Abstract
Many proteins have been implicated in the physiological function of telomerase, but specific roles of telomerase-associated proteins other than telomerase reverse transcriptase (TERT) remain ambiguous. To gain a more comprehensive understanding of catalytically active enzyme composition, we performed affinity purification of epitope-tagged, endogenously assembled Tetrahymena telomerase. We identified and cloned genes encoding four telomerase proteins in addition to TERT. We demonstrate that both of the two new proteins characterized in detail, p65 and p45, have essential roles in the maintenance of telomere length as part of a ciliate telomerase holoenzyme. The p65 subunit contains an La motif characteristic of a family of direct RNA-binding proteins. We find that p65 in cell extract is associated specifically with telomerase RNA, and that genetic depletion of p65 reduces telomerase RNA accumulation in vivo. These findings demonstrate that telomerase holoenzyme proteins other than TERT play critical roles in RNP biogenesis and function.
Keywords: Tetrahymena, telomerase, telomere, ribonucleoprotein
Telomerase is a ribonucleoprotein (RNP) reverse transcriptase specialized for addition of telomeric repeats to chromosome 3′-ends. The telomerase reverse transcriptase protein (TERT) contains active-site motifs characteristic of viral reverse transcriptases and other motifs shared only among TERTs (for review, see Kelleher et al. 2002). The telomerase RNA subunit provides a template sequence for telomeric repeat synthesis and also functions in determining enzyme catalytic properties (for review, see Blackburn 2000; Harrington 2003). TERT and telomerase RNA from a few species, if coexpressed in specific cell lysates, can reconstitute a properly templated reverse transcriptase activity (Weinrich et al. 1997). For Tetrahymena TERT and telomerase RNA assembled in rabbit reticulocyte lysate, the activity of the minimal recombinant enzyme has notable differences from endogenous enzyme activity in substrate specificity and elongation processivity (Collins and Gandhi 1998; Hardy et al. 2001). These discrepancies could derive from altered protein or RNA folding, differences in the modification state of TERT or telomerase RNA, or differential association of additional RNP factors.
Biochemical fractionation of cell extract suggests that physiologically assembled telomerase RNPs possess a substantial mass in excess of TERT and telomerase RNA (for review, see Collins 1999). Many proteins that associate with telomerase complexes have been identified using biochemical, genetic, and candidate approaches (for review, see Ford et al. 2002; Harrington 2003). Owing to the scarcity of endogenous telomerase complexes, however, it has been difficult to determine whether individual telomerase-associated proteins interact with a common RNP or with distinct, heterogeneous RNP populations.
One important biological role for telomerase-associated proteins is the assembly of telomerase RNA into stable RNP, a role evidently not fulfilled by TERT itself. In vertebrate cells, the telomerase RNA H/ACA motif is essential for precursor RNA 3′-end processing and mature RNA accumulation (Mitchell et al. 1999a). This motif interacts with dyskerin, NHP2, NOP10, and GAR1 (Mitchell et al. 1999b; Dragon et al. 2000; Pogacic et al. 2000), proteins also associated with a large family of H/ACA small nucleolar (sno) RNPs that catalyze the posttranscriptional, site-specific conversion of uridine to pseudouridine (for review, see Kiss 2002). In Saccharomyces cerevisiae, Sm proteins associate with telomerase RNA to enhance RNA accumulation (Seto et al. 1999). To date, none of these proteins implicated in cellular telomerase RNP assembly is unique to telomerase RNP, complicating the analysis of their telomerase biogenesis roles in specific. In the ciliate Euplotes aediculatus, telomerase affinity purification by template-complementary oligonucleotide hybridization identified two telomerase components, TERT and p43 (Lingner and Cech 1996). Euplotes p43 contains an La motif (Aigner et al. 2000), a defining characteristic of the eukaryotic La protein and additional La motif-containing proteins with direct RNA-binding activities (for review, see Wolin and Cedervall 2002). Indeed, recombinant p43 can bind telomerase RNA directly and antibodies against p43 substantially immunodeplete telomerase RNA and activity from Euplotes nuclear extracts (Aigner et al. 2000, 2003). A role for p43 in telomerase nuclear retention has been proposed based on changes in the efficiency of telomerase RNA electroelution from macronuclei when p43 is targeted for RNAi (Möllenbeck et al. 2003).
Telomerase-associated proteins also play biologically important roles in coupling telomerase RNP to telomere substrates. In S. cerevisiae, mutations in the genes encoding Est1p, Est3p, and Cdc13p can impair telomerase-dependent telomere length maintenance without affecting enzyme activity in cell extract (Lingner et al. 1997). Est1p and Est3p were shown by immunoprecipitation to interact with catalytically active telomerase RNP (Steiner et al. 1996; Hughes et al. 2000). Current models suggest that Est1p either recruits telomerase to a telomeric G-strand 3′-overhang or binds there independently to stimulate the activity of prebound telomerase RNP (Evans and Lundblad 1999; Taggart et al. 2002). Human and Schizosaccharomyces pombe EST1 proteins sharing limited conservation with the budding yeast Est1p have recently been identified using a bioinformatics approach. At least a subset of the human EST1 proteins can interact with active telomerase and affect telomere biology (for review, see Lundblad 2003; Vega et al. 2003).
The ciliate Tetrahymena thermophila is a favorable model system for the study of telomeres and telomerase. Tetrahymena have a silent, diploid germ-line micronucleus containing five pairs of mitotically segregated chromosomes. The transcriptionally active macronucleus, in contrast, has ∼20,000 chromosomes and thus ∼40,000 telomeres. Fully half of these derive from amplification of a palindromic chromosome encoding ribosomal RNA (rDNA). The macronuclear genome also contains ∼45 copies each of ∼200 additional chromosomes created during a developmentally programmed process of fragmentation and amplification. The large number of telomeres obliges an abundance of telomerase that can be readily detected in cell extracts (Greider and Blackburn 1985). In addition to this biochemical advantage, T. thermophila is highly amenable to genetic and molecular genetic analysis (for review, see Turkewitz et al. 2002). The macronucleus divides amitotically during vegetative growth, producing daughter cells with different allele frequencies at each locus. In the process of phenotypic assortment, a locus disrupted by integration of a selectable marker can be enriched relative to the wild-type locus by culture growth under selective pressure to reach one of two endpoints: a complete gene knockout if the targeted gene is not essential, or a function-limiting gene dose knockdown in the case of essential genes (for review, see Karrer 2000).
Even in ciliate systems, telomerase purification has proven to be a difficult task. Telomeres and telomerase are most abundant in spirotrichous ciliates such as Euplotes. Affinity purification from E. aediculatus extracts using template-complementary oligonucleotide hybridization was successful in the identification of TERT and p43 but was also noted to induce a change in RNP conformation or composition (Lingner and Cech 1996). Human and Tetrahymena cell extracts subjected to a similar purification strategy were even more clearly disrupted by this approach, with substantial changes in telomerase RNP mass and composition (Schnapp et al. 1998; data not shown). Conventional chromatography strategies are also disruptive, because of the extent of purification required. The Tetrahymena p80/p95 complex identified in this manner by cofractionation with telomerase RNA (Collins et al. 1995) is not required for telomerase catalytic activity and negatively regulates telomere length in vivo (Miller and Collins 2000). We have now developed a more rapid and gentle affinity purification strategy that preserves the integrity of telomerase holoenzyme. Here, we exploit the combined biochemical and genetic advantages of Tetrahymena as a telomerase model system to isolate telomerase-associated proteins and characterize their roles in enzyme function.
