SUMMARY
A substantial amount of mitochondrial energy is required for cell cycle progression. However, the mechanisms coordinating the mitochondrial respiration with G2/M transition, a critical step in cell division, remains to be elucidated. Here we show that a fraction of cell cycle CyclinB1/Cdk1 proteins localizes into the matrix of mitochondria and phosphorylates a cluster of mitochondrial proteins including the complex I (CI) subunits in the respiratory chain. The CyclinB1/Cdk1-mediated CI subunit phosphorylation enhances CI activity, whereas deficiency of such phosphorylation in each of the relevant CI subunits results in impairment of CI function. Mitochondria-targeted CyclinB1/Cdk1 increases mitochondrial respiration with enhanced oxygen consumption and ATP generation, which provides cells with efficient bioenergy for G2/M transition and shortens overall cycling time. Thus, CyclinB1/Cdk1-mediated phosphorylation of mitochondrial substrates allows cells to sense and respond to an increased energy demand for G2/M transition, and subsequently to up-regulate mitochondrial respiration for a successful cell cycle progression.
Keywords: CyclinB1/Cdk1, Mitochondrial respiration, Phosphorylation, Cell cycle, G2/M-checkpoint
INTRODUCTION
Cell cycle progression depends on highly ordered events controlled by Cyclins and Cyclin-dependent kinases (CDKs) (Hartwell and Weinert, 1989). CyclinB1/Cdk1 complex specifically regulates entry into mitosis (Hunt, 1989; Nurse, 1990). Through its cytoplasmic, nuclear, and centrosomal localization, CyclinB1/Cdk1 synchronizes the critical events of mitosis such as nuclear envelope breakdown and centrosome separation (Takizawa and Morgan, 2000). Accumulating evidence suggests that mitochondrial respiration is linked with cell cycle regulators; CyclinD1 coordinates mitochondrial bioenergetics in G1 progression (Sakamaki et al., 2006), CyclinE controls the formation of high energy-charged mitochondria in the G1/S transition (Mitra et al., 2009), and CyclinB1/Cdk1 is involved in the integration of mitochondrial fission with the onset of G2/M transition (Taguchi et al., 2007). However, the mechanism underlying the G2/M phase-regulated mitochondrial bioenergetics needs to be further investigated.
G1/S and G2/M checkpoints are energy-sensitive and require pronounced bioenergy supply for de novo synthesis of biomasses needed for cell cycle phase transitions (Sweet and Singh, 1995, 1999). In proliferating mammalian cells, mitochondrial ATP is generated via oxidative phosphorylation (OXPHOS) machinery (electron transportation chain), which is composed of 5 multi-subunit complexes; Complex I – Complex V (CI-CV). CI is the largest complex, with 46 subunits, and is the major entry point of electrons into OXPHOS. A functional CI is required not only for overall mitochondrial respiration (Petrosillo et al., 2009; Roessler et al., 2010), but also for a successful cell cycle progression (Owusu-Ansah et al., 2008).
In this study, we detected a fraction of CyclinB1/Cdk1 proteins located in the matrix of mitochondria and found an increased influx of mitochondrial CyclinB1/Cdk1 to be associated with elevated mitochondrial bioenergetics in G2/M transition, and further identified a cluster of CI subunits of OXPHOS as novel CyclinB1/Cdk1 substrates. Our results showed that the CyclinB1/Cdk1-mediated phosphorylation of CI subunits upregulates CI enzymatic activity to enhance overall mitochondrial respiration during G2/M transition, indicative of mitochondrial CyclinB1/Cdk1 as an important coordinator orchestrating mitochondrial bioenergetics with a successful G2/M progression for cell division.
RESULTS
The Presence of CyclinB1/Cdk1 in Mitochondria Is Enhanced at G2/M Transition
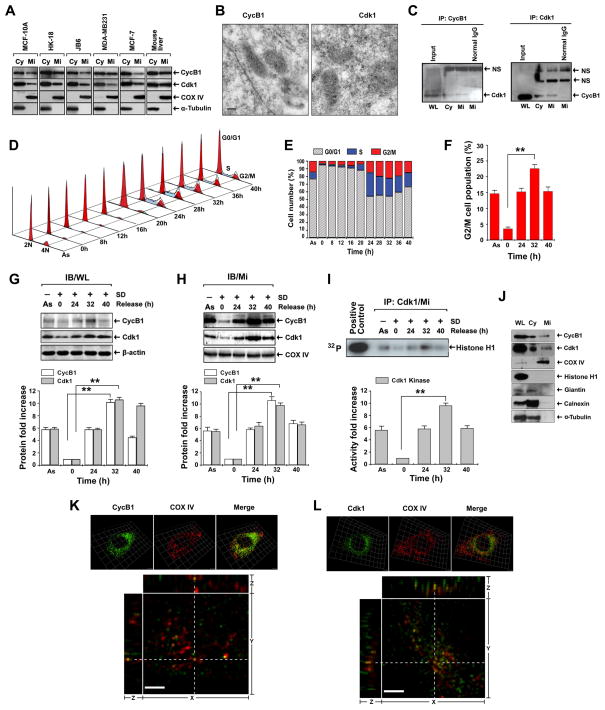
CyclinB1/Cdk1 protein was detected in the mitochondria from an array of human and mouse cell lines: human breast epithelial MCF-10A cells, human skin keratinocytes HK18, mouse skin epithelial cells JB6, human breast cancer MDA-MB-231 and MCF-7 cells, as well as mouse liver tissues (Figure 1A). MCF-10A cell line was used for all further experiments. The presence of CyclinB1 and Cdk1 in mitochondria was further confirmed by immuno-gold labeling electron microscopy (Figure 1B). The co-localization of CyclinB1/Cdk1 in mitochondria was observed with electron microscopy by double labeling technique using different sizes of gold particles (Figure S1), and by co-immunoprecipitation analysis showing that CyclinB1 and Cdk1 formed a complex in the mitochondria (Figure 1C), suggesting CyclinB1/Cdk1 complex formed in the mitochondria is enzymatically active.
Figure 1. Mitochondrial CyclinB1/Cdk1 Is Actively Correlated with G2/M Transition.
(A) Immunoblotting (IB) analysis of CyclinB1 (CycB1) and Cdk1 of cytosolic (Cy) and mitochondrial (Mi) fractions isolated from an array of human and mouse cell lines and mouse liver tissue (α-Tubulin, cytosolic marker; COX IV, mitochondrial marker). MCF-10A cells were used in all further experiments.
(B) Mi CyclinB1 and Cdk1 detected by immunoelectron microscopy with gold-labeled antibodies (dot arrows, 10 nm gold-particles for CyclinB1 and Cdk1; scale bar, 250 nm).
(C) Immunoprecipitation (IP) of CyclinB1 followed by IB of Cdk1 or reverse using whole cell lysates (WL), cytosolic (Cy)and mitochondrial (Mi) fractions (NS, non-specific binding).
(D, E, F) Cell cycle distribution after release from G0/G1 synchronization (As, asynchronous cells) with G2/M population peakedat 32 h (F) (mean ± SD; n = 3; **p < 0.01).
(G, H) IB of CyclinB1 and Cdk1 in whole cell lysates (WL, G) and Mi fraction (H) at indicated time intervals after release from G0/G1 synchronization.
(I) Kinase assay of Mi Cdk1 isolated by IP after release from G0/G1 synchronization (Histone H1, control substrate; commercial Cdk1, positive control). The lower panels show estimated fold changes of protein expression (G, H) and Cdk1 kinase activity in autoradiography (I) using Image software (mean ± SD; n = 3; **p < 0.01).
(J) CyclinB1 and Cdk1 in WL, Cy and Mi fractions isolated from G2/M-peaked cells were detected by IB together with different markers (Histone H1, nuclear protein; Cox IV, Mi marker, Giantin, Golgi apparatus membrane marker; Calnexin, endoplasmic reticulum membrane marker, α-Tubulin, Cy marker).
(K, L) Mitochondrial localization of CyclinB1 and Cdk1 detected by immunofluorescence using 3D structured illumination super-resolution microscopy. Upper panels, fluorescence images of immunostained mitochondrial CyclinB1 (K) and Cdk1 (L) isolated from G2/M-enriched cells with COX IV as control. Lower panels, the raw fluorescence images were also deconvoluted with a proprietary software package to enhance the resolutions. Yellow dots from merged images represent co-localization of CyclinB1/Cdk1 and COX IV from orthogonal views (xy-, yz- and xz- planes). See also Figures S1–S4, Movies S1 & S2.
