Abstract
Overexpression of human epidermal growth factor receptor 2 (HER2) is associated with tumor aggressiveness and poor prognosis in breast cancer. With the availability of therapeutic antibodies against HER2, great strides have been made in the clinical management of HER2 overexpressing breast cancer. However, de novo and acquired resistance to these antibodies presents a serious limitation to successful HER2 targeting treatment. The identification of novel epitopes of HER2 that can be used for functional/region-specific blockade could represent a central step in the development of new clinically relevant anti-HER2 antibodies. In the present study, we present a novel computational approach as an auxiliary tool for identification of novel HER2 epitopes. We hypothesized that the structurally and linearly evolutionarily conserved motifs of the extracellular domain of HER2 (ECD HER2) contain potential druggable epitopes/targets. We employed the PROSITE Scan to detect structurally conserved motifs and PRINTS to search for linearly conserved motifs of ECD HER2. We found that the epitopes recognized by trastuzumab and pertuzumab are located in the predicted conserved motifs of ECD HER2, supporting our initial hypothesis. Considering that structurally and linearly conserved motifs can provide functional specific configurations, we propose that by comparing the two types of conserved motifs, additional druggable epitopes/targets in the ECD HER2 protein can be identified, which can be further modified for potential therapeutic application. Thus, this novel computational process for predicting or searching for potential epitopes or key target sites may contribute to epitope-based vaccine and function-selected drug design, especially when x-ray crystal structure protein data is not available.
Introduction
Human epidermal growth factor receptor 2 (HER2) is one of four members of the EGF receptor family of receptor tyrosine kinases that mediate cell proliferation, differentiation and survival [1]. Overexpression of HER2, resulting from amplification of the ErbB2 gene, is observed in approximately 20% of breast cancers, and amplification of HER2 significantly correlates with increased disease aggressiveness and thereby with poor patient outcome [2], [3], [4], [5], [6]. Overexpression of HER2 can be detected in the early stages of breast cancer, and it is maintained in the progression to metastatic disease [7], [8], indicating that HER2 has an important effect on breast cancer progression. As a result, HER2 has become a critical therapeutic target in the treatment of breast cancer patients.
Trastuzumab, a monoclonal antibody directed against the extracellular domain of HER2, which consists of four domains (domain I, II, III and IV) [9], is currently the first choice of treatment for HER2-positive breast cancer patients, as it improves overall survival and reduces the risk of disease recurrence when administered in combination with chemotherapy (for review see [10]). Nevertheless, not all HER2 positive patients benefit from Trastuzumab treatment [11] and around 15% of breast cancer patients relapse after an initial response to trastuzumab-based therapy, suggesting that de novo or acquired resistance to trastuzumab has developed [12]. Thus, additional therapeutic agents are necessary in the treatment of HER2-positive breast cancer patients, with the aim of improving survival.
Pertuzumab is another humanized monoclonal antibody that binds to the extracellular domain II of HER2, the dimerization arm [13], thereby blocking signaling transduction that results from dimerization with other members of the EGFR family [14]. Although pertuzumab had low clinical efficacy when used alone, it has an excellent effect in HER2-positive breast cancer patients when used in combination with trastuzumab [15], [16], [17]. Pertuzumab administrated in combination with trastuzumab and docetaxel significantly prolongs the progression-free survival without increased cardiac toxic side effects in metastatic breast cancer patients [18], [19]. These data suggest that an additive or perhaps synergistic effect can be achieved using several antibodies directed against different epitopes of the same protein (HER2) [19].
Another therapeutic strategy to block HER2 makes use of small molecule tyrosine kinase inhibitors (TKIs), such as the dual EGFR/HER2 TKI lapatinib [20], [21]. Unfortunately, as it is the case for other molecular targeted therapies, the clinical responses to lapatinib tend to be short-lived. However, several lines of evidence suggest continued dependence of HER2+ breast cancers on HER2 signaling network after progression on anti-HER2 therapy (reviewed in [22]), providing a rationale for multilayered HER2 blockade. Therefore, searching for additional epitopes/targets of HER2 is needed to broaden clinical selection and improve the efficacy of anti-HER2 treatment. Currently, the use of three-dimensional (3D) structural data combined with some experimental approaches such as pepscan, phage display, or mutagenesis scanning, are the gold standard of epitope-based vaccine design [23], [24]. But 3D data are not available for all proteins, and experimental approaches are expensive and time-demanding techniques. Thus, computational processes that could function as a compensational approach to predictably identify some desirable epitopes or functional targets for rational vaccine or drug design are badly needed.
In the present study, we report a novel computational process used to predict functional motifs of HER2, which matched very well with epitopes of trastuzumab and pertuzumab (HER2's antibodies).
Materials and Methods
Data collection and protein BLAST (basic local alignment search tool)
The human HER2 amino acid sequence with accession number P04626 was compared to the UNIPROT database using BLAST via the web service (http://www.uniprot.org/) with default setting.
Structural conserved motifs scanning in PROSITE database
According to the domain organization of the ErbB family, the extracellular domain region of HER2 has ∼620 residues [25]. The human extracellular domain of HER2 (1-620) amino sequence was uploaded in the PROSITE Scan online service (http://prosite.expasy.org/scanprosite/). The databases used for structurally conserved motifs scanning included UniProtKB/Swiss-Prot, splice variants and UniProtKB/TrEMBL databases. The searching results with low level scores were also allowed to show in the output. The other parameters used the default setting.
Searching for fingerprints in the PRINTS database
The amino sequence of the extracellular domain of HER2 (ECD HER2) was aligned for fingerprints in web service (http://www.bioinf.man.ac.uk/cgi-bin/dbbrowser/fingerPRINTScan/FPScan_fam.cgi) with default settings.
