Abstract
The colinearity of genes in Hox clusters suggests a role for chromosome structure in gene regulation. We reveal programmed changes in chromatin structure and nuclear organization upon induction of Hoxb expression by retinoic acid. There is an early increase in the histone modifications that are marks of active chromatin at both the early expressed gene Hoxb1, and also at Hoxb9 that is not expressed until much later. There is also a visible decondensation of the chromatin between Hoxb1 and Hoxb9 at this early stage. However, a further change in higher-order chromatin structure, looping out of genes from the chromosome territory, occurs in synchrony with the execution of the gene expression program. We suggest that higher-order chromatin structure regulates the expression of the HoxB cluster at several levels. Locus-wide changes in chromatin structure (histone modification and chromatin decondensation) may establish a transcriptionally poised state but are not sufficient for the temporal program of gene expression. The choreographed looping out of decondensed chromatin from chromosome territories may then allow for activation of high levels of transcription from the sequence of genes along the cluster.
Keywords: Chromatin decondensation, chromosome territory, differentiation, gene expression, histone modification, nuclear structure
To understand how chromatin regulates gene expression it is necessary to analyze not only nucleosome structure and modifications but also higher-order chromatin condensation and nuclear organization. Many correlations between transcription and nucleosome modifications have been found. For example, active β-globin loci are enriched in hyperacetylated histones H3 and H4, and H3 dimethylated on lysine 4 (met2H3-K4), whereas the silent domain flanking the chicken locus has hypoacetylated histones and H3 dimethylated on K9 (met2H3-K9; Litt et al. 2001; Bulger et al. 2003; Kim and Dean 2003). How histone modifications influence transcription is still debated. They can act as binding sites for specific regulatory proteins (Fischle et al. 2003) and may directly affect higher-order chromatin structure and condensation (Tse et al. 1998; Wolffe and Hayes 1999; Carruthers and Hansen 2000; Wang et al. 2001).
Chromatin organization at and beyond the level of the 30-nm chromatin fiber is poorly understood. Cytologically, chromatin decondensation is seen at transcriptionally active regions in the polytene chromosomes of dipteran insects (Andersson et al. 1984) and when transcriptional regulators are artificially targeted to sites in the mammalian genome (Tumbar et al. 1999; Tsukamoto et al. 2000; Muller et al. 2001; Ye et al. 2001; Nye et al. 2002). It is not clear whether chromatin decondensation accompanies the induction of endogenous mammalian gene expression.
There is also a complex relationship between transcription and the position of genes within chromosome territories (CTs). Some studies show inactive genes located in the interior of CTs and active genes concentrated at the territory periphery (Kurz et al. 1996; Dietzel et al. 1999), but active genes can be transcribed from inside of CTs (Mahy et al. 2002b). Moreover, regions with a high density of coordinately expressed genes locate in loops that extend outside of CTs in expressing cells but not in nonexpressing cells (Volpi et al. 2000; Williams et al. 2002). Localization outside of CTs also occurs at genomic regions with a high-density of broadly expressed genes (Mahy et al. 2002a). Hence, it has been suggested that there is a correlation between high levels of transcriptional activity and localization outside of CTs (Mahy et al. 2002a).
It is not known whether chromatin decondensation and CT organization are just consequences of the changes in histone modifications accompanying gene activation, or whether they are independent levels of chromatin structure over and above the level of the nucleosome. At the mammalian β-globin locus, altered nuclear organization (movement away from heterochromatin) correlates with general histone acetylation and the LCR appears to be responsible for localizing the region outside of CTs prior to gene activation. These studies suggest that nuclear reorganization is upstream of histone modifications in a pathway to chromatin opening and that it is necessary to create a transcriptionally poised state that precedes gene expression (Schubeler et al. 2000; Ragoczy et al. 2003).
To understand the relationships between gene expression, nucleosome modification, chromatin decondensation, and nuclear organization, we have studied the developmentally regulated Hoxb gene cluster. Establishing correct spatial and temporal domains of Hox expression is important for anteroposterior (AP) patterning (Krumlauf 1994; Popperl et al. 1995; Studer et al. 1998). This is partly achieved through the colinearity of gene order along the chromosome, with the time and place of gene activation (Fig. 1A; for review, see Kmita and Duboule 2003). The mechanisms for vertebrate Hox gene regulation are largely unknown, but cis-acting regulatory elements (see Marshall et al. 1994; Popperl et al. 1995; Maconochie et al. 1997; Oosterveen et al. 2003), as well as higher-order silencing mechanisms (Kondo and Duboule 1999) are involved. A progressive transition from an inactive (closed) to an active (open) chromatin state, propagated through the cluster from 3′ to 5′, has been proposed (Van der Hoeven et al. 1996; Kondo and Duboule 1999; Roelen et al. 2002). These models are inferred from transgenic reporter approaches (see Marshall et al. 1994; Maconochie et al. 1997) and from rearrangements that reposition Hox genes within a cluster (Van der Hoeven et al. 1996; Kondo et al. 1998).