Results
Transgene expression of epitope-tagged Tetrahymena TERT
Transgenes expressing epitope-tagged TERT were integrated into the Tetrahymena genome by replacement of the nonessential β-tubulin 1 (BTU1) coding region. Growth of the Tetrahymena strain CU522 shows enhanced sensitivity to taxol, because of an amino acid substitution in the β-tubulin protein encoded at BTU1. Disruption of BTU1 does not compromise viability because a second gene also encodes β-tubulin, but in strain CU522, BTU1 disruption confers increased resistance to taxol (Gaertig et al. 1994). Of the several epitope tags screened for use in large-scale TERT affinity purification, the most effective was a C-terminal tandem affinity purification (TAP) tag (Rigaut et al. 1999). This epitope tag consists of two terminally located protein A domains that bind to IgG, an adjacent TEV protease cleavage site to allow elution from IgG resin, and a calmodulin-binding peptide that binds to calmodulin resin in the presence of calcium and elutes with EGTA (see Supplementary Fig. 1A). The TAP tag has been used in previous studies for the purification of other ribonucleoprotein complexes (Puig et al. 2001; Dragon et al. 2002).
We targeted a transgene encoding TAP-tagged TERT (TERT-TAP) for homologous recombination into the BTU1 locus, placing TERT expression under control of the endogenous BTU1 promoter (Gaertig et al. 1999). Correct integration specificity was verified by Southern blot hybridization (data not shown). The resulting strain expressed both TERT-TAP from the BTU1 locus and wild-type TERT from the endogenous TRT locus, as monitored by immunoblotting with a polyclonal antibody specific for TERT (Fig. 1A). The immunoblot signal from TERT-TAP is substantially amplified by binding of the TERT antibody to the protein A domains of the TAP tag. To determine whether TERT-TAP was produced at a physiologically appropriate level, we incubated total cell extract with TEV protease to remove the TERT-TAP protein A domains before immunoblot analysis (Supplementary Fig. 1B). Using this method, we determined that epitope-tagged TERT in this and all other TERT transgene strains accumulated to a level less than or equal to endogenous TERT in wild-type cells.
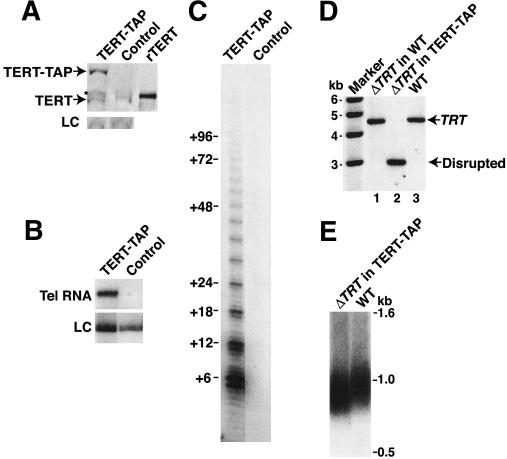
Figure 1.
TERT-TAP is expressed and catalytically active. (A) Whole-cell extract from TERT-TAP or a control strain was normalized by total protein concentration and analyzed by immunoblot for TERT. Control cells in panels A, B, and C expressed both wild-type TERT and a tagged TERT differing from TERT-TAP by substitution of the Flag peptide epitope for both protein A domains. Recombinant TERT (rTERT) expressed in E. coli was used as an immunoblot standard. A nonspecific cross-reacting polypeptide is shown as a loading control (LC), with equal loading also verified by staining of the immunoblot membrane (not shown). In the TERT-TAP sample, a minor TERT-TAP degradation product (*) migrates with slightly slower mobility than wild-type TERT (see Supplementary Fig. 1 for additional data). (B,C) Extracts normalized by total protein concentration from TERT-TAP cells and control cells were bound to IgG resin. Beads were assayed for telomerase RNA by Northern blot (B) and were assayed independently for telomerase activity (C). The activity assay product ladder is annotated to indicate product lengths from processive synthesis of telomeric repeats. Prior to RNA precipitation for Northern blot analysis, a truncated form of purified recombinant telomerase RNA was added as a loading control (LC). (D) Genomic DNA was digested with HindIII and analyzed by Southern blot from untransformed wild-type cells (WT); wild-type cells transformed with the TRT gene disruption cassette, phenotypically assorted and then released from selection (ΔTRT in WT); or TERT-TAP cells similarly transformed with the TRT gene disruption cassette, assorted and then released from selection (ΔTRT in TERT-TAP). The probe recognizes the wild-type and disrupted TRT loci with equivalent hybridization potential. (E) Macronuclear telomere length was assayed in wild-type cells or TERT-TAP cells disrupted for the wild-type TRT gene. Genomic DNA was digested with HindIII, resolved on a nondenaturing agarose gel, and assayed by Southern blot to detect rDNA telomeres. The hybridization probe recognizes the 357-bp subtelomeric region of rDNA chromosomes.
To determine whether TERT-TAP was assembled into a catalytically functional telomerase RNP, we assayed for cofractionation of telomerase RNA and activity with TERT-TAP after purification on IgG resin. Both telomerase RNA and activity were recovered on IgG resin from extracts of strains expressing TERT-TAP but not from control strains expressing TERT C-terminally fused to a TAP tag variant containing a Flag epitope in place of the protein A domains (Fig. 1B,C). This result indicates that TERT-TAP associates with telomerase RNA in enzymatically functional telomerase RNP complexes. Notably, upon purification of the epitope-tagged TERT, we failed to detect any association with coexpressed wild-type TERT even when the wild-type TERT was much more abundant (Supplementary Fig. 2). This result suggests that Tetrahymena TERT is monomeric in the active telomerase RNP purified from cell extract.
To determine whether TERT-TAP telomerase RNPs were functional in vivo as well as in vitro, we disrupted the endogenous TRT gene to eliminate the expression of wild-type TERT protein. We used phenotypic assortment to test whether TERT-TAP could substitute genetically for wild-type TERT during vegetative growth (see Supplementary Fig. 3). Because TRT is an essential gene in wild-type cells, it is impossible to completely replace the endogenous TRT locus with a transgene conferring paromomycin resistance (Miller and Collins 2000). Instead, under maximum selective pressure, a combination of wild-type and disrupted loci is observed. This genotype reverted to predominantly wild-type TRT in the absence of selection (Fig. 1D, lane 1). In contrast, in strains containing the TERT-TAP transgene, the endogenous TRT locus could be fully replaced with a transgene encoding paromomycin resistance. No back-assortment to wild-type TRT was detected even after multiple generations of growth in the absence of selective pressure (Fig. 1D, lane 2). Immunoblot analysis confirmed that TRT knockout cells with the TERT-TAP transgene do not express wild-type TERT protein (Supplementary Fig. 1B). Moreover, these cells expressed TERT-TAP protein at the normal physiological level of the endogenous TERT protein (Supplementary Fig. 1B). These experiments demonstrate that TERT-TAP fulfills the essential functions of wild-type TERT in vivo. Cells expressing TERT-TAP instead of wild-type TERT had no growth defect and no apparent change in telomerase activity assayed in cell extract (data not shown). Cells expressing only TERT-TAP maintained macronuclear telomere lengths within the variability between wild-type strains (Fig. 1E; data not shown), confirming that the tagged protein functions in telomerase-dependent telomere length maintenance. All subsequent experiments shown here were performed using the TERT-TAP transgene strain lacking wild-type TERT.
