Abstract
Polyamines are small molecules associated with a wide variety of physiological functions. Bacterial pathogens have developed subtle strategies to exploit polyamines or manipulate polyamine-related processes to optimize fitness within the host. During the transition from its innocuous E. coli ancestor, Shigella, the aetiological agent of bacillary dysentery, has undergone drastic genomic rearrangements affecting the polyamine profile. A pathoadaptation process involving the speG gene and the cad operon has led to spermidine accumulation and loss of cadaverine. While a higher spermidine content promotes the survival of Shigella within infected macrophages, the lack of cadaverine boosts the pathogenic potential of the bacterium in host tissues. Enteroinvasive E. coli (EIEC) display the same pathogenicity process as Shigella, but have a higher infectious dose and a higher metabolic activity. Pathoadaption events affecting the cad locus have occurred also in EIEC, silencing cadaverine production. Since EIEC are commonly regarded as evolutionary intermediates between E. coli and Shigella, we investigated on their polyamine profile in order to better understand which changes have occurred along the path to pathogenicity. By functional and molecular analyses carried out in EIEC strains belonging to different serotypes, we show that speG has been silenced in one strain only, favouring resistance to oxidative stress conditions and survival within macrophages. At the same time, we observe that the content of spermidine and putrescine, a relevant intermediate in the synthesis of spermidine, is higher in all strains as compared to E. coli. This may represent an evolutionary response to the lack of cadaverine. Indeed, restoring cadaverine synthesis decreases the expression of the speC gene, whose product affects putrescine production. In the light of these results, we discuss the possible impact of pathoadaptation events on the evolutionary emergence of a polyamine profile favouring to the pathogenic lifestyle of Shigella and EIEC.
Introduction
Escherichia coli is not only a harmless commensal of the human and animal intestine, but also a major cause of morbidity and mortality [1], [2]. As in many other bacterial pathogens, the evolution of E. coli towards pathogenic phenotypes has been determined mainly by two mechanisms: the acquisition of virulence genes and the loss or modification of genes of the core genome [3]. E. coli acquires virulence determinants by horizontal gene transfer as parts of plasmids, bacteriophages, transposons or pathogencity islands, and this process plays a crucial role in the colonization of a new host environment and in the successful establishment of a pathogenic lifestyle [4], [5]. Genome analyses clearly confirm that also the loss of genes by means of so-called pathoadaptive mutations has strongly contributed to the emergence of pathogenic E. coli strains [6], [7].
On the basis of specific virulence factors and pathogenicity processes, pathogenic E. coli have been subdivided into different pathotypes, including intestinal and extraintestinal strains [1]. Among intestinal pathogenic E. coli, enteroinvasive E. coli (EIEC) are intracellular pathogens causing a severe enteric syndrome in humans, mainly in developing countries [5]. The pathogenesis of EIEC strains is based on their capacity to invade cells of the colonic epithelium, replicate intracellularly and spread to adjacent cells causing inflammatory destruction of the intestinal epithelial barrier. The mechanism is similar to that of Shigella, the causative agent of bacillary dysentery [8] and EIEC strains are therefore included in the same pathotype as Shigella [1]. However, EIEC do not display the full set of characters that define Shigella nor they underwent the extensive gene decay observed in Shigella [9]. Molecular analyses confirm that EIEC strains are widely distributed among E. coli phylogenetic groups and generally correspond to bioserotypes found in a dozen of E. coli serogroups [10], [11], [12]. Many EIEC strains have Shigella-like features: they can be Lac−, non-motile, low-level indole-producing, and (at least in some cases) they carry the typical Shigella O-antigen. On the other hand, in contrast to Shigella, EIEC have a higher infectious dose and a higher metabolic activity, retaining the ability to catabolize substrates widely used by E. coli [10], [13], [14].
It is widely acknowledged that the major event that gave rise to the Shigella/EIEC pathotype has been the acquisition of a large virulence plasmid (pINV) coding for the invasion plasmid antigens (Ipa proteins), for their Type III secretion system (T3SS), and for many other genes involved in invasion, survival and intracellular spread [15], [16], [17], [18]. The expression of pINV genes is affected by multiple environmental stimuli acting through a regulatory cascade which involves nucleoid proteins, specific regulators and sRNAs encoded by both, pINV and the chromosome [19], [20], [21]. A complementary yet significant step towards the pathogenic lifestyle has been the inactivation of several chromosomal genes which negatively interfere with the expression of virulence factors required for survival within the host. This process, commonly referred to as pathoadaptation, has been mainly studied in Shigella spp. [6], [7]. The antivirulence loci identified so far in Shigella encode a broad spectrum of functions, confirming that adaptation to the new host environments is the result of a long and ordered process targeting core genome determinants.
Among pathoadaptive mutations, a paradigmatic case is represented by those affecting the polyamine content of Shigella [22], [23]. Polyamines are small polycationic molecules found in almost all cells and are associated with a wide variety of physiological functions, including translation, gene regulation, stress resistance, cell proliferation, and differentiation [24], [25]. Considerable evidence has recently built up that bacteria have evolved mechanisms to turn polyamines to their own advantage in order to increase their fitness within the host. In particular, major polyamines (cadaverine, spermidine, putrescine) have been shown to optimize virulence processes and intracellular survival rates in several human pathogens [26], [27]. In this respect, Shigella exhibits a peculiarity since polyamines have antagonistic effects on the invasive process: while higher spermidine levels correlate with an increased survival of Shigella during the infection of macrophages [22], the lack of cadaverine production increases the pathogenic potential of the bacterium in host tissues [23]. The higher level of spermidine is determined by the absence of spermidine acetyltransferase (SAT), the enzyme (encoded by the speG gene) which converts spermidine into its inert form, N-acetylspermidine. Cadaverine, whose synthesis depends on the cad gene [28], normally blocks the release of Shigella into the cytoplasm of the infected cells and inhibits the migration of polymorphonuclear leukocytes across the intestinal epithelium [6], [29]. The lack of functional cad and speG genes is the result of a convergent evolution, which banks on inactivating events ranging from point mutations to large deletions [23], [30], [31].
Besides the acquisition of the pINV plasmid, EIEC and Shigella share additional pathoadaptive mutations, e.g. the lack of the OmpT protease [32] and the loss of cadaverine production [33]. Another typical pathoadaptive mutation observed in Shigella is the requirement for exogenous nicotinic acid, due to inactivation of the nad genes [34]. It has been shown recently that, as opposed to Shigella, the inability to synthesize nicotinic acid is not a generalized feature among EIEC strains [35]. This agrees with the view that EIEC may represent an evolutionary intermediate in the transition towards a full-blown phenotype, with some mutational events still confined to Shigella.
In the present study, we have investigated on the polyamine profile in EIEC in order to understand whether, besides cad genes, also other genes involved in polyamine metabolism had been lost during pathoadaptation. Moreover, we asked if and how the loss of cadaverine may have affected the expression of other genes involved in the synthesis of polyamines. The results we present reveal that EIEC have an intermediate polyamine content as compared to E. coli and Shigella and that cadaverine negatively affects the synthesis of putrescine. All together our observations contribute to reconstruct the evolutionary events that have determined the emergence in Shigella of a polyamine profile well suited to an invasive lifestyle.
Materials and Methods
Bacterial strains and plasmids
Bacterial strains are listed in Table 1. E. coli ULS655 carries a deletion of the cadC gene [36]. Strain ULS86, carrying a deletion of the entire cadBA operon, and strains EIEC HN280 DG and 53638 DG, carrying a speG deletion, were constructed using the one-step method of gene inactivation [37] by transforming MG1655 pKD46, HN280 pKD46 and 53638 pKD46 with amplicons obtained using the plasmid pKD13 as template and the oligo pair caf/car (cadBA deletion) or dgf/dgr (speG deletion) (Table S1). Under the experimental conditions used, no differences in growth rate were observed between EIEC strains and their speG derivatives.
