Abstract
Dendritic cells (DCs) are major antigen-presenting cells that play a key role in initiating and regulating innate and adaptive immune responses. DCs are critical mediators of tolerance and immunity. The functional properties of DCs decline with age. The purpose of this study was to define the age-associated molecular changes in DCs by gene array analysis using Affymatrix GeneChips. The expression levels of a total of 260 genes (1.8%) were significantly different (144 down-regulated and 116 upregulated) in monocyte-derived DCs (MoDCs) from aged compared to young human donors. Of the 260 differentially expressed genes, 24% were down-regulated by more than 3-fold, suggesting that a large reduction in expression occurred for a notable number of genes in the aged. Our results suggest that the genes involved in immune response to pathogens, cell migration and T cell priming display significant age-related changes. Furthermore, downregulated genes involved in cell cycle arrest and DNA replication may play a critical role in aging-associated genetic instability. These changes in gene expression provide molecular based evidence for age-associated functional abnormalities in human DCs that may be responsible for the defects in adaptive immunity observed in the elderly.
Introduction
Aging is characterized by a progressive decline in immune responses, resulting in an increased susceptibility to infections and impaired response to vaccination [1]–[2]. Paradoxically, aging is associated with chronic inflammatory state [3]. and an increased incidence of diseases associated with inflammation including atherosclerosis, Alzheimer's disease, and arthritis [4]. The molecular mechanisms responsible for this paradox are not well characterized.
Dendritic cells (DCs) play a critical role in bridging innate and adapotive immune response [5]–[6]. Upon antigen capture, immature DCs become activated and migrate to the lymphoid organs where they acquire co-stimulatory molecules, toll-like receptors (TLRs) and chemokine receptors, and prime lymphocytes to initiate an adaptive immune response. In aged humans, several functions of DCs are compromised including phagocytosis, uptake of antigens, and migration [5]. There is an aberrant cytokine secretion by various DC subsets, including increased basal levels of pro-inflammatory cytokines [7]–[8], but their response to foreign antigens upon stimulation is decreased [7], [9]. In contrast, the immunogenicity to self-antigens as a result of epigenetic changes is increased [10]–[11], suggesting a loss of peripheral self-tolerance. Interferon type I and type III secretion by plasmacytoid dendritic cells and monocyte-derived DCs (MoDCs) and the capacity of DCs to prime naïve T cell subsets are also impaired in aged humans [12]–[13].
In this investigation, we have compared differential gene expression patterns in MoDCs from young and aged subjects using a fold-change cutoff of ≥1.5. We have (1) identified the gene expression patterns from aged MoDCs, (2) demonstrated that expression changes associated with aging occur in genes involved in immune response, cell cycle and response to oxidative stress, and (3) provided candidate genes for further studies. These age-associated changes in gene expression patterns in MoDCs provide molecular bases for age-associated functional abnormalities in immune responses, which may play an important role in the progressive decline in adaptive response in aging.
Materials and Methods
Blood donors
Blood was collected from healthy aged and young donors. Young donors ranged from 20–30 years of age and elderly donors were between 75–90 years of age. Description of cohort used for microarray analysis is provided in Table 1 while Table 2 provides description of donors for PCR and cell cycle analysis. Aged healthy subjects are of middle class social state and living independently. Each donor was requested to discontinue any vitamins, minerals and antioxidants one week prior to blood draw. This study was approved by the Institutional Review Board of the University of California, Irvine. All participants signed their written informed consent forms.
Table 1. MoDCs donor characteristics for microarray data.
| Donor Array ID | Age | Sex |
| Young 1 | 22 | Female |
| Young 2 | 25 | Female |
| Young 3 | 27 | Female |
| Young 4 | 20 | Male |
| Aged 1 | 84 | Female |
| Aged 2 | 77 | Female |
| Aged 3 | 78 | Female |
| Aged 4 | 87 | Female |
| Aged 5 | 81 | Male |
Table 2. MoDCs donor characteristics for qRT-PCR and cell cycle.
| Donor qRT-PCR ID | Age | Sex |
| Y1-p* | 28 | Male |
| Y2-p* | 26 | Male |
| Y3-p* | 32 | Female |
| Y4-p* | 23 | Female |
| Y5-p* | 25 | Female |
| Y6-p | 22 | Male |
| Y7-p | 24 | Male |
| Y8-p | 27 | Female |
| Y9-p | 25 | Female |
| Y10-p | 29 | Female |
| A277-p* | 76 | Male |
| A164-p* | 94 | Female |
| A268-p* | 95 | Female |
| A146-p* | 85 | Male |
| A276-p* | 85 | Female |
| A163-p | 87 | Female |
| A103-p | 81 | Male |
| A244-p | 84 | Female |
| A83-p | 72 | Male |
| A276-p | 85 | Female |
*Cell cycle analysis of DNA content in DCs from these aged and young donors.
Preparation of human monocyte-derived dendritic cells
MoDCs were prepared essentially as described [7], [14]. Briefly, monocytes were purified from the PBMCs by positive selection using CD14 magnetic beads (Stemcell Sep, Vancouver, BC). The purity of the isolated CD14+ monocytes was >90% as determined by flow cytometry (FACS). To induce differentiation of DCs, purified CD14+ monocytes were cultured in RPMI 1640 10% fetal bovine serum. 50 ng/ml supplemented with human rGM-CSF and 10 ng/ml human rIL-4 (Peprotech, Rocky Hill, NJ). Half of the medium was replaced every 2 days and MoDCs were collected after 6 days. The purity of the MoDCs was >95% as determined by the expression of CD14− CD11c+ and HLA-DR+.
RNA extraction and purification
Total RNA was extracted from MoDCs of aged and young subjects using the TRI Reagent kit (Molecular Research Center Inc., Cincinnati, OH, USA), following the manufacturer's protocol. The integrity of intact total RNA was verified with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Real-time quantitative reverse transcriptase PCR
qRT-PCR was performed in MoDCs from ten young and ten aged donors (Table 2, Donor qRT-PCR). 1 µg of total RNA was reverse transcribed to cDNA by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random hexamers as primers following the manufacturer's instructions. The cDNA was amplified by PCR in a 20-µL reaction mixture containing 1 µL of 20 µM primers, 2 µL of SYBR Green PCR Master Mix (Applied Biosystems) and 4 µL of cDNA (50x diluted from original synthesized cDNA) or water as a negative control.