Affinity purification of epitope-tagged TERT reveals additional telomerase proteins
Extracts from TERT-TAP cells were used for telomerase affinity purification with only minor changes from the standard TAP protocol (Puig et al. 2001). Telomerase activity (Fig. 2A), telomerase RNA (data not shown), and TERT protein (Fig. 2B) were recovered and enriched with each specific binding and elution step. Importantly, telomerase activity in the purified samples retained the high repeat addition processivity of the authentic cellular holoenzyme. By SDS-PAGE, four major polypeptides in addition to TERT were evident by silver stain (Fig. 2C) or Coomassie stain (data not shown) in several variations of purification conditions and with numerous extract preparations. These polypeptides were not recovered in purifications performed in parallel from extracts of control strains (data not shown). Notably, p80 and p95 were not detected in the purified material either by silver stain (Fig. 2C) or immunoblot (data not shown; see Discussion).
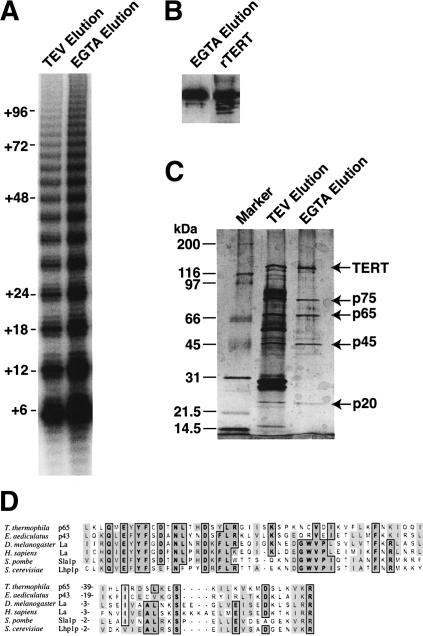
Figure 2.
Four major proteins copurify with TERT-TAP. (A) Telomerase activity was assayed in the TEV protease and EGTA elutions of the affinity purification. The activity assay product ladder is annotated to indicate product lengths from processive synthesis of telomeric repeats. (B) TERT recovered in an aliquot of the EGTA elution was examined by immunoblot in comparison with 5 ng of recombinant TERT standard (rTERT). (C) Aliquots of the TEV protease and EGTA elutions were resolved by SDS-PAGE, and proteins were visualized by silver stain. The molecular weights of proteins in the Marker lane are indicated at left, and the most abundant polypeptides in the EGTA elution are indicated at right. (D) The La motif in p65 was aligned with La motifs in other proteins. Identical residues are boxed and shaded dark. Similar residues are lightly shaded. The numbering at left in the lower alignment panel indicates the number of amino acids not shown between conserved sequences at the end of the top panel and the beginning of the bottom panel.
Peptide sequences were obtained from proteolytic digests of each protein and used to design primers for degenerate PCR. PCR products were obtained for which translation in silico corroborated additional peptide sequence not used in primer design. Subsequently, RT–PCR and RACE were used to obtain full-length cDNA sequences, and PCR was used to obtain the sequences of the macronuclear genomic loci (see Supplemental Material). Each full-length gene is predicted to encode a protein with molecular weight closely matching the apparent molecular weight by SDS-PAGE. For chronological reasons, we began our analysis of telomerase holoenzyme proteins with p65 and p45.
We designated the p45 gene TAP45, for TERT- or telomerase-associated protein of 45 kDa. The protein encoded by the full-length cDNA exhibits no apparent homology to other known proteins when examined using BLAST searches of the GenBank database. We termed the p65 gene TAP65. BLAST searches of GenBank with p65 revealed limited homology to other proteins within an N-terminal region representing a divergent La motif (Fig. 2D). This motif, strikingly also found in Euplotes p43, was originally discovered in the eukaryotic La protein and has subsequently been found in additional direct RNA-binding proteins (for review, see Wolin and Cedervall 2002). In its most characterized role, La binds to the 3′ polyuridylate tract of primary transcripts of RNA polymerase III and to other small RNA precursors, protecting them from exonucleolytic degradation and promoting subsequent assembly to mature RNP. Both the La motif and a putative RNA recognition motif (RRM) C-terminal to the La motif were detected in a p65 query of the Prosite database (http://us.expasy.org/prosite). Sequence homology between Tetrahymena p65 and Euplotes p43 was contained almost exclusively within the La motif, not the adjacent putative RRM. The full-length forms of the two proteins share ∼20% amino acid identity and ∼40% amino acid similarity overall.
Endogenous p65 and p45 are components of telomerase holoenzyme
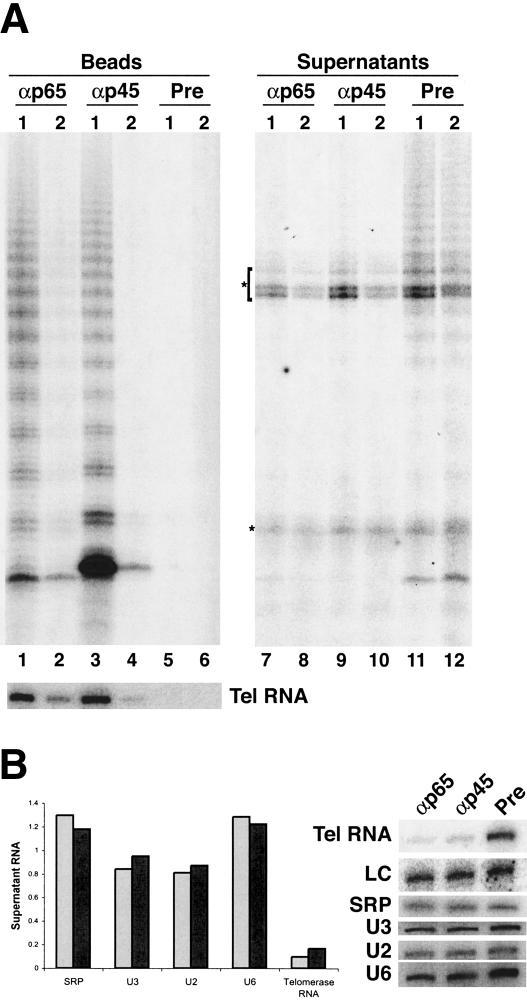
Because no extensive purification protocol, including our affinity purification, can recover all of the tagged protein, telomerase RNA, or telomerase activity in the starting material, it was important to determine using whole-cell extract whether p65 and p45 associate with a large percentage of the total telomerase RNP pool. We thus performed immunodepletion studies with polyclonal antibodies raised and affinity-purified against full-length p65 and p45 overexpressed using synthetic genes in Escherichia coli (R. Prathapam, K.L. Witkin, and K. Collins, unpubl.). These antibodies recognized their cognate proteins specifically (Supplementary Fig. 4). We subjected wild-type Tetrahymena whole-cell extract to serial depletions in successive, 1-h incubations with bead-immobilized antibody. Antibody specific for p65 or p45 recruited telomerase activity and telomerase RNA to beads (Fig. 3A, lanes 1–4), whereas preimmune serum (Fig. 3A, lanes 5–6) or affinity-purified antibody raised against an unrelated human protein (data not shown) did not. Depletion with p65 or p45 antibody reduced telomerase activity in an aliquot of the supernatant to a barely detectable (after round 1) or undetectable (after round 2) level (Fig. 3A, lanes 7–10), whereas preimmune serum (Fig. 3A, lanes 11–12) or another control antibody (data not shown) did not. Northern blot hybridization revealed that telomerase RNA remaining in the second-round immunodepletion supernatant was reduced to <10% of the amount of telomerase RNA in the control antibody supernatant (Fig. 3B). In contrast, other small RNAs including SRP RNA, U3 snoRNA, U2 small nuclear (sn) RNA, and U6 snRNA remained in the supernatant after immunodepletion with either antibody. Notably, none of the antibodies depleted a nonspecific radiolabeling activity evident in the whole-cell extracts (Fig. 3A, products marked by an asterisk). Using either p65 or p45 antibody, the majority of telomerase activity and RNA could be removed from supernatants and recovered on antibody beads. This finding demonstrates that both proteins are stable telomerase RNP components, and furthermore that the predominant pool of telomerase RNP contains both p65 and p45 bound together to each molecule of telomerase RNA.