Table 1. Bacterial strains used in this study.
| Bacterial strains | Serotype | Relevant Features | Defective gene in the cad locus | Source or reference |
| EIEC | ||||
| HN280 | O135 | Lac−, pINV (260 kb), pCRY (160 kb), LDC−b | cadC | Prosseda et al., 2006 |
| 4608 | O143 | Lac+, pINV (250 kb), LDC- | cadC | WRAIRc |
| 53638 | O144 | Lac−, pINV (250 kb), pCRY (140 kb), LDC− | cadC | WRAIRc |
| 13.80 | O124 | Lac−, pINV (240 kb), LDC− | cadC cadBA | IPCd |
| 6.81 | O115 | Lac−, pINV (250 kb), pCRY (160 kb), LDC− | cadC cadBA | IPCd |
| HN280 DG | O135 | HN280 derivative ΔspeG | cadC | This study |
| 53638 DG | O144 | 53638 derivative ΔspeG | cadC | This study |
| S.flexneri | ||||
| M90T | 5 | pINV (213 kb), pCRY (2.1 and 6.1 kb), LDC− | cadC cadBA | Sansonetti et al., 1982 |
| E. coli K-12 | ||||
| MG1655 | E. coli K12, F− ilvG rfb-50 rph-1; LDC+ | ATCCe | ||
| ULS655 | MG1655 derivative ΔcadC, Kmr, LDC− | cadC | Casalino et al., 2010 | |
| ULS86 | MG1655 derivative ΔcadBA, Kmr, LDC− | cadBA | This study | |
| DH10b | F− recA1 ΔlacX74 galK16 araD139 rpsL | Invitrogen-Life Technologies, Inc |
All the strains are designed by their original laboratory name.
LDC, lysine decarboxylase.
WRAIR, Walter Reed Army Institute of Research.
IPC, Institut Pasteur Collection.
ATCC, American Type Culture Collection.
Plasmid pCC55 is a pACYC184 derivative carrying the cadC gene of MG1655 [33]. pULS37 and pULS13 are pACYC184 derivatives containing the E. coli speG gene under the control of its regulatory region or of the Ptac promoter, respectively [22]. Plasmids containing the ynfB-speG locus of EIEC strains (pG13.80, pG6.81, pG4608, pG53630, and pG280) were constructed by cloning into pGEM-T Easy amplicons obtained using the pgf/ygt oligo pair (Table S1) and the corresponding EIEC genomic DNA as template. DH10b was used as recipient in cloning experiments.
Bacterial cells were routinely grown at 37°C in Luria-Bertani (LB) broth. When required cells were grown in polyamine free M9 complete medium (M9 minimal medium supplemented with 10 µg/ml thiamine, 0.2% glucose, 0.5% casamino acids and 10 µg/ml nicotinic acid). Solid media contained 1.6% agar. When required, antibiotics were included at the following concentrations: ampicillin (Ap), 100 µg/ml; chloramphenicol (Cm) 30 µg/ml; kanamycin (Km) 30 µg/ml, streptomycin (Sm) 50 µg/ml; tetracycline, (Tc) 5 µg/ml.
General molecular procedures
DNA purification, transformation, restriction, electrophoresis, amplification and purification of DNA fragments were carried out as described previously [22], [38]. Pfu Taq DNA polymerase was used to obtain longer transcripts and high fidelity. All oligonucleotides used in this study are listed in Table S1 and have been designed mainly on the basis of the genomic sequence of the Escherichia coli K-12 MG1655 [39].
Real time PCR
Total RNA purification and cDNA synthesis was performed as previously described [40]. Real time PCR was performed using a 30 µl reaction mix containing 2 µl cDNA. At least three wells were run for each sample. The amount of speG and speC transcripts was analysed using the 2−ΔΔCt method [41] and the results were indicated as n-fold increase relative to the reference sample. The ΔCt-values in the Student's t test have been considered to determine whether datasets of relative gene expression were significantly different from those in a chosen calibrator. Primers for the nusA transcript, used as endogenous control, and for the above-mentioned transcripts were experimentally validated for suitability to the 2−ΔΔCt method. The following oligos (Table S1) were used: rgf/rgr for speG, scf/scr for speC, and nusAF/nusAR for nusA.
Primer extension
Total RNA from EIEC strains HN280, 13.80 and 4608 grown to OD600 0.6 was extracted by a modified hot-phenol method and quantified spectrophotometrically as described [42]. The primer (peg) was 5′-end labelled with [γ-32P]dATP using T4 polynucleotide kinase and hybridized with 50 µg total RNA as previously described [43]. Reverse transcription experiments were carried out and the resulting cDNAs were run on denaturing 6% polyacrylamide gels, along with a sequencing ladder that was generated by using the same primer and HN280 and 4608 DNAs as templates. Sequencing reactions were performed with [α-32P]dATP.
Polyamine quantification
Bacteria were grown in M9 complete medium to OD600 0.7-0.8, centrifuged and resuspended in PBS. Cells were disrupted by sonication and polyamines were extracted from the lysate with 3% percloric acid containing 5 mM 1,6-diaminehexane as a polyamine internal standard. After derivatization with dansylchloride, the simultaneous fluorimetric determination of intracellular polyamines was performed by reverse-phase high-performance liquid chromatography and polyamines were quantified as described previously [44]. The polyamine concentration in total cellular homogenates was normalized to cell number and expressed as nmol/108 cells.
Bacterial sensitivity to oxidative stress
The susceptibility to oxidative stress was evaluated by analysing the survival of EIEC HN280 and 53638 strains and their derivatives after exposure to H2O2 in liquid cultures. To this end, 15 ml of bacterial cultures grown in M9 complete medium to OD600 0.7 were centrifuged and the pellets were suspended in 1 ml PBS. An equal volume of PBS containing 20 mM H2O2 was added and the mix was incubated at 37°C for 1 h. The reaction was stopped by adding catalase to 0.1 mg/ml. The number of bacteria surviving oxidative stress was quantified by plating aliquots on LB agar. Survival percentages were calculated by comparison with the corresponding untreated strains.
Ornithine decarboxylase activity assays
Monitoring of ornithine decarboxylase (L-ornithine decarboxylase, ODC) activity was performed according to the Ngo method [45]. Bacterial cultures were grown overnight, diluted in fresh M9 complete medium and allowed to grow to OD600 0.7–0.8. 10 ml for each culture were centrifuged and pellets suspended in 1 ml PBS. 200 U of DNase and 10 µg/ml of RNase A were added to the samples and, after 30 minutes on ice, sonicated. Lysates were centrifuged at 14000 rpm for 15 minutes and supernatants were filtered (0.22 µm) and then dialyzed against 150 mM phosphate buffer. 100 µl aliquots were incubated at 37°C for 30 minutes in 400 µl reaction mixture containing 3 mM L-ornithine, 75 nM pyridoxal phosphate (PLP), 1.5 mM EDTA and 2.5 mM β-mercaptoethanol. Reactions were stopped by adding 20 µl 70% perchloric acid. Samples were centrifuged at 14000 rpm for 15 minutes and 200 µl of the supernatant were derivatized with dansyl chloride [46]. 10 µl of each sample were used to perform the TLC assay on Silica gel. The TLC separation was performed using chloroform:triethylamine (25∶2) as solvent. Finally, bands corresponding to putrescine were scraped from the plate and, after ethyl acetate extraction, fluorescence was measured on supernatants using a multilabel counter. Protein concentrations were determined by the Bradford method.
Cultures of macrophages and bacterial infection
The murine macrophage-like J774 cells (ATCC, Manassas, VA) were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine and penicillin-streptomycin at 37°C in a humidified 5% CO2 atmosphere. For bacterial infection, 4×105 cells per well were seeded in 12-well tissue culture plates and grown overnight. Before infection, host cells were cultured with serum-free, antibiotic-free medium for 1 hour. In order to produce a competitive infection, J774 cells were simultaneously infected with HN280 pACYC184 and HN280 pULS13 at a multiplicity of infection of 100 and centrifuged 10 min at 700 g. After a 30 min incubation at 37°C (time 0) cells were extensively washed with PBS and fresh cell culture medium containing 10 µg/ml of gentamicin was added to kill extracellular bacteria. The cell/bacteria mixture was further incubated at 37°C for the indicated time. To determine the number of intracellular bacteria, the cells were washed once with PBS and lysed by adding 0.5 ml of 1% Triton X-100 in PBS to each well for 5 min. Samples were mixed, diluted and plated onto LB agar to determine the number of CFU recovered from the lysate. The number of intracellular bacteria at different time points was compared to bacteria recovered at time zero. To calculate the competitive index (C.I.), the ratios of strains HN280 pULS13/HN280 pACYC184 recovered from the infected cultures were determined and normalized by dividing by the corresponding ratio in the initial inoculum.
Nucleotide sequence accession numbers
DNA from plasmids pG13.80, pG6.81, pG4608, pG53630, and pG280 was used as template for sequencing the ynfB-speG locus. The sequence data were compared to known nucleotide and protein sequences using the BLAST server (National Center of Biotechnology Information, Bethesda, Md.). The ynfB-speG sequences were deposited at GeneBank under accession numbers KJ825879; KJ825880; KJ825881; KJ825882; and KJ825883.