Initial denaturation of DNA was carried out at 95°C for 10 min; forty amplification cycles were performed, each cycle consisting of denaturation (95°C, 30 s) and annealing with extension (65°C, 1 min). Each sample was amplified in triplicate, and results were normalized with GAPDH gene expression as ‘housekeeper.’ A 4-log absolute standard curve (synthesized by Ziren Research LLC, Irvine, CA) [15] dilution series was run using each primer pair at optimal concentration, and amplification efficiencies were calculated. The fold changes of differential expression between healthy young donors and aged subjects were calculated using the ratios of each gene of interest to GAPDH. For a list of primer sequences, see Table S3.
Gene array processing and statistical analysis
MoDC samples from the four young and five aged donors (Table 1) were examined for gene expression profile by a method previously described [16] using 9 HG-U133A_2 Gene Array chips (Affymetrix, Santa Clara, CA, USA, http://www.affymetrix.com). “All microarray procedures were performed at the Gene Array Technology Facility, University of California, Irvine. Each chip contains 22,277 25-mer probe sets corresponding to 18450 unique transcripts and 14500 genes. A 10 µg aliquot of total RNA from each sample was processed and applied to a HG-U133A_2 Gene Array chip according to the manufacturer's protocol (http://www.affymetrix.com/support/technical/manual/expression_ manual.affx). Microarray Command Console V3.1 (Affymetrix) was used for gene expression image analysis and quality control. Affymetrix default settings were used, and statistical parameters such as background, noise, and spike-in controls were found to be within acceptable limits. Ratios of 3′ to 5′ mRNA transcripts of constitutively expressed internal housekeeping gene controls, human β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were also measured to be within the recommended Affymetrix guidelines. Expression Console Software (Affymetrix) was used to produce CEL files. Web-based Genetic Analysis Software (www. Genesifter.net, Geospiza, Seattle, WA, USA) was used to analyze the data. By uploading each CEL file to Genesifter's website, normalized gene expression levels were obtained using GC-RMA. Initial analysis focused on removing genes that did not meet our criteria for differential expression by applying t-tests followed by the Benjamini-Hochberg method for false discovery rate (FDR) adjustment” [16]–[17]. “Genes were accepted for further analysis if they changed by more than 1.5 fold and had an adjusted p≤0.05 (by t-test) with an FDR <5%. Lists of genes with expression changes meeting our criteria were filtered by the Gene Ontology (GO) program by performing biological function analyses. Degree of enrichment for biological processes and molecular functions of interest were assessed by analyzing z-scores generated by the GO program or by applying a p value <0.05. The z-score statistically rates a GO category by evaluating whether the number of genes included in the category is either greater or less than that expected by chance [18], using as criteria either z-scores >2.0 (genes are over-represented in the corresponding GO category) or <−2.0 (under-represented). Hierarchical clustering analysis [19] was performed using the program CLUSTER from Genesifter” [16]. Microarray data measured in HG-U133A_2 Gene Array chips of mRNA samples of DCs from 4 young and 5 aged donors are submitted to GEO, accession number: GSE 58015.
Cell cycle analysis of DNA content by FACS in DCs from aged and young donors
Cell cycle analysis of DNA content was performed in MoDCs from five young and five aged donors (Table 2, Donor qRT-PCR). MoDCs about (5×105) were washed with Phosphate Buffered Saline (PBS) and incubated in ice-cold 70% ethanol/30% PBS at 4°C for 1 h. Cells were then washed once with PBS, resuspended in 0.5 ml PBS plus 0.5 ml DNA extraction buffer (0.2 M Na2HPO4, 0.4 µM citric acid, pH 7.8), incubated for 5 min at room temperature. Finally, cells were resuspended in 1 ml of staining solution [20 µg/ml RNase A (Qiagen), 20 µg/ml propidium iodide (Sigma) in PBS], and incubated for 1 h in the dark at room temperature. Ten thousand cells were acquired on FACS Calibur and analysis was done using the FlowJo cell cycle program.
Results
Global characteristics of age-associated gene expression changes in human MoDCs
In this study, we analyzed the differential expression of genes in MoDCs from four young and five aged (Table 1) healthy human donors using Affymetrix GeneChip HG U133A 2.0 microarrays. Of the 14,500 well-characterized genes measured, 260 genes (1.8%) displayed significant donor age-associated expression changes. Although the total numbers of up-regulated and down-regulated genes were approximately similar (1.24∶1.0 down:up ratio), 24% of the down-regulated genes displayed differential changes more than three-fold, indicating that the levels of some genes were highly reduced in MoDCs from aged donors as compared to young subjects (Fig. 1A, B, Table S1).
Figure 1. Clustering of Gene expression changes in young and aged MoDCs.
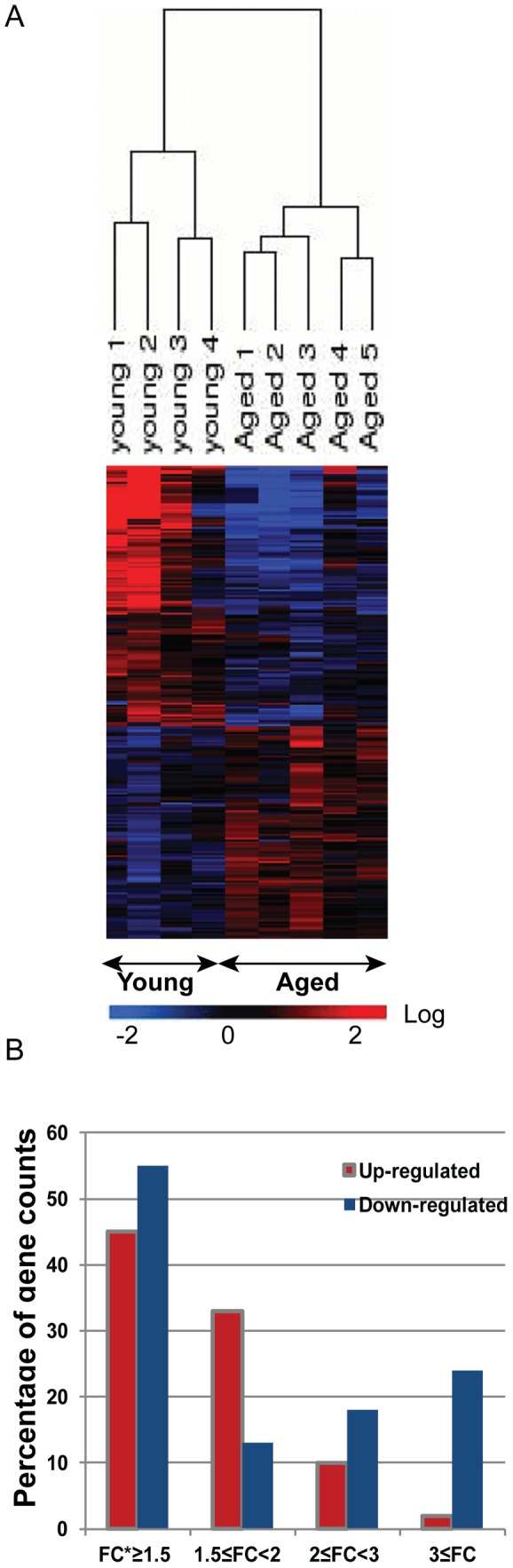
(A) Hierarchical clustering heat map of 260 genes. with significant expression changes in young and aged MoDCs. Each column represents an individual donor; each row refers to a gene. Gene expression changes with respect to median changes across age are denoted by: red, up-regulated (ratio ≥1.5); blue, down-regulated (ratio <1/1.5); and black, unchanged. (B) Distribution of 260 genes by expression size, as percentages of total numbers of up- or down-regulated genes. *:FC = Fold change.