Figure 3.
Endogenous p65 and p45 are associated with telomerase activity and RNA. Whole-cell extract was subject to immunodepletion with antibodies against p65 (αp65) and p45 (αp45) or with preimmune serum (Pre). (A) A small aliquot of extract samples following first (1) or second (2) round immunodepletion (Supernatants) and a 30-fold higher relative amount of each bead-bound sample (Beads) were assayed for telomerase activity. Asterisks indicate telomerase-independent background from radiolabeling of nucleic acids in the crude extract. Telomerase RNA associated with an aliquot of each antibody bead sample was detected by Northern blot. (B) The extent of supernatant RNA depletion was quantitated following the second round of immunodepletion by comparing p65 (αp65), p45 (αp45), and preimmune (Pre) antibody depletions to control for any RNA degradation over the incubation in crude extract. A single blot was probed for telomerase RNA, recombinant truncated telomerase RNA added prior to precipitation of RNA samples as a loading control (LC) and endogenous control RNAs (at right). Hybridization signal intensity was quantitated by PhosphorImager analysis to determine the specificity of immunodepletion (at left) using antibodies against p65 (light gray) and p45 (dark gray). Recovery of control RNAs normalized to the preimmune immunodepletion control ranged from 80% to 120% in the single experiment shown, reflecting small irreproducible differences in sample recovery.
p65 associates specifically with telomerase RNA
To further confirm the association of p65 and p45 with active telomerase RNP, we created Tetrahymena strains expressing an epitope-tagged version of each protein. Proteins fused at their C termini to a TAP tag were expressed from transgenes integrated into the CU522 macronuclear genome at the BTU1 locus as described for TERT-TAP above. We confirmed the intended integration events by Southern blot (data not shown). In each case, the tagged protein accumulated at a much lower level than TERT-TAP and likely for this reason was unable to substitute genetically for the wild-type protein. Although p65-TAP and p45-TAP expression levels were too low to permit detection of associated proteins after affinity purification from cell extract, purification could recover telomerase activity substantially enriched over any background recovered from wild-type control extract (Fig. 4A). The relative recovery of telomerase activity is not a meaningful comparison in this experiment, because individual TAP-tagged proteins are expected to bind IgG resin at different efficiencies and are present at different levels in the starting extract as well.
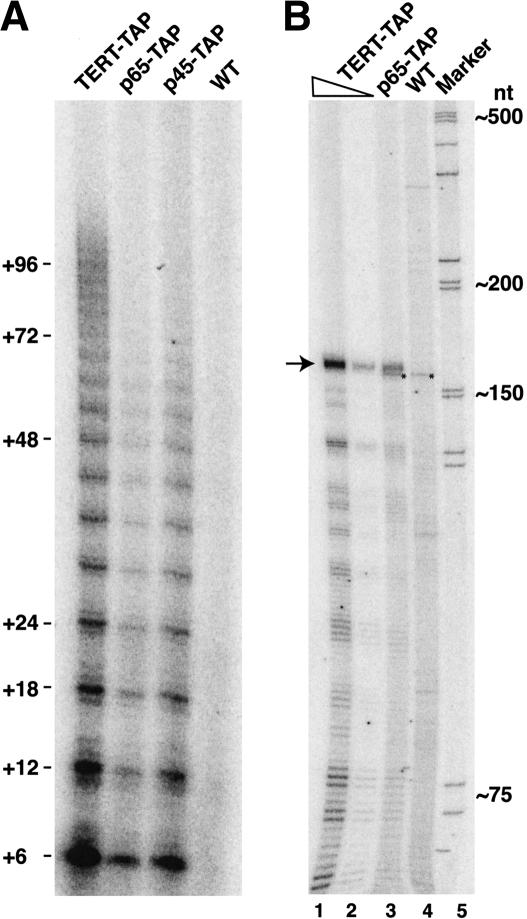
Figure 4.
Epitope-tagged p45 and p65 copurify active telomerase RNP. (A) Whole-cell extracts from TERT-TAP, p65-TAP, p45-TAP, and wild-type strains normalized by total protein concentration were incubated with IgG resin. Specifically bound complexes were eluted from the resin with TEV protease and assayed for telomerase activity. (B) RNAs recovered after the complete TAP purification protocol from TERT-TAP or p65-TAP cell extracts were radiolabeled by a single 3′-nucleotide addition. Because p65-TAP was expressed at a lower level than TERT-TAP, 90% (lane 1) and 10% (lane 2) of the TERT-TAP purification were loaded in separate lanes for better comparison to 100% of the p65-TAP purification (lane 3). RNAs were resolved by denaturing gel electrophoresis. The arrow indicates the predominant RNA species copurified with TERT-TAP and p65-TAP that also comigrated with a purified telomerase RNA standard (data not shown). The asterisk indicates a background RNA present in the control purification from wild-type cells (lane 4). The Marker lane contains end-labeled DNA ladder.
The presence of an La motif in p65 raises the intriguing possibility that it could be the Tetrahymena La protein, playing a general role in binding RNA polymerase III transcripts including telomerase RNA. We used two sequential steps of affinity purification of p65-TAP to investigate whether p65 is preferentially associated with telomerase RNA, as expected of a telomerase-specific protein, or whether like La it associates with the large variety of RNA species. We compared the spectrum of RNAs that copurified with TERT-TAP and p65-TAP from whole-cell extracts, also monitoring any background RNAs recovered in a parallel mock purification from extract of a control strain. RNA was precipitated from the purified samples, radiolabeled by addition of a single 3′-nucleotide, then resolved by denaturing gel electrophoresis (Fig. 4B). The TERT-TAP-associated RNA profile (Fig. 4B, lanes 1–2 loaded at two different concentrations) matched the p65-TAP associated RNA profile (Fig. 4B, lane 3) and was composed of a single predominant species not present in the wild-type extract control purification (Fig. 4B, lane 4). The RNA associated with TERT-TAP and p65-TAP migrated consistent with an expected telomerase RNA size of 159 nt and comigrated with a recombinant telomerase RNA standard (data not shown). Some RNA degradation occurred during the 3′-radiolabeling protocol, resulting in a variable ladder of smaller RNA products in all samples including the purified RNA standard. The similarity of the TERT- and p65-associated RNA profiles suggests that both proteins are preferentially associated with telomerase RNA.
Telomerase p65 and p45 are encoded by essential genes
The genes encoding TERT and telomerase RNA are essential for Tetrahymena viability (Miller and Collins 2000). To determine whether the genes encoding p65 and p45 are similarly essential, we performed gene disruption of TAP65 and TAP45 loci in a manner identical to the TRT gene disruption described above. In strains taken to maximal phenotypic assortment and then released from selective pressure for an extended interval of growth, analysis of TAP65 (Fig. 5A) and TAP45 (Fig. 5B) loci revealed that both wild-type and disrupted genes remained present. The persistence of the wild-type loci through phenotypic assortment indicates that both p65 and p45 are encoded by essential genes (see Supplementary Fig. 3).
Figure 5.