Comparison of structurally conserved motifs with the epitopes of HER2's antibodies based on the crystal 3D structure
The crystal structure of the extracellular domain of the human HER2 complexed with trastuzumab Fab (PDB ID: 1N8Z) and the structure of HER2-pertuzumab complex (PDB ID: 1S78) were obtained from NCBI (http://www.ncbi.nlm.nih.gov/structure/?term=HER2). Using Cn3D software, the structurally conserved fragments of ECD HER2 were superposed and labeled on the two crystal structures. Furthermore, as supplementary reference to our prediction, FoldX (http://foldx.crg.es/) were introduced with in silico mutagenesis 'repair module', which allowed to predict the amino acids involved in the binding of HER2 to the antibodies by alanine mutation based on 'force field' statistical considerations [26].
Results
The HER2 amino acid sequence was highly conserved in mammals
We hypothesized that functional structure motifs could be conserved and maintained during evolution. In order to determine the level of homology in the HER2 amino acid sequence among mammals, available orthologs of HER2 were searched by BLAST in the UniProt database. The result showed that the HER2 protein sequence indeed was highly homologous across species with over 80% similarity in mammals, including horse (Equus caballus), pig (Sus scrofa), mouse (Mus musculus), rat (Rattus norvegicus), and cow (Bos taurus) (Table 1). This indicated that some meaningful sequences/structures of ECD HER2 protein existed among these conserved amino acid sequences.
Table 1. The identity of human HER2 protein in the UniPort database.
| Enter accession number | Organism | Protein names | Length | Identity |
| P04626 | Homo sapiens (Human) | Receptor tyrosine-protein kinase erbB-2 | 1255 | 100% |
| G3QPD2 | Gorilla gorilla gorilla (Lowland gorilla) | Uncharacterized protein | 1258 | 99% |
| K7DJM0 | Pan troglodytes (Chimpanzee) | V-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (Avian) | 1255 | 99% |
| H2QCV1 | Pan troglodytes (Chimpanzee) | Uncharacterized protein | 1247 | 99% |
| G1QK40 | Nomascus leucogenys (Hylobates leucogenys) | Uncharacterized protein | 1255 | 99% |
| P04626-4 | Homo sapiens (Human) | Isoform 4 of Receptor tyrosine-protein kinase erbB-2 | 1240 | 99% |
| I0FM22 | Macaca mulatta (Rhesus macaque) | Receptor tyrosine-protein kinase erbB-2 isoform a | 1255 | 98% |
| G7NI22 | Macaca mulatta (Rhesus macaque) | Receptor tyrosine-protein kinase erbB-2 | 1255 | 98% |
| P04626-5 | Homo sapiens (Human) | Isoform 5 of Receptor tyrosine-protein kinase erbB-2 | 1225 | 100% |
| F6WAF2 | Callithrix jacchus (White-tufted-ear marmoset) | Receptor tyrosine-protein kinase erbB-2 isoform a (Uncharacterized protein) | 1255 | 96% |
| G7PUM4 | Macaca fascicularis (Cynomolgus monkey) | Receptor tyrosine-protein kinase erbB-2 | 1231 | 98% |
| H2NU95 | Pongo abelii (Pongo pygmaeus abelii) | Uncharacterized protein | 1256 | 97% |
| F7CBG7 | Callithrix jacchus (White-tufted-ear marmoset) | Uncharacterized protein | 1239 | 96% |
| F6UJQ9 | Callithrix jacchus (White-tufted-ear marmoset) | Uncharacterized protein | 1225 | 97% |
| F6VNY4 | Equus caballus (Horse) | Uncharacterized protein | 1258 | 94% |
| Q49LT4 | Felis catus (Cat) (Felis silvestris catus) | Epidermal growth factor receptor type 2 | 1260 | 94% |
| F1PIQ9 | Canis familiaris (Dog) (Canis lupus familiaris | Receptor tyrosine-protein kinase erbB-2 | 1260 | 93% |
| H0X3G4 | Otolemur garnettii (Garnett's greater bushbaby) | Uncharacterized protein | 1256 | 93% |
| H9BB15 | Felis catus (Cat) (Felis silvestris catus) | Erb2 | 1260 | 93% |
| G1SZL0 | Oryctolagus cuniculus (Rabbit) | Uncharacterized protein | 1255 | 93% |
| M3WCC2 | Felis catus (Cat) (Felis silvestris catus) | Uncharacterized protein | 1260 | 93% |
| H9BNW8 | Ursus americanus (Euarctos americanus) | V-erb-b2 erythroblastic leukemia viral oncogene-like protein 2 | 1260 | 92% |
| G1LTV6 | Ailuropoda melanoleuca (Giant panda) | Uncharacterized protein | 1263 | 92% |
| O18735 | Canis familiaris (Dog) (Canis lupus familiaris) | Receptor tyrosine-protein kinase erbB-2 (EC 2.7.10.1) (Proto-oncogene c-ErbB-2) (p185erbB2) (CD antigen CD340) | 1259 | 92% |
| I3M2X8 | Spermophilus tridecemlineatus (Ictidomys tridecemlineatus) | Uncharacterized protein | 1259 | 93% |
| M3YU62 | Mustela putorius furo (Mustela furo) | Uncharacterized protein | 1260 | 92% |
| F1MCQ7 | Bos taurus (Bovine) | Uncharacterized protein | 1257 | 92% |
| W8FW17 | Tupaia chinensis (Chinese tree shrew) | ERBB2 | 1255 | 92% |
| A8WED5 | Canis familiaris (Dog) (Canis lupus familiaris) | HER-2 | 1242 | 93% |
| M3X794 | Felis catus (Cat) (Felis silvestris catus) | Uncharacterized protein | 1260 | 92% |