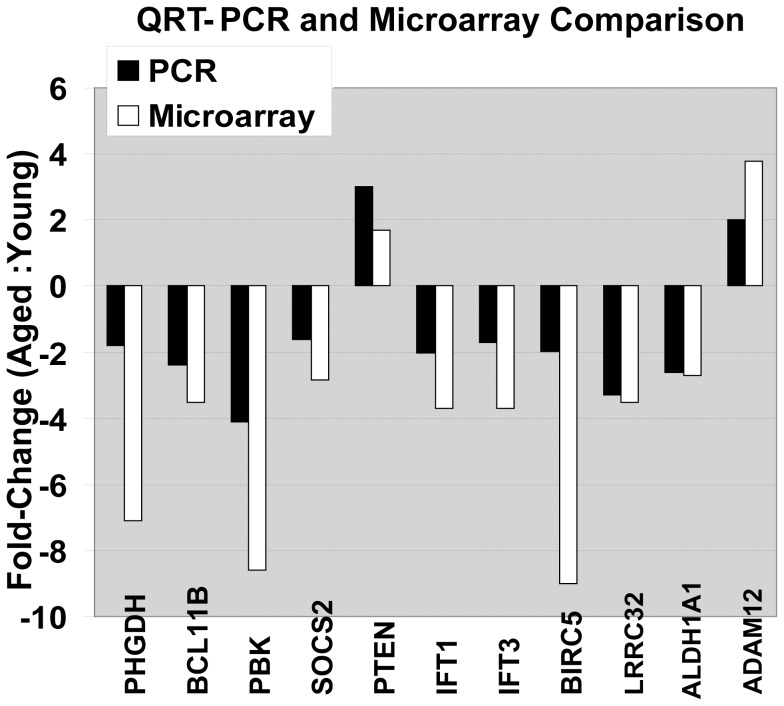
In addition, we carried out real-time quantitative RT-PCR (qRT-PCR) analysis in triplicate for 10 selected genes using cDNA from MoDCs from ten young and ten aged individuals. (Table 1B). The age-related changes in gene expression measured by qRT-PCR were in agreement with microarray data for those specific genes (Fig. 2). The Pearson correlation coefficient between the qRT-PCR and microarray data was 0.64, (Fig. 2; for gene ID and primer sequences see Table S3).
Figure 2. Validation of microarray data with real-time quantitative RT-PCR.
qRT-PCR validation. Black bars represent microarray hybridizations, while white bars represent values from qRT-PCR. Ratio of expression for each gene (aged group to young group) is shown as fold change. The Pearson correlation coefficient between the qRT-PCR and microarray data was 0.64.
Functional association of expression changes in MoDCs from aged donors
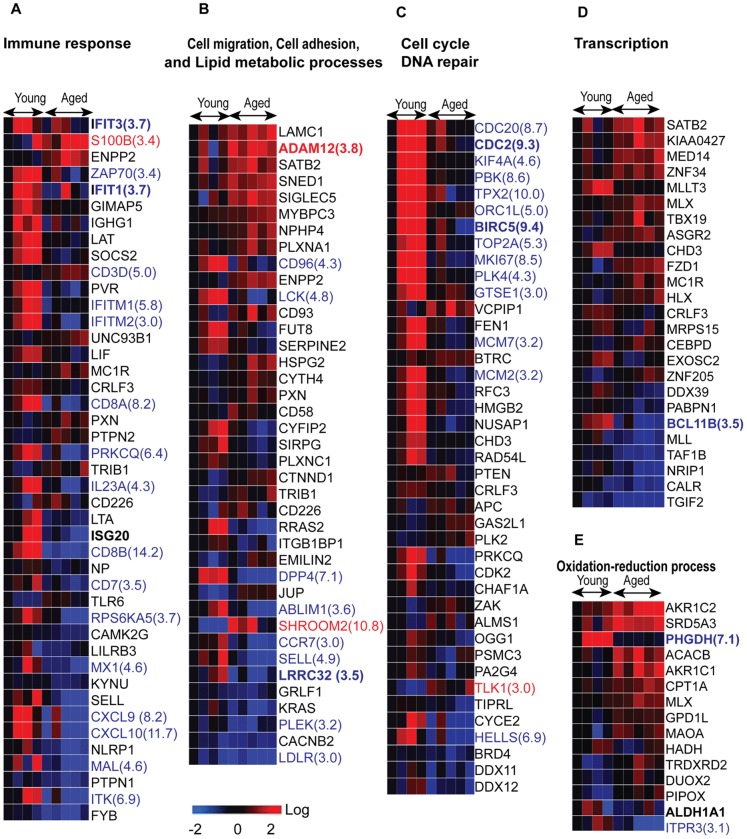
Age-associated expression of 209 genes filtered by Gene Ontology and involved in specific cellular processes were either suppressed or induced. Expression levels of many of these genes were highly reduced (>3 fold), with down-regulated genes being over-represented in several categories including immune response, cell migration and cell cycle (Fig. 3A–C, Table 3; Table S2). Genes with altered expression involved in immune response included: interferon-stimulated genes ISG20, MX1, CXCL10, and CXCL9; IFN-induced transmembrane proteins IFITM1 and IFITM2; and IFN-induced proteins with tetratricopeptide repeats IFIT1 and IFIT3. Similarly modulated genes involved in DC cell migration and regulation of T cell activation included IL23, CCR7, and DDP4; those involved in the cell surface receptor-linked signaling pathway included CD8A, CD8B, SIRPG, ITK and LCK; and finally genes involved in the toll-like receptor signaling pathway included RPS6KA5, TLR6 and Unc93B1. The downregulated genes involved in cell cycle and DNA repair included CDC20, CDC2, ORC1 (ORC1L), BIRC5, MCM7, MCM2, CDK2, PBK, TPX2, ZAK, PSMC3, ZAK, APC, FEN1, RFC3, OGG1, CHD3, and DSCC1. Other downregulated cell cycle genes, such as TPX2, BIRC5, KIF4A, and GTSE1 play key roles in spindle assembly and are required for mitosis and for response to DNA damage repair. That many genes are highly down-regulated (≥3-fold) may indicate that aged-donor DCs are less likely to respond to an immune challenge and that the integrity of these cells may be impaired.