The exquisite control of CyclinB1/Cdk1 activity peaking at metaphase is necessary for a successful G2/M transition. To investigate whether the mitochondrial abundance of CyclinB1/Cdk1 changes in correspondence with their total cellular protein levels within cell cycle progression, cells were synchronized at G0/G1 phase by serum deprivation (SD) for 48 h (Davis et al., 2001). After being released by switching them to the normal medium, mitochondrial and cellular CyclinB1 and Cdk1 were examined along with the cell cycle progression. The fluorescence-activated cell sorting (FACS) revealed that the G2/M population peaked at 32 h after release from G0/G1 synchronization (Figure 1D–F), which was paralleled with the maximal enhancement of CyclinB1 and Cdk1 proteins (Figure 1G, H). Consistently, the maximal kinase activity of mitochondrial Cdk1 was detected at the same time point (Figure 1I).
The purity of the mitochondrial preparations was studied with immunoblotting using markers from several subcellular components. COX IV, a mitochondrial resident protein, was detected exclusively in the mitochondrial preparation, whereas Histone H1, a nuclear protein, was only detected in whole lysate but not in other fractions. A Golgi apparatus marker, giantin; an endoplasmic reticulum marker, calnexin; and a cytoskeleton marker, α-tubulin were detected in cytoplasmic fraction, but were absent in mitochondrial fraction, indicating a high purity of the mitochondrial preparations (Figure 1J).
The matrix localization of CyclinB1/Cdk1 was further studied by immunofluorescence assays (IFAs) with double staining using antibodies to CyclinB1 or Cdk1 along with COX IV. The results of three dimensional deconvolution fluorescence microscopy (Figure 1K, 1L, upper panels) showed that CyclinB1 and Cdk1 were co-localized with COX IV in G2/M-enriched cells at 32 h. Using the structured illumination super-resolution fluorescence microscopy, we further demonstrated that CyclinB1 and Cdk1 localized in the very proximity of COX IV. The average distance between the COX IV and CyclinB1 or Cdk1 was calculated less than 350 nm which was significantly below the lower limit of the size of a mitochondrion (600 ~ 1000 nm) (Kennady et al., 2004). Furthermore, some merged dots emerged from CyclinB1/Cdk1 and COX IV co-localization were also shown on orthogonal views (xy-, yz- and xz- planes in Figure 1K, 1L, lower panels; Movies S1 & S2). The images in Figure 1K–L are representative z-sections showing the co-localization of COXIV and CyclinB1/Cdk1 complex in the mitochondria. To achieve optimal resolution for observing mitochondrial localization of CyclinB1/Cdk1, we used thin optical sectioning to achieve 30–40 z-sections along the Z direction across a whole cell. The CyclinB1 and Cdk1 staining in the nucleus was not visible on the representative z-sections in Figure 1K–L, but could be seen on the image of the same cell with 0.5 μm up in the z-section (Figure S2). The centrosome-like structures stained by anti-CyclinB1 or anti-Cdk1 antibodies were shown in Figure S3 & S4. As the z-value increased from Z=0, the centrosome-like structures gradually disappeared (Figures S3 & S4). Collectively, the data obtained from different approaches indicate that a fraction of CyclinB1/Cdk1 proteins localizes in mitochondria, and an increased mitochondrial influx of CyclinB1/Cdk1 is associated with G2/M transition.
Potential Mitochondrial Targets of CyclinB1/Cdk1
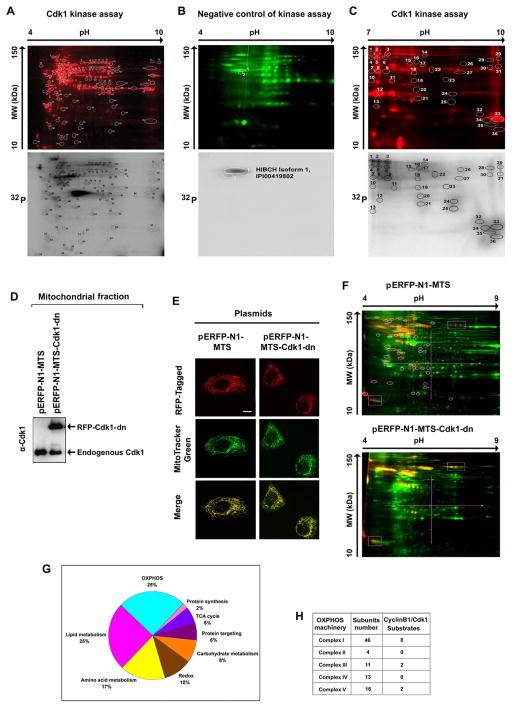
A repertoire of mitochondrial proteins are phosphorylated during mitosis (Dephoure et al., 2008). Cdk1 belongs to the serine/threonine (S/T) kinase family, which catalyzes the transfer of a phosphate from ATP to proline (P)-oriented S or T residues. Its substrates contain either an optimal (S/T*-P-x-K/R; x, any residue) or a minimal (S/T*-P) Cdk1 consensus motif. To identify the potential CyclinB1/Cdk1 targets in mitochondria, we performed an in vitro CyclinB1/Cdk1 kinase assay, in which the commercial Cdk1 was incubated with mitochondrial proteins isolated from G0/G1 cells that possessed low endogenous CyclinB1/Cdk1 activity. Triplicate Cdk1 kinase assay products were analyzed by 2D gel analysis with pH 4–10 gel strips, which generated about 110 spots with incorporated [γ-32P] ATP (Figure 2A), among which only 2 spots (one protein) were phosphorylated by endogenous mitochondrial kinases other than Cdk1; detected here by the negative control kinase assay, in which Cdk1 activity was blocked by the treatment of Cdk1 inhibitor, RO-3306 (Figure 2B). Further 2D gel analysis using pH 7–10 gel strips revealed 36 proteins with incorporated [γ-32P] ATP (Figure 2C). Mass spectrometry of the total 146 spots showed that Cdk1 potentially phosphorylates 52 different proteins after exclusion of the spots corresponding to the same proteins. The results of Scansite search (http://scansite.mit.edu) indicated that all of the 52 substrates contain at least one minimal consensus site, and 19 targets (including 8 CI subunits) contain an optimal Cdk1 consensus phosphorylation motif (Table S1).
Figure 2. Potential Mitochondrial Substrates of Cdk1.
(A) Mitochondrial (Mi) proteins (1 mg) isolated from G0/G1 cells were incubated with commercial Cdk1 in the presence of [γ-32P] ATP, of which 50 μg was separated by 2-D gel electrophoresis (pH 4–9) after labeled with Cy5 (upper panel). The phosphorylated spots were detected by autoradiography (lower panel) and the circled spots were extracted and identified by mass spectrometry and the potential Cdk1 -phosphorylated Mi proteins are listed in Table S1.
(B) A negative control of (A) with the absence of commercial Cdk1 and presence of the Cdk1 inhibitor RO-3306.
(C) Cdk1-Mi target proteins detected with the same Cdk1 kinase assay as (A) with electrophoresis in the range pH 7–10.
(D) Immunoblotting analysis of Cdk1 in Mi fractions isolated from cells transfected with mitochondria-targeted dominate negative mutant Cdk1 and empty vectors.
(E) Representative images of mitochondria-targeted (MTS) Cdk1-wt-RFP, Cdk1-dn-RFP and empty vector MTS-RFP.
(F) Mi proteins extracted from G2/M-peaked cells transfected with mitochondria-targeted empty vector (pERFP-N1-MTS, upper panel) or mutant Cdk1 (pERFP-N1-MTS-Cdk-dn, lower panel) were labeled with Cy5 (green), separated by 2-D gel and phosphorylated proteins were stained with Pro-Q Diamond dye (red). Spots absent in the Mi profile of cells with mitochondria-targeted mutant Cdk1 compared with the vector control transfectants (circled) were extracted and analyzed by mass spectrometry. The potential Cdk1 Mi targets detected by these experiments were listed in Table S3.
(G) Summary of potential Cdk1-targeted mitochondrial proteins detected by in vitro (A–C) and in vivo (E–F) Cdk1 kinase assays.
(H) Distribution of potential MiCdk1 targets in the subunits of complexes. See also Tables S1–S4.