| D2I2F8 | Ailuropoda melanoleuca (Giant panda) | Putative uncharacterized protein | 1236 | 92% |
| G3SLF2 | Loxodonta africana (African elephant) | Uncharacterized protein | 1257 | 90% |
| K7GS43 | Sus scrofa (Pig) | Uncharacterized protein | 1231 | 91% |
| S7MLI8 | Myotis brandtii (Brandt's bat) | Receptor tyrosine-protein kinase erbB-2 | 1227 | 91% |
| G3H5Y0 | Cricetulus griseus (Cricetulus barabensis griseus) | Receptor tyrosine-protein kinase erbB-2 | 1256 | 89% |
| S9YT31 | Camelus ferus (Wild Bactrian camel) | Receptor tyrosine-protein kinase erbB-2 | 1253 | 90% |
| F1RWM5 | Sus scrofa (Pig) | Uncharacterized protein | 1234 | 88% |
| Q8K3F9 | Rattus norvegicus (Rat) | Neu protooncoprotein | 1259 | 88% |
| F1LRR9 | Rattus norvegicus (Rat) | Receptor tyrosine-protein kinase erbB-2 | 1257 | 88% |
| P06494 | Rattus norvegicus (Rat) | Receptor tyrosine-protein kinase erbB-2 | 1257 | 88% |
| W5PQJ8 | Ovis aries (Sheep) | Uncharacterized protein | 1251 | 89% |
| Q60553 | Mesocricetus auratus (Golden hamster) | Receptor tyrosine-protein kinase erbB-2 | 1254 | 88% |
| P70424 | Mus musculus (Mouse) | Receptor tyrosine-protein kinase erbB-2 | 1256 | 88% |
| H0V6A0 | Cavia porcellus (Guinea pig) | Uncharacterized protein | 1236 | 88% |
| J3QLU9 | Homo sapiens (Human) | Receptor tyrosine-protein kinase erbB-2 | 1055 | 99% |
| G1PRB8 | Sarcophilus harrisii (Sarcophilus laniarius) | Uncharacterized protein | 1244 | 85% |
| G3WQQ2 | Sarcophilus harrisii (Sarcophilus laniarius) | Uncharacterized protein | 1255 | 81% |
| F7C962 | Monodelphis domestica (Gray short-tailed opossum) | Uncharacterized protein | 1257 | 80% |
The table only shows resemblance to HER2 higher than 80%.
The ECD HER2 protein sequence contained three evolutionally conserved structural motifs
To determine the structurally conserved motifs in the ECD HER2 protein, the protein sequence from M1 to E620 was uploaded in Prosite ExPASy. We obtained three hits (Table 2).
Table 2. The structurally conserved motifs of ECD HER2 detected by PROSITE Scan.
| Predicted Conserved motifs | Amino Acid Residues | Predicted domain (condition) | Contact region |
| F1 (L244-K311; PDB sequence number.) | LHCPALVTYNTDTFESMPNPEGRYTFGASCVTACPYNYLSTDVGSCTLVCPLHNQEVTAEDGTQRCEK | Sushi (Disulfide 246C-x-293C and 277C-x-309C) | Heterodimerization region and Pertuzumab binding region. |
| F2 (N549-E558; PDB sequence number.) | NGSVTCFGPE | NHL | Trastuzumab interacting region. |
| F3 (D570-E598; PDB sequence number.) | DPPFCVARCPSGVKPDLSYMPIWKFPDEE | ZF-THAP type, degenerate | Trastuzumab interacting region. |
The amino acids in bold present involved residues in epitopes of antibodies: trastuzumab and pertuzumab.
The first hit, Fragment 1 (F1), was predicted as a type of Sushi domain by the presence of four conserved cysteine residues (C246-C293 and C277-C309), forming two disulfide bounds [27], which covered amino acid residues from L244 to K311 according to the PDB sequence number.
The second hit, Fragment 2 (F2), was predicted as a NHL domain, defined by amino acid homological presenting in three proteins: NCL-1, HT2A, and LIN-41[28], which was located from N549 to E558 (PDB sequence number). Finally, the third hit, Fragment 3 (F3), was predicted as a degenerated zinc finger THAP-type domain (C2H2) [29], [30], which embraced 29 amino residues from D570 to E598 (PDB sequence number) (Table 2). The zinc finger HTAP degenerate domain mapped to the N-terminal location of a sequence-specific DNA-binding factor [31], [32].
Three structurally conserved fragments are located in functional domains of ECD HER2
In order to explore the superposition of those structurally conserved fragments with functional domains of ECD HER2, the predicted fragments were superposed on the crystal structure of ECD HER2 (Figure 1) [33]. F1 (L244-K311) was completely located at center of domain II (G200-R329), which formed the exposed dimerization arm of HER2, suggesting that the predicted fragment F1 involved in HER2 dimerization and hereby could affect the activation of HER2. F2 (N549-E558) and F3 (D570-E598) were located at domain IV (W499-N607) which was the arm extending to domain II and further mediating the activation of HER2. Trastuzumab worked well in clinic via its interaction with this domain [33]. The results indicated that these two predicted fragments are matched with HER2's functional domains and probably they were critical sites for HER2's activation. (To keep consistency with the crystal structure study [33], we used the PDB residues number here, rather than the sequential residues number of HER2.)
Figure 1. The location of the three structurally conserved motifs (F1, F2 and F3) on ECD HER2 crystal structure.