Figure 3. Heat maps of expression changes of genes in cell processes affected by donor age.
(A–F) Heat maps of the genes by functional group (for detailed gene information, see Table S2, worksheets A, B). Expression of genes in red text were increased, and of genes in blue decreased, by ≥3 fold. Each column represents a donor; each row refers to a gene.
Table 3. Numbers of genes (total in list, up- and down-regulated) and associated z-scores >|2.0| in selected functional categories that contained significantly over-represented differentially expressed genes in naïve aged-donor DCs compared to young.
| Ontology | List | Up | Down | z-up | z-down |
| cellular process | 209 | 91 | 118 | 2.28 | 2.88 |
| I. signaling transduction | 93 | 41 | 52 | 1.22 | 2.57 |
| 1. response to stress | 61 | 18 | 42 | −0.34 | 4.1 |
| 2. cell surface receptor linked signaling pathway | 54 | 18 | 36 | 0.34 | 3.84 |
| 3. immune system process | 57 | 15 | 42 | 0.15 | 7.08 |
| 4. immune response | 46 | 8 | 38 | −0.26 | 9.52 |
| 5. defense response | 34 | 8 | 26 | −0.22 | 9.01 |
| 6. locomotion | 37 | 14 | 23 | 1.6 | 3.48 |
| 7. cell migration | 22 | 10 | 12 | 2.28 | 2.0 |
| 8. chemotaxis | 19 | 7 | 10 | 0.35 | 2.42 |
| 9. antigen receptor-mediated signaling pathway | 8 | 0 | 8 | −0.84 | 8.63 |
| II. localization | 93 | 56 | 37 | 3.25 | 1.38 |
| 1.transport | 68 | 39 | 29 | 2.22 | −0.47 |
| 2. cellular localization | 41 | 20 | 21 | 2.1 | 1.53 |
| 3. protein localization | 31 | 15 | 16 | 2.08 | 1.1 |
| 4. transmembrane transport | 24 | 16 | 8 | 3.69 | 0.32 |
| 5. cell adhesion | 23 | 16 | 7 | 2.42 | 0.9 |
| 6. cell morphogenesis | 25 | 15 | 6 | 3.66 | 0.93 |
| III. cell cycle | 42 | 7 | 35 | −1.44 | 5.28 |
| 1. cell cycle arrest | 18 | 1 | 17 | −1.6 | 4.1 |
| 2. DNA replication | 10 | 0 | 10 | −1.7 | 3.72 |
| IV. oxidation-reduction process | 16 | 13 | 3 | 2.09 | −1.6 |
In contrast, expression levels of genes involved in the processes of oxidation-reduction and cell adhesion were modestly increased (Fig. 3B, E, Table 3; Table S2). The genes involved in oxidation-reduction included AKR1C2, AKR1C1, SRD5A3, ACACB, CPT1A, MAOA, DUOX2; and those involved in cell adhesion include HSPG2, JUP, CD93, LAMC1, CTNND1, PXN, SNED1, CYTH4 and SIGLEC5. These results indicate that aged DCs may be in a state of chronic oxidative stress. In addition, PHGDH, (a gene encoding a metabolic enzyme that plays a critical role in serine biosynthesis in the phosphorylation pathway) [20] was downregulated (Fig 3E).
More cells were arrested in G1 phase in MoDCs from aged donors as compared to MoDCs from young donors
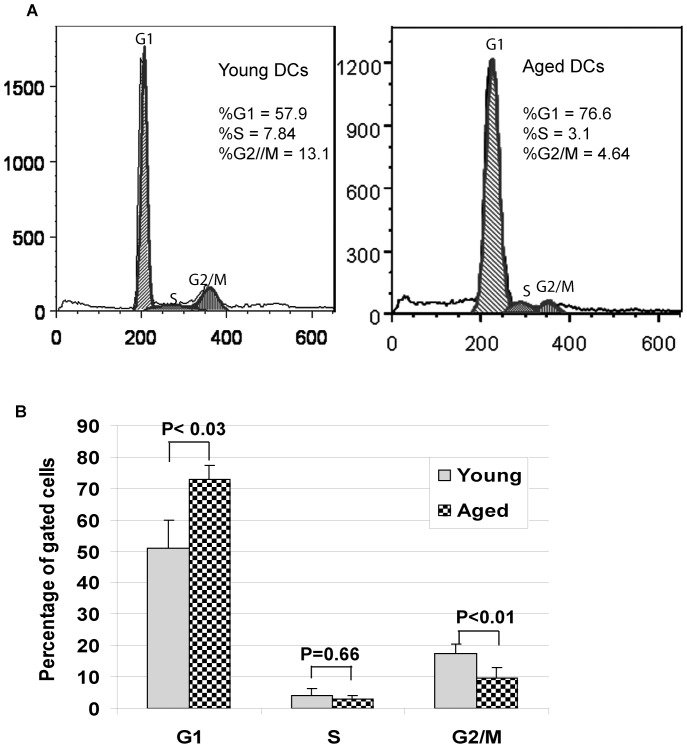
Cell cycle analysis of DNA content in DCs from aged and young donors demonstrated significantly increased number of cells in G1 phase (73% vs 51%, p<0.03) from old donors compared to young donors. In contrast, fewer cells were in G2/M phase (9.8% vs 17.4%, p<0.01). These results indicate that more cells were arrested in G1 phase in aged DCs than in young (Fig. 4), a profile reminiscent of senescent cells.