Figure 1.
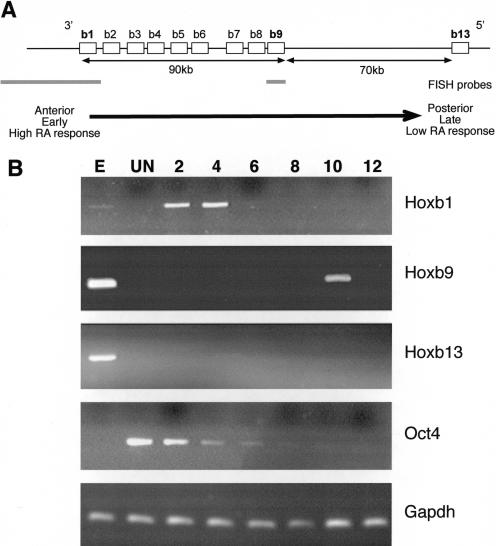
Organization and differential activation of the Hoxb locus in ES cells. (A) Chromosomal organization of genes (open boxes) in the murine Hoxb complex. 5′ and 3′ indicate the direction of the transcription. The distance between genes is indicated below the map, as is the position of FISH probes (gray bars) for Hoxb1 and Hoxb9. The arrow indicates the colinear properties of the Hoxb cluster with respect to spatial expression (anterior to posterior), time of expression (early to late), and sensitivity to RA. Modified from Hunt and Krumlauf (1992). (B) Differential expression of Hoxb genes in OS25 ES cells. RT–PCR analysis of Hoxb1, 9, and 13 in undifferentiated (UN) cells and in cells induced to differentiate with RA for 2 to 12 d. Differentiation was confirmed by the loss of Oct4 expression. Total RNA from E11.5 embryo (E) and analysis of Gapdh were used as positive controls.
Given the interest in the control of Hox gene expression, it is surprising that little is known about their chromatin structure. By using retinoic acid (RA) to induce regulated expression of Hoxb genes in mouse embryonic stem (ES) cells, we have performed chromatin immunoprecipitation (ChIP) to track histone modifications, and fluorescence in situ hybridization (FISH) to analyze chromatin condensation and nuclear organization. Within 4 d of induction, there are increases in histone modifications associated with transcriptional activity (AcH3-K9 and met2H3-K4) at both the early expressed Hoxb1 and also at genes that will not be transcribed until later in the developmental program (Hoxb9). At this stage there is also a dramatic decondensation of the HoxB cluster, which is not a simple consequence of the increase in histone acetylation. There is then a choreographed extrusion of genes outside of CTs that is coincident with the temporal program of gene expression. We propose a two-step model for how chromatin structure regulates expression from HoxB. First a locus-wide change in chromatin structure (histone modification and chromatin decondensation) may create a transcriptionally poised state. Then a progressive 3′ to 5′ change in higher-order chromatin structure, which is manifest as the movement outside of CTs, is necessary to allow for the programmed expression of genes along the cluster.