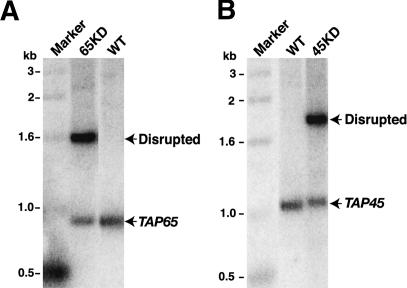
The genes encoding p45 and p65 are essential. Cells were transformed with a construct designed to disrupt the endogenous gene encoding either p65 (TAP65 locus) or p45 (TAP45 locus). (A) Genomic DNA from wild-type cells and from cells transformed with the TAP65 disruption construct (65KD), maximally assorted and released from selective pressure was digested with BbsI and SacI and then analyzed by Southern blot. The probe recognizes wild-type and disrupted TAP65 loci with equivalent hybridization potential. (B) Genomic DNA from wild-type cells and from cells transformed with the TAP45 disruption construct (45KD), maximally assorted and released from selective pressure was digested with SpeI and SacI and then analyzed by Southern blot. The probe recognizes wild-type and disrupted TAP45 loci with equivalent hybridization potential.
Short telomeres in p65 and p45 knockdown strains
We used the maximally assorted gene knockdown strains to examine the impact of limiting p65 or p45 function on the physiological role of telomerase in telomere length maintenance. Half of all macronuclear telomeres have precisely the same subtelomeric sequence, derived from the palindromic rDNA chromosome ends. To examine telomere length in p65 and p45 knockdown strains, purified genomic DNA was digested with HindIII, resolved by denaturing gel electrophoresis, and probed for the terminal fragment of the macronuclear rDNA chromosome using an oligonucleotide complementary to the subtelomeric region on the telomeric repeat AC-rich strand. Previous studies using numerous experimental controls have demonstrated that telomere length maintenance is not affected by the presence of selective drug alone (Miller and Collins 2000; data not shown). All cultures were maintained at a selective drug concentration that permitted a growth rate comparable to that of wild-type cells grown in the absence of drug selection.
We found that telomeres were dramatically shorter in p65 and p45 knockdown strains (Fig. 6A, lanes 2–3) than in wild-type cells (Fig. 6A, lane 1). Each hybridizing DNA fragment contains a precise 357 nt of subtelomeric sequence and a variable length of telomeric repeat array. We estimate that under these assortment conditions, the telomeric repeat tract in p65 and p45 knockdown strains was predominantly ∼150–350 bp. In contrast, the telomeric repeat tract in wild-type cells was predominantly ∼350–550 bp. Genomic DNA digested with the frequently cutting restriction enzyme MseI and probed using a telomeric repeat oligonucleotide confirmed the substantial telomere loss on rDNA and other chromosome ends in p65 and p45 knockdown strains (data not shown).
Figure 6.
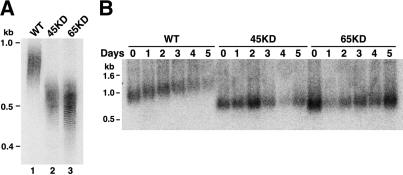
p65 and p45 are required for telomere maintenance in the macronucleus. (A) Genomic DNA from wild-type, p45 knockdown, and p65 knockdown cells was digested with HindIII before Southern blot analysis to detect rDNA telomeres. DNA was resolved on a denaturing acrylamide gel. Markers were detected by hybridization to the same membrane to calculate telomere length as precisely as possible. (B) Wild-type, p45 knockdown, and p65 knockdown strains were grown under maximum drug selection for 24 h followed by 5 d of continuous log-phase growth in the absence of selection. Cells were harvested each day from 0 to 5 d after release from selection for telomere length analysis as in A, except that DNA was electrophoresed on a nondenaturing agarose gel to resolve telomere lengths >1 kb. Markers were detected by ethidium bromide staining of a gel slice removed before the remainder of the gel was transferred to membrane and should be considered as an approximation.
In wild-type strains, macronuclear telomere length increases by 3–10 bp per generation when cultures are kept in continuous log-phase growth at 30°C (Larson et al. 1987). We examined whether this telomere length regulation was affected in p65 and p45 knockdown strains. Cultures maintained under selection (day 0) were released into drug-free media and maintained in log-phase growth for 10 d (∼80–100 doublings total). Telomere length was assayed for rDNA as described above, except that nondenaturing gel electrophoresis was used to resolve longer telomeres (Fig. 6B). In wild-type cells, telomere length increased from day 0 to day 5 as expected (Fig. 6B, WT lanes). In contrast, telomere elongation was marginal in p65 and p45 knockdown strains. By 10 d of growth in the absence of selection, telomere length in the knockdown strains had increased detectably but still remained less than telomere length in wild-type cells at day 0 (data not shown). Analysis of rDNA and other telomeres using genomic DNA digested with MseI and probed with a telomeric repeat oligonucleotide confirmed these altered telomere length dynamics (data not shown).
A telomerase RNA accumulation defect in p65 knockdown strains
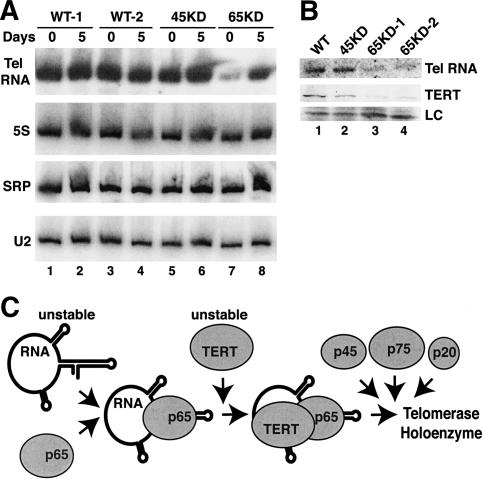
One mechanism that could account for the defect in telomere maintenance in the p65 and p45 knockdown strains is a failure in accumulation of the telomerase RNP. Total RNA prepared from cultures at days 0 and 5 of the time course described above was analyzed for telomerase RNA by Northern blot hybridization. Telomerase RNA accumulation was unaffected by continuous log-phase growth in two independent wild-type cell cultures (Fig. 7A, lanes 1–4). No change in telomerase RNA level was detected in p45 knockdown cultures under selective pressure (Fig. 7A, lane 5) or after 5 d of continuous culture in the absence of selection (Fig. 7A, lane 6). In contrast, telomerase RNA accumulation in the p65 knockdown strain was inhibited dramatically under selective pressure (Fig. 7A, lane 7) and only partially recovered in 5 d of continuous culture after release from selection (Fig. 7A, lane 8).
Figure 7.
Telomerase RNA and TERT levels decrease in p65 knockdown strains. (A) Total RNA was prepared from equal numbers of wild-type cells in log-phase growth under no drug selection (WT-1) or from wild-type and knockdown strain cells in log-phase growth under the maximum sublethal drug selection (WT-2, 45KD, 65KD). These same cell cultures were also grown for an additional 5 d of continuous log-phase growth in the absence of selection. RNA levels were analyzed by Northern blot for telomerase, 5S, SRP, and U2 RNAs. (B) Whole-cell extracts from wild-type, p45 knockdown, and two independent p65 knockdown strains normalized by total protein concentration were used for Northern blot analysis of telomerase RNA (top) or immunoblot analysis of TERT (middle). A nonspecific cross-reacting polypeptide is shown as a loading control (LC) for the immunoblot (bottom), with equal loading also verified by staining of the immunoblot membrane (data not shown). (C) This model depicts the proposed role for p65 in establishing the level of telomerase RNA accumulation, with the subsequent incorporation of TERT, p45, p75, and p20 to form a biologically active telomerase RNP holoenzyme.