Figure 4. Cell cycle phases are different between MoDCs from aged and young donors.
(A) Distribution of aged and young MoDCs existing in the various phases of the cell cycle as determined by FACS. (B) Statistical analysis and percentages of MoDCs from young and aged donors in each phase of the cell cycle (n = 5, each group). Donors' information sees Table 2.
Discussion
Aging is characterized by a progressive decline in the adaptive immune response and chronic inflammation. The functionality of dendritic cells, both for orchestrating the immune response and for maintaining immune tolerance, is compromised with age [5]. Identifying the functions and signaling pathways of genes that exhibit substantial (≥1.5-fold) expression changes in MoDCs from healthy aged human donors provides insights into the molecular mechanisms underlying immunosenescence.
Expression levels of genes belonging to several critical functional groups, most notably immune defense response against viruses, were reduced in MoDCs from aged donors. Type I Interferons play a critical role in defense against viral infections as they not only interfere with viral replication but also lead to further activation of DCs, macrophages via the induction of set of genes known as interferon-stimulated genes. Our data demonstrates substantial down-regulation (by about 3-fold) in the expression of several interferon-stimulated genes, such as IFITM1, IFITM2, IFIT1, IFIT3, ISG20, and MX (Fig. 3A, Table S2), in MoDCs from aged subjects. IFIT and IFITM each denote a genetically and functionally distinct family of IFN-stimulated genes that have antiviral activities against a range of human viruses [21]–[22]. IFIT1 binds to the free 5′-ppp moiety on RNA of a number of viruses, including influenza A virus, and inhibits infection by forming a complex with IFIT2 and IFIT3 that sequesters viral nucleic acid [22]. Therefore, decreased IFIT1 and IFIT3 gene expression and decreased expression of IFITM1 and IFITM2 suggest that in aging there is an impairment of both IFN-I production and IFN-induced signaling mechanisms. ISG20 is an interferon-inducible 3′-5′ exonuclease that inhibits replication of several human and animal RNA viruses [23]–[24]. Mx1 (Myxoma resistance protein 1 or human MXA) is a member of a family of large GTPases that are induced exclusively by IFN-I and have broad antiviral activity against several viruses, including influenza A [25]. We have previously reported that both plasmacytoid dendritic cells and MoDCs from aged donors secrete reduced levels of type I interferons in response to influenza virus [12]. Our present data suggests that aged DCs are compromised not only with respect to the production of type I interferon but also with regard to the signaling response to IFN-I that contribute to the increased susceptibility of aged subjects to viral infections.
DCs sense and capture pathogens via the expression of pathogen recognition receptors (PRRs), such as Toll-like receptors (TLR), which are critical for the development of adaptive immunity [26]–[27]. Our data show age-related alterations in several critical genes in the TLR signaling pathway (Fig 3A, Table S2), including upregulation of the gene encoding Unc93B1; this factor interacts with TLRs -3, -7, and -9 and is involved in the production of pro-inflammatory cytokines [28]. In contrast, expression of the SOCS2 gene [29], which acts as a negative regulator of TLR-induced DC activation, was down-regulated. Defects in negative-feedback loops regulating inflammation lead to increased inflammation-sensitive autoimmune diseases [30]. Thus, the alterations in Unc93B1 and SOCS2 may result in an enhancement of TLR activation-associated inflammation [31]–[32]. This is consistent with our previous studies where we have reported an increase in inflammatory cytokine, TNF-a and IL-6 secretion from MoDCs of elderly in response to Lipopolysaccharide [7] More recently, we have also demonstrated that MoDCs from elderly are deficient in the production of anti-inflammatory cytokine, IL-10 which controls inflammation [12]. The T cells primed by these inflammatory DCs also display reduced IL-10 production. Furthermore, the DCs from elderly were also found to be defective in maintaining tolerance in that they displayed enhanced basal level of activation and increased reactivity to self-antigens [10]. Several genes which are involved in the tolerizing function of DCs appear downregulated in the elderly [Figure 3]. These include AldhA1 which codes for RALDH enzymes which plays an important role in maintenance of mucosal tolerance by DCs [33] (Fig. 2, 3). A reduction in the expression of KYNU is also observed in the DCs from aged (Figure 3). KYNU encodes of Kynureninase and is involved in tryptophan metabolism which is one of the pathways in DCs involved in the generation of T regulatory cells [34]. Furthermore, we have observed reduced expression of LRRC33 which is also important for generation of regulatory T cells [35] (Fig. 2,3). Aged DCs also express increased levels of metalloproteinases such as ADAM12 (Fig. 2, 3). We have reported that this enhances the permeability of airway epithelial cells barrier leading to increased airway inflammation [36].
Dendritic cells (DCs) are critical for adaptive immunity and tolerance, and the migration and accurate positioning of DCs is indispensable for the priming of T cells and immune surveillance. Several genes involved in these processes were downregulated (Fig. 3A, B, and Table S2). CCR7, with an approximately 3-fold reduction in expression, was prominent; its product is a receptor required for the migration of DCs and T cells to the areas of lymph nodes where T cell priming occurs [37]–[38]. The protein encoded by ITGB1BP1 (integrin beta 1 binding protein 1), which binds to small GTPase Cdc42, also plays an important role in controlling cell adhesion and migration [39], and antigen-uptake by DCs [40]. Cdc42 by itself is also a major regulator of endocytosis in DCs [41]. Our functional studies support these hypotheses as we have previously reported impairment in DC antigen uptake and migration [7] in the aged subjects. Dipeptidylpeptidase 4 (DPP4/CD26), co-localizes with membrane-bound adenosine deaminase on human DCs and enhances the ability of the latter to stimulate T-cell proliferation [42]. CD58 (lymphocyte function-associated antigen 3, LFA-3), is significant in that its molecular interaction with CD2 counter-receptors mediates the ability of monocytes to augment T cell activation [24]. Protein products of the genes ITK, LCK, and SIPG are also involved in T cell-mediated immunity. Of these, ITK is a member of the family of Tec kinases, and is known to have an important role in CD4+ T cell differentiation [43]. Therefore, the reduced expression of ITK in aged DCs may explain decreased priming ability and T cell proliferation in aged humans. Therefore, downregulation of DPP4, CD58 and ITK would be consistent with our observation of impaired capacity of aged MoDCs to prime T cells [11].