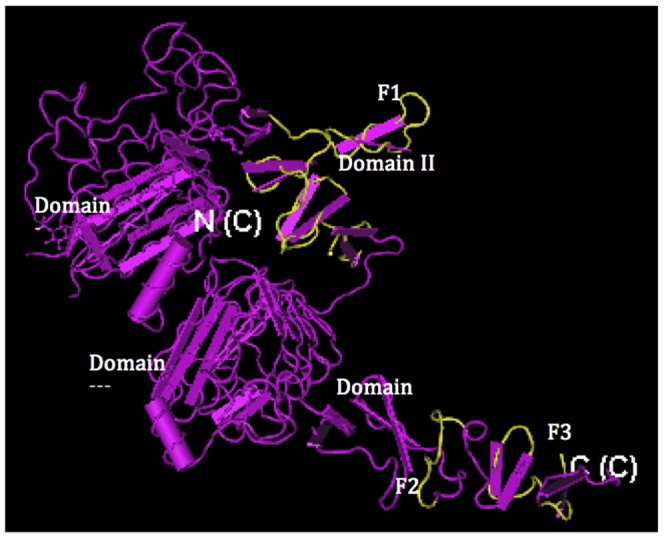
Purple: ECD HER2 receptor; Yellow: structurally conserved motifs (F1, F2 and F3). N(C) and C(C) are ECD HER2's N-terminal and C-terminal, respectively.
The structurally conserved fragments matched well with epitopes of HER2 antibodies
To investigate whether the predicted structurally conserved motifs/fragments could be key regions for the biological function(s) of HER2, they were compared with epitopes of HER2 antibodies based on the HER2: trastuzumab [33]/HER2: pertuzumab [14] co-crystal structures, respectively. The result showed that the conserved F1 did not locate in the interaction surface with trastuzumab, whereas the F2 and F3 were involved in the interface of trastuzumab binding (Figure 2A). According to Cho et al., three loops in the domain IV of HER2 are involved in the HER2-Trastuzumab binding surface [33] (Figure 2). We found the two residues of F2 P557 and E558 were located in loop 1 (P557-D561). The four residues of F3 D570, P571, P572 and F573 were positioned in loop 2 (D570-F573), and the residues of F3 K593, F594, P595, D596, E597 and E598 were found in loop 3 (K593-P603) (Figure 2B). The results showed the predicted F2 and F3 partly consisted of the epitope of trastuzumab, implying the predicted F2 and F3 probably are the pivotal regions for HER2's activation.
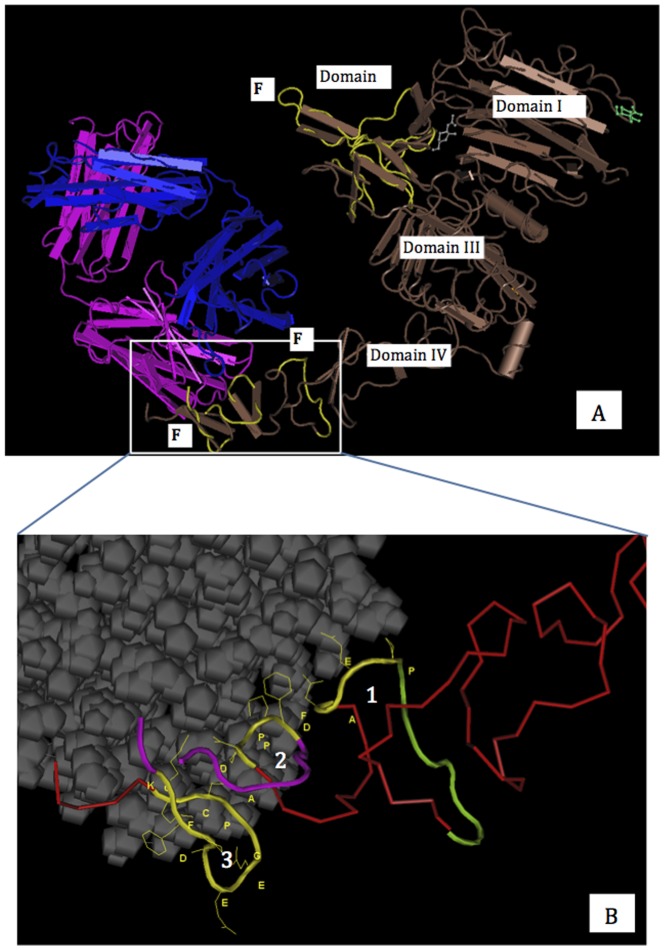
Figure 2. Comparison of structurally conserved motifs of ECD HER2 with the epitope of trastuzumab.
A: The location of F1, F2 and F3 in the tube worm representation of the HER2-trastuzumab complex. HER2 is colored dark brown and trastuzumab is in chain form: light chain in bright purple and heavy chain in blue. The structurally conserved motifs (F1, F2 and F3) are highlighted in yellow. B: Enlarged view of binding interface between ECD HER2 and trastuzumab. Trastuzumab is shown in grey, with a space-fill protein backbone and side chains style. The numbers indicted in the figure show the three loop regions (highlighted in yellow) mediating the interaction with trastuzumab. F2 is shown in light green and F3 is in bright purple. The overlapped residues between F2, F3 and interaction loops are P557 and E558 (F2, on loop 1 (P557-D561)), D570, P571, P572 and F573 (F3, on loop 2 (D570-F573)), K593, F594, P595, D596, E597 and E598 (F3, on loop 3 (K593-P603)). According to the crystal structure of HER2-trastuzumab complex [33], there are eight residues invisible in the F3 region. The numbers shown in the figure are consistent with PDB data rather than sequential numbering for HER2 residues.
The structural conserved F1 was completely embedded in the center of domain II (in blue, Figure 3A), which has been revealed as the interface with pertuzumab [14]. Furthermore, we found that the directly interacted residues of HER2 with pertuzumab, H245, Y252, F257, K311 and H296, and important residues of HER2, H245, V286, S288, L295, H296 and K311, for affinity with pertuzumab [14] were located in the predicted conserved F1 (Figure 3B, showed in dark green). This result suggested that the predicted conserved F1 contained pertuzumab's epitopes.