The preferential interaction of p65 with telomerase RNA in extract (Fig. 4B) would predict a specific telomerase RNA accumulation defect in the p65 knockdown strain. To test this hypothesis, we reprobed the same Northern blot membrane to detect additional small RNAs. Steady-state levels of 5S rRNA, SRP RNA, and U2 snRNA (Fig. 7A) and also U6 snRNA and U3 snoRNA (data not shown) were not affected by growth conditions in any strain. These RNAs share properties with telomerase RNA including transcription by RNA polymerase III (5S, SRP, U6) or a nuclear site of function (U2, U6, U3). We also investigated whether TERT levels decreased in p65 knockdown strains in parallel with the decrease in telomerase RNA. We compared the levels of telomerase RNA and TERT in whole-cell extracts of the wild-type and knockdown strains. Telomerase RNA detected by Northern blot hybridization (Fig. 7B, top) and wild-type TERT detected by immunoblot (Fig. 7B, middle) were both reduced in extracts from two independently generated p65 knockdown strains (Fig. 7B, lanes 3–4), in comparison to extracts from wild-type or p45 knockdown strains (Fig. 7B, lanes 1–2). These findings provide genetic evidence that p65 functions in vivo to promote telomerase RNA accumulation, playing a crucial and apparently specific, physiological role in telomerase holoenzyme RNP assembly.
Discussion
Telomerase holoenzyme purification
To accomplish a rapid and gentle affinity purification of telomerase RNP, we expressed TAP-tagged Tetrahymena TERT in substitution of the wild-type protein. Telomerase RNPs that incorporate the tagged TERT were shown to have full catalytic activity in vitro and in vivo. Four major proteins copurified with TERT and telomerase RNA, with several less abundantly recovered proteins also coenriched in the purified material (Fig. 2C). Our studies do not address the composition of any hypothetical telomerase RNP lacking TERT or the potential association of developmentally stage-specific holoenzyme proteins.
A conserved role for p65
In yeast and mammalian systems, telomerase proteins other than TERT confer telomerase RNA stability and promote RNP accumulation in vivo. In vertebrate telomerase RNAs, an H/ACA motif serves as the binding site for cooperative assembly of dyskerin, NHP2, NOP10, and GAR1 (Mitchell et al. 1999b; Dragon et al. 2000; Pogacic et al. 2000); the yeast telomerase RNA precursor instead assembles with Sm proteins (Seto et al. 1999). Our findings here reveal that a role for holoenzyme proteins in determining steady-state telomerase RNA accumulation is universal, even though the mechanism of their interaction with telomerase RNA is not. Ciliate telomerase RNAs (∼150 nt) are much smaller than yeast (>1 kb) or vertebrate (∼450 nt) telomerase RNAs and are transcripts of RNA polymerase III rather than RNA polymerase II (for review, see Collins 1999). Nonetheless, telomerase RNA accumulation in Tetrahymena requires the holoenzyme protein p65. By extension, we anticipate that telomerase RNA accumulation in Euplotes will require p43.
Several lines of evidence suggest that p65 functions directly to increase the cellular stability of telomerase RNA (Fig. 7C). In Tetrahymena, overexpression of transgene-encoded telomerase RNA or TERT does not increase the absolute level of subunit accumulation; instead, it competitively reduces endogenous subunit expression to maintain the same overall level of accumulation (Yu et al. 1990; D. Cunningham and K. Collins, unpubl.). This finding suggests that a third factor establishes the steady-state level of telomerase RNP. We propose that p65 is this factor. The presence of an La motif and putative RRM indicate that p65 should bind telomerase RNA directly. The dramatic reduction in telomerase RNA accumulation in p65 gene knockdown strains supports this conclusion. Although TERT accumulation is also reduced in these strains, we believe that TERT is destabilized as a secondary effect of telomerase RNA depletion. This would be consistent with the observation in S. cerevisiae that accumulation of the yeast TERT Est2p is reduced in the absence of telomerase RNA (Taggart et al. 2002). Results from other Tetrahymena telomerase RNP purification strategies reveal that telomerase RNP can be isolated lacking TERT but that TERT is not stable independent of telomerase RNA (K.L. Witkin and K. Collins, unpubl.). Together, these findings suggest that telomerase RNA is destabilized in the absence of p65, and that TERT is degraded in the absence of stable telomerase RNA (Fig. 7C).
Although the La motif is a hallmark feature of the eukaryotic La protein, it is shared by a larger family of proteins distinct from La (for review, see Wolin and Cedervall 2002). Pairwise comparisons of the sequences of Tetrahymena p65, Euplotes p43, and authentic La proteins did not reveal substantial homology beyond the La motif. La itself can associate with the human telomerase RNA transcribed by RNA polymerase II (Ford et al. 2001). However, our experimental evidence suggests that p65 is an La-motif, telomerase-specific protein instead of ciliate La. First, whereas telomerase RNA level decreased in p65 knockdown strains, the accumulation of other RNA polymerase III transcripts was unaffected. Second, the purification of p65-TAP readily recovered telomerase RNA but not other RNA polymerase III transcripts. We note that BLAST searches of the Tetrahymena macronuclear genome (http://www.tigr.org/tdb/e2k1/ttg) can yield at least one region other than the TAP65 locus with potential to encode an La motif. Whether this region produces an mRNA transcript and corresponding protein, and if so whether the protein represents ciliate La, remain to be determined.
Other telomerase holoenzyme protein roles
In addition to directing telomerase RNA folding and RNP stability, telomerase holoenzyme proteins must also coordinate RNP interactions with intranuclear storage sites (Fang and Cech 1995; Wong et al. 2002) and with telomeres (for review, see Vega et al. 2003). Genetic criteria demonstrate that p45 is required for telomere length maintenance but not for cellular accumulation of telomerase RNA or TERT, suggesting that p45 acts at a step after stable RNP biogenesis (Fig. 7C). Telomerase holoenyzme proteins including p45 could be required for catalytic activation. Because knockdown strains retain some wild-type telomerase enzyme even at maximal phenotypic assortment, we cannot reliably assay the activity of an RNP lacking p45 by using p45 knockdown cell extract. Further analysis will be required to determine the precise role of p45 in telomerase function and to investigate roles for p75 and p20 in telomerase-dependent telomere length maintenance.
Does the set of five proteins and telomerase RNA recovered by TERT-TAP affinity purification represent a complete telomerase holoenzyme? Telomerase activity in Tetrahymena extract fractionates at ∼270 kDa by glycerol gradient sedimentation or ∼500 kDa by gel filtration (Collins and Greider 1993; Wang and Blackburn 1997). The RNP recovered through TERT-TAP affinity purification would be consistent with these mass estimates. A strikingly similar fractionation has been reported for active telomerase in extracts of E. aediculatus, comparing both glycerol gradient sedimentation and gel filtration (Lingner and Cech 1996; Aigner et al. 2003). This suggests that the telomerase-associated proteins identified here could also be components of a Euplotes telomerase holoenzyme, with yet additional proteins joining to form larger holoenzyme complexes detected in mated Euplotes crassus undergoing developmentally programmed chromosome fragmentation (Greene and Shippen 1998).