In addition to immune defense, our data show that the expression levels of many genes involved in chromatin organization, cell cycle arrest, DNA replication and repair and regulation of transcription were reduced by more than 3-fold in aged-donor cells (Fig. 3C, Table S2). Genes involved in cell cycle arrest included CDC20, CDC2, ORC1 (ORC1L), BIRC5, MCM7, MCM2, CDK2, PBK, TPX2, ZAK; those involved in DNA replication and repair included PSMC3, HELLS, MLL, FEN1, RFC3, OGG1, CHD3, and DSCC1. Cellular degeneration, uncontrolled cell proliferation and genomic instability are major hallmarks of aging [44]–[45]. CDC2, a cyclin-dependent kinase that normally drives cells into mitosis, is also the ultimate target of pathways that mediate rapid arrest in G2 in response to DNA damage [46]. CCNE2 (CYCE2) is a CDK2 partner in the late G1 and S phases of the mammalian cell cycle [47]. ORC1L (ORC1), a component of ORC (origin of replication complex), is required for efficient cell-cycle progression (transition from G1 to S phase) and DNA replication [48]. Some of these genes encode microtubule-associated proteins such as TPX2 [24], [49]. BIRC5 [50] and GTSE1 play a key role in spindle assembly and are required for mitosis and for responding to DNA damage. GTSE1 is also required for cell migration and for microtubule-dependent disassembly of focal adhesions [51]. KIF4A is associated with chromosomes during mitosis and mediates the interaction between mitotic chromatin and microtubules [52]. FEN1 is a structure-specific nuclease with 5′-flap endonuclease and 5′-3′ exonuclease activities and is involved in DNA replication and repair [53]. The RFC3 gene encodes DNA replication factor C subunit 3, a protein that maintains genome stability by inhibiting cell cycle progression in the presence of DNA damage or incomplete replication [54]. HELLS encodes a helicase involved in DNA repair, while the protein encoded by MLL mediates modifications to chromatin associated with epigenetic transcriptional activation [55]–[56].CHD3 encodes chromodomain helicase DNA binding protein 3, a member of a family of chromatin remodeling proteins involved in repression of gene expression [57]. We have previously demonstrated that type I and type III interferon promoter genes display increased association with the repressor histone, H3K9me3 in the unstimulated state in MoDCs from the elderly while their association with activator histone, H3K4me3 is reduced on activation with influenza virus [12].
In contrast to DNA repair genes, three genes important in cell cycle regulation, PTEN, ZAK and APC, were elevated. PTEN expression has both a potent inhibitory effect on DNA synthesis and the ability to block cell cycle progression in G1 [58]. Overexpression of PTEN results in decreased PKB/Akt phosphorylation and cyclin E/cdk2 activity, and inhibits S-phase entry in MCF-7 cells [59]. We have also observed an increased expression of PTEN at both mRNA and protein levels, and a reduced degree of AKT phosphorylation in aged MoDCs [7]. A greater percentage of cells from aged donors were observed to be in G1, along with a lower percentage of these cells in M phase (Fig. 4). Notably, cellular senescence is a state of stable cell cycle arrest [60] that occurs not only in normally proliferating cells, but also in arrested quiescent or terminally differentiated cells [61]. The significant changes that were observed in cell cycle-associated genes, despite the fact that DCs are terminally differentiated, suggests that these genes may be playing a very important role in the maintenance of cellular architecture, chromatin structure, cell cycle arrest and DNA repair, all of which are fundamentally important for the maintenance of DC cell integrity, genomic stability and basic functions.
Genes involved in transcription regulation that were downregulated in aged-donor MoDCs included CALR, TAF1B, PABPN1, CEBPD and Bcl11b (Fig 3C, D). Calreticulin (CALR) acts as an important modulator of the regulation of gene transcription mediated by nuclear hormone receptors [62]. TAF1B and PABPN1 are essential for the initiation of transcription by RNA polymerase I. Investigating the roles these genes in the age-related changes in DC cellular integrity, may provide new insights into the age-associated functional decline of DCs.
The present study utilizes monocyte derived DCs which display both similarities and differences than DCs found in circulation or tissues. Both circulating and in vitro–differentiated MoDCs share the main functional properties of APCs by priming naive T cells and inducing immune tolerance. They also display similar phenotypic expression of DC markers such as low expression of CD14, CD40, CD80 and high expression of CD11c and MHCII [63] Even though both DCs are of myeloid lineage. MoDCs are more homogenous population compared to peripheral DCs, which are divided into two major DC populations expressing CD1c and CD14. Genome-wide expression profiling has demonstrated that MoDC are more similar to inflammatory DCs which are DC derived from monocytes in an inflammatory setting than circulatory DCs. Nevertheless, studies of both are equally relevant as inflammatory DCs have been demonstrated to play a major role in protection against lung infection of influenza virus [64]. The study of MoDCs thus provides insight into the age-associated changes in the functions of monocytes, inflammatory DCs and some shared functions of circulatory DCs.
By comparing expression changes associated with donor age, we observed that such changes in DCs were greater than those observed in aged CD8 + T cells [16] (Fig. S1). A substantially larger number of genes, that are involved in immune responses and cellular integrity, were down-regulated over 3-fold (24% vs. 4%) in MoDCs. Therefore, despite the fact that donor age altered fewer genes in MoDCs as compared to aged CD8+ T cells, the greater magnitude in expression of downregulated genes nevertheless may result in significant alterations in MoDC functions.
In summary, by utilizing microarray gene analysis of MoDCs from young and aged donors, we have identified distinctive age-related gene expression patterns in MoDCs from aged donors. Genes with expression changes associated with donor age appear to play important roles in fundamental cellular processes such as immune response, cellular and DNA integrity, and oxidation-reduction. These changes may alter the functionality of aged human MoDCs, and impair adaptive immunity and exacerbate inflammation in the elderly.
Supporting Information
Percentages of up-and down-regulated genes from aged-donor DCs compared to aged-donor CD8+ T cells.
(TIF)
A list of 260 genes with expression changes in DCs associated with donor age and meeting the criteria of p <0.05 (student's t -test), FDR <5%, and fold change ≥1.5.
(XLS)
Classification by cellular function of genes displayed in Figure 3 . These are genes with age-associated expression changes in cells derived from 4 young and 5 aged individual donors. Expression level means, standard errors, p-statistics and per-array values are tabulated.
(XLS)
Sequences of primers used for qRT-PCR.
(XLS)
Acknowledgments
We thank Seung-Ah Chung for gene target preparation and processing for microarray analysis at Genomics High-Throughput Facility, UCI.
Funding Statement
The authors have no support or funding to report.
