Abstract
Eradication of Staphylococcus aureus biofilms after rifampin treatment was tested in a mouse model of device-related infection by using biophotonic imaging. Following treatment, the bioluminescent signals decreased to undetectable levels, irrespective of the age of the biofilm. After the final treatment, the signals rebounded in a time-dependent manner and reached those for the untreated mice. Readministration of rifampin was unsuccessful in eradicating reestablished infections, with the rifampin MICs for such bacteria being increased and with the bacteria having point mutations in the rpoB gene.
Many antibiotics are unable to completely eradicate biofilm infections, and cessation of treatment often results in the reestablishment of the biofilm. To better understand and control biofilms on indwelling medical devices, new approaches to drug efficacy screening in vivo are essential. The development of novel strategies, especially in vivo, have been severely impeded by the absence of longitudinal monitoring systems which are quantitative and which can be used in real time, permitting the assessment of antibiotic activity in the same animal. Recently, we demonstrated such a continuous method for real-time monitoring of biofilms using bioluminescent bacteria grown on Teflon catheters to investigate the interaction of biofilm bacteria with antibiotics during a chronic infection in a mouse model (12). In the present study, we investigated the effectiveness of rifampin against biofilms at different stages during growth and further tested the efficacy of this antibiotic against biofilms that reestablished following the termination of treatment.
Staphylococcus aureus ATCC 12600 was engineered for bioluminescence by inserting a modified complete lux operon, as described previously (11), and was designated S. aureus Xen 29. The experimental infection in a murine model was established with precolonized catheter carrying 104 CFU of S. aureus Xen 29 as described previously (11, 12). At various times after implantation of the catheters, groups of mice (n = 47) were treated with rifampin by administering 30 mg/kg of body weight intraperitoneally in 0.1 ml of saline. This dose of rifampin was previously shown to produce the maximum bactericidal effect against Xen 29 in the mouse model of biofilm infection (12). Antibiotic was given to mice twice daily for a total of 4, 5, or 6 days, starting on either day 7 or day 23 after catheter implantation. A second group of animals (n = 15) served as an untreated infection control group by being implanted with infected catheters but being treated with saline. Yet a third group of animals (n = 4) served as a negative control group, being implanted with sterile catheters. Total photon emissions from defined regions of interest within the images of each mouse were quantified by using Living Image software (Xenogen Corp., Alameda, Calif.), as described previously (12). The bacteria recovered from catheters were screened for resistance to rifampin that developed during therapy by plating the sonicated fluid onto Mueller-Hinton agar containing rifampin at various concentrations, as described previously (12). Bacteria that showed increased resistance to rifampin were further tested by the broth dilution method to determine the MIC (12). Two isolates, recovered from catheters with 23-day-old established biofilms and subsequently treated for 4 days with rifampin, were further characterized. One of these isolates (mutant I) was recovered 35 days postimplantation (i.e., 8 days after rifampin treatment), and the other (mutant II) was recovered 49 days postimplantation (i.e., 22 days after rifampin treatment). Rifampin resistance in S. aureus has previously been reported to be due to mutations in the rpoB gene (22), which encodes the β subunit of RNA polymerase. Thus, PCR was used to amplify the rifampin resistance-determining (Rif) regions of the rpoB gene directly from bacterial cells of rifampin-resistant strains (22). The rpoB gene fragment was amplified with the oligonucleotide primers SArpoF (5′-GAG TTG TAC GTG AAA GAA TG-3′) and SArpoR (5′-CAT GTG TAT CTA AAT CAA C-3′). Mutations conferring rifampin resistance were determined by DNA sequencing of the PCR fragment with primer SArpoF (Sequetech, Mountain View, Calif.).