Results
The polyamine content of EIEC strains
Enteroinvasive Escherichia coli (EIEC) do not represent a homogeneous group of pathogenic E. coli. Indeed, EIEC strains differ widely with respect to biochemical features, serotype, and plasmid content. Molecular phylogenetic analyses show that during evolution EIEC have derived several times independently from E. coli [10], [12]. In analogy to Shigella, all EIEC have lost the ability to synthetize cadaverine [33]. In a previous study [22] we have shown that Shigella has also lost speG, the gene encoding spermidine acetylase (SAT, an enzyme catalyzing the synthesis of N-acetylspermidine from spermidine), and that the lack of SAT induces a beneficial accumulation of intracellular spermidine favouring bacterial survival under oxidative stress conditions.
Since EIEC and Shigella display essentially the same pathogenicity mechanism and belong to the same pathotype, we asked whether they also share the same polyamine profile. To this end, we analysed a set of five invasive EIEC strains (HN280, 13.80, 4608, 6.81, and 53638) with different serotypes and different geographic origin (Table 1). To ascertain if and to what extent the polyamine content of these strains had been modified as compared to E. coli, their commensal ancestor, we assayed, by means of HPLC, the intracellular level of the major polyamines (spermidine, putrescine, cadaverine, spermine, N-acetylspermidine and N-acetylspermine) when cells were grown in a polyamine-free medium. The polyamine profiles obtained have been compared also with those of S. flexneri M90T (Table 1), which has lost cadaverine and N-acetylspermidine during the evolutionary transition from non-pathogenic E. coli [22], and of E. coli K-12 MG1655.
As reported in Table 2, N-acetylspermidine, the inert form of spermidine obtained by spermidine acetylation (Fig. 1), is still present in all EIEC strains. Strain 13.80 shows a level of N-acetylspermidine comparable to E. coli K-12 MG1655. In strains 53638, 6.81, and 4608 N-acetylspermidine is 3.0- to 3.3-fold higher than in E. coli K-12 MG1655, while strain HN280 exhibits a 1.8-fold reduction. Statistical analysis shows that these differences are significant (Table 2). As far as intracellular spermidine is concerned, all EIEC strains attain a higher level as compared to E. coli. In particular, the increase in strains 13.80, HN280 and 53638 (1.2- to 2.8-fold) is significant (p<0.05), and the higher levels found in strains 4608 and 6.81 are slightly below the significance threshold (p = 0.051 and 0.058 respectively). When referencing the spermidine levels of EIEC to Shigella, a significant reduction shows up in all EIEC (1.9- to 2.3-fold; p<0.05) except in strain 53638, where the level remains comparable. It is worth stressing that Shigella spp. has faced a complete loss of the speG gene product [22]. Interestingly, also the level of putrescine is significantly increased (2.4- to 4.5-fold, p<0.01) in EIEC as compared to E. coli while it is comparable to that observed in Shigella. In agreement with previous findings [23], [33], we observe that cadaverine is absent from EIEC and Shigella spp. Finally, endogenous spermine and acetylspermine are not found in EIEC and Shigella (data not shown), which is not surprising since it has been reported also in E. coli [24], commonly considered as the commensal ancestor of both Shigella and EIEC [16].
Table 2. Analysis of polyamine content in different EIEC and S. flexneri strains.
| Strain | NSPD | PUT | CAD | SPD | |
| E. coli K-12 | |||||
| MG1655 | 5.06±0.41 | 4.59±0.38 | 11.2±0.69 | 3.14±0.35 | |
| EIEC | |||||
| 13.80 | 5.43±0.43 | 12.23±1.03** | nd | 3.89±0.17* | |
| HN280 | 2.80±0.27* | 11.04±1.00** | nd | 4.68±0.22* | |
| 4608 | 16.64±1.22** | 15.20±1.12** | nd | 3.90±0.12 | |
| 6.81 | 15.88±1.34** | 14.98±1. 16** | nd | 3.85±0. 13 | |
| 53638 | 15.23±1.43** | 20.88±1.99** | nd | 8.90±0.74** | |
| S. flexneri | |||||
| M90T | nd | 12.33±2.10** | nd | 8.96±0. 68** |
Values reported are in nmol per 108 cells and represent the average ± standard deviations. E. coli K-12 MG1655 has been used as reference. N-SPD: Acetyl spermidine; PUT: Putrescine; CAD: Cadaverine; SPD: spermidine. Student's t tests were performed comparing PUT, SPD, NSPD and CAD concentration in EIEC and Shigella strains with the polyamine concentration detected in E. coli K12 MG1655.
*denotes 0.05>p≥0.01;
**denotes p<0.01.
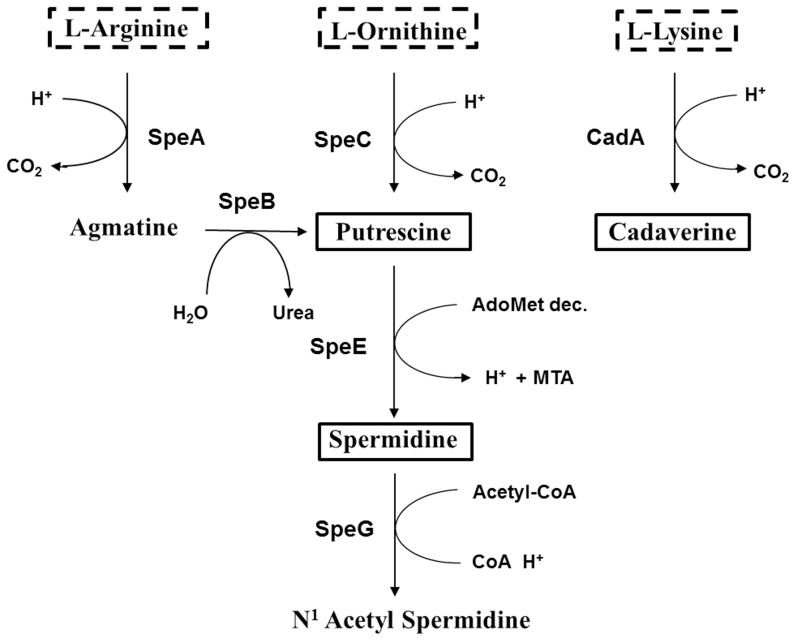
Figure 1. Major steps of the polyamine metabolism in E. coli.
The diagram depicts the pathway of polyamine biosynthesis in E. coli. Data are drawn from Ecocyc database [39]. Dotted boxes: precursor aminoacids. AdoMet dec: S-Adenosyl-L-Methioninamine. MTA: S-Methyl-5′-Thioadenosine.
Altogether, our observations indicate that in EIEC, the levels of intracellular putrescine is increased and the levels of spermidine tend to be higher as compared to E. coli K-12 MG1655. Moreover, N-acetylspermidine is still present in all strains, in contrast with Shigella. This suggests that the polyamine content of EIEC strains has settled at a level intermediate between Shigella and E. coli K-12 and that the lack of N-acetylspermidine, with the consequent spermidine accumulation, is a trait limited to Shigella spp.