A point mutation generated by replacing an aspartate (D) residue by an asparagine (N) at the position 146 (D146N) of Cdk1 results in a dominant negative (dn) Cdk1 mutant (van den Heuvel and Harlow, 1993). To generate a mitochondria-targeted Cdk1-dn to explore Cdk1-regulated mitochondrial functions, we constructed the plasmid pERFP-N1-MTS-Cdk1-dn linking a mitochondrial targeting sequence (MTS) composed of 29 amino acid residues derived from the subunit VIII of the human cytochrome C oxidase (Table S2) to the RFP-tagged dominant negative Cdk1. The pERFP-N1-MTS was used as an empty vector control with mitochondria-targeted ERFP (Figure 2D & 2E). Mitochondrial phosphoproteins in G2/M cells transfected with both constructs were profiled by 2D gel analysis. Compared with the mock transfectants (Figure 2F, upper panel), a group of mitochondrial phosphoproteins was apparently absent or decreased in the Cdk1-dn transfectants (Figure 2F, lower panel). Mass spectrometry analysis of the spots detected in Figure 2F revealed that 15 of the in vivo CyclinB1/Cdk1-phosphorylated mitochondrial proteins (Table S3) were among the list of proteins detected with the in vitro kinase assay (Table S1). Thus, our results of phosphoproteomics identified a specific cluster of mitochondrial proteins that were potentially phosphorylated by CyclinB1/Cdk1, including the elements of OXPHOS, metabolism of lipids, amino acids and carbohydrates, tricarboxylic acid cycle, and redox balance. Among these 52 potential mitochondrial substrates, summarized according to their functional category in Figure 2G, 12 targets belonged to the OXPHOS machinery (mitochondrial respiration chain) including the 5 major respiratory complexes (CI, CII, CIII, CIV and CV) (Figure 2H). Intriguingly, 8 of the identified OXPHOS targets were CI subunits and they all contained the optimal Cdk1 consensus phosphorylation motif (Table S4) determined by Scansite search, suggesting a role of CyclinB1/Cdk1 in regulation of mitochondrial ATP generation at G2/M transition by phosphorylation of the CI subunits.
CyclinB1/Cdk1 Is Localized in Mitochondrial Matrix and Phosphorylates the CI Subunits in the Mitochondrial Respiration Chain
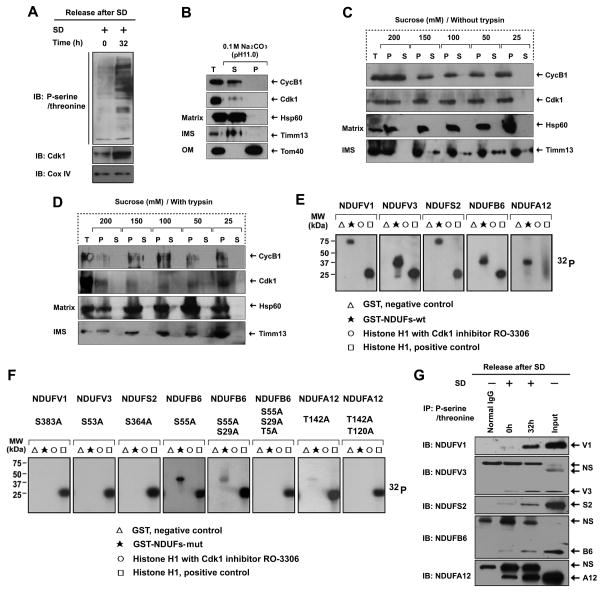
Consistent with our observation that the mitochondrial influx of CyclinB1/Cdk1 proteins and Cdk1 kinase activity were increased on G2/M phase (Figure 1G–1I), the overall mitochondrial phosphoprotein levels were also markedly enhanced in G2/M-enriched cells (32 h) compared to the G0/G1 cells (0 h) (Figure 3A). The phosphorylation of mitochondrial inner membrane resident CI subunits by Cdk1 implied that CyclinB1/Cdk1 might localize into the mitochondrial matrix, inner membrane, or intermembrane space (IMS), where the complex may freely access its subunits. To determine the sub-mitochondrial localization of CyclinB1/Cdk1, the sub-mitochondrial fractions were prepared using the alkaline extraction method with the soluble proteins recovered in the supernatants. Timm13 subunit of the TIM (the translocase of inner membrane) complex, which resides in the inner membrane space; and Tom40 subunit of the TOM (the translocase of outer membrane) complex, residing on the outer membrane, was utilized as the markers of sub-mitochondrial compartments. Unlike Tom40 in the pellets, the presence of CyclinB1/Cdk1 in the supernatant excluded the possibility of its OM localization (Figure 3B). The results also indicated that CyclinB1/Cdk1 are either soluble proteins localizing into the matrix like Hsp60; or integral membrane proteins localizing into the inner membrane like Timm13 (Figure 3B). To further specify the sub-mitochondrial localization of CyclinB1 or Cdk1, mitoplasting was performed by diluting mitochondria in hypotonic buffers with decreasing concentrations of the osmoticum sucrose from 200 mM to 25 mM. The outer membrane began to rupture at 150 mM of sucrose, while the inner membrane remained intact until the final concentration at 25 mM of sucrose (Figure 3C). In combination with mitoplasting, protease protection assay was performed using trypsin to digest exposed proteins following outer membrane rupture. Similar to Hsp60 and unlike Timm13, CyclinB1 and Cdk1 were protected from trypsin digestion, indicating their mitochondrial matrix localization (Figure 3D).
Figure 3. Mitochondrial CyclinB1/Cdk1 localizes in the Matrix and Phosphorylate CI Subunits at G2/M Transition.
(A) Immunoblotting analysis of mitochondrial (Mi) phospho-serine/threonine proteins isolated from G0/G1 (0 h) and G2/M(32 h) cells after release from G0/G1 synchronization.
(B) Sub-mitochondrial localization of CyclinB1 and Cdk1 detected by alkaline extraction (Antonyuk et al.). Matrix proteins were separated from integral membrane proteins by extracting mitochondria with sodium carbonate (pH 11), then the total input (T), soluble matrix proteins (S), and membrane vesicle pellets (P) were immunoblotted for CyclinB1, Cdk1, Tom40 (an outer membrane protein), TIMM13 (an inter-space protein), and HSP60 (a matrix protein).
(C, D) Sub-mitochondrial localization of CyclinB1 and Cdk1 detected via mitoplasting and protease protection assay (Antonyuk et al.). Mitochondria were incubated in gradient hypotonic sucrose buffer as indicated to digest the outer membrane of mitochondria with or without soybean trypsin. The total (T), pellet (P), and supernatant (S) fractions were subjected to IB analysis with indicated antibodies.
(E, F) Five GST-fused human wild type CI subunits (E) and their mutants (F) in the indicated potential Cdk1 phosphorylation sites (note, multiple mutations created in NDUFB6 and NDUFA12) were synthetized and tested as substrates in kinase assay with commercial Cdk1.
(G) Mitochondrial proteins from G0/G1 and G2/M cells were extracted by IP using a phospho-serine/threonine antibody followed by IB using antibodies to each of the CI subunits (normal IgG, control for the IP reaction; NS, non -specific band). See also Figure S5 & Table S4.
To validate the phosphorylation of aforementioned 8 CI subunits by CyclinB1/Cdk1 complex, we constructed these CI subunits as GST fusion proteins (Table S5), and expressed them in E. coli (Figure S5). Among them, 5 of the synthesized CI subunits were expressed as soluble proteins and can be phosphorylated by in vitro Cdk1 kinase assay (Figure 3E). These results were repeated and confirmed by generating phosphorylation defective forms of these 5 CI subunits through substitutions of Serine/Threonine (S/T) residues with Alanine (A) on either Cdk1 optimal or minimal consensus motifs (T383 on NDUFV1, S105 on NDUFV3, S364 on NDUFS2, S55/S29/T5 on NDUFB6 and T142/T120 on NDUFA12). The mutation of Cdk1 consensus motifs severely diminished their phosphorylation (Figure 3F). Consistent with the increased influx and activity of CyclinB1/Cdk1 in mitochondria, the phosphorylation levels of the 5 endogenous CI subunits were also increased in G2/M transition compared to G0/G1 phase (Figure 3G). Collectively, these data demonstrate that during G2/M transition, an increased influx of CyclinB1/Cdk1 into the mitochondrial matrix promotes phosphorylation of CI subunits, which may facilitate electron transport through the mitochondrial respiration chain.