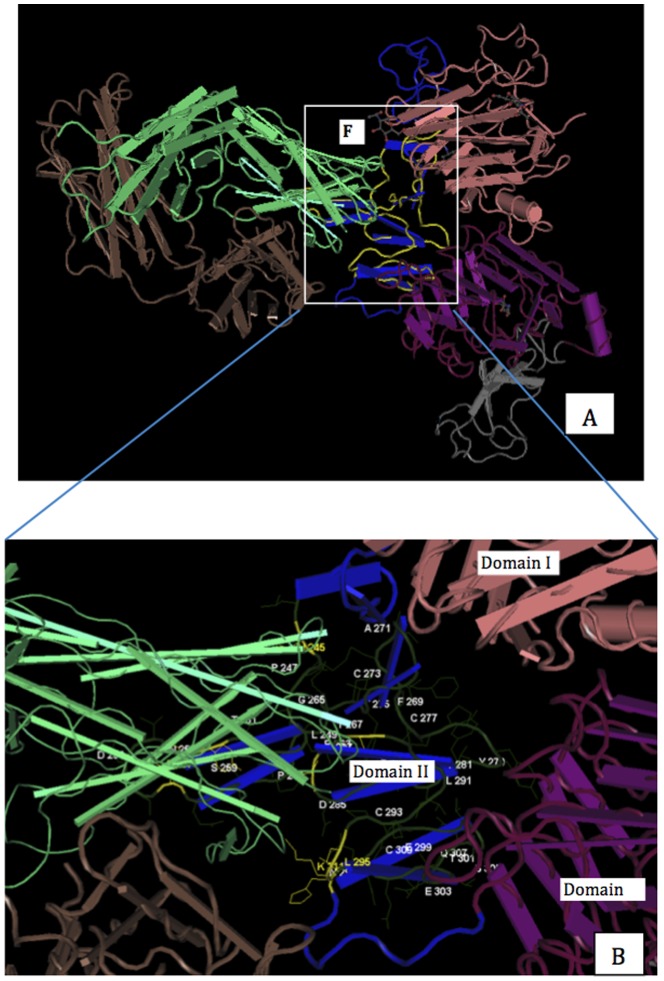
Figure 3. Comparison of structurally conserved motifs of ECD HER2 with the epitope of pertuzumab Fab.
A: The location of F1 in the tube worm representation of the HER2-pertuzumab complex. According to Franklin, M.C. et al. [14], ECD HER2 is colored according to domain: domain I in pink, domain II in blue, domain III in dark purple and domain IV in grey. Pertuzumab is shown as a chain: light chain in green and heavy chain in dark grey. The highlighted in yellow shows the location of structurally conserved F1. B: The enlarged view shows the binding interface between ECD HER2 and pertuzumab. Domain II is shown in blue, and the F1 is shown as a tube worm with wire side-chains in dark green. The residues involving in the interaction with pertuzumab [14] are highlighted in yellow. The numbers shown in the figure are consistent with PDB data rather than sequential numbering for HER2 residues.
All these structural comparisons results between the predicted conserved fragments and epitopes of HER2's antibodies implied that the structurally/functionally conserved fragments of HER2 probably contain more potentially powerful epitopes/target sites.
The ECD HER2 protein sequence contained some conserved fingerprints
For purpose of combination of linear feature and structural characterization of epitopes to reduce the prediction error, the PRINTS database, another deterministic pattern, was introduced. And then, 9 top fingerprints were significantly screened from the ECD HER2 sequence with p-value<0.01 (Table 3). Among them, the fingerprints ANATRNSFRASE 280, BACEL 290, and TRANSYNTHGLU 316 overlapped with F1; the fingerprints 4FE4SFRDOXIN 577 partly overlapped with F2; the fingerprints ALPHATUBULIN 600 and 4FE4SFRDOXIN 595 overlapped with F3 (The position number is consistent with ECD HER2 amino acid number).
Table 3. The linearly conserved motifs (fingerprints) of ECD HER2 searched by PRINTS.
| Fingerprint Name | P-value | Position | Sequence | Matched fingerprints with structurally conserved motifs |
| ALPHATUBULIN | 9.2×10−6 | 439 | GAYSLTLQGLGIS | |
| 600 | CPSGVKPDLSYMPI * | F3 | ||
| ANATRNSFRASE | 4.1×10−5 | 79 | EVQGYVLIAHNQVRQVPLQRLRIV | |
| 280 | ESMPNPEGRYTFGASCVTACPYNY * | F1 | ||
| ONCOGENEAML1 | 110 | DNYALAVLDNGDPLNNTTPVTG* | ||
| 0.00021 | 374 | LAFLPESFDGDPASNTAPLQ | ||
| FMOXYGENASE | 125 | NTTPVTGASPGGLRELQ | ||
| 0.00048 | 354 | RAVTSANIQEFAGCKKIF | ||
| BASICPTASE | 236 | CHEQCAAGCTG | ||
| 0.00052 | 529 | VNCSQFLRGQECVEEC | ||
| 4FE4SFRDOXIN | 577 | FGPEADQCVACA* | F2 | |
| 0.0013 | 595 | FCVARCPSGVKP * | F3 | |
| BACE1 | 113 | ALAVLDNGDPLNNTTPVTGAS | ||
| 0.0023 | 290 | TFGASCVTACPYNYLSTDVG | F1 | |
| TRNASYNTHGLU | 0.0033 | 177 | NQLALTLIDTNRSR* | |
| 316 | PLHNQEVTAED * | F1 | ||
| ANIONEXCHNGR | 0.0020 | 11 | LLLALL | |
| 251 | DCLACLHF* |
The fingerprints overlapped with structurally conserved motifs, but not present in the interface of HER2: antibodies were showed in italics. Underlined residues present key sites involved in epitopes of trastuzumab and pertuzumab.
*indicates that this motif affected the interaction energy between HER2 and pertuzumab and Fab trastuzumab in 1S78 and 1N8Z 3D structures, respectively.