Our TERT-TAP affinity purification did not recover p80 and p95, two proteins characterized by co-fractionation with telomerase RNA through multiple steps of conventional chromatography (Collins et al. 1995). While purified recombinant p80 and p95 bind to telomerase RNA and telomeric repeat substrate DNA in vitro (Gandhi and Collins 1998), they are not important for catalytic activity in vitro or in vivo and at the physiological level are negative regulators of telomere length (Miller and Collins 2000). It has been suggested that p80 and p95 are not associated with active telomerase because the two proteins more effectively coimmunoprecipitate each other than they coimmunoprecipitate TERT, telomerase RNA or telomerase activity from cell extracts (Mason et al. 2001). Such results are difficult to interpret conclusively given that the p80/p95 complex is in substantial molar excess of telomerase RNP (K.L. Witkin and K. Collins, unpubl.). However, in addition, immunopurification of TERT does not obligatorily copurify p80/p95 (Mason et al. 2001). We find that under some conditions—for example, RNP purification using 2′-O-methyl RNA oligonucleotides complementary to the template region—p80 and p95 are recovered with telomerase RNA to a much greater extent than is TERT (K.L. Witkin and K. Collins, unpubl.). The nucleic acid binding properties of p80/p95 may promote an interaction with misfolded telomerase RNP in cell lysate under purification conditions that are less gentle than used here for purification of TAP-tagged RNP. Disruptive conditions may allow the formation of an RNP complex that does not reflect a physiologically functional state of telomerase RNP. We note that even under optimized conditions, whether using crude Tetrahymena extract or extract enriched for telomerase, antibodies against p80 or p95 immunoprecipitated a much smaller fraction of the total enzyme pool than did antibodies against p45 or p65 shown here. This comparison indicates that p80 and p95 are not stable holoenzyme components in the same manner as are p45 and p65. The litmus test of immunodepletion remains to be performed for p20 and p75, if appropriate antibody reagents can be produced. Finally, we note that neither p65 nor p45 has appreciable sequence homology to full-length Est1p or the motifs conserved among EST1 proteins (for review, see Lundblad 2003). The ongoing efforts in Tetrahymena genome sequencing, assembly, and annotation will be helpful in determining whether this ciliate encodes a potential Est1p ortholog.
Materials and methods
Tetrahymena growth and transformation
Cells were grown in 2% proteose peptone, 0.2% yeast extract, 10 μM FeCl3 with 250 μg/mL ampicillin and streptomycin and 1.25 μg/mL Fungizone (amphotericin B). When indicated, cells were starved in 10 mM Tris-HCl (pH 7.5) for 12–24 h. All transformations were performed on starved cells by particle bombardment (Cassidy-Hanley et al. 1997). For transformations into the BTU1 locus of CU522, cells were allowed to recover for 2–5 h at 30°C then were selected in 20–30 μM paclitaxel (Sigma). Cells were passaged to progressively higher paclitaxel concentrations for 2–3 d until cells were growing in 50 μM paclitaxel, after which they were passaged daily in 50 μM paclitaxel for 5 d. Transgenes encoding p45-TAP and p65-TAP were transformed into CU522 strains partially assorted for p45 and p65 gene disruptions. For TRT gene knockout, cells were transformed and assorted as described previously (Miller and Collins 2000). Cells were subjected to maximal selective pressure by stepwise passaging to the maximum tolerated paromomycin concentration, continued passaging at this drug concentration for 16 d (∼100 doublings), then passaging for 8 d(∼50 doublings) in the absence of selection. For TAP45 and TAP65 gene knockdowns, cells were grown for ∼150 doublings in the presence of paromomycin and 100 doublings after release from drug selection.
Transformation constructs
The C-terminal TAP tag was fused to the synthetic TERT gene (Collins and Gandhi 1998) by subcloning of PCR-amplified tag sequence from the vector pRSAB1234 (a gift from Erin O'Shea, University of California, San Francisco). The entire TERT-TAP coding region was cloned into pBICH3 (a gift from Jacek Gaertig, University of Georgia, Athens) with NsiI and BamHI, inserting the sequence between the BTU1 promoter and 3′-flanking genomic sequence. Expression constructs for p65-TAP and p45-TAP were constructed as for TERT-TAP except that the wild-type genomic loci were used as the coding regions. The TRT gene disruption construct, as described (Miller and Collins 2000), consisted of a neomycin-resistance expression cassette designed to recombine into the TRT locus, replacing a segment of the gene. The TAP45 gene disruption construct contains the same neomycin-resistance expression cassette cloned between an ∼520-bp and an ∼580-bp segment of the TAP45 gene, designed to replace the encoded protein from V150 to E215. Similarly, the TAP65 disruption construct contains the neomycin-resistance cassette cloned between an ∼600-bp and an ∼735-bp segment of the TAP65 gene, replacing the region encoding G181 to Q302.
Protein, RNA, DNA, and activity assays
To make whole-cell extracts, starved cells were lysed with 0.2% NP-40 in T2MG buffer (20 mM Tris-HCl at pH 8.0, 1 mM MgCl2, 10% glycerol) in the presence of protease inhibitors and β-mercaptoethanol, and then centrifuged at 130,000 × g for 1 h. Affinity-purified rabbit polyclonal antibody raised against a peptide from the Tetrahymena TERT motif AB-loop region was used for immunoblot detection (Collins and Gandhi 1998). Activity assays were performed by direct primer extension in reaction buffer with 50 mM Tris-acetate (pH 8.0), 10 mM spermidine, 5 mM β-mercaptoethanol, 2 mM MgCl2, 0.4 mM dTTP, 0.3 μM [α-32P]dGTP (800 Ci/mmole, NEN), and 0.4 μM DNA primer (G4T2)3. Samples from affinity purification were washed or diluted into T2MG to make up half of the activity assay reaction volume, such that elution buffer made minimal contribution to the final reaction conditions. Assays were incubated for 1 h at 30°C.
Total RNA was prepared by standard methods (Ausubel et al. 1996). For RNA 3′-end labeling, RNA isolated from the affinity purification EGTA elution was incubated with 0.4 μM cordycepin (5000 Ci/mmole; PerkinElmer Lifesciences) and 120 U of poly(A) polymerase (USB) for 20 min at 30°C, extracted with phenol/chloroform and then resolved by electrophoresis on a 9% polyacrylamide denaturing gel. Small RNA Northern blots were probed with 5′-end-labeled DNA oligonucleotides complementary to the intended RNA target. Genomic DNA isolation was performed as described previously (Ahmed et al. 1998). Southern blots to assess integration events were analyzed with random hexamer-primed probes corresponding to the genomic region immediately 5′ of the integrated transgene. Telomeric Southern blot hybridization was performed as described (Miller and Collins 2000), except that DNA was separated on a 4% acrylamide or 0.7% agarose gel.
Affinity purification
For large-scale purification, 800 mL of extract was used corresponding to ∼200 L of starved cells at 2–4 × 105 cells/mL. Extracts were adjusted to 0.1 M NaCl and incubated in batch with IgG agarose (Sigma) for 1–2 h at room temperature. Resin was washed three times in wash buffer (WB: 20 mM Tris-HCl at pH 8.0, 2 mM MgCl2, 0.2 mM EGTA, 0.1 M NaCl, 0.1% NP-40, 10% glycerol, 1 mM DTT, 0.1 mM PMSF) and twice in TEV protease elution buffer (10 mM Tris-HCl at pH 8.0, 0.1 M NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM DTT). Elution was accomplished by incubation in elution buffer for 1–1.5 h mixing at room temperature with recombinant TEV protease. The elution was adjusted to 3 mM CaCl2 and mixed with 3 volumes of calmodulin binding buffer (CBB: 10 mM Tris-HCl at pH 8.0, 0.1 M NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10 mM β-mercaptoethanol), then incubated with calmodulin affinity resin (Stratagene) for 1 h at 4°C. Resin was washed three times in CBB then eluted with three sequential additions of calmodulin elution buffer (10 mM Tris-HCl at pH 8.0, 0.1 M NaCl, 1 mM magnesium acetate, 1 mM imidazole, 0.1% NP-40, 10 mM EGTA, 10 mM β-mercaptoethanol). For large-scale purifications, samples were precipitated with tricholoacetic acid. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose, stained with Ponceau S, excised, and digested with Lys-C protease. Peptides were separated by HPLC and sequenced by Edman degradation.