CyclinB1/Cdk1-mediated CI Phosphorylation Is Required for CI function
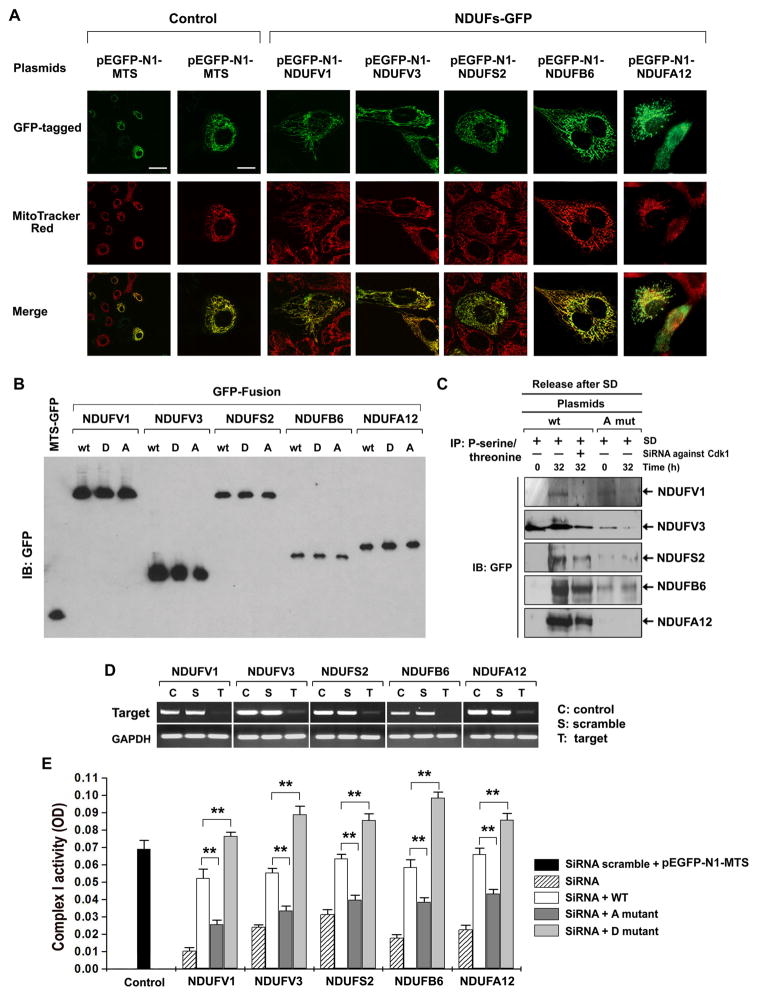
CI as the major entry point of electrons into the respiratory chain plays a fundamental role in mitochondrial ATP generation (Janssen et al., 2006). To investigate CyclinB1/Cdk1-mediated regulation of CI activity, we constructed plasmids expressing the GFP-tagged CI subunits, each with 3 different forms, i.e., wild type (wt, Table S6), phosphorylation-mimic D form (S/T to D, Table S7), and phosphorylation-defective A form (S/T to A, Table S7). The unambiguous mitochondrial localization of each GFP-tagged CI subunit was identified by confocal microscopy (Figure 4A) and the transfection efficiency was analyzed by immunoblotting (Figure 4B). As expected, in vivo phosphorylation of each subunit at G2/M phase was inhibited in cells transfected with Cdk1 siRNA (Figure 4C, lane 3), which arrested cells at the G2/M phase (Figure S5). The phosphorylation defective mutant subunits showed no phosphorylation in both G0/G1 and G2/M phases (Figure 4C, lanes 4–5). To address whether the phosphorylation of CI subunits is required for CI activity, we tested rescue ability of wt and phosphorylation-defective CI subunits by knocked-down the expression of each of 5 endogenous CI subunits with siRNAs targeting their 5′- or 3′-UTRs (Table S8) without affecting the expression of exogenous subunits (Figure 4D). The siRNA treatments resulted in 65–82% reduction in CI activity when compared to control cells transfected with scrambled siRNA-treated empty vector pEGFP-N1-MTS (Figure 4E). Reconstituting each of CI subunits with their wt or D forms significantly rescued the demolished CI activities, whereas the phosphorylation-defective A form mutants failed to restore CI activities, indicating that CI activity is dependent on theCdk1 -dependent phosphorylation of the CI subunits.
Figure 4. Cdk1-mediated Phosphorylation of CI Subunits Is Required for CI Activity.
(A) Representative images of mitochondria (Mi)-localized CI subunits of transfected cells co-stained with GFP and Mito Tracker Red (Control, empty MTS-GFP vector, scale bar: left = 30 μm, right =10 μm).
(B) Expression of wild type or mutant forms of each CI subunit in transfected cells tested by IB of GFP (MTS-GFP, empty vector; A, alanine mutation; D, aspartate mutation.
(C) Phosphorylated proteins were extracted by IP with phospho-serine/threonine antibody from wild type and A mutant CI transfected cells at 0 h and 32 h after release from synchronization, followed by IB for GFP. Additional cells transfected with wild type CI and treated with Cdk1 siRNA were included as controls.
(D) RT-PCR analysis of siRNA-mediated inhibition on each of CI subunits.
(E) CI activity rescued by wild type but not mutant CI subunits. CI activity was measured in cells co-transfected with wild type or mutant CI subunits along with siRNAs against each of the endogenous CI subunits (See also Table S8). SiRNA scramble + MTS-EGFP, a non-target control; A mutant, phosphorylation-defective form; D mutant, constitutive phosphorylation-mimic form; one unit of OD is equal to 29.3 μmoles/min/mg of proteins (mean ± SD, n = 3; **p < 0.01). See also Figure S6, Tables S6, S7 & S8.
CyclinB1/Cdk1 Coordinates CI Activity with G2/M Transition
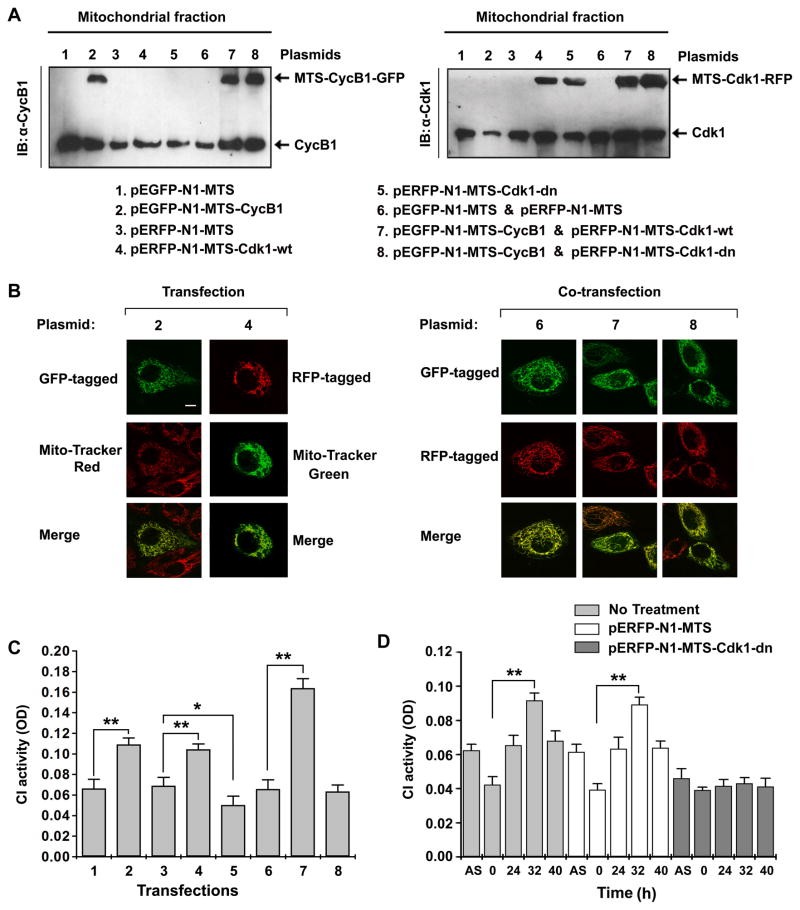
To determine whether mitochondrial CyclinB1/Cdk1 is sufficient to induce CI activity at G2/M transition, we analyzed a correlation between CyclinB1/Cdk1 abundance and CI activity in vitro. To mimic the peak levels of mitochondrial CyclinB1/Cdk1 at G2/M transition, we transfected cells with the mitochondrial targeted sequence (MTS)-oriented CyclinB1/GFP and/or Cdk1/RFP constructs (Table S2 & S4). The GFP- or RFP-tagged and mitochondria-targeted CyclinB1 and Cdk1 proteins were confirmed by immunoblotting (Figure 5A) and confocal microscopy (Figure 5B). As shown in Figure 5C, compared with the empty vector transfectants (Lanes 1, 3, & 6), the CI activity was enhanced by 174%, 169%, and 242% in cells expressing mitochondrial CyclinB1 alone (Lane 2), Cdk1 alone (Lane 4), and CyclinB1/Cdk1 (Lane 7), respectively; while mitochondrial mutant Cdk1-dn alone or in combination with CyclinB1 did not affect the CI activity (Lanes 5 & 8), suggesting a positive coordination between mitochondrial CyclinB1/Cdk1 and CI activity.