Genetic and functional analysis of the speG gene in EIEC
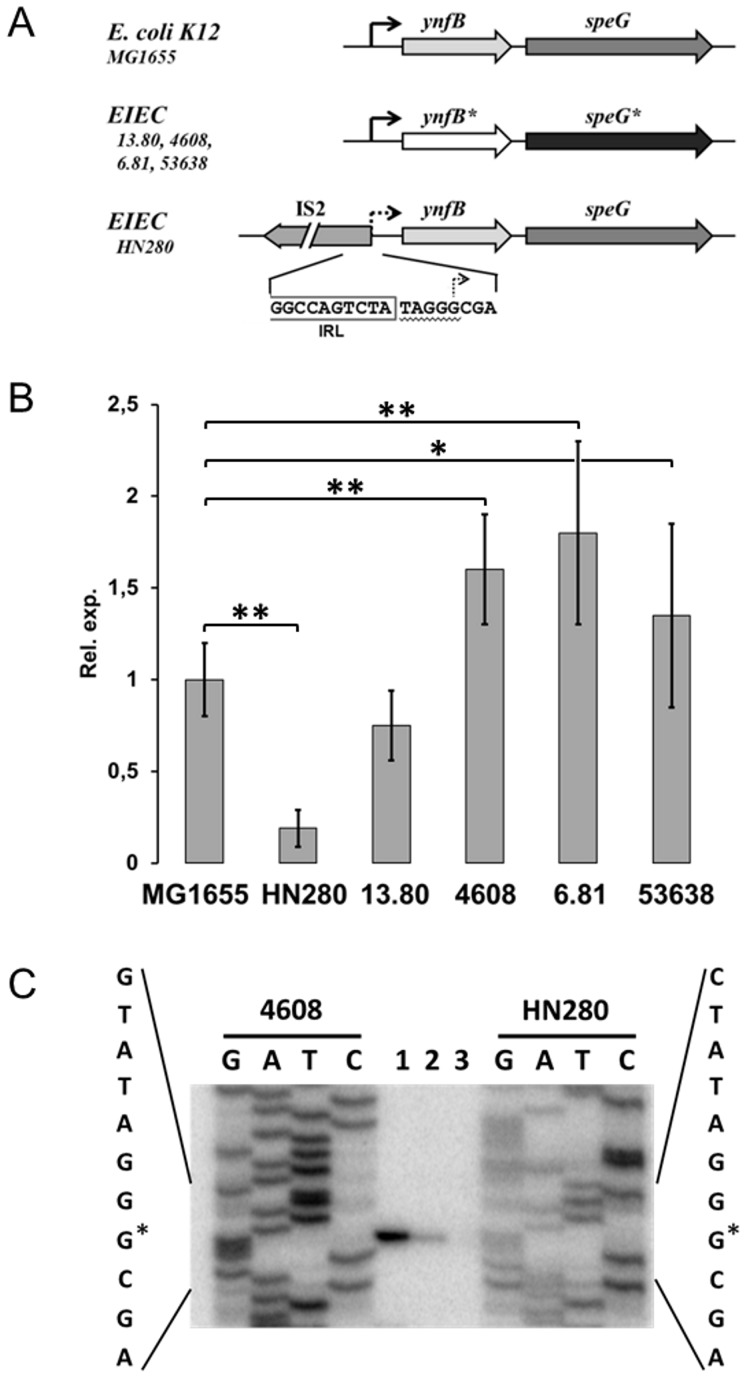
To understand whether the different levels of N-acetylspermidine observed in our EIEC strains (Table 2) depend on mutations or rearrangements in the speG gene promoter, we cloned the corresponding operon (ynfB-speG) into pGEM-T Easy, obtaining plasmids pG280, pG13.80, pG4608 pG6.81 and pG53638. PCR amplification, performed using a primer pair (pgf/ygt) flanking the entire operon, shows that only HN280 gives rise to a product larger than the amplicon of the control strain (MG1655), suggesting the presence of an IS element. Sequence analysis confirms that HN280, which exhibits a reduced level of N-acetylspermidine (Table 2), harbours an IS2 element inserted within the promoter at position -4 from the transcription start site (+1) (Fig. 2A). Moreover, sequence data (Table S2) reveal that EIEC strains other than HN280 carry a TA transversion within the ynfB-speG promoter (position -28), as well as an AC transversion upstream (position -111) the transcription start site. Since these transversions are found in all strains having an N-acetylspermidine level higher than, or comparable to, the E. coli K-12 MG1655 control strain, it is possible to speculate that these point mutations are not responsible for the different expression of speG.
Figure 2. Molecular and functional analysis of the ynfB-speG operon in EIEC.
(A) Genetic organization of the ynfB-speG operon in EIEC strains. The operon at the top is based on the E. coli K-12 MG1655 sequence as reported in (National Center of Biotechnology Information, Bethesda, Md.). The right-pointing arrow marks the transcription start site (+1). The asterisk (*) indicates genes carrying non-synonymous mutations. The nucleotide sequence at the bottom refers to the IS2 insertion into the ynfB-speG promoter of EIEC HN280 (IRL: left-end inverted repeat of IS2. Underlined: direct repeat at the IS2 insertion site). (B) Relative speG transcription in EIEC strains as monitored by in vivo Real-Time PCR using MG1655 as a control. Strains were grown at 37°C in M9 complete medium. At least three wells were run for each sample and the error bars display the calculated maximum (RQMax) and minimum (RQMin) expression levels that represent standard error of the mean expression level (RQ value). * denotes 0.05>p≥0.01; ** denotes p<0.01. (C) Primer extension analysis of the transcripts generated under the control of the ynfB-speG promoter of EIEC strains HN280, 13.80, and 4608. The autoradiograph is the result of a typical experiment performed with the peg primer on RNA extracted from strain 4608 (lane 1), 13.80 (lane 2), and HN280 (lane 3). Lanes G, A, T, and C show the sequencing ladder generated with the same primer. The asterisk marks the transcription start site (+1).
Also in the case of the ynfB gene, located upstream speG and encoding a protein of unknown function, a non-synonymous mutation, responsible for a I14F substitution, is present in all strains. Two additional mutations, determining a T25A and a M49I substitution in the YnfB protein, are present only in EIEC 13.80 (Table S2). Except for the presence of the IS2 element, the HN280 ynfB-speG sequence perfectly matches that of E. coli K-12 MG1655. Altogether, these data suggest that in EIEC the ynfB-speG operon did not undergo the severe rearrangements/deletions seen in Shigella [22]. However, the presence of an IS2 element in the HN280 ynfB-speG promoter could represent a symptom of nascent pathoadaptive evolution of the operon.
In order to correlate the amount of N-acetylspermidine observed in our EIEC strains (Table 2) with the expression of the speG gene, we monitored the transcription of speG in its native background by means of Real Time PCR. As compared to the control, speG transcription is strongly activated in EIEC strains 4608, 6.81, and 53638. While the speG transcript of strain 13.80 attains a level comparable to that of the control, speG transcription in HN280 is severely (5-fold) reduced (Fig. 2B). A possible explanation for this reduction is that the presence of the IS2 element very close to the transcription start site hampers the transcription of the ynfB-speG operon. In order to verify this hypothesis, we analysed, by means of primer extension experiments, ynfB-speG transcription in strains HN280, 13.80 and 4608, chosen as representative of the diverse levels of speG expression. The results (Fig. 2C) fully agree with the Real Time PCR analysis (Fig. 2B) and confirm that the transcription start site of the EIEC 4608 and 13.80 ynfB-speG operon corresponds to the one previously predicted in E. coli K-12 [39]. No signal is detected for strain HN280 (Fig. 2C, lane 3), indicating that the IS2 insertion destroys the promoter and suggesting that the residual expression observed in Real Time PCR assays (Fig. 2B) may be due to read-through from unspecific upstream sites.
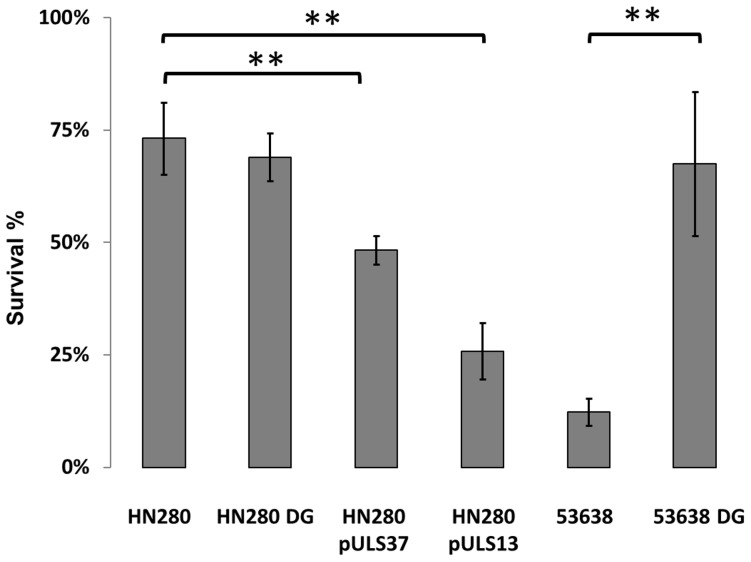
To understand whether the IS2-mediated silencing of speG confers on EIEC HN280 a selective advantage in response to oxidative stress, we compared the survival of HN280 (wt), the derivatives carrying a plasmid with speG under the control of its native promoter (pULS37) or of the Ptac promoter (pULS13) and, as a control, the HN280 derivative carrying the deletion of the entire speG gene (HN280DG). While HN280 and HN280DG respond in a similar manner to the presence of H2O2, restoration of SpeG activity in HN280 reduces resistance to oxidative stress, and the reduction is directly correlated to speG expression since it is further enhanced when speG is under the control of a strong promoter (pULS13) (Fig. 3). Additional evidence about the capacity of SpeG to negatively affect resistance to H2O2 was obtained by studying an EIEC strain (53638) which harbours a functional speG gene. The data in figure 3 evidence that under oxidative stress a 53638 speG defective derivative (53638 DG) has a higher survival rate as compared to the parental strain.