Results
RA induces a temporal program of Hoxb gene expression in ES cells
In vivo the activation of Hox gene expression by retinoids is mediated by cis-acting RA response elements (RAREs; for review, see Gavalas and Krumlauf 2000). Hox genes also show a colinear temporal response to RA ex vivo. In embryonal carcinoma cells, genes at the 3′ extremity of the clusters respond earlier, and at lower RA concentrations, than do more 5′ genes (Fig. 1A; Simeone et al. 1990, 1991; Papalopulu et al. 1991). RA also induces Hox expression in ES cells (Papalopulu et al. 1991). We induced expression of the HoxB cluster with RA in wild-type ES cells but obtained a nonhomogeneous response as revealed by the expression of SSEA-1 antigen in residual undifferentiated ES cells (Brown et al. 1993; data not shown). Because studies of nuclear organization rely on analysis at the single cell level, it was important to have a more homogenous population of cells, before and after differentiation. Therefore, we chose to use OS25 ES cells that allow for G418 selection of undifferentiated (Sox2 expressing) cells, and then selection against undifferentiated (Oct4 expressing) cells with Ganciclovir after differentiation (Billon et al. 2002). The expression of alkaline phosphatase and SSEA-1 markers, and Oct4, confirmed the undifferentiated state of cells grown in the presence of LIF and G418 (Billon et al. 2002; data not shown). No expression of Hoxb genes was detected in these cells (Fig. 1B). Differentiation, induced by withdrawing LIF and adding RA, resulted in the complete loss of alkaline phosphatase and SSEA-1 markers and extinction of Oct4 expression (Fig. 1B). After 2 to 4 d of differentiation (day 2 through 4), a robust pulse of Hoxb1 expression was detected, but there was no expression of the more 5′ Hoxb9 and Hoxb13. A pulse of Hoxb9 expression was detected at day 10, at which time Hoxb1 had been silenced (Fig. 1B). No Hoxb13 expression was detected during the time course of the experiment. Therefore, we are able to recapitulate the sequence of expression from Hoxb in this ex vivo system, enabling us to follow the chromatin structure of the complex during induction.
Changes in histone H3 modification at Hoxb1 and Hoxb9
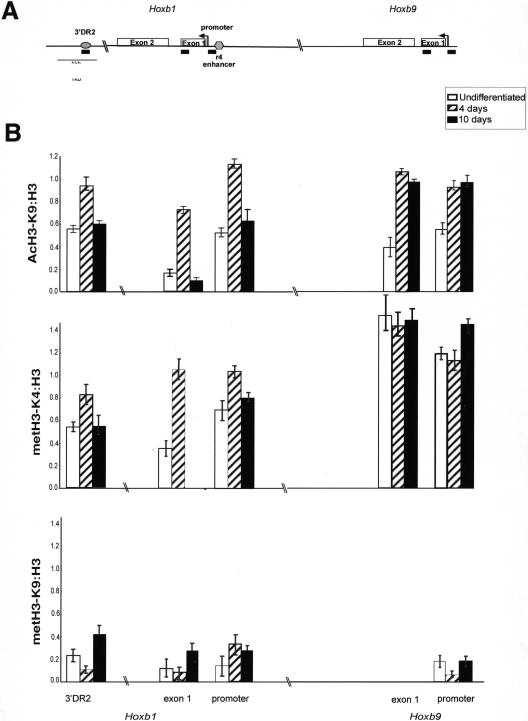
Transcriptionally active genes are generally characterized by high levels of histone acetylation and metH3-K4 (for review, see Zhang and Reinberg 2001; Lachner et al. 2003). We used ChIP and quantitative real-time PCR to examine changes in H3 modification at Hoxb1 and Hoxb9 before differentiation (neither gene expressed) and at day 4 (Hoxb1 expressed) and day 10 (Hoxb9 expressed) of differentiation. We also examined the r4 enhancer upstream of Hoxb1, which is required for its rhombomere 4 restricted expression (Popperl et al. 1995), and the RARE (3′DR2) located 1.2 kb 3′ of Hoxb1 that mediates the early response to RA (Marshall et al. 1994; Fig. 2A). The presence of nucleosomes in these regions was confirmed by ChIP with antibody that detects the C terminus of H3 (Verreault et al. 1996). By day 4, increased H3 acetylated at K9 (AcH3-K9) was detected at all sites in Hoxb1 and Hoxb9 that were examined, even though only Hoxb1 is expressed at this time. Levels of met2H3-K4 also increased at Hoxb1 by day 4 of differentiation, and surprisingly, constitutively high levels of this histone modification were found at exon 1 and the promoter of the silent Hoxb9 both before and after differentiation. AcH3-K9 and met2H3-K4 were lost from Hoxb1 by day 10, when the gene has been silenced, but they persist at Hoxb9 (Fig. 2B).
Figure 2.