Figure 5. Mitochondrial Cdk1 Is Required for G2/M-associated CI Activation.
(A) IB of mitochondrial fraction isolated from cells for mitochondria-targeted CyclinB1 with wild type or dominant negative mutant Cdk1 (plasmids are indicated on the bottom. pEGFP-N1-MTS and pERFP-N1-MTS were empty vector controls for MTS-Cylcin B1 and MTS-Cdk1 respectively; see also Table S2 for detailed information on the plasmid constructs).
(B) Representative images of mitochondria-localized CyclinB1 (2), Cdk1 (4), or co-localized wild type and mutant Cdk1 with CyclinB1(6, 7, 8).
(C) CI activity measured with asynchronous cells at 48 h following transfection of above indicated plasmids (mean ± SD; n = 3; *p < 0.05; **p < 0.01).
(D) CI activity measured in control (no treatment) and in cells with empty vector (pERFP-N1-MTS) or Cdk1-dn mutant (pERFP-N1-MTS-Cdk1-dn) at indicated times after release from G0/G1 synchronization (mean ± SD;n = 3, ** p < 0.01).
The endogenous effects of mitochondrial CyclinB1/Cdk1 on CI function were examined in synchronous cells. Consistent with the timing of maximal mitochondrial CyclinB1/Cdk1 at 32 h after synchronization (Figure 1H–1J), the CI activity was at its peak value in G2/M (32 h) cells that were left untreated or mock transfected with the empty vector (pERFP-N1-MTS). To further confirm that the increase of CI activity at G2/M transition was CyclinB1/Cdk1-dependent, we utilized Cdk1-dn construct to mimic low mitochondrial CyclinB1/Cdk1 activity. This G2/M-related enhancement of CI activity was completely absent in synchronous cells expressing mitochondrial mutant Cdk1-dn (Figure 5D).
CyclinB1/Cdk1 Enhances Mitochondrial Respiration for G2/M Transition
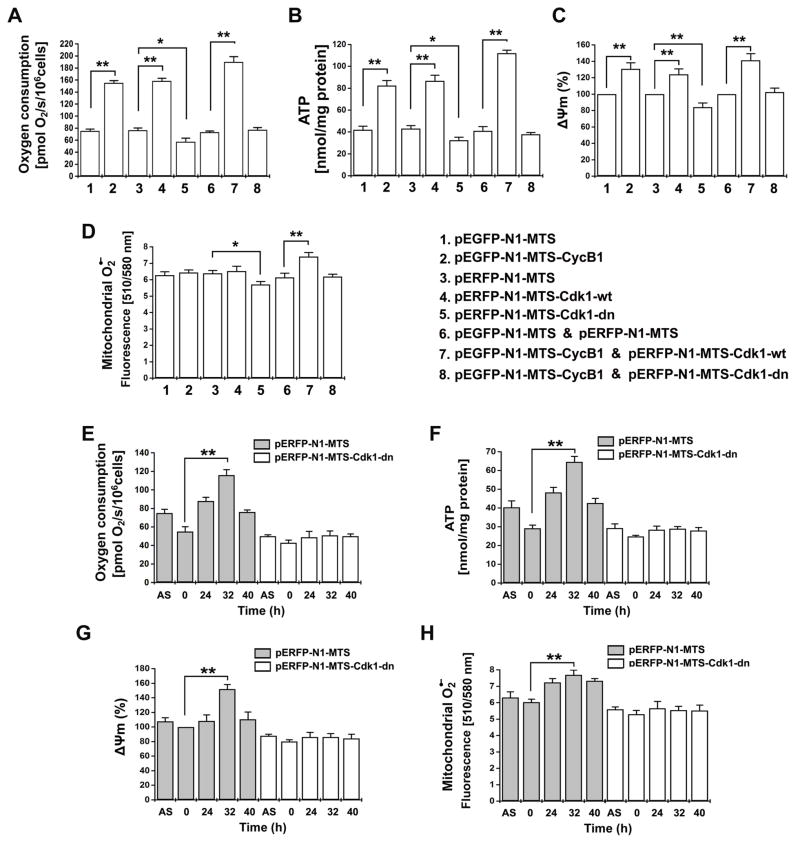
ATP is generated by OXPHOS through coupling of electron transport with proton (H+) pumping in the respiration chain, which creates mitochondrial membrane potential (Δψm) and increases oxygen consumption and reactive oxygen species (ROS) formation. To investigate whether CyclinB1/Cdk1-mediated phosphorylation is required for the mitochondrial respiration in G2/M transition, we tested the mitochondrial functions in cells transfected with MTS-CyclinB1, MTS-Cdk1-wt, MTS-Cdk1-dn, alone or in combination with each other. Similar to the altered CI activity (Figure 5C–D), the mitochondrial oxygen consumption, ATP generation and membrane potential (Δψm) were markedly increased in accordance with mitochondrial abundance of CyclinB1/Cdk1 (Figure 6A–C). Mitochondria-targeted CyclinB1 (Lane 2), Cdk1 (Lane 4) as well as CyclinB1/Cdk1 (Lane 7) all enhanced mitochondrial functions compared with empty vector transfected control cells (Figure 6A–C, Figure S6). Interestingly, a substantial increase in the mitochondrial superoxide (O2•−) levels was only observed in cells co-transfected with MTS-CyclinB1 and MTS-Cdk1 (Figure 6D, lane 7). On the other hand, overexpression of mutant MTS-Cdk1-dn in mitochondria suppressed all of the tested mitochondrial functions (Lane 5, Figure 6A–D) when compared to control cells (Lane 3, Figure 6A–D). The Cdk1-dn-mediated mitochondrial suppression could be rescued to levels of mock control cells by co-transfection of CyclinB1 (Lane 8, Figure 6A–D), supporting that CyclinB1 is able to remedy the inhibitory effect of Cdk1-dn (van den Heuvel and Harlow, 1993).
Figure 6. Mitochondrial Cdk1 Is Required for G2/M-associated Mitochondrial Respiration.
(A–D) Mitochondrial oxygen consumption, ATP generation, Δψm, and O2·− levels were measured in cells at 48 h after transfection with the indicated plasmids.
(E–H) Mitochondrial oxygen consumption, ATP generation, Δψm, and O2·− levels were measured in cells with empty vector (pERFP-N1-MTS) or Cdk1-dn mutant (pERFP-N1-MTS-Cdk1-dn) at indicated times after release from G0/G1 synchronization (mean ± SD; n = 3; *p <0.05; **p <0.01). See also Figures S7 & S8.
The mitochondrial activity was also tested in synchronized cells transfected with mock or MTS-Cdk1-dn vectors to gain insight on the regulation of mitochondrial activity in conjunction with G2/M transition. Consistent with the in vitro results showing a positive correlation between CyclinB1/Cdk1 abundance and mitochondrial respiration (Figure 6A–D), the endogenous mitochondrial activity was increased at G2/M-enriched population (32 h) in the cells transfected with empty vector (Figure 6E–H, Figure S7). The oxygen consumption rate and ATP generation were more than doubled (Figure 6E, F); the Δψm and the superoxide levels were increased significantly (Figure 6G, H). However, such boost of G2/M-associated mitochondrial activities was absent in cells expressing the mitochondria-targeted inactive Cdk1 (Cdk1-dn). Collectively, CyclinB1/Cdk1-mediated enhancement of mitochondrial activity and ATP generation is a predominant feature of the G2/M progression of the cell cycle.