Characterization of the two rifampin-resistant isolates, mutants I and II, revealed that both have a single base pair change, although at different positions, in the Rif region of the rpoB gene. Mutant I had a TCT to CCT codon change that resulted in an amino acid change of serine to proline at position 464, while mutant II had a AGC to AAC codon change that resulted in an amino acid change of aspartic acid to asparagine at position 471 (the substituted bases are underlined). Although the presence of other mutations in nonsequenced regions of the rpoB gene—or, indeed, other genes—cannot be excluded, it is likely that these mutations are at least partially accountable for the rifampin resistance. S. aureus isolates from the untreated group of mice showed no differences in susceptibilities to rifampin or mutations in the rpoB gene throughout the 50-day study period.
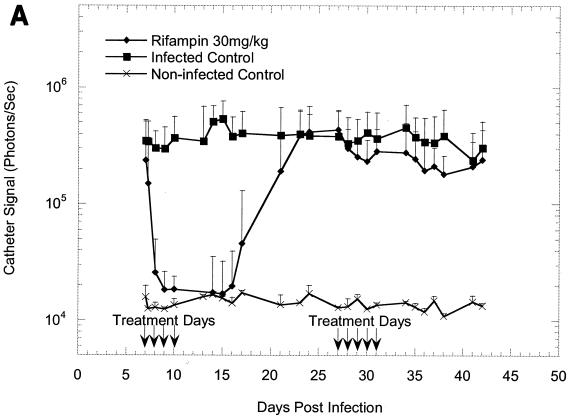
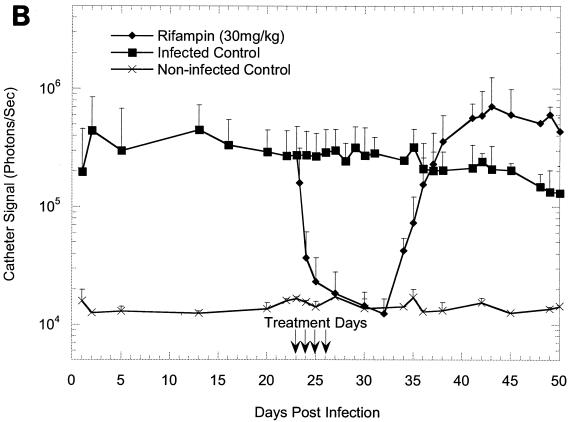
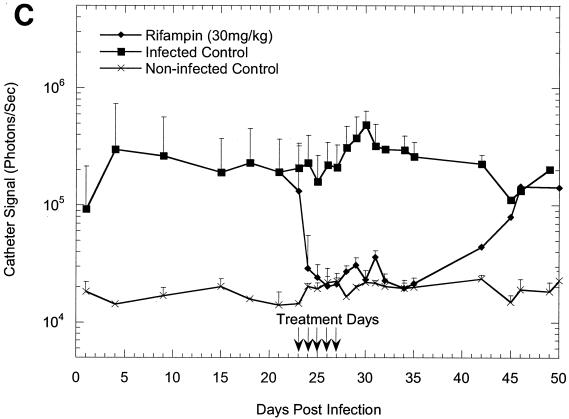
The effectiveness of rifampin administration to mice implanted with precolonized catheters under a variety of different conditions is shown in Fig. 1A to C. Following implantation, the bioluminescence signals increased exponentially over 24 h, as shown in previous studies (11, 12), reaching approximately 105 photons/s/catheter and remaining at this level until the termination of the study, thus allowing the efficacy of antibiotic treatment to be effectively monitored. The kinetics of the rifampin-induced reduction in bioluminescence on a 23-day-old biofilm was similar to that seen on a 7-day-old biofilm (Fig. 1A and B), indicating that the age of a biofilm does not affect its sensitivity to rifampin. Once the signal was reduced, it remained undetectable for the next 7 to 8 days in groups of animals treated for 5 days but gradually increased and reached the levels for the untreated animals by 18 days after the final treatment (Fig. 1C). In contrast, bacterial regrowth began approximately 5 days after the final treatment in groups to which rifampin treatment was given only for four consecutive days (Fig. 1A and B). The bioluminescent signal intensity seen from a 23-day-old biofilm after four consecutive days of rifampin treatment increased beyond that seen for the untreated control group after 36 days and reached 106 photons/s/catheter (Fig. 1B).