Some of both linear and structural conserved regions were beyond the epitopes of HER2's antibodies
To further explore the match between the linear and structural conserved regions and epitopes of HER2's antibodies (trastuzumab and pertuzumab), they were compared. Some linear and structural conserved overlapped regions, including fingerprints ALPHATUBULIN 600, ANTRNSFRASE 280, 4FE4SRDOXIN 595, BACE1 290 and TRNASYNTHGLU 316, did not present in the interface of HER2:antibodies (Table 3. shaded fingerprints). The result showed that the predicted structurally conserved fragments contained some linearly conserved segments, suggesting that those segments could be other efficient epitopes or drug target sits. In order to evaluate the effect of those conserved motifs listed in Table 3 on binding energies to pertuzumab and Fab trasutumab, FoldX was introduced as a supplementary reference. The result showed that some amino acids in the overlapped regions of structurally and linearly conserved motifs were predictively involved in the interaction with trastuzumab and pertuzumab (Table 3), implying our prediction performance was reliable.
Discussion
Great achievements have been obtained with antibody-based therapies in the treatment of cancer [34]. Trastuzumab and pertuzumab, two antibodies targeting the HER2 protein, are good examples of antibody-based therapies. Clinical studies have shown that these two monoclonal antibodies, which are directed against different epitopes of HER2, display an additive/synergistic anti-tumor effect when they are used in combination with docetaxel in the treatment of metastatic breast cancer, even though some patients developed resistance [15], [16], [19]. Hence, it is conceivable that using several antibodies toward the same molecular target results in an additive/synergistic effect, since the antibodies are immunologically generated towards different parts of the antigen. Therefore, more efficient antibodies against HER2 could provide more options for treatment and greater benefits for HER2-positive breast cancer patient than those presently in use.
The identification of the most efficient epitopes/target sites, as the first step for epitope-based vaccine/drug design, is still an unsolved problem. In the present study, we hypothesized that evolutionary structurally conserved fragments of the ECD of HER2 contain efficient druggable epitopes/targets.
We used the PROSITE database that can detect biologically meaningful inter-domains, based on manually derived alignments and extensive manually curated documentation [35], to predict structurally conserved motifs. We also introduced the PRINTs database which is a compendium of protein motifs with a series of conserved regions of aligned sequences, since the linear segment is another essential part of protein epitopes [36]. We found that all predicted fragments were located in the important dimerization arms regions of HER2: within domain II and IV. Comparing the structurally conserved fragments of HER2 with epitopes of trastuzumab and pertuzumab, respectively, we further found that the predicted fragments were involved in the formation of epitopes for trastuzumab and pertuzumab, which are the most efficient known antibodies against HER2. Thus, our results showed that the evolutionary structurally conserved fragments of HER2 contain novel epitopes/targets, not being targeted by trastuzumab or pertuzumab, which strongly supports our hypothesis.
Furthermore, considering that the linear conserved motifs most probably have a meaningful role in the formation of druggable epitopes, we combined the two kinds of motifs and compared the results generated. Some of these segments were not presented in the epitopes of trastuzumab and pertuzumab, but included in the structurally conserved fragments. The result implies that these segments could be potential new epitopes/targets of ECD HER2 for peptide-based vaccine and drug design. Hence, this novel computational process provides a new complementary approach for efficiently selecting critical regions/sites as epitope for drug targeting.
Numerous studies have shown that HER2 is involved in many fundamental cellular processes, including cell migration, cell survival and cell proliferation and differentiation (for review [1]), suggesting that the basal function of HER2 is probably shared in mammals. Indeed, our results confirmed that the homology of HER2 amino acid sequence is very high in mammal species with over 80% similarity. Thus, it is reasonable to suggest that some similar functional inter-structures exist in the HER2 protein. Three-conserved inter-structure motifs of ECD HER2 (named as F1, F2 and F3) were identified as best hits using the PROSITE Scan. The first two, F1 and F2, corresponding to the Sushi and NHL domains in the ECD of HER2, respectively, are linked to protein-protein interaction, protein-binding modules and cell adhesion, all of which are coherent with the function of HER2, a growth factor receptor protein [37], [38], [39], [40], [41].
However, the third domain, F3, was predicted as a degenerated zinc finger HTAP domain, which has DNA-binding capability. It is rather intriguing that the trans-membrane part of HER2 should contain a DNA-binding domain, even if HER2 reportedly can function as a transcriptional regulator [42]. Studies have demonstrated that HER2 is involved in COX2
[43], and ribosomal RNA gene [44] transcriptional regulation and HER2 can be located to the nucleus, where transcriptional activity takes place reasonably [42]. In addition, a truncated HER2 has also been reported to be located to nucleus and to contribute to acquired resistance to HER2 kinase inhibitors [45], [46]. Taken as a whole these lines of evidence suggest that a HER2 may function as a transcriptional regulator, in which case a DNA-binding domain is not a remarkable finding.
We found that all identified fragments, based on their predicted structural features, can be rationalized to match the activation of HER2. Hence, this prediction approach probably is a believable method for drug design, especially when one selects a protein or activity of a protein to inhibit.
To add bio-function assessment to the predictive model, in the present study, the correspondence between localization of predicted fragments and HER2's functional domains was studied using co-crystal structure of ECD HER2: antibody.
The X-ray crystal structure of ECD HER2 has revealed that the domain II of HER2, plays a key role in the activation of the receptor, through which contacting and forming dimerization with other ErbB family members [33], [47]. According to our hypothesis, some of evolutionally conserved fragments should be included in domain II of HER2. Indeed, F1, as predicted as Sushi domain, located in the center of domain II and completely overlapped with interaction surface with pertuzumab [14]. Furthermore, Franklin et al. revealed by a crystallographic study that the residues H245, Y252, F257, H296 and K311 interact with pertuzumab directly, and the residues H245, H296, K311, V286, S288 and L295 significantly mediated the affinity of HER2 to pertuzumab. Without exception, we also found that all these pivotal residues located in the predicted F1, as expected. This further confirms that the structurally conserved motif F1 probably has more efficient, potential epitopes/targets.