Figure 3. Effect of speG on the sensitivity to oxidative stress.
The sensitivity to H2O2 was assayed on EIEC strain HN280 (wild type), on its speG-deleted derivative HN280DG, on HN280 complemented with a plasmid carrying speG under the control of its own promoter (pULS37) or of the Ptac promoter (pULS13), on strain 53638 (wild type), and on its speG defective derivative 53638 DG. Survival in the presence of H2O2 was measured after exposure of strains, grown in M9 complete medium, to H2O2 for 1 h at 37°C. The survival percentage is relative to growth without exposure to H2O2. Error bars indicate the standard deviations relative to at least three independent experiments. ** denotes p<0.05.
Finally, to verify whether the lack of the speG gene confers a selective advantage during infection in EIEC, we performed an in vitro competitive assay analyzing the survival within macrophages of strain HN280 transformed with a plasmid harbouring/lacking a functional copy of speG. To this end, HN280 pULS13 and HN280 pACYC184 were grown to OD600 0.6–0.7, mixed and used to infect a murine macrophage cell line (J774). Bacterial survival was monitored during two hours after infection by lysing the macrophages and plating appropriate dilutions on LB plates. The resulting competitive index (ratio of survival rates HN280 pULS13/HN280 pACYC184) corresponds to 0.52 at 1 h and 0.41 at 2 h, indicating that the absence of speG enhances survival in macrophages, in agreement with our previous findings in Shigella [22]. All together these observations confirm that also in EIEC the absence of SpeG activity confers an increased capability on the bacterium to defy antagonistic host environments.
Interplay between spermidine and cadaverine production
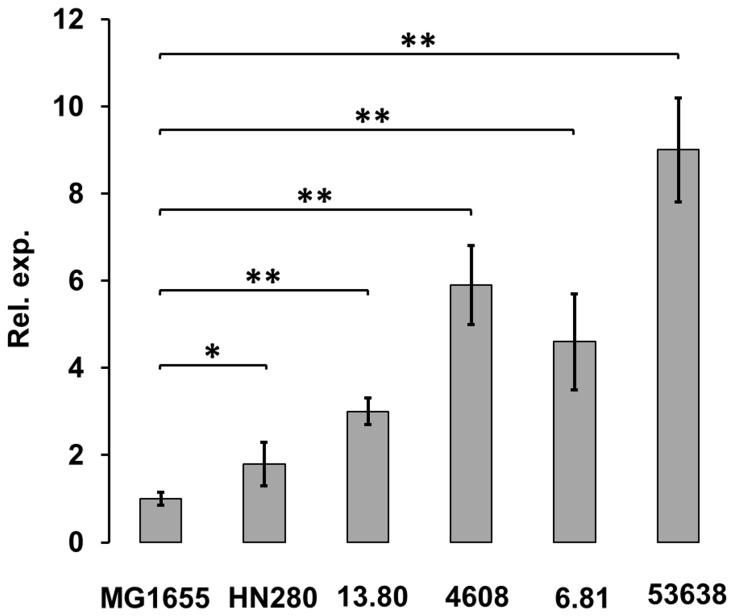
Putrescine is found to be approximately 2 to 4-fold higher in EIEC than in E. coli K-12 MG1655 (Table 2). Putrescine production (Fig. 1) results from direct ornithine decarboxylation, mediated by the SpeC decarboxylase, and from arginine decarboxylation followed by agmantine ureohydrolyzation determined by the SpeA and SpeB proteins, respectively [24]. To verify whether the increased level of putrescine in EIEC might depend on increased transcription of speA, speB or speC, we monitored mRNA levels in their native backgrounds by Real Time PCR. While no difference is observed in speA and speB transcription between EIEC and MG1655 strains (data not shown), speC transcription is always higher, the increase spanning from 2- to 9-fold. (Fig. 4).
Figure 4. Expression of speC in EIEC strains.
The in vivo speC transcription was monitored by Real-Time PCR and referred to the level observed in the MG1655 control. Strains were grown at 37°C in M9 complete medium. At least three wells were run for each sample and the error bars display the calculated maximum (RQMax) and minimum (RQMin) levels that represent standard error of the mean expression level (RQ value); * denotes 0.05>p≥0.01; ** denotes p<0.01.
To further clarify this issue, we considered that, despite their genetic and biochemical heterogeneity, all EIEC strains lack lysine decarboxylase and are therefore unable to produce cadaverine [33]. In E. coli, the synthesis of cadaverine depends mainly on the gene products of the cad locus, which includes three genes, cadA, cadB and cadC [28]. The cadBA operon, encoding lysine decarboxylase (CadA) and a lysine cadaverine antiporter (CadB), is submitted to the control of CadC, an integral inner membrane protein which acts both, as signal sensor and as a positive transcriptional regulator. Besides positively regulating the cadBA operon, CadC has been shown to negatively interfere with the synthesis of the argine-dependent acid-resistance system [36]. A previous analysis [33] of the major event triggering the loss of cadaverine synthesis in the five EIEC strains used in this study has highlighted that in strains HN280, 4608 and 53638 the cadBA operon is integer and that its silencing depends exclusively on the absence of a functional CadC. On the contrary, more severe rearrangements have occurred in EIEC 13.80, where the entire cad locus has been lost, and in EIEC 6.81, where the cadC gene is inactivated by an IS1 insertion and the cadBA operon is affected by a frameshift mutation in the cadB gene [33] (Table 1).
Taking advantage of these cad-silencing mutations, we verified whether the expression of the speC gene is affected by cadaverine or by the presence/absence of CadC. To this end, we introduced a functional cadC gene (plasmid pCC55) into the five EIEC strains studied and compared the polyamine levels of strains containing or lacking a functional cadBA operon. The results (Table 3) indicate that in strains HN280, 4608, 53638 (where the ectopic cadC gene is able to restore cadaverine production because of the presence of an integer cadBA operon) the amount of intracellular putrescine is severely reduced (about 3-fold). A similar reduction in the putrescine level is observed also in the E. coli K-12 control strain ULS655 (MG1655ΔcadC) when cadaverine synthesis is restored by the introduction of pCC55. On the contrary, in strains 13.80 and 6.81, which do not contain a functional cadBA operon, the level of putrescine is only slightly affected by the presence of CadC. These findings rule out that the CadC regulator is involved in the control of the speC gene and strongly suggest that the absence of cadaverine in EIEC has led to increased synthesis of putrescine.
Table 3. Analysis of the polyamines levels in EIEC strains complemented or not with a functional copy cadC gene.
| Strain | cadC | NSPD | PUT | CAD | SPD | |
| ULS655 | - | 5.47±0.30 | 6.75±0.33 | 0 | 2.92±0.23 | |
| ULS655 | + | 4.38±0.56 | 3.40±0.17** | 21.15±1.02** | 3.12±0.30 | |
| 13.80 | − | 5.21±0.26 | 11.46±0.55 | 0 | 3.03±0.19 | |
| 13.80 | + | 7.08±0.33* | 13.14±0.66* | 0 | 2.96±0.15 | |
| HN280 | − | 3.18±0.12 | 13.64±0.68 | 0 | 4.27±0.15 | |
| HN280 | + | 4.88±0.24* | 4.60±0.21** | 15.15±0.75** | 3.99±0.15 | |
| 4608 | − | 16.82±0.84 | 14.80±0.74 | 0.00 | 3.22±0.13 | |
| 4608 | + | 15.24±0.76 | 3.85±0.18** | 19.27±0.96** | 2.04±0.09** | |
| 6.81 | − | 16.73±0.84 | 15.71±0.78 | 0 | 3.96±0.14 | |
| 6.81 | + | 18.90±0.93* | 18.87±0.94* | 0 | 4.03±0.20 | |
| 53638 | − | 14.19±0.71 | 21.92±1.01 | 0 | 9.08±0.45 | |
| 53638 | + | 20.28±0.99* | 7.82±0.39** | 21.91±1.05** | 5.29±0.33** | |
Values reported are in nmol per 108 cells and represent the average ± standard deviations. E. coli ULS655 (MG1655 cadC) has been used as reference. N-SPD: Acetyl spermidine; PUT: Putrescine; CAD: Cadaverine; SPD: spermidine. cadC+: strain carrying the pCC55 plasmid; cadC -: strain carrying the pACYC184 vector. Student's t tests were performed comparing PUT, SPD, NSPD and CAD concentrations in CadC complemented EIEC strains with the polyamine concentration detected in the corresponding wt (cadC −) strains.