FIG. 1.
In vivo bioluminescence monitoring of S. aureus Xen 29 in the mouse model of biofilm infection during treatment with rifampin. (A) Effects of 4 days of rifampin treatment of a 7-day-old biofilm or 5 days of rifampin treatment of a reestablished biofilm; (B) effects of 4 days of rifampin treatment of a 23-day-old biofilm; (C) effects of 5 days of rifampin treatment of a 23-day-old biofilm. The total number of photons detected over the infected catheter per second was determined with an IVIS camera and plotted with respect to time. Each mouse was implanted with two catheters, and each datum point is the mean ± standard deviation for four to six catheters. Arrows indicate the days of antibiotic administration. Data are the averages of the results from three to four separate experiments.
To determine whether bioluminescence imaging can be used to monitor the efficacy of rifampin therapy on recovering biofilm infections (in real time), the antibiotic was readministered for five consecutive days to a group of animals (n = 16) after 16 days following cessation of the initial therapy (Fig. 1A). Notably, retreatment of the animals with reestablished biofilm infections had no effect on bioluminescence, indicating the presence of a rifampin-resistant population. Indeed, as discussed above, staphylococci recovered from the catheters in animals with recurring infections following rifampin treatments were confirmed to be resistant to rifampin by MIC (5 to 10 μg/ml) and genetic analyses. Our results are consistent with those of previous studies reporting the rapid emergence of rifampin-resistant staphylococci when rifampin is used as monotherapy for the treatment of device-related infections (1, 15).
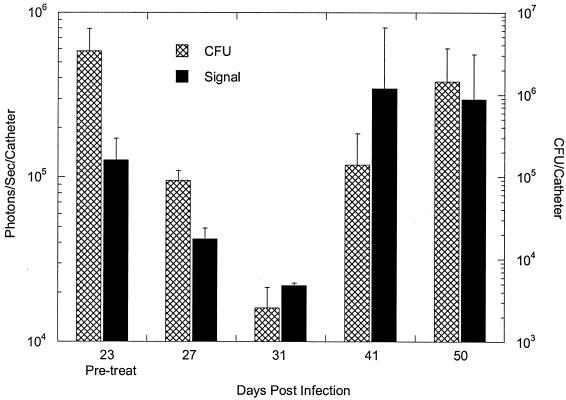
To assess the correlation between bioluminescence and the traditional colony-counting method, the mice were first imaged to determine the total photon counts emitted from implanted catheters, followed by extraction of the bacteria from the catheters for CFU determinations. We previously established that the numbers of CFU extracted from catheters correspond to the light signal, and other investigators (4, 6-9, 11-13, 16, 17) have also shown that a strong correlation exists between the numbers of viable cells from tissue homogenates and bioluminescence. Similarly, in the present study, a strong correlation (R2 = 0. 98) between the mean bioluminescence and the number of CFU was found for 23-day-old biofilms treated with rifampin for 4 days and untreated biofilms (Fig. 1B and 2). Similar results were obtained with the other treated groups, except in the study in which rifampin treatment of a 23-day-old biofilm was initiated for 5 days (Fig. 1C), in which the correlation coefficient between the numbers of CFU and the light signal appeared to be moderately low (R2 = 0.67) on day 50 (probably due to a sampling error). With the exception of the data for this time point, the data indicated that measurement of a change in photon flux from bioluminescence imaging is a convenient method for the monitoring of treatment of biofilm infections in living mice and quantification of the relative differences in therapeutic efficacy.
FIG. 2.
Correlation between bioluminescence and traditional CFU determination methods (CFU) after 4 days of rifampin treatment of a 23-day-old biofilm. Viable counts are reported as the numbers of CFU per catheter, and bioluminescence is represented as the numbers of photons per second per catheter, obtained with an IVIS camera. Each datum point is the mean ± standard deviation for three to four catheters. Mice were first imaged for bioluminescence, followed by removal and extraction of the bacteria from the catheters for CFU determinations.