Results from X-ray structure analysis have shown that domain IV of HER2 is another important domain for activation of HER2, being involved in receptor-receptor contact [48], [49], suggesting that the domain IV should contain a receptor-receptor contacting structure. Our prediction showed that a NHL structure in F2, which has been proposed to be a protein-protein interaction structure [39], [41], exists in domain IV. Furthermore, the X-ray crystal structure of the ECD HER2 complex with trastuzumab has also revealed that domain IV of HER2 is the domain of epitope of trastuzumab, by its' loop 1 (P557-D561), loop 2 (D570-F573), and loop 3 (K593-P603) to contact trastuzumab [33]. Similarity, our predicted fragments F2 and F3 partly formed the epitope of trastuzumab, which meet our expectations.
Since the linear peptides are the basis of forming epitopes, we introduced the PRINTScan tool to scan the linear conserved segment and compared these with structurally conserved motifs. We found that some fingerprints also appeared in the rest residues of the structurally conserved motifs (highlighted in Table 2). These results indicate that the overlapped regions in structurally and linearly conserved motifs could be potentially efficient epitopes/targets for a new vaccine or drug design.
By and large, one can distinguish three main mechanism of trastuzumab resistance: (1) functional bypass of blockade by either up-regulation of downstream signaling or equivalent alternate pathways, (2) failure to elicit immune-mediated killing of tumor cells, and (3) hindrance of antibody binding to HER2. With respect to the latter, our study may provide cues for novel molecular mechanisms of resistance by pinpointing important biological epitopes of ECD HER2. Conversely, the approach presented here may provide the means to identify potentially druggable epitopes/targets that can bypass known impediments to antibody binding to HER2, such as expression of a constitutively active, truncated form of HER2 that lacks the ECD, and consequently the binding site of trastuzumab, or masking of trastuzumab cognate epitopes by steric hindrance of HER2 by cell surface proteins [45], [50], [51].
In conclusion, by comparing the structurally conserved motifs of ECD HER2 protein with epitopes of trastuzumab and pertuzumab, we confirmed our hypothesis that evolutionally structural conserved motifs of the ECD HER2 protein contain potentially druggable epitopes/targets. Furthermore, on the basis of a comparison between the structurally conserved motifs and the linearly conserved motifs, we proposed that some segments being potentially more important biological epitopes of ECD HER2 can be further modified for potential therapeutic application. In the present study, we also provide a novel procedure to predict or search for potential epitopes or key target sites in proteins, which may contribute to the design of epitope-based vaccines and drugs. In particular, when there is no X-ray crystal structure available, it is easy to narrow the searching region by means of this process, sharply reducing the interesting residues.
Acknowledgments
We are grateful to Henrik J. Ditzel professor, Center for Medical Biotechnology Institute of Medical Biology, University of Southern Denmark, for his comments and advice on this work.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data may be found within the paper.
Funding Statement
The study was financially supported by the National Natural Science Foundation of China and the Danish National Research Foundation, and by National Science and Technology Ministry of China (No. 2010ZX09401-306-2-22). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103: 211–225. [DOI] [PubMed] [Google Scholar]
- 2. Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, et al. (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244: 707–712. [DOI] [PubMed] [Google Scholar]
- 3. Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, et al. (1997) HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 15: 2894–2904. [DOI] [PubMed] [Google Scholar]
- 4. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, et al. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235: 177–182. [DOI] [PubMed] [Google Scholar]
- 5. Owens MA, Horten BC, Da Silva MM (2004) HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer 5: 63–69. [DOI] [PubMed] [Google Scholar]
- 6. Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, et al. (2004) HER-2 testing in breast cancer using parallel tissue-based methods. JAMA 291: 1972–1977. [DOI] [PubMed] [Google Scholar]
- 7. Latta EK, Tjan S, Parkes RK, O'Malley FP (2002) The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 15: 1318–1325. [DOI] [PubMed] [Google Scholar]
- 8. Carlsson J, Nordgren H, Sjostrom J, Wester K, Villman K, et al. (2004) HER2 expression in breast cancer primary tumours and corresponding metastases. Original data and literature review. British journal of cancer 90: 2344–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, et al. (1992) Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 89: 4285–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Esteva FJ, Yu D, Hung MC, Hortobagyi GN (2010) Molecular predictors of response to trastuzumab and lapatinib in breast cancer. Nat Rev Clin Oncol 7: 98–107. [DOI] [PubMed] [Google Scholar]
- 11. Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, et al. (2002) Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 20: 719–726. [DOI] [PubMed] [Google Scholar]
- 12. Nahta R, Esteva FJ (2006) HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res 8: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harbeck N, Beckmann MW, Rody A, Schneeweiss A, Muller V, et al. (2013) HER2 Dimerization Inhibitor Pertuzumab - Mode of Action and Clinical Data in Breast Cancer. Breast Care (Basel) 8: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, et al. (2004) Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer cell 5: 317–328. [DOI] [PubMed] [Google Scholar]
- 15. Portera CC, Walshe JM, Rosing DR, Denduluri N, Berman AW, et al. (2008) Cardiac toxicity and efficacy of trastuzumab combined with pertuzumab in patients with [corrected] human epidermal growth factor receptor 2-positive metastatic breast cancer. Clin Cancer Res 14: 2710–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baselga J, Gelmon KA, Verma S, Wardley A, Conte P, et al. (2010) Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. J Clin Oncol 28: 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cortes J, Fumoleau P, Bianchi GV, Petrella TM, Gelmon K, et al. (2012) Pertuzumab monotherapy after trastuzumab-based treatment and subsequent reintroduction of trastuzumab: activity and tolerability in patients with advanced human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol 30: 1594–1600. [DOI] [PubMed] [Google Scholar]