*denotes 0.05>p≥0.01;
**denotes p<0.01.
Cadaverine negatively interferes with SpeC expression
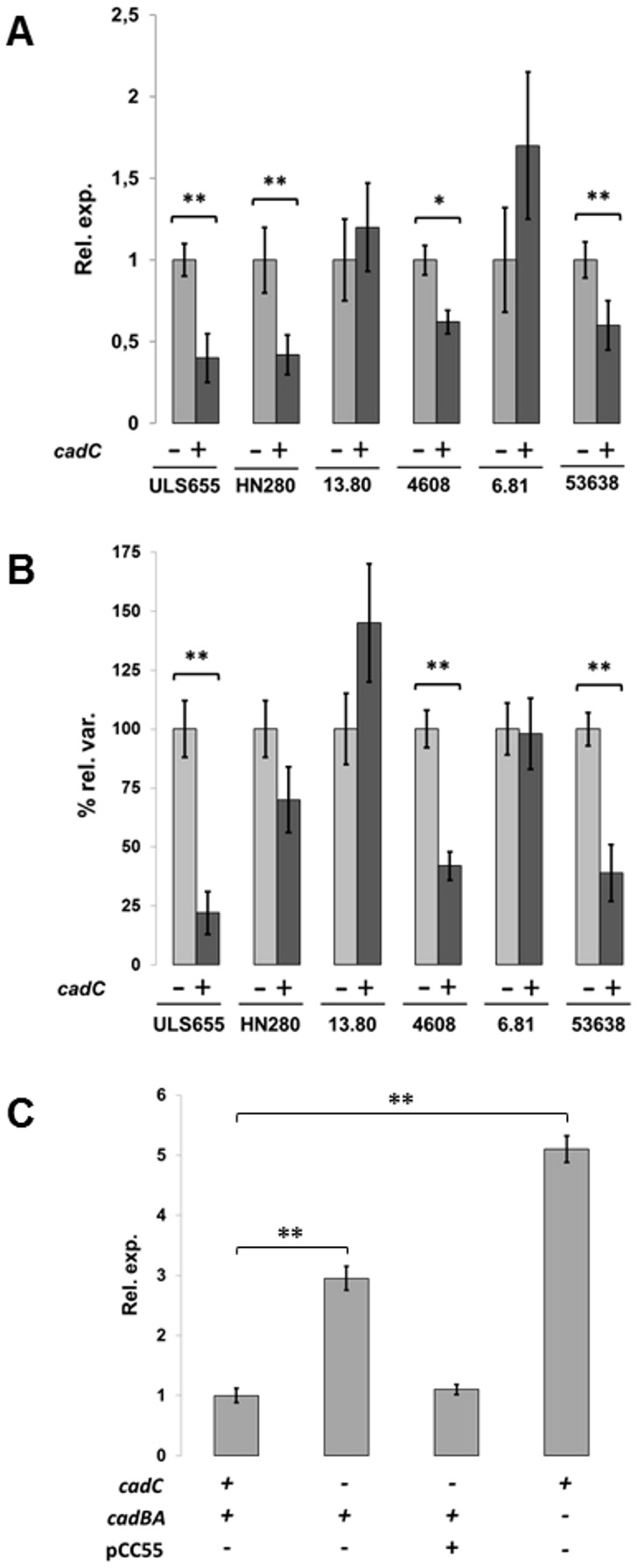
To find out how strictly the expression of SpeC depends on cadaverine, we analysed speC transcription in EIEC strains and in the ULS655 control strain complemented with a functional cadC gene (pCC55). Comparative analysis performed by Real Time PCR (Fig. 5A) indicates that speC transcription is strongly reduced in ULS655 and in EIEC HN280, 4608 and 53638 when cadaverine synthesis is restored by the introduction of a functional cadC gene [33]. On the contrary, the presence of CadC does not affect significantly speC expression in EIEC strains 13.80 and 6.81, which lack an integer cadBA operon. To ascertain whether the observed transcriptional regulation is primarily responsible for SpeC expression and putrescine production, we estimated relative ornithine decarboxylase activity in total cell extracts from EIEC strains HN280, 4608, 53638, 13.80, and 6.81. As shown in Fig. 5B ornithine decarboxylase activity is decreased only in strains which produce cadaverine (HN280, 4608 and 53638) after cadC-complementation. This nicely matches with the reduced transcriptional regulation of the speC gene in the same strains, shown in Fig. 5A. Furthermore, these data are consistent with the polyamine abundance reported in Table 3 and strongly stress the key role played by cadaverine in modulating the level of putrescine.
Figure 5. Cadaverine interferes with speC expression.
Experiments were performed using strains lacking only the cadC gene (EIEC HN280, 4608 and 53638 and E. coli K-12 ULS655) or the entire cad operon (EIEC 13.80 and 6.81). Strains were transformed with pCC55 (a pACYC184 derivative containing a functional cadC gene) or pACYC184. (A) Transcription of speC in the presence or absence of cadaverine as monitored by Real-Time PCR. At least three wells were run for each sample and the error bars display the calculated maximum (RQMax) and minimum (RQMin) levels that represent standard error of the mean expression level (RQ value). * denotes 0.05>p≥0.01; ** denotes p<0.01 (B) Ornithine decarboxylase (ODC) activity in the presence or absence of cadaverine. ODC activity was measured in total cell extracts by assaying putrescine production. The synthesized putrescine was made fluorescent through chemical modification and separated by TLC. The fluorimetric data were normalized against the total protein content of each sample. Results are shown as variation (percentage) in strains carrying pCC55 (cadC) vs the corresponding wt strain carrying the backbone construct. * denotes 0.05>p≥0.01; ** denotes p<0.01. (C) Transcription of speC in E. coli K-12 (MG1655); ** denotes p<0.01. Transcription in strains producing or lacking cadaverine was monitored by Real-Time PCR as in panel A.
In order to check whether the inhibitory effect of cadaverine on speC expression also applies to polyamine metabolism in E. coli, and to rule out incidental contributions from undefined factors possibly present in the EIEC background, we studied the role of cadaverine in an E. coli K-12 background. Exploiting the one-step gene inactivation method [37], we first constructed an E. coli MG1655 derivative defective in the cadBA operon (ULS86, Table 1). Then, we introduced the pCC55 plasmid into ULS655 (cadC) or in ULS86 (cadBA). The expression of the speC gene was monitored by Real Time PCR. The results (Fig. 5C) show that in a cadC defective background (ULS655) the level of speC transcription is 3-fold higher as compared to wt (MG1655) and to cadC complemented ULS655 strain, and that in a cadBA defective background (ULS86 pCC55) the increase is even higher (5-fold). These data strongly support the hypothesis that the higher level of putrescine observed in EIEC strains is a consequence of the evolutionary loss of the cad operon.
Discussion
Polyamines are small polycationic molecules found in eukaryotic and prokaryotic cells, associated with a wide variety of biological functions. Putrescine, cadaverine, spermidine and spermine are the major polyamines in bacteria [24]. Their intracellular content is regulated by the concerted action of biosynthesis and uptake processes, as well as degradation and efflux mechanisms. Synthesis usually depends on the decarboxylation of precursor aminoacids (or other intermediates) which are then converted into functional polyamines. Trafficking relies mainly on uptake and exchange processes mediated by specific ABC transporters and antiporters [25]. In recent years, it has become increasingly evident that, in addition to core physiological functions, polyamines play an active role in bacterial virulence [22], [23], [47], [48], [49]. Several strategies have been developed by bacterial pathogens to exploit polyamines or manipulate polyamine-related processes to optimize bacterial fitness within the host. Some bacterial pathogens are able to utilize polyamines to favour their survival within the host or to alter the immune response of the host. In other cases, polyamines are crucial to promote the expression of virulence factors, as T3SS, or to activate biofilm formation [26], [27].
The essential role played by polyamines in bacterial virulence is nicely exemplified by Shigella, an intracellular pathogen which belongs to the E. coli species and causes a severe enteric syndrome in humans [22]. Shigella evolved from its innocuous ancestor, E. coli, through several steps, which include a gain of functions facilitating the intracellular survival and loss of functions hampering the full expression of an invasive phenotype [7], [11], [16]. While the acquisition of the large virulence plasmid (pINV) by the Shigella/EIEC pathotype has induced, in a single step, the capacity to enter and multiply inside the highly specialized intracellular environment of the human intestinal mucosa [5], [8], [17], the loss of antivirulence functions (pathoadaptive mutations) has acted progressively to increase the pathogenic potential of these strains [6], [7]. Among pathoadaptive mutations, a paradigmatic case is represented by the inactivation of genes involved in the biosynthesis of polyamines. In particular, as compared to the commensal E. coli, Shigella has completely lost cadaverine [23], [30], [31] and N-acetylspermidine, (the inert form of spermidine), and displays a marked accumulation of spermidine [22].