Mitochondrial CyclinB1/Cdk1 Facilitates G2/M Progression
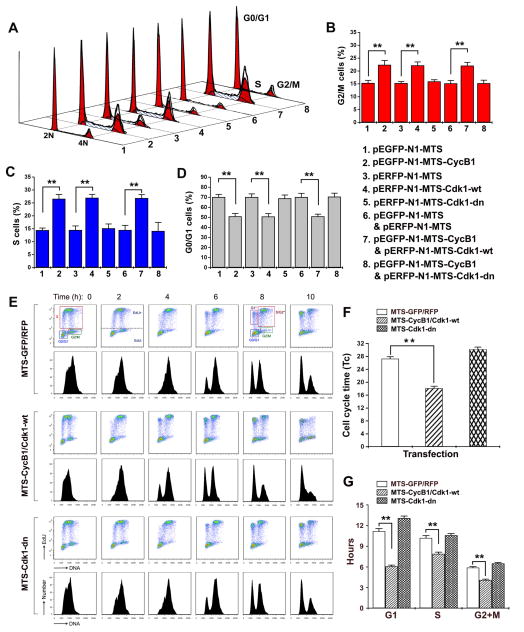
Previous studies have shown that mitochondrial bioenergetics has key roles in cell cycle progression, mainly in the progression of G1 phase (Schieke et al., 2008; Schulz et al., 2006) and in S phase (Mitra et al., 2009). A recent publication also showed that mitochondrial oxidative phosphorylation is activated in S and G2/M phases in tumor cells (Bao et al., 2013). To investigate a potential role of mitochondrial CyclinB1/Cdk1 in mitochondrial metabolism in G2/M progression, we examined the cell population kinetics of the MCF-10A cells that were transfected with MTS-linked CyclinB1 and/or Cdk1 (Figure 7). Cell populations constitutively overexpressing the mitochondrial wt CyclinB1/Cdk1 showed a decreased number of cells in G1 and increased number of cells in S and G2/M phases compared to vector control or mutant CyclinB1/Cdk1 expressing cells (Figure 7A–D). This change in the cell cycle distribution is consistent with accelerated transit through G1 phase and is expected, given that G1 phase of the cell cycle is the most responsive to changes in cellular metabolic events. We then measured the potential doubling time (Tc) and the duration of each cell cycle phase, and found that the duration of G1 was on average ~5 hrs shorter in cells transfected with wt mitochondrial CyclinB1/Cdk1. Interestingly, the duration of S and G2/M phases were also shortened by ~2 hrs each (Figure 7E–F, Table S9), even though this did not reflect on the population distribution (Figure 7B–C). Although shorter, the S and G2/M phases showed an increased population of cells owing to the larger alterations in G1 phase and the overall cell cycle durations. To further investigate whether the cells progressed through G2/M phase faster when mitochondrial CyclinB1/Cdk1 is enhanced, we performed a pulse-chase labeling using a novel thymidine analogue, ethynyl deoxyuridine (EdU) to label the population of cells undergoing DNA synthesis (Terry and White, 2006). Tracking of the EdU-positive population of cells as they progressed through the S and G2/M phases and accumulated in G1 phase allowed the visualization of the cell cycle captured over a 22-hr window. The results revealed that the labeled S phase cells progressed through the G2/M phase and appeared in the G1 phase as fast as 4 hrs in cells with wt mitochondrial CyclinB1/Cdk1 compared to 6 hrs in vector control and mutant CyclinB1/Cdk1 transfected cells, indicating that G2/M phase is accelerated due to the enforced enhancement of mitochondrial CyclinB1/Cdk1 (Figure 7G). The results of DNA content analysis and EdU analysis indicate that mitochondrial energy supply is a rate-limiting event for cell cycle progression. Taken together, these results show that CyclinB1/Cdk1-mediated up-regulation of mitochondrial bioenergetics is required for a successful progression through G2/M transition and the factors that control commitment to the cell cycle, such as those present at the onset of G2/M, may do so by regulating the energy supply in mammalian cells.
Figure 7. Mitochondrial Cdk1 Enhances G2/M Transition and Overall Cycle Progression.
(A–D) Cell population kinetics in cells after 48 h transfection with indicated mitochondria-targeted CyclinB1, Cdk1, CyclinB1/Cdk1 and control vectors; a representative plot (A), and percentage in G2/M(B), S (C) and G0/G1(D)phases (mean ± SD; n = 3; **p <0.01).
(E–F). Cell cycle time (Tc, in G) and each phase (G1, S, and G2/M in H) progression analysis were performed based on the flow cytometry analysis of the DNA content. (Mean ± SD; n = 3; *p <0.05; **p <0.01). See also Figure S9 & Table S9.
(G) Cell cycle analysis with EdU pulse-chase labeling. Cells were pulsed with EdU for 30 min following 24 h transfection with indicated constructs on the left. The EdU-positive population was followed over time as it progressed through cell cycle phases. Scatter plot histograms of EdU-labeled cells were stained for DNA content (X-axis) and EdU (Y-axis). The lower figures in each panel show the mean fluorescence intensity of the EdU labeled nuclei. The time points were indicated in h after the EdU pulse. For all time points, unique gates displaying the following populations were drawn: G0/G1, S, and G2/M. For 6, 8, and 10-h time points, EdU- labeled G1*, S/G2*, and G2/M* populations are shown.
DISCUSSION
The synchronization of cytoplasmic and nuclear events by temporally and spatially regulated activity of CyclinB1/Cdk1 for cell cycle G2/M transition has been thoroughly elucidated over decades (Barnes et al., 2001; Baumann, 2012; Coudreuse and Nurse, 2010; Draetta and Beach, 1988; Lenormand et al., 1999; Nurse, 2002; Wittenberg and Reed, 1988). Accumulating evidence indicates that not only mitochondrial morphology but also respiration is altered in cell cycle progression (Garedew et al., 2012; Owusu-Ansah et al., 2008). Notably, the ATP production in G2/M phase of colon cancer cells is predominantly dependent on mitochondrial respiration while G1 phase cells rely more on glycolysis for their energy needs during cell cycle, and the most active mitochondrial oxidative phosphorylation is found to occur in G2/M phase (Bao et al., 2013). The present study defines a novel role for CyclinB1/Cdk1 in mitochondria, by which CyclinB1/Cdk1 improves mitochondrial respiration and ATP generation to meet the increased energy demand for G2/M transition. We show that the mitochondrial abundance as well as the kinase activity of CyclinB1/Cdk1 was dramatically increased when cells are in G2/M transition. Our data further support that through phosphorylation and activation of 5 subunits of the CI in the OXPHOS, CyclinB1/Cdk1 is able to enhance CI activity and mitochondrial ATP output. Through phosphorylation and activation of its mitochondrial targets, including the 5 subunits of the CI in the OXPHOS machinery, CyclinB1/Cdk1 enhances CI activity and ATP output, as to ensure a sufficient energy supply for the critical cell division processes such as the spindle formation and centrosome separation (Nakada et al., 2010).
It has been well established that the sub-cellular redistribution of cell cycle regulators is required for checkpoint regulation during cell cycle (Gavet and Pines, 2010). Although CyclinB1 and Cdk1 are expressed in late S and G2 phases of the mammalian cell cycle, they are activated at late G2 phase (Hartwell and Weinert, 1989). The activation of CyclinB1/Cdk1 complex immediately triggers its rapid accumulation in the nucleus via a 40-fold increase in nuclear import, which is also dependent on Cdk1 activity (Gavet and Pines, 2010). The redistribution of CyclinB1/Cdk1 to the mitochondrial matrix demonstrates a unique mechanism coordinating the mitotic events in other cellular compartment and mitochondrial activity for G2/M progression. Although CyclinB1/Cdk1 was detected in the mitochondria of asynchronous cells in different cell cycle phases, the timing of mitochondrial influx of CyclinB1/Cdk1 was consistent with the accumulation of G2/M cells, importantly, the kinase activity of Cdk1 was also maximized at the G2/M (Figure 1), indicating that CyclinB1/Cdk1-mediated mitochondrial bioenergetics is integrated into the overall process of G2/M transition.
Reversible protein phosphorylation plays a key role in the regulation of various mitochondrial functions (Pagliarini, 2006). A large number of Cdk-phosphorylated proteins have been detected in mitotic HeLa cells (Dephoure et al., 2008). Among this profile, Drp-1 together with other mitochondrial proteins was identified to be the target of Cdk1-mediated phosphorylation in early mitosis to stimulate mitochondrial fission (Taguchi et al., 2007). In addition, CI subunit NDUFV3 is identified as a Cdk1 substrate during HeLa cell mitosis (Dephoure et al., 2008), indicating that the CyclinB1/Cdk1 targets a multitude of mitochondrial proteins during cell division. However, the functions of the mitochondrial phosphoproteins, especially those involved in cell cycle-related mitochondrial bioenergetics, remain unknown. CyclinB1/Cdk1 mitochondrial targets identified in this study include those involved in electron transport (CI-CV), tricarboxylic acid cycle, amino acid and lipid metabolism, and redox balance (Figure 2G–H, Table S1). CI not only functions as the major entry point for electrons into the respiratory chain, but also the largest complex in the OXPHOS system. CI dysfunction impairs electron transport, leading to major defects in overall mitochondrial energy metabolism (Janssen et al., 2006). We found that the optimal CyclinB1/Cdk1 phosphorylation motifs are predominantly present in the CI subunits and CyclinB1/Cdk1-mediated CI phosphorylation enhances CI activity synchronously with G2/M transition. Furthermore, mitochondrial matrix-localization of CyclinB1/Cdk1 complex evidently brings it in close proximity to the CI that resides on the inner membrane, implicating the availability of CyclinB1/Cdk1 for a quick response to enhanced ATP demands of the cell via phosphorylation of its mitochondrial targets. Therefore, Cdk1-mediated increase in the mitochondrial ATP output is likely a result of up-regulated phosphorylation of CI subunits and subsequently enhanced CI activity at G2/M phase.