To determine the activity of the antibiotic on the biofilm infections, conventional methods would typically require that infected animals be killed at each sampling point and that the foreign body be removed to estimate the pathogen burden, determined by measurement of the number of CFU recovered from the biofilm. This method can have the problem of antibiotic carryover and other artifacts due to handling, and these problems positively bias the data for the groups treated with antibiotics (5, 14). Additionally, efficient and real-time estimation of pharmacodynamic parameters, such as postantibiotic effect and delayed regrowth, factors that are critical in drug discovery programs, is not possible in a living animal by present methodologies based on determination of CFU counts. The main advantage of the present method was the in situ detection of staphylococci without destruction of the biofilm structure by detachment procedures. The detachment may result in the conversion of the organisms from sessile to planktonic forms, with possible phenotypic changes (2). Therefore, bioluminescence-based monitoring of biofilm bacteria provides additional information about the microbial physiological state, independently of the culturability of these bacteria. In addition, the experiment can be performed in the presence of the animal's host defense mechanisms.
The excellent bactericidal activity of rifampin against young or mature biofilms is probably due to its activity against bacteria at all phases of growth, including actively growing, semidormant, nongrowing, and intracellular bacteria (3, 18, 21). Because of its potency, rifampin appears to be effective against biofilm-embedded organisms that are recognized to be heterogeneous in the physiological or metabolic state (10, 19, 20).
Before we applied biophotonic imaging to the measurement of biofilms, no convenient method that allows noninvasive, real-time monitoring of the efficacies of therapeutic agents and the relapse of infection in a living animal following the termination of treatment regimens was available. Using a recombinant bioluminescent S. aureus strain and a low-light imaging system, were we able not only to visualize the entire course of infection but also to determine the precise effects of different antibiotic treatment regimens and subsequent unique patterns of disease relapse, all using a minimal number of animals. We also demonstrated the therapeutic failure of the antibiotic in eradicating reestablished biofilms on the implanted device in vivo. In this respect, our model mimics the clinical picture, in which relapse and the recalcitrant nature of the infection to treatment are commonly seen, phenomena that probably contribute significantly to the frequent failure of antibiotic therapy for biofilm infections in humans. We believe that our approach is ideally suited for evaluation of the in vivo efficacies of antimicrobial compounds at an early stage in antibiofilm drug discovery.
Acknowledgments
We thank E. Reynolds and C. Dalesio (Graphics and Communications, Xenogen Corporation) for assistance with drawings.
REFERENCES
- 1.Brandt, C. M., M. S. Rouse, B. M. Tallan, N. W. Laue, W. R. Wilson, and J. M. Steckelberg. 1995. Effective treatment of cephalosporin-rifampin combinations against cryptic methicillin-resistant β-lactamase-producing coagulase-negative staphylococcal experimental endocarditis. Antimicrob. Agents Chemother. 39:1815-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. R., and P. Williams. 1985. The influence of environment on envelope properties affecting survival of bacteria in infections. Annu. Rev. Microbiol. 39:527-556. [DOI] [PubMed] [Google Scholar]
- 3.Chaisson, R. E. 2003. Treatment of chronic infections with rifamycins: is resistance likely to follow? Antimicrob. Agents Chemother. 47:3037-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 5.Contag, C. H., S. D. Spilman, P. R. Contag, M. Oshiro, B. Eames, P. Dennery, D. K. Stevenson, and D. A. Benaron. 1997. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol. 66:523-531. [DOI] [PubMed] [Google Scholar]