- 18.Sendur MA, Aksoy S, Zengin N (2012) Pertuzumab plus trastuzumab in metastatic breast cancer. N Engl J Med 366 : 1348; author reply 1349–1350. [DOI] [PubMed] [Google Scholar]
- 19. Baselga J, Cortes J, Kim SB, Im SA, Hegg R, et al. (2012) Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 366: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, et al. (2006) Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med 355: 2733–2743. [DOI] [PubMed] [Google Scholar]
- 21. Kaufman B, Trudeau M, Awada A, Blackwell K, Bachelot T, et al. (2009) Lapatinib monotherapy in patients with HER2-overexpressing relapsed or refractory inflammatory breast cancer: final results and survival of the expanded HER2+ cohort in EGF103009, a phase II study. Lancet Oncol 10: 581–588. [DOI] [PubMed] [Google Scholar]
- 22. Garrett JT, Arteaga CL (2011) Resistance to HER2-directed antibodies and tyrosine kinase inhibitors: mechanisms and clinical implications. Cancer Biol Ther 11: 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gershoni JM, Roitburd-Berman A, Siman-Tov DD, Tarnovitski Freund N, Weiss Y (2007) Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs 21: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clementi N, Mancini N, Castelli M, Clementi M, Burioni R (2013) Characterization of epitopes recognized by monoclonal antibodies: experimental approaches supported by freely accessible bioinformatic tools. Drug Discov Today 18: 464–471. [DOI] [PubMed] [Google Scholar]
- 25. Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, et al. (2003) An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell 12: 541–552. [DOI] [PubMed] [Google Scholar]
- 26. Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, et al. (2005) The FoldX web server: an online force field. Nucleic Acids Research 33: W382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reid KB, Day AJ (1989) Structure-function relationships of the complement components. Immunology Today 10: 177–180. [DOI] [PubMed] [Google Scholar]
- 28. Slack FJ, Ruvkun G (1998) A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci 23: 474–475. [DOI] [PubMed] [Google Scholar]
- 29. Pavletich NP, Pabo CO (1991) Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 252: 809–817. [DOI] [PubMed] [Google Scholar]
- 30. Lee MS, Gippert GP, Soman KV, Case DA, Wright PE (1989) Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 245: 635–637. [DOI] [PubMed] [Google Scholar]
- 31. Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, et al. (2003) The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci 28: 66–69. [DOI] [PubMed] [Google Scholar]
- 32. Bessiere D, Lacroix C, Campagne S, Ecochard V, Guillet V, et al. (2008) Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways. J Biol Chem 283: 4352–4363. [DOI] [PubMed] [Google Scholar]
- 33. Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, et al. (2003) Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 421: 756–760. [DOI] [PubMed] [Google Scholar]
- 34. Ben-Kasus T, Schechter B, Sela M, Yarden Y (2007) Cancer therapeutic antibodies come of age: targeting minimal residual disease. Mol Oncol 1: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, et al. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Research 34: W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivanciuc O, Schein CH, Garcia T, Oezguen N, Negi SS, et al. (2009) Structural analysis of linear and conformational epitopes of allergens. Regul Toxicol Pharmacol 54: S11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Vega S, Iwamoto T, Nakamura T, Hozumi K, McKnight DA, et al. (2007) TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J Biol Chem 282: 30878–30888. [DOI] [PubMed] [Google Scholar]
- 38. Wei XQ, Orchardson M, Gracie JA, Leung BP, Gao BM, et al. (2001) The sushi domain of soluble IL-15 receptor alpha is essential for binding IL-15 and inhibiting inflammatory and allogenic responses in vitro and in vivo. Journal of Immunology 167: 277–282. [DOI] [PubMed] [Google Scholar]
- 39. Loer B, Bauer R, Bornheim R, Grell J, Kremmer E, et al. (2008) The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat Cell Biol 10: 422–428. [DOI] [PubMed] [Google Scholar]
- 40. Souri M, Kaetsu H, Ichinose A (2008) Sushi domains in the B subunit of factor XIII responsible for oligomer assembly. Biochemistry 47: 8656–8664. [DOI] [PubMed] [Google Scholar]
- 41. Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK (2003) Model of the brain tumor-Pumilio translation repressor complex. Genes Dev 17: 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie Y, Hung MC (1994) Nuclear localization of p185neu tyrosine kinase and its association with transcriptional transactivation. Biochem Biophys Res Commun 203: 1589–1598. [DOI] [PubMed] [Google Scholar]
- 43. Wang SC, Lien HC, Xia W, Chen IF, Lo HW, et al. (2004) Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 6: 251–261. [DOI] [PubMed] [Google Scholar]
- 44. Li LY, Chen H, Hsieh YH, Wang YN, Chu HJ, et al. (2011) Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res 71: 4269–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, et al. (2007) Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst 99: 628–638. [DOI] [PubMed] [Google Scholar]
- 46. Xia W, Liu Z, Zong R, Liu L, Zhao S, et al. (2011) Truncated ErbB2 expressed in tumor cell nuclei contributes to acquired therapeutic resistance to ErbB2 kinase inhibitors. Mol Cancer Ther 10: 1367–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, et al. (2003) The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell 11: 495–505. [DOI] [PubMed] [Google Scholar]
- 48. Berezov A, Chen J, Liu Q, Zhang HT, Greene MI, et al. (2002) Disabling receptor ensembles with rationally designed interface peptidomimetics. The Journal of biological chemistry 277: 28330–28339. [DOI] [PubMed] [Google Scholar]
- 49. Saxon ML, Lee DC (1999) Mutagenesis reveals a role for epidermal growth factor receptor extracellular subdomain IV in ligand binding. The Journal of biological chemistry 274: 28356–28362. [DOI] [PubMed] [Google Scholar]
- 50. Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ (2006) Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nature clinical practice Oncology 3: 269–280. [DOI] [PubMed] [Google Scholar]
- 51. Spector NL, Blackwell KL (2009) Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 27: 5838–5847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data may be found within the paper.