References
- 1. McGlauchlen KS, Vogel LA (2003) Ineffective humoral immunity in the elderly. Microbes and Infection 5: 1279–1284. [DOI] [PubMed] [Google Scholar]
- 2. Wick G, Grubeck-Loebenstein B (1997) The aging immune system: Primary and secondary alterations of immune reactivity in the elderly. Experimental Gerontology 32: 401–413. [DOI] [PubMed] [Google Scholar]
- 3.Cevenini E, Monti D, Franceschi C (2013) Inflamm-ageing. Current Opinion in Clinical Nutrition & Metabolic Care 16: : 14–20 10.1097/MCO.1090b1013e32835ada32813. [DOI] [PubMed] [Google Scholar]
- 4. McGeer PL, McGeer EG (2004) Inflammation and the Degenerative Diseases of Aging. Annals of the New York Academy of Sciences 1035: 104–116. [DOI] [PubMed] [Google Scholar]
- 5. Banchereau J, Steinman RM (1998) Dendritic cells and the control of immunity. Nature 392: 245–252. [DOI] [PubMed] [Google Scholar]
- 6. Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, et al. (2000) Immunobiology of Dendritic Cells. Annual Review of Immunology 18: 767–811. [DOI] [PubMed] [Google Scholar]
- 7. Agrawal A, Agrawal S, Cao JN, Su H, Osann K, et al. (2007) Altered innate immune functioning of dendritic cells in elderly humans: a role of phosphoinositide 3-kinase-signaling pathway. J Immunol 178: 6912–6922. [DOI] [PubMed] [Google Scholar]
- 8. Panda A, Qian F, Mohanty S, van Duin D, Newman FK, et al. (2010) Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J Immunol 184: 2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Agrawal A, Gupta S (2011) Impact of aging on dendritic cell functions in humans. Ageing Res Rev 10: 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agrawal A, Tay J, Ton S, Agrawal S, Gupta S (2009) Increased reactivity of dendritic cells from aged subjects to self-antigen, the human DNA. J Immunol 182: 1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agrawal A, Tay J, Yang GE, Agrawal S, Gupta S (2010) Age-associated epigenetic modifications in human DNA increase its immunogenicity. Aging (Albany NY) 2: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prakash S, Agrawal S, Cao J-n, Gupta S, Agrawal A (2013) Impaired secretion of interferons by dendritic cells from aged subjects to influenza. AGE 35: 1785–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sridharan A, Esposo M, Kaushal K, Tay J, Osann K, et al. (2011) Age-associated impaired plasmacytoid dendritic cell functions lead to decreased CD4 and CD8 T cell immunity. AGE 33: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jonuleit H, Kühn U, Müller G, Steinbrink K, Paragnik L, et al. (1997) Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. European Journal of Immunology 27: 3135–3142. [DOI] [PubMed] [Google Scholar]
- 15. Zhou YH, Raj VR, Siegel E, Yu L (2010) Standardization of Gene Expression Quantification by Absolute Real-Time qRT-PCR System Using a Single Standard for Marker and Reference Genes. Biomark Insights 5: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao JN, Gollapudi S, Sharman EH, Jia Z, Gupta S (2010) Age-related alterations of gene expression patterns in human CD8+ T cells. Aging Cell 9: 19–31. [DOI] [PubMed] [Google Scholar]
- 17. Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375. [DOI] [PubMed] [Google Scholar]
- 18.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, et al.. (2003) MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 4. [DOI] [PMC free article] [PubMed]
- 19. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Furuta S (2008) An essential role for de novo biosynthesis of L-serine in CNS development. Asia Pac J Clin Nutr 17: 312–315. [PubMed] [Google Scholar]
- 21. Diamond MS, Farzan M (2013) The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pichlmair A, Lassnig C, Eberle C-A, Gorna MW, Baumann CL, et al. (2011) IFIT1 is an antiviral protein that recognizes 5[prime]-triphosphate RNA. Nat Immunol 12: 624–630. [DOI] [PubMed] [Google Scholar]
- 23. Espert L, Degols G, Lin YL, Vincent T, Benkirane M, et al. (2005) Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol 86: 2221–2229. [DOI] [PubMed] [Google Scholar]
- 24. Gollob JA, Ritz J (1996) CD2-CD58 Interaction and the Control of T-Cell Interleukin-12 Responsiveness. Annals of the New York Academy of Sciences 795: 71–81. [DOI] [PubMed] [Google Scholar]
- 25. Haller O, Staeheli P, Kochs G (2007) Interferon-induced Mx proteins in antiviral host defense. Biochimie 89: 812–818. [DOI] [PubMed] [Google Scholar]
- 26. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, et al. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akira S, Takeda K (2004) Toll-like receptor signalling. Nat Rev Immunol 4: 499–511. [DOI] [PubMed] [Google Scholar]
- 28. Kim YM, Brinkmann MM, Paquet ME, Ploegh HL (2008) UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452: 234–238. [DOI] [PubMed] [Google Scholar]
- 29. Alexander WS, Hilton DJ (2004) The Role of Suppressors of Cytokine Signaling (SOCS) Proteins in Regulation of the Immune Response. Annual Review of Immunology 22: 503–529. [DOI] [PubMed] [Google Scholar]
- 30. Gilli F, Navone ND, Perga S, Marnetto F, Caldano M, et al. (2011) Loss of braking signals during inflammation: a factor affecting the development and disease course of multiple sclerosis. Arch Neurol 68: 879–888. [DOI] [PubMed] [Google Scholar]
- 31. McBerry C, Gonzalez RM, Shryock N, Dias A, Aliberti J (2012) SOCS2-induced proteasome-dependent TRAF6 degradation: a common anti-32. inflammatory pathway for control of innate immune responses. PLoS One 7: e38384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fukui R, Saitoh S, Kanno A, Onji M, Shibata T, et al. (2011) Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity 35: 69–81. [DOI] [PubMed] [Google Scholar]
- 33. Molenaar R, Knippenberg M, Goverse G, Olivier BJ, de Vos AF, et al. (2011) Expression of Retinaldehyde Dehydrogenase Enzymes in Mucosal Dendritic Cells and Gut-Draining Lymph Node Stromal Cells Is Controlled by Dietary Vitamin A. The Journal of Immunology 186: 1934–1942. [DOI] [PubMed] [Google Scholar]
- 34. Mellor AL, Munn DH (2004) Ido expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol 4: 762–774. [DOI] [PubMed] [Google Scholar]