The silencing of genes involved in the biosynthesis of cadaverine is a key factor in the optimization of the pathogenicity process of Shigella, since secreted cadaverine blocks the release of the bacterium into the cytoplasm of infected cells by stabilizing the endosomal membrane and negatively affects Shigella-induced proinflammatory events by inhibiting PMN migration to the infection site [6]. The increased spermidine content of Shigella depends on the lack of a functional speG gene, i.e. on the absence of spermidine acetyltransferase (SAT), the enzyme which converts spermidine into N-acetylspermidine [50]. A distinct advantage results: higher spermidine levels have been shown [22] to increase survival within macrophages during the initial step of the infection process. Enteroinvasive E. coli (EIEC) share with Shigella the same infective process and, as for genetic and phenotypic features, are considered evolutionary intermediates between the harmless E. coli and the harmful Shigella [10], [12]. Similarly to Shigella, also EIEC have acquired the pINV virulence plasmid and have undergone pathoadaptation starting from their ancestor [5]. While the lack of cadaverine has been extensively analysed in EIEC [33], [36], so far no data concerning the presence of the other polyamines were available.
The results we obtained in this study indicate that the polyamine content of EIEC is intermediate between E. coli and Shigella. Indeed, intracellular putrescine is significantly increased in EIEC while spermidine tends to be higher as compared to E. coli K-12. However, N-acetylspermidine is still present in most strains we have analysed, indicating that the loss of speG as pathoadaptive mutation is an emerging, albeit not fully acquired, trait of EIEC. In particular, in four out of five EIEC strains we have analysed, the speG gene is expressed at a level comparable or higher than in the E. coli K-12 control, whereas only one EIEC strain (HN280) displays a severe reduction of N-acetylspermidine (Table 2). We have shown that, a few bases upstream the transcription start site, the promoter of the HN280 ynfB-speG operon harbours an IS2 element which is likely responsible for silencing the speG gene (Fig. 2A). The residual speG transcription in HN280 is likely the result of read-through transcription, since primer extension analysis does not reveal any signal in correspondence to the expected transcription start base (Fig. 2C).
As for the increased expression of speG in EIEC stains 4608, 6.81 and 53638 (Fig. 2B), our observations indicate that it does not depend on the presence of two transversions within the promoter region (positions -28 and -111), since these base changes (Table S2) are found also in EIEC strain 13.80, where speG transcription is comparable to that observed in the E. coli K-12 control (Fig. 2B). Therefore, it is reasonable to assume that the level of speG transcription detected in these strains may be due to the increased spermidine biosynthesis elicited by the higher putrescine content. In particular, this is evidenced by EIEC 53638 where putrescine and spermidine attain the highest levels among the strains investigated.
It is not surprising that the lack of speG (and the consequent accumulation of spermidine) is not a common feature of EIEC strains, since EIEC do not constitute a homogeneous group of pathogenic E. coli. Indeed, phylogenetic studies of housekeeping genes reveal that EIEC do not belong to a single E. coli cluster and have several distinct evolutionary origins, i.e they emerged in several independent events from multiple ancestral E. coli [4], [10], . EIEC possess many Shigella-like features, however they do not have the full set of characters that define Shigella strains and are not included in any of the three Shigella clusters. In contrast to Shigella, EIEC have a high metabolic activity since they still retain the ability to catabolize substrates widely utilized by E. coli [14]. It is still an open question whether EIEC strains should be regarded as ancestral forms which are developing towards Shigella or as forms that have acquired a Shigella-type adaptation to the human host but are better equipped to face the challenges of the outer environment.
Interestingly, the evolutionary analysis of the polyamine content of EIEC we present in this study underscores the interplay between cadaverine and the expression of the speC gene, encoding ornithine decarboxylase. As one of the major activities in the polyamine pathway this enzyme is responsible for an essential step, the conversion of the L-ornithine into putrescine (Fig. 1). Our data show that the absence of cadaverine in EIEC, depending on the silencing of the cad operon brought about by convergent evolution [29], is responsible for increased speC transcription and, consequently, increased putrescine production. Though the molecular mechanisms adopted by cadaverine to negatively interfere with speC expression deserve further investigation to be fully clarified, the crucial role of cadaverine shows up. Indeed, restoring cadaverine synthesis (by introducing a functional cadC gene) in EIEC strains harboring a functional cadBA operon reduces speC expression and intracellular putrescine (Fig. 5C).
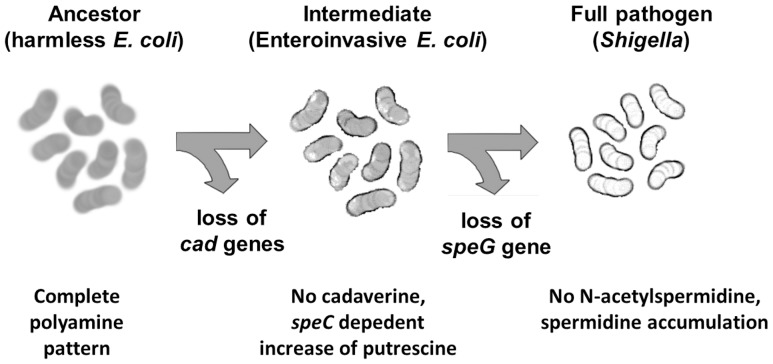
On the basis of our observations, it is reasonable to speculate (Fig. 6) that during the transition from a commensal ancestor (E. coli) towards an enteroinvasive pathogenic phenotype (EIEC) the modification of the polyamine profile might have been triggered by the loss of cadaverine. It is known that cadaverine negatively interferes with the invasive process [23] and it is likely that it has been lost during the initial steps of the pathoadaptation process. In turn, the lack of cadaverine may have induced an increased putrescine level, as we have observed in our EIEC collection and reproduced in an E. coli K-12 background. Since putrescine is a relevant intermediate in the synthesis of spermidine and, consequently, of N-acetyspermidine, an increase in putrescine may well have caused higher levels of both, spermidine and N-acetylspermidine. The inactivation of the speG gene would represent the last step, favouring the accumulation of spermidine (and the elimination of N-acetylspermidine) and enhancing the survival in oxidative environments. In this view, the loss of speG-function has already occurred in Shigella spp. and, banking on our data, could be regarded as an ongoing process in EIEC. In both microorganisms, the lack of speG increases the resistance to oxidative stress and confers higher survival within macrophages. Altogether, our observations agree well with the hypothesis that EIEC are an intermediate step during the transition of E. coli towards a full-blown Shigella phenotype, and further clarify mechanisms and strategies adopted by these bacterial pathogens during the infectious processes.
Figure 6. A model of the pathoadaptive evolution of the polyamine profile in EIEC and Shigella spp.
In this view the loss of cad genes marks the transition from the polyamine profile of the commensal E. coli ancestor to that of enteroinvasive E. coli (EIEC), characterized by lack of cadaverine and increased putrescine content. The loss of the speG gene, occurring in a successive evolutionary step and inducing spermidine accumulation (and, consequently, higher resistance to oxidative stress), gives rise to the Shigella-type polyamine profile.
Supporting Information
Oligos used in this study.
(DOC)
List of the point mutations (transitions and transversion) found in the EIEC ynfB-speG locus and its promoter.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All sequence files are available from the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank; accession numbers: KJ825879, KJ825880, KJ825881, KJ825882, KJ825883).