For immunodepletion, affinity-purified rabbit polyclonal antibodies raised against full-length p45 and p65 and control rabbit polyclonal antibodies were prebound to Protein A Sepharose (Amersham Biosciences) and then incubated with whole-cell extract adjusted to 50 mM NaCl for 1 h at 4°C for each round of immunodepletion. Antibody resin was subsequently washed in WB as above except for the presence of 50 mM NaCl.
GenBank accession numbers
The accession numbers for the sequences reported here are AY280525 (TAP45) and AY280524 (TAP65).
Acknowledgments
We thank Jacek Gaertig for Tetrahymena strain CU522 and the pBICH3 targeting construct, Sharleen Zhou for peptide sequencing, and the Lemaux lab for generous use of their biolistic transformation system. Thanks to Ramadevi Prathapam for invaluable help growing Tetrahymena, Michael Miller for Tetrahymena advice, and Neal Freedman and members of the Collins lab for helpful discussions on the manuscript. This work was supported by NIH grant GM54198 to K.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1201704.
References
- Ahmed S., Sheng, H., Niu, L., and Henderson, E. 1998. Tetrahymena mutants with short telomeres. Genetics 150: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner S., Lingner, J., Goodrich, K.J., Grosshans, C.A., Shevchenko, A., Mann, M., and Cech, T.R. 2000. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 19: 6230–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner S., Postberg, J., Lipps, H.J., and Cech, T.R. 2003. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42: 5736–5747. [DOI] [PubMed] [Google Scholar]
- Ausubel F., Brent, R., Kingston, R., Moore, D., Seidman, F., Smith, J., and Struhl, K. 1996. Current protocols in molecular biology. John Wiley & Sons, Inc., New York.
- Blackburn E.H. 2000. The end of the (DNA) line. Nat. Struct. Biol. 7: 847–850. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D., Bowen, J., Lee, J.H., Cole, E., VerPlank, L.A., Gaertig, J., Gorovsky, M.A., and Bruns, P.J. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. 1999. Ciliate telomerase biochemistry. Annu. Rev. Biochem. 68: 187–218. [DOI] [PubMed] [Google Scholar]
- Collins K. and Gandhi, L. 1998. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. 95: 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. and Greider, C.W. 1993. Nucleolytic cleavage and non-processive elongation catalyzed by Tetrahymena telomerase. Genes & Dev. 7: 1364–1376. [DOI] [PubMed] [Google Scholar]
- Collins K., Kobayashi, R., and Greider, C.W. 1995. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell 81: 677–686. [DOI] [PubMed] [Google Scholar]
- Dragon F., Pogacic, V., and Filipowicz, W. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20: 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad, V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120. [DOI] [PubMed] [Google Scholar]
- Fang G. and Cech, T.R. 1995. Telomerase RNA localized in the replication band and spherical subnuclear organelles in hypotrichous ciliates. J. Cell Biol. 130: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Shay, J.W., and Wright, W.E. 2001. The La antigen associates with the human telomerase ribonucleoprotein and influences telomere length in vivo. RNA 7: 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Wright, W.E., and Shay, J.W. 2002. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene 21: 580–583. [DOI] [PubMed] [Google Scholar]
- Gaertig J., Thatcher, T.H., Gu, L., and Gorovsky, M.A. 1994. Electroporation-mediated replacement of a positively and negatively selectable β-tubulin gene in Tetrahymena thermophila. Proc. Natl. Acad. Sci. 91: 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., Gao, Y., Tishgarten, T., Clark, T.G., and Dickerson, H.W. 1999. Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nature Biotech. 17: 462–465. [DOI] [PubMed] [Google Scholar]
- Gandhi L. and Collins, K. 1998. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes & Dev. 12: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E.C. and Shippen, D.E. 1998. Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes & Dev. 12: 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn, E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Hardy C.D., Schultz, C.S., and Collins, K. 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276: 4863–4871. [DOI] [PubMed] [Google Scholar]
- Harrington L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194: 139–154. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Evans, S.K., Weilbaecher, R.G., and Lundblad, V. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10: 809–812. [DOI] [PubMed] [Google Scholar]
- Karrer K.M. 2000. Tetrahymena genetics: Two nuclei are better than one. In Tetrahymena thermophila (eds. D.J. Asai and J.D. Forney), pp. 127–186. Academic Press, San Diego.
- Kelleher C., Teixeira, M.T., Forstemann, K., and Lingner, J. 2002. Telomerase: Biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27: 572–579. [DOI] [PubMed] [Google Scholar]
- Kiss T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Larson D.D., Spangler, E.A., and Blackburn, E.H. 1987. Dynamics of telomere length variation in Tetrahymena thermophila. Cell 50: 477–483. [DOI] [PubMed] [Google Scholar]
- Lingner J. and Cech, T.R. 1996. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. 93: 10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Cech, T.R., Hughes, T.R., and Lundblad, V. 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. 94: 11190–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. 2003. Telomere replication: An EST fest. Curr. Biol. 13: R439–R441. [DOI] [PubMed] [Google Scholar]
- Mason D.X., Autexier, C., and Greider, C.W. 2001. Tetrahymena proteins p80 and p95 are not core telomerase components. Proc. Natl. Acad. Sci. 98: 12368–12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.C. and Collins, K. 2000. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol. Cell 6: 827–837. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Cheng, J., and Collins, K. 1999a. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.R., Wood, E., and Collins, K. 1999b. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555. [DOI] [PubMed] [Google Scholar]
- Möllenbeck M., Postberg, J., Paeschke, K., Rossbach, M., Jonsson, F., and Lipps, H.J. 2003. The telomerase-associated protein p43 is involved in anchoring telomerase in the nucleus. J. Cell. Sci. 116: 1757–1761. [DOI] [PubMed] [Google Scholar]
- Pogacic V., Dragon, F., and Filipowicz, W. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20: 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Séraphin, B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Schnapp G., Rodi, H.-P., Rettig, W.J., Schnapp, A., and Damm, K. 1998. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 26: 3311–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Zaug, A.J., Sobel, S.G., Wolin, S.L., and Cech, T.R. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180. [DOI] [PubMed] [Google Scholar]
- Steiner B.R., Hidaka, K., and Futcher, B. 1996. Association of the Est1 protein with telomerase activity in yeast. Proc. Natl. Acad. Sci. 93: 2817–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart A.K., Teng, S.C., and Zakian, V.A. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026. [DOI] [PubMed] [Google Scholar]
- Turkewitz A.P., Orias, E., and Kapler, G. 2002. Functional genomics: The coming of age for Tetrahymena thermophila. Trends Genet. 18: 35–40. [DOI] [PubMed] [Google Scholar]
- Vega L.R., Mateyak, M.K., and Zakian, V.A. 2003. Getting to the end: Telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 4: 948–959. [DOI] [PubMed] [Google Scholar]
- Wang H. and Blackburn, E.H. 1997. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 16: 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich S.L., Pruzan, R., Ma, L., Ouellette, M., Tesmer, V.M., Holt, S.E., Bodnar, A.G., Lichsteiner, S., Kim, N.W., Trager, J.B., et al. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17: 498–502. [DOI] [PubMed] [Google Scholar]
- Wolin S.L. and Cedervall, T. 2002. The La protein. Annu. Rev. Biochem. 71: 375–403. [DOI] [PubMed] [Google Scholar]
- Wong J.M., Kusdra, L., and Collins, K. 2002. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4: 731–736. [DOI] [PubMed] [Google Scholar]
- Yu G., Bradley, J.D., Attardi, L.D., and Blackburn, E.H. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132. [DOI] [PubMed] [Google Scholar]