ MIC ≤ 4 μg/ml) but not resistant (MIC ≥ 8 μg/ml) to 5FC, by inference, the majority of clade I isolates are FURR/FURS heterozygotes, representing a pool of isolates from which resistance may rapidly develop through homozygosis to FURR/FURR. As the FUR1 gene is a component of a salvage pathway and nonessential, the conversion from FURS/FURR to FURR/FURR could occur and be maintained with or without selective pressure. This may account for the high rates with which resistance to 5FC develops in select cases during therapy. Presumably, resistant strains develop more readily from clade I strains. A study to test this hypothesis in vitro is under way in our laboratory. The discovery of the mutation responsible for both resistance and decreased susceptibility could lead to the development of rapid tests for resistance and the identification of isolates likely to become resistant during therapy.
The mechanism by which the substitution of cysteine for arginine at amino acid position 101 of the Fur1 protein confers resistance is beyond the scope of this work. Some insights, however, may be drawn from previous studies. It has been shown that 5FC-resistant C. albicans isolates have undetectable UPRTase activity (25). Crystallography studies of the T. gondii UPRTase enzyme have shown that the Arg126 residue plays a role in the interactions at the interface between FUR1 monomers in the formation of the dimers necessary for the function of the enzyme (23). The Arg101-to-Cys101 mutation found in FURR of C. albicans corresponds to Arg126 of T. gondii (Fig. 3). We hypothesize, therefore, that replacement of arginine with cysteine in the C. albicansFur1 disrupts dimerization, which prevents the formation of the active enzyme and results in decreased UPRTase activity and 5FC resistance.
FIG. 3.
Alignment of protein sequences of the known and predicted FUR1 gene products for a number of fungi and the parasite T. gondii. The sequences were aligned using the ClustalW multiple alignment editor. The arginine residue at position 101 in sensitive C. albicans strains is in boldface type. Note that this residue is conserved. Identical residues in all sequences are denoted by asterisks, while conservative replacements are denoted by either two stacked solid circles (based on similar functional groups) or a single solid circle (based on similar effects on secondary structure).
The discovery of the mating type locus of C. albicans (5) and subsequent description of fusion and mating (6, 8, 9, 10, 11) have provided a possible mechanism for recombination in natural C. albicans populations. Our initial data showing that a decrease in susceptibility and resistance to 5FC were exclusive characteristics of clade I, and the data presented here showing that a mutant allele of the FUR1 gene was dispersed throughout one clade but absent in non-clade I strains, suggest that if mating does occur, it happens only between members of the same clade. These results are consistent with studies suggesting that the population structure of C. albicans is mainly clonal (4, 17). The results presented here reinforce previous concerns (18, 24) that not all strains, or clades, of C. albicans are equal, and that representative strains should be selected from each of the five C. albicans clades in future studies of drug resistance or phenotypes of clinical relevance.
Acknowledgments
This work was funded by National Institutes of Health grant AI39735 to D.R.S. A.R.D. is supported by Wellcome Trust Medical Microbiology Research Fellowship 064466.
REFERENCES
- 1.Blignaut, E., C. Pujol, S. R. Lockhart, S. Joly, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals new clade in South Africa. J. Clin. Microbiol. 40:826-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuenca-Estrella, M., T. M. Diaz-Guerra, E. Mellado, and J. L. Rodriguez-Tudela. 2001. Flucytosine primary resistance in Candida species and Cryptococcus neoformans. Eur. J. Clin. Microbiol. Infect. Dis. 20:276-279. [DOI] [PubMed] [Google Scholar]
- 3.Defever, K. S., W. L. Whelan, A. L. Rogers, E. S. Beneke, J. M. Veselenak, and D. R. Soll. 1982. Candida albicans resistance to 5-fluorocytosine: frequency of partially resistant strains among clinical isolates. Antimicrob. Agents Chemother. 22:810-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gräser, Y., M. Volovsek, J. Arrington, G. Schönian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 6.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 7.Jund, R., and F. Lacroute. 1970. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J. Bacteriol. 102:607-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels, and D. R. Soll. 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 2:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 11.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Normark, S., and J. Schönebeck. 1973. In vitro studies of 5-fluorocytosine resistance in Candida albicans and Torulopsis glabrata. Antimicrob. Agents Chemother. 2:114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller, M. A., S. A. Messer, L. Boyken, H. Huynh, R. J. Hollis, and D. J. Diekema. 2002. In vitro activities of 5-fluorocytosine against 8,803 clinical isolates of Candida spp.: global assessment of primary resistance using National Committee for Clinical Laboratory Standards susceptibility testing methods. Antimicrob. Agents Chemother. 46:3518-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polak, A., and M. Grenson. 1973. Evidence for a common transport system for cytosine, adenine and hypoxanthine in Saccharomyces cerevisiae and Candida albicans. Eur. J. Biochem. 32:276-282. [DOI] [PubMed] [Google Scholar]
- 16.Polak, A., and H. J. Scholer. 1975. Mode of action of 5-fluorocytosine and mechanisms of resistance. Chemotherapy 21:113-130. [DOI] [PubMed] [Google Scholar]
- 17.Pujol, C., J. Reynes, F. Renaud, M. Raymond, M. Tibayrenc, F. J. Ayala, F. Janbon, M. Mallie, and J. M. Bastide. 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90:9456-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pujol, C., M. A. Pfaller, and D. R. Soll. 2003. Flucytosine resistance is restricted to a single genetic glade of Candida albicans. Antimicrob. Agents Chemother. 48:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pujol, C., M. Pfaller, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans bloodstream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 40:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rex, J. H. M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, F. C. Odds, M. G. Sinald, T. J. Walsh, and A. L. Barry. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro:in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24:235-247. [DOI] [PubMed] [Google Scholar]
- 22.Scherer, S., and D. A. Stevens. 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher, M. A., D. Carter, D. M. Scott, D. S. Roos, B. Ullman, and R. G. Brennan. 1998. Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding. EMBO J. 17:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soll, D. R., and C. Pujol. 2003. Candida albicans clades. FEMS Immunol. Med. Mycol. 1623:1-7. [DOI] [PubMed] [Google Scholar]
- 25.Whelan, W. L., and D. Kerridge. 1984. Decreased activity of UMP pyrophosphorylase associated with resistance to 5-fluorocytosine in Candida albicans. Antimicrob. Agents Chemother. 26:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]