Funding Statement
RC is supported by fellowships of the Istituto Pasteur Fondazione Cenci Bolognetti. MLDM was supported by a post-doc grant from Università Roma Tre. This work was supported by grants from Ministero della Ricerca e dell'Istruzione (PRIN 2012/WWJSX8K and FIRB), and by grants of Sapienza Università di Roma. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kaper JB, Nataro JP, Mobley HL (2004) Pathogenic Escherichia coli . Nat Rev Microbiol 2: 123–140. [DOI] [PubMed] [Google Scholar]
- 2. Leimbach A, Hacker J, Dobrindt U (2013) E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 358: 3–32. [DOI] [PubMed] [Google Scholar]
- 3. Ochman H, Moran NA (2001) Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292: 1096–1099. [DOI] [PubMed] [Google Scholar]
- 4. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, et al. (2013) Recent advances in understanding enteric pathogenic Escherichia coli . Clin Microbiol Rev 26: 822–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bliven KA, Maurelli AT (2012) Antivirulence genes: insights into pathogen evolution through gene loss. Infect Immun 80: 4060–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prosseda G, Di Martino ML, Campilongo R, Fioravanti R, Micheli G, et al. (2012) Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model. Res Microbiol 163: 399–406. [DOI] [PubMed] [Google Scholar]
- 8. Sansonetti PJ (2006) Shigellosis: an old disease in new clothes? PLoS Med 3: e354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng Y, Chen Z, Liu SL (2011) Gene decay in Shigella as an incipient stage of host adaptation. PlosOne 6: e277754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR (2004) Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp . Infect Immun 72: 5080–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pupo GM, Lan R, Reeves PR (2000) Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA 97: 10567–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng J, Yang J, Jin Q (2009) The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli . Infect Genet Evol 9: 147–152. [DOI] [PubMed] [Google Scholar]
- 13. Moreno AC, Ferreira LG, Martinez MB (2009) Enteroinvasive Escherichia coli vs. Shigella flexneri: how different patterns of gene expression affect virulence. FEMS Microbiol Lett 301: 156–163. [DOI] [PubMed] [Google Scholar]
- 14. Silva RM, Toledo RF, Trabulsi LR (1980) Biochemical and cultural characteristics of invasive Escherichia coli . J Clin Microbiol 11: 441–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sansonetti PJ, Hauteville H, Formal SB, Toucas M (1982) Plasmid-mediated invasiveness of Shigella-like Escherichia coli . Ann Microbiol (Inst Pasteur) 132A: 351–355. [PubMed] [Google Scholar]
- 16. Lan R, Reeves PR (2002) Escherichia coli in disguise: molecular origins of Shigella . Microbes Infect 4: 1125–1132. [DOI] [PubMed] [Google Scholar]
- 17. Parsot C (2005) Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol Lett 252: 11–18. [DOI] [PubMed] [Google Scholar]
- 18. Scribano D, Petrucca A, Pompili M, Ambrosi C, Bruni E, et al. (2014) Polar localization of PhoN2, a periplasmic virulence-associated factor of Shigella flexneri, is required for proper IcsA exposition at the old bacterial pole. PLoS One 9: e90230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prosseda G, Fradiani PA, Di Lorenzo M, Falconi M, Micheli G, et al. (1998) A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli . Res Microbiol 149: 15–25. [DOI] [PubMed] [Google Scholar]
- 20. Prosseda G, Falconi M, Nicoletti M, Casalino M, Micheli G, et al. (2002) Histone-like protein and Shigella invasivity regulon. Res Microbiol 153: 461–468. [DOI] [PubMed] [Google Scholar]
- 21. Tran CN, Giangrossi M, Prosseda G, Brandi A, Di Martino ML, et al. (2011) A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri . Nucleic Acids Res 39: 8122–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barbagallo M, Di Martino ML, Marcocci L, Pietrangeli P, De Carolis E, et al. (2011) A new piece of the Shigella pathogenicity puzzle: spermidine accumulation by silencing of the speG gene. PLoS One 6: e27226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A (1998) “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp and enteroinvasive Escherichia coli . Proc Natl Acad Sci USA 95: 3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tabor CW, Tabor H (1985) Polyamines in microorganisms. Microbiol Rev 49: 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Igarashi K, Kashiwagi K (2010) Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42: 39–51. [DOI] [PubMed] [Google Scholar]
- 26. Shah P, Swiatlo E (2008) A multifacet role for polyamines in bacterial pathogens. Mol Microbiol 68: 4–16. [DOI] [PubMed] [Google Scholar]
- 27. Di Martino ML, Campilongo R, Casalino M, Micheli G, Colonna B, et al. (2013) Polyamines: emerging players in bacteria-host interactions. Int J Med Microbiol 303: 484–91. [DOI] [PubMed] [Google Scholar]
- 28. Meng SY, Bennett GN (1992) Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol 174: 2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prosseda G, Latella MC, Barbagallo M, Nicoletti M, Al Kassas R, et al. (2007) The two-faced role of cad genes in the virulence of pathogenic Escherichia coli . Res Microbiol 158: 487–493. [DOI] [PubMed] [Google Scholar]
- 30. Day WA, Fernandez RE, Maurelli AT (2001) Pathoadaptive mutation that enhance virulence: genetic organization of the cadA region of Shigella spp. Infect Immun 69: 7471–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Casalino M, Latella MC, Prosseda G, Ceccarini P, Grimont F, et al. (2005) Molecular evolution of the lysine decarboxylase-defective phenotype in Shigella sonnei . Int J Med Microbiol 294: 503–512. [DOI] [PubMed] [Google Scholar]
- 32. Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, et al. (1993) The absence of a surface protease, OmpT, determines the intracellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol Microbiol 9: 459–468. [DOI] [PubMed] [Google Scholar]
- 33. Casalino M, Latella MC, Prosseda G, Colonna B (2003) CadC is the preferential target of a convergent evolution driving enteroinvasive Escherichia coli towards a lysine decarboxylase-defective phenotype. Infect Immun 71: 5472–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prunier AL, Schuch R, Fernández RE, Mumy KL, Kohler H, et al. (2007) nadA and nadB of Shigella flexneri 5a are antivirulence loci responsible for the synthesis of quinolinate, a small molecule inhibitor of Shigella pathogenicity. Microbiology 153: 2363–2372. [DOI] [PubMed] [Google Scholar]
- 35. Di Martino ML, Fioravanti R, Barbabella G, Prosseda G, Colonna B, et al. (2013) Molecular evolution of the nicotinic acid requirement within the Shigella/EIEC pathotype. Int J Med Microbiol 303: 651–661. [DOI] [PubMed] [Google Scholar]
- 36. Casalino M, Prosseda G, Barbagallo M, Iacobino A, Ceccarini P, et al. (2010) Interference of the CadC regulator in the arginine-dependent acid resistance system of Shigella and enteroinvasive E. coli . Int J Med Microbiol 300: 289–295. [DOI] [PubMed] [Google Scholar]
- 37. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giangrossi M, Prosseda G, Tran CN, Brandi A, Colonna B, et al. (2010) A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri . Nucleic Acids Res 38: 3362–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, et al. (2011) EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Research 39: D583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prosseda G, Mazzola A, Di Martino ML, Tielker D, Micheli G, et al. (2010) A temperature-induced narrow DNA curvature range sustains the maximum activity of a bacterial promoter in vitro. Biochemistry 49: 2778–2785. [DOI] [PubMed] [Google Scholar]
- 41. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 42. Prosseda G, Latella MC, Casalino M, Nicoletti M, Michienzi S, et al. (2006) Plasticity of the Pjunc promoter of ISEc11, a new insertion sequence of the IS1111 family. J Bacteriol 188: 4681–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Carolis E, Posteraro B, Florio AR, Colonna B, Prosseda G, et al. (2011) Analysis of heat-induced changes in protein expression of Stenotrophomonas maltophilia K279a reveals a role for GroEL in the host-temperature adaptation. Int J Med Microbiol 301: 273–281. [DOI] [PubMed] [Google Scholar]
- 44. Matés JM, Márquez J, García-Caballero M, Núñez de Castro I, Sánchez-Jiménez F (1992) Simultaneous fluorometric determination of intracellular polyamines separated by reversed-phase high-performance liquid chromatography. Agents Actions 36: 17–21. [DOI] [PubMed] [Google Scholar]
- 45. Ngo TT, Brillhart KL, Davis RH, Wong RC, Bovaird JH, et al. (1987) Spectrophotometric Assay for Ornithine Decarboxylase. Anal Biochem 160: 290–293. [DOI] [PubMed] [Google Scholar]
- 46. Madhubala R (1998) Methods in Molecular Biology: Polyamine protocols. : Humana Press Inc; 79: 131p. [DOI] [PubMed] [Google Scholar]
- 47. Chaturvedi R, Asim M, Hoge S, Lewis ND, Singh K, et al. (2010) Polyamines impair immunity to Helicobacter pylori by inhibiting L-arginine uptake required for nitric oxide production. Gastroenterology 139: 1686–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Joshi GS, Spontak JS, Klapper DG, Richardson AR (2011) ACME encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jelsback L, Thomsen LE, Wallrodt I, Jensen PR, Olsen JE (2012) Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLosOne 7: e36149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fukuchi J, Kashiwagi K, Yamagishi M, Ishihama A, Igarashi K (1995) Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli . J Biol Chem 270: 18831–18835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligos used in this study.
(DOC)
List of the point mutations (transitions and transversion) found in the EIEC ynfB-speG locus and its promoter.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All sequence files are available from the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank; accession numbers: KJ825879, KJ825880, KJ825881, KJ825882, KJ825883).