- 35. Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, et al. (2009) GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A 106: 13445–13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash S, Agrawal S, Vahed H, Ngyuen M, Benmohamad L, et al.. (2014) Dendritic cells from aged subjects contribute to chronic airway inflammation by activating bronchial epithelial cells under steady state. Mucosal Immunol. [DOI] [PMC free article] [PubMed]
- 37. Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, et al. (1999) CCR7 Coordinates the Primary Immune Response by Establishing Functional Microenvironments in Secondary Lymphoid Organs. Cell 99: 23–33. [DOI] [PubMed] [Google Scholar]
- 38. Braun A, Worbs T, Moschovakis GL, Halle S, Hoffmann K, et al. (2011) Afferent lymph-derived T cells and DCs use different chemokine receptor CCR7-dependent routes for entry into the lymph node and intranodal migration. Nat Immunol 12: 879–887. [DOI] [PubMed] [Google Scholar]
- 39. Bouvard D, Millon-Fremillon A, Dupe-Manet S, Block MR, Albiges-Rizo C (2006) Unraveling ICAP-1 function: toward a new direction? Eur J Cell Biol 85: 275–282. [DOI] [PubMed] [Google Scholar]
- 40. Nobes C, Marsh M (2000) Dendritic cells: New roles for Cdc42 and Rac in antigen uptake? Current biology: CB 10: R739–R741. [DOI] [PubMed] [Google Scholar]
- 41. Chi X, Wang S, Huang Y, Stamnes M, Chen JL (2013) Roles of rho GTPases in intracellular transport and cellular transformation. Int J Mol Sci 14: 7089–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhong J, Rao X, Deiuliis J, Braunstein Z, Narula V, et al. (2013) A Potential Role for Dendritic Cell/Macrophage-Expressing DPP4 in Obesity-Induced Visceral Inflammation. Diabetes 62: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boucheron N, Ellmeier W (2012) The role of Tec family kinases in the regulation of T-helper-cell differentiation. Int Rev Immunol 31: 133–154. [DOI] [PubMed] [Google Scholar]
- 44. Maslov AY, Vijg J (2009) Genome instability, cancer and aging. Biochimica et Biophysica Acta (BBA) - General Subjects 1790: 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu B, Yip R, Zhou Z (2012) Chromatin remodeling, DNA damage repair and aging. Curr Genomics 13: 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stark GR, Taylor WR (2006) Control of the G2/M transition. Mol Biotechnol 32: 227–248. [DOI] [PubMed] [Google Scholar]
- 47. Lauper N, Beck AR, Cariou S, Richman L, Hofmann K, et al. (1998) Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene 17: 2637–2643. [DOI] [PubMed] [Google Scholar]
- 48. Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, et al. (2012) The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gruss OJ, Wittmann M, Yokoyama H, Pepperkok R, Kufer T, et al. (2002) Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat Cell Biol 4: 871–879. [DOI] [PubMed] [Google Scholar]
- 50. Cheung CH, Cheng L, Chang KY, Chen HH, Chang JY (2011) Investigations of survivin: the past, present and future. Front Biosci 16: 952–961. [DOI] [PubMed] [Google Scholar]
- 51. Scolz M, Widlund PO, Piazza S, Bublik DR, Reber S, et al. (2012) GTSE1 Is a Microtubule Plus-End Tracking Protein That Regulates EB1-Dependent Cell Migration. PLoS One 7: e51259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee YM, Lee S, Lee E, Shin H, Hahn H, et al. (2001) Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem J 360: 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng L, Jia J, Finger LD, Guo Z, Zer C, et al. (2011) Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Research 39: 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ellison V, Stillman B (2003) Biochemical Characterization of DNA Damage Checkpoint Complexes: Clamp Loader and Clamp Complexes with Specificity for 5′ Recessed DNA. PLoS Biol 1: e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. von Eyss B, Maaskola J, Memczak S, Mollmann K, Schuetz A, et al. (2012) The SNF2-like helicase HELLS mediates E2F3-dependent transcription and cellular transformation. EMBO J 31: 972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, et al. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117. [DOI] [PubMed] [Google Scholar]
- 57. Zhang H, Rider SD, Henderson JT, Fountain M, Chuang K, et al. (2008) The CHD3 Remodeler PICKLE Promotes Trimethylation of Histone H3 Lysine 27. Journal of Biological Chemistry 283: 22637–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moon SK, Kim HM, Kim CH (2004) PTEN induces G1 cell cycle arrest and inhibits MMP-9 expression via the regulation of NF-kappaB and AP-1 in vascular smooth muscle cells. Arch Biochem Biophys 421: 267–276. [DOI] [PubMed] [Google Scholar]
- 59. Hlobilkova A, Knillova J, Svachova M, Skypalova P, Krystof V, et al. (2006) Tumour suppressor PTEN regulates cell cycle and protein kinase B/Akt pathway in breast cancer cells. Anticancer Res 26: 1015–1022. [PubMed] [Google Scholar]
- 60. Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, et al. (2011) Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16ink4a . Cell Cycle 10: 457–468. [DOI] [PubMed] [Google Scholar]
- 61. Campisi J (2011) Cellular senescence: putting the paradoxes in perspective. Curr Opin Genet Dev 21: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Michalak M, Burns K, Andrin C, Mesaeli N, Jass GH, et al. (1996) Endoplasmic Reticulum Form of Calreticulin Modulates Glucocorticoid-sensitive Gene Expression. Journal of Biological Chemistry 271: 29436–29445. [DOI] [PubMed] [Google Scholar]
- 63. MacDonald KPA, Munster DJ, Clark GJ, Dzionek A, Schmitz J, et al. (2002) Characterization of human blood dendritic cell subsets. Blood 100: 4512–4520. [DOI] [PubMed] [Google Scholar]
- 64. Neyt K, Lambrecht BN (2013) The role of lung dendritic cell subsets in immunity to respiratory viruses. Immunological Reviews 255: 57–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Percentages of up-and down-regulated genes from aged-donor DCs compared to aged-donor CD8+ T cells.
(TIF)
A list of 260 genes with expression changes in DCs associated with donor age and meeting the criteria of p <0.05 (student's t -test), FDR <5%, and fold change ≥1.5.
(XLS)
Classification by cellular function of genes displayed in Figure 3 . These are genes with age-associated expression changes in cells derived from 4 young and 5 aged individual donors. Expression level means, standard errors, p-statistics and per-array values are tabulated.
(XLS)
Sequences of primers used for qRT-PCR.
(XLS)