Identification of new mitochondrial CyclinB1/Cdk1 substrates will be critical for further revealing the mechanistic insights of cell division (Ubersax et al., 2003). In this study, proteomics approach identified a group of novel Cdk1 targets in mitochondria of G2/M cells. However, our approach failed to identify some of the known mitochondrial CyclinB1/Cdk1 substrates including Drp-1 (Taguchi et al., 2007), Bad (Konishi et al., 2002), Bcl-2 (Furukawa et al., 2000), Mcl-1 (Harley et al., 2010) and Bcl-xl (Schmitt et al., 2007); potentially due to cell type, context and stimuli dependent nature of interaction of Cdk1 with these substrates. Cdk1 interacts with Bcl-2 in M phase arrested cells (Pathan et al., 2001); with BAD in apoptotic neurons (Konishi et al., 2002); and with Mcl-1 during mitotic arrest (Harley et al., 2010), showing that none of these interactions have been found in G2/M phase cells. Furthermore, although Bcl-xl localizes into the outer membrane of the mitochondria, its interaction with Cdk1 was identified in the nucleus (Schmitt et al., 2007). It has been shown that CyclinB1/Cdk1 phosphorylates mitochondrial p53 in colon cancer (Nantajit et al., 2010) and survivin in yeast (O’Connor et al., 2002); however we did not detect these in the G2/M phase mitochondria in MCF10A cells. Nevertheless, a recent study found that MnSOD (SOD2) can be phosphorylated and activated by CyclinB1/Cdk1 in the mitochondria in MCF10A cells (Candas et al., 2013), which was, indeed, detected in our profile of in vitro CyclinB1/Cdk1 targets (Table S1). The targeting of MnSOD by Cdk1 may be beneficial to cells for coping with the increased superoxide levels resulting from the mitochondrial CyclinB1/Cdk1-mediated enhancement of oxidative phosphorylation, suggesting a potential network of CyclinB1/Cdk1-phosphorylated elements in the regulation of mitochondrial metabolic activity to meet an “extra” cellular fuel demand for a normal cell division.
The mechanisms causing CyclinB1/Cdk1 mitochondrial influx are largely unknown. Two types of mitochondrial targeting signals have been described; pre-sequences and internal signals (Neupert and Herrmann, 2007; Truscott et al., 2003). Pre-sequences direct precursor proteins into the mitochondrial matrix, inner membrane or intermembrane space via the TOM complex on the outer mitochondrial membrane. The internal targeting information, best characterized for members of the carrier family of the mitochondrial inner membrane, is hidden in the mature protein amongst other amino acid residues important for folding and function. The internal targeting signals can spread throughout the length of the protein. The proteins with mitochondria-targeting sequences (MTS)are transferred into the mitochondria via their interactions with chaperon proteins that can be recognized by the TOM complex (Neupert and Herrmann, 2007; Truscott et al., 2003). Prediction of mitochondrial localization of cellular proteins can be performed based on the presence of an N-terminal pre-sequence (Claros and Vincens, 1996). In this study, we utilized Mitoprot database to search for pre-sequences within CyclinB1 and Cdk1. Mitoprot revealed no pre-sequences in Cdk1; however, an N-terminal 42-residue region of CyclinB1 that is known to be responsible for its cytoplasmic localization and nuclear shuttling (Pines and Hunter, 1994), was found to contain MTS and thus may be responsible for CyclinB1/Cdk1 influx into mitochondria. Like many other mitochondrial proteins, CyclinB1/Cdk1 may be relocated to mitochondria via internal signals within the protein sequence and chaperon proteins (Chacinska et al., 2009).
Cellular proliferation represents significant energetic commitments and presumably requires an increase in energy. The significance of CyclinB1/Cdk1 in G2/M cell cycle transition and the importance of mitochondria in cellular ATP generation have been well-appreciated over decades. Here, we reveal a mechanism by which cells can sense and deal with the increased cellular fuel demand at the G2/M transition via CyclinB1/Cdk1-mediated CI activation and mitochondrial ATP generation, which represents a unique synchronization between mitochondrial activity and G2/M transition to ensure a sufficient energy supply for cell cycle progression.
EXPERIMENTAL PROCEDURES
CyclinB1/Cdk1 in vitro Kinase Assay with 2D gels
CyclinB1/Cdk1 kinase assay was performed with established methods (Pan and Hurwitz, 1993). In the presence of phosphatase and protease inhibitors, 1 mg of mitochondrial proteins was incubated with 800 units of CyclinB1-Cdk1 in 1200 μL of 1 × kinase assay reaction buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, 0.01% Brij 35, pH 7.5) supplemented with 100 μM of ATP and 100 μCi/μmol of [γ-32P] ATP at 30°C for 2 h (Atherton-Fessler et al., 1993). In vitro kinase assay in the absence of CyclinB1/Cdk1 was set as negative control. Analysis by two dimensional (2D) SDS-PAGE was performed after 2 h of reaction. The protocol of CyclinB1/Cdk1 in vitro kinase assay with mitochondria-localized immuno-captured CyclinB1/Cdk1 is presented in the supplementary information (SI).
Two-dimensional Fluorescence Gel Electrophoresis
One milligram of mitochondrial proteins was used for CyclinB1/Cdk1 kinase assay and then precipitated using 20% TCA before labeling with 400 pmol of Cy2 (GE Healthcare), followed by electrophoresis and staining with Pro-Q Diamond solution (Molecular Probes) and CyDye and imaging by Typhoon 9410 laser scanner (Amersham). For 2D-DIGE analysis with [γ-32P] ATP-incorporated phosphorylation, the gel was exposed to phosphor-screen for at least 12 h and the image was acquired using the Phosphostorage mode on Typhoon 9410 Scanner.
CI Activity Assay
Mitochondrial CI activity was determined by measuring rotenone sensitive NADH:cytochrome c reductase (NCCR) activity (Martinvalet et al., 2008). Mitochondrial fractions were rinsed once with hypotonic buffer and the pellet was sonicated and the assay was performed at 30°C using triplicates with 5 μg of mitochondrial proteins in 100 μL of potassium phosphate buffer. The oxidation rate of NADH was determined using Spectra Max M2e at the wavelength of 340 nm for 3 min. The assay was also performed in the presence of 5 μM of rotenone to determine the rotenone-insensitive CI activity. For CI rescuing experiments, cells were transfected with CI subunit-specific siRNA’s (10–30 nM) by Lipofectamine™ RNAi MAX for 48 h and then continuously transfected with the wild type or mutant CI subunit for an additional 48 h using Lipofectamine 2000. CI activity was calculated by subtracting the rotenone-insensitive activity from the total activity of NADH-mediated oxidation rate, and was presented as optical density value (OD value).
Oxygen Consumption
Oxygen consumption was determined following reported methods with a Clark-type oxygen electrode (Digital Model 20; Rank Brothers, Cambridge, UK) interfaced with Pico software to provide a real-time display of oxygen concentration (PicoLog Recorder and Technology, Interworld Electronics, Point Roberts, UK). Cells were gently scraped off the culture plates and 1×107 cells were suspended into 200 μl of respiration solution (20 mm HEPES, pH 7.1, 250 mm sucrose, 10 mm MgCl2, 2 mm phosphate) and permeabilization of plasma membrane was carried out by digitonin (25 μg/ml). Respiration was initiated with the addition of CI substrate glutamate/malate at final concentration of 12.5 mM.