- 6.Contag, P. R., I. N. Olomu, D. K. Stevenson, and C. H. Contag. 1998. Bioluminescent indicators in living mammals. Nat. Med. 4:245-247. [DOI] [PubMed] [Google Scholar]
- 7.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis, K. P., J. Yu, C. Bellinger-Kawahara, D. Joh, M. J. Hawkinson, G. Xiao, T. F. Purchio, M. G. Caparon, M. Lipsitch, and P. R. Contag. 2001. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamblin, M. R., D. A. O'Donnell, N. Murthy, C. H. Contag, and T. Hasan. 2002. Rapid control of wound infections by targeted photodynamic therapy monitored by in vivo bioluminescence imaging. Photochem. Photobiol. 75:51-57. [DOI] [PubMed] [Google Scholar]
- 10.Huang, C. T., F. P. Yu, G. A. McFeters, and P. S. Stewart. 1995. Nonuniform spatial patterns of respiratory activity within biofilms during disinfection. Appl. Environ. Microbiol. 61:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadurugamuwa, J. L., L. Sin, E. Albert, J. Yu, K. Francis, M. DeBoer, M. Rubin, C. Bellinger-Kawahara, T. R. Parr Jr., Jr., and P. R. Contag. 2003. Direct continuous method for monitoring biofilm infection in a mouse model. Infect. Immun. 71:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadurugamuwa, J. L., L. V. Sin, J. Yu, K. P. Francis, R. Kimura, T. Purchio, and P. R. Contag. 2003. Rapid direct method for monitoring antibiotics in a mouse model of bacterial biofilm infection. Antimicrob. Agents Chemother. 47:3130-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuklin, N. A., G. D. Pancari, T. W. Tobery, L. Cope, J. Jackson, C. Gill, K. Overbye, K. P. Francis, J. Yu, D. Montgomery, A. S. Anderson, W. McClements, and K. U. Jansen. 2003. Real-time monitoring of bacterial infection in vivo: development of bioluminescent staphylococcal foreign-body and deep-thigh-wound mouse infection models. Antimicrob. Agents Chemother. 47:2740-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, S. H., and A. Camilli. 2000. Novel approaches to monitor bacterial gene expression in infected tissue and host. Curr. Opin. Microbiol. 3:97-101. [DOI] [PubMed] [Google Scholar]
- 15.Lucet, J. C., M. Herrmann, P. Rohner, R. Auckenthaler, F. A. Waldvogel, and D. P. Lew. 1990. Treatment of experimental foreign body infection caused by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2312-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocchetta, H. L., C. J. Boylan, J. W. Foley, P. W. Iversen, D. L. LeTourneau, C. L. McMillian, P. R. Contag, D. E. Jenkins, and T. R. Parr, Jr. 2001. Validation of a noninvasive, real-time imaging technology using bioluminescent Escherichia coli in the neutropenic mouse thigh model of infection. Antimicrob. Agents Chemother. 45:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siragusa, G. R., K. Nawotka, S. D. Spilman, P. R. Contag, and C. H. Contag. 1999. Real-time monitoring of Escherichia coli O157:H7 adherence to beef carcass surface tissues with a bioluminescent reporter. Appl. Environ. Microbiol. 65:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson, E., H. Hanberger, and L. E. Nilsson. 1997. Pharmacodynamic effects of antibiotics and antibiotic combinations on growing and nongrowing Staphylococcus epidermidis cells. Antimicrob. Agents Chemother. 41:107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wentland, E. J., P. S. Stewart, C. T. Huang, and G. A. McFeters. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12:316-321. [DOI] [PubMed] [Google Scholar]
- 21.Widmer, A. F., A. Gaechter, P. E. Ochsner, and W. Zimmerli. 1992. Antimicrobial treatment of orthopedic implant-related infections with rifampin combinations. Clin. Infect. Dis. 14:1251-1253. [DOI] [PubMed] [Google Scholar]
- 22.Wielders, C. L., A. C. Fluit, S. Brisse, J. Verhoef, and F. J. Schmitz. 2002. mecA gene is widely disseminated in Staphylococcus aureus population. J. Clin. Microbiol. 40:3970-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]