ATP Assay
Cellular ATP content was determined in cell lysates using a luciferase-based ATP assay kit (Molecular Probes). After designated treatments, cells were rinsed twice with cold PBS, 40 μL of cold 0.5% (W/V) trichloroacetic acid (TCA) were added and incubated on ice with shaking for 20 min. Cells were supplemented with 140 μL of 250 mM Tris-Acetate (pH 7.75) per sample and 10 μL of this cell suspension was mixed with 90 μL of ATP Assay Solution. ATP levels were determined using a Turner TD20/20 Luminometer (Promega). Normalization was performed with the protein concentration in the cell lysate.
Detection of Mitochondrial Superoxide
Cells (7 × 103) were seeded in 96-well plates and grown for 24 h before experiments. After designated treatments, cells were incubated at 37°C for 10 min with 5 μM of MitoSOX solution (Molecular Probes), washed three times with PBS (pH 7.4) and resuspended in 100 μL of PBS (pH 7.4). Mitochondrial superoxide was measured at excitation/emission wavelengths of 510/580 nm using Spectra Max M2e plate reader (Molecular Devices).
Statistical Analysis
Data are presented as mean ± SD. Student’s t test was used to evaluate two group comparisons and a value of p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Grete Adamson at Electron Microscopy Laboratory, University of California Davis School of Medicine for technical assistance in mitochondrial analysis, and Dr. Haiying Hang at Institute of Biophysics, Chinese Academy of Sciences, for analysis of cell cycle data. This work was supported by NIH grants CA133402, CA152313 and Department of Energy Office of Science DE-SC0001271 (JL)and NIH Intramural Research Program of NIDDK(LW, TF).
Footnotes
References
- Antonyuk SV, Han C, Eady RR, Hasnain SS. Structures of protein-protein complexes involved in electron transfer. Nature. 2013;496:123–126. doi: 10.1038/nature11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton-Fessler S, Parker LL, Geahlen RL, Piwnica-Worms H. Mechanisms of p34cdc2 regulation. Mol Cell Biol. 1993;13:1675–1685. doi: 10.1128/mcb.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Mukai K, Hishiki T, Kubo A, Ohmura M, Sugiura Y, Matsuura T, Nagahata Y, Hayakawa N, Yamamoto T, et al. Energy management by enhanced glycolysis in G1-phase in human colon cancer cells in vitro and in vivo. Mol Cancer Res. 2013;11:973–985. doi: 10.1158/1541-7786.MCR-12-0669-T. [DOI] [PubMed] [Google Scholar]
- Barnes EA, Kong M, Ollendorff V, Donoghue DJ. Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J. 2001;20:2214–2223. doi: 10.1093/emboj/20.9.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K. Cell cycle: Order in the pericentriolar material. Nat Rev Mol Cell Biol. 2012;13:749. doi: 10.1038/nrm3471. [DOI] [PubMed] [Google Scholar]
- Candas D, Fan M, Nantajit D, Vaughan AT, Murley JS, Woloschak GE, Grdina DJ, Li JJ. CyclinB1/Cdk1 phosphorylates mitochondrial antioxidant MnSOD in cell adaptive response to radiation stress. J Mol Cell Biol. 2013;5:166–175. doi: 10.1093/jmcb/mjs062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- Davis PK, Ho A, Dowdy SF. Biological methods for cell-cycle synchronization of mammalian cells. Biotechniques. 2001;30:1322–1326. 1328, 1330–1321. doi: 10.2144/01306rv01. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draetta G, Beach D. Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell. 1988;54:17–26. doi: 10.1016/0092-8674(88)90175-4. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Iwase S, Kikuchi J, Terui Y, Nakamura M, Yamada H, Kano Y, Matsuda M. Phosphorylation of Bcl-2 protein by CDC2 kinase during G2/M phases and its role in cell cycle regulation. J Biol Chem. 2000;275:21661–21667. doi: 10.1074/jbc.M906893199. [DOI] [PubMed] [Google Scholar]
- Garedew A, Andreassi C, Moncada S. Mitochondrial dynamics, biogenesis, and function are coordinated with the cell cycle by APC/C CDH1. Cell Metab. 2012;15:466–479. doi: 10.1016/j.cmet.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. 2010;189:247–259. doi: 10.1083/jcb.200909144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hunt T. Embryology. Under arrest in the cell cycle. Nature. 1989;342:483–484. doi: 10.1038/342483a0. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA. Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Kennady PK, Ormerod MG, Singh S, Pande G. Variation of mitochondrial size during the cell cycle: A multiparameter flow cytometric and microscopic study. Cytometry A. 2004;62:97–108. doi: 10.1002/cyto.a.20091. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Lehtinen M, Donovan N, Bonni A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell. 2002;9:1005–1016. doi: 10.1016/s1097-2765(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Lenormand JL, Dellinger RW, Knudsen KE, Subramani S, Donoghue DJ. Speedy: a novel cell cycle regulator of the G2/M transition. EMBO J. 1999;18:1869–1877. doi: 10.1093/emboj/18.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell. 2008;133:681–692. doi: 10.1016/j.cell.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantajit D, Fan M, Duru N, Wen Y, Reed JC, Li JJ. Cyclin B1/Cdk1 phosphorylation of mitochondrial p53 induces anti-apoptotic response. PLoS One. 2010;5:e12341. doi: 10.1371/journal.pone.0012341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annual review of biochemistry. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nurse PM. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Nurse PM. Nobel Lecture. Cyclin dependent kinases and cell cycle control. Biosci Rep. 2002;22:487–499. doi: 10.1023/a:1022017701871. [DOI] [PubMed] [Google Scholar]
- O’Connor DS, Wall NR, Porter AC, Altieri DC. A p34(cdc2) survival checkpoint in cancer. Cancer Cell. 2002;2:43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Hurwitz J. Reconstitution of cyclin-dependent cdc2 and cdk2 kinase activities in vitro. J Biol Chem. 1993;268:20433–20442. [PubMed] [Google Scholar]
- Pathan N, Aime-Sempe C, Kitada S, Basu A, Haldar S, Reed JC. Microtubule-targeting drugs induce bcl-2 phosphorylation and association with Pin1. Neoplasia. 2001;3:550–559. doi: 10.1038/sj.neo.7900213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo G, Matera M, Moro N, Ruggiero FM, Paradies G. Mitochondrial complex I dysfunction in rat heart with aging: critical role of reactive oxygen species and cardiolipin. Free Radic Biol Med. 2009;46:88–94. doi: 10.1016/j.freeradbiomed.2008.09.031. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler MM, King MS, Robinson AJ, Armstrong FA, Harmer J, Hirst J. Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance. Proc Natl Acad Sci U S A. 2010;107:1930–1935. doi: 10.1073/pnas.0908050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, Jiao X, Li A, Zhang X, Lu Y, et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, McCoy JP, Jr, Finkel T. Coordination of mitochondrial bioenergetics with G1 phase cell cycle progression. Cell Cycle. 2008;7:1782–1787. doi: 10.4161/cc.7.12.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Beauchemin M, Bertrand R. Nuclear colocalization and interaction between bcl-xL and cdk1(cdc2) during G2/M cell-cycle checkpoint. Oncogene. 2007;26:5851–5865. doi: 10.1038/sj.onc.1210396. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Thierbach R, Voigt A, Drewes G, Mietzner B, Steinberg P, Pfeiffer AF, Ristow M. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J Biol Chem. 2006;281:977–981. doi: 10.1074/jbc.M511064200. [DOI] [PubMed] [Google Scholar]
- Sweet S, Singh G. Accumulation of human promyelocytic leukemic (HL-60) cells at two energetic cell cycle checkpoints. Cancer Res. 1995;55:5164–5167. [PubMed] [Google Scholar]
- Sweet S, Singh G. Changes in mitochondrial mass, membrane potential, and cellular adenosine triphosphate content during the cell cycle of human leukemic (HL-60) cells. J Cell Physiol. 1999;180:91–96. doi: 10.1002/(SICI)1097-4652(199907)180:1<91::AID-JCP10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12:658–665. doi: 10.1016/s0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- Terry NHA, White RA. Flow cytometry after bromodeoxyuridine labeling to measure S and G2+M phase durations plus doubling times in vitro and in vivo. Nat Protocols. 2006;1:859–869. doi: 10.1038/nprot.2006.113. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Brandner K, Pfanner N. Mechanisms of protein import into mitochondria. Curr Biol. 2003;13:R326–337. doi: 10.1016/s0960-9822(03)00239-2. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI. Control of the yeast cell cycle is associated with assembly/disassembly of the Cdc28 protein kinase complex. Cell. 1988;54:1061–1072. doi: 10.1016/0092-8674(88)90121-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.