Histone H3 modifications upon induction of HoxB in ES cells. (A) Map of Hoxb and the elements analysed by ChIP. Exons of Hoxb1 and Hoxb9 are represented by two open bars. The arrows denote the transcription start sites of Hoxb1 and Hoxb9. The 3′ RARE (3′DR2; Marshall et al. 1994) and the r4 enhancer (Popperl et al. 1995) are indicated by the filled bars. The position of the PCR products detected by ChIP are indicated by black bars underneath. (B) Quantification of ChIP by real-time PCR. The graphs show the mean levels of product amplified from samples after ChIP with antibodies recognizing AcH3-K9, met2H3-K4, and met2H3-K9 (after subtraction of mock IP levels) normalized relative to those obtained by ChIP with antibody recognizing the C terminus of H3. The analysis was carried out on chromatin prepared from undifferentiated ES cells (open bars) and from cells induced to differentiate with RA for 4 d (hatched bars) and 10 d (filled bars).
We conclude that RA rapidly induces histone modifications (AcH3-K9 and met2H3-K4), which are considered as marks of active chromatin, at not only the 3′ end of the cluster where gene expression is induced at an early stage but also at more 5′ regions (Hoxb9). At these later sites, the changes in histone modifications precede transcriptional activation by many days. The subsequent loss of these histone modifications from Hoxb1 parallels the cessation of Hoxb1 expression. However, at no stage did we detect substantial levels of met2H3-K9 (Fig. 2B), suggesting that this marker of gene repression is not important for the switch on or off of Hoxb expression in this system.
Chromatin decondensation at Hoxb
It has been suggested that an “opening” of chromatin structure may contribute to the regulation of Hox clusters (Van der Hoeven et al. 1996; Kondo and Duboule 1999; Roelen et al. 2002). Cytological chromatin decondensation accompanies transcriptional activation in some reporter systems (Tumbar et al. 1999; Tsukamoto et al. 2000; Muller et al. 2001). To determine if there are similar levels of chromatin decondensation accompanying induction of Hoxb, we measured the distance (d) between Hoxb1 and Hoxb9 hybridization signals in nuclei before and after RA induction. At probe separations of <2 Mb, there is a linear relationship between the meansquare interphase distances between them (d2) and their separation in kilobases (van den Engh et al. 1992). This analysis has been used to show that different parts of the human genome have different levels of chromatin compaction (Yokota et al. 1997).
Hoxb1 and Hoxb9 are 90 kb apart (Fig. 1A), and in nuclei of undifferentiated ES cells, they could barely be separated from one another. After day 2 of differentiation, they were readily separable in both MAA and pFa-fixed cells (Fig. 3A). Some recondensation of this region was then seen by day 10. The distribution of d values after day 2 conforms to a Rayleigh distribution (Sachs et al. 1995) (SD/〈d〉 = 0.56 and median/〈d〉 = 0.9) indicating that the decondensed chromatin describes a random walk path.
Figure 3.
Decondensation of Hoxb. (A) FISHof Hoxb1 (red) and Hoxb9 (green) to MAA-fixed nuclei from undifferentiated (UN) cells, from cells at 2 or 10 d of differentiation, and from undifferentiated cells treated with TSA. Nuclei were counterstained with DAPI (blue). Bar, 5μm. Below it, each image the mean-square interprobe distance—d2 (μm2) ± S.E.M.—is shown (n = 50 cells). The value for cells at 2 d, fixed with pFa, is also shown. (B) FISH of Pax6 (red) and Rcn (green) probes on cells before and after (2 d) differentiation.
Decondensation is not a general consequence of differentiation. The interphase separation between two control genes (Rcn and Pax6), the expression of which is not induced by RA and which are separated by 300 kb on MMU2, is not significantly different before and after 2 d of differentiation (P = 0.46; Fig. 3B).
The visual decondensation of Hoxb could represent a specific change in its higher-order chromatin structure or could just be a passive consequence of the increase in histone acetylation at the locus (Fig. 2) and its influence on folding of the chromatin fiber. To distinguish between these two possibilities, we experimentally increased histone acetylation in undifferentiated ES cells, by inhibiting the action of histone deacetylases (HDACs) with trichostatin A (TSA), in the absence of RA. Expression of Hoxb was determined by real-time RT–PCR. During the normal RA-induced differentiation program, there is an ∼10-fold increase in the levels of Hoxb1 mRNA by day 4. In contrast, only a very small (less than twofold) increase in Hoxb1 mRNA was detected after exposure of undifferentiated ES cells to TSA, and there was no expression of Hoxb9 (data not shown). TSA pretreatment does not prevent the normal execution of the program of RA-induced Hoxb expression because the correct sequence of Hoxb1 and Hoxb9 expression was seen at appropriate times after exposure of TSA-treated undifferentiated cells to RA (data not shown).
ChIP and real-time PCR showed that, in undifferentiated cells, TSA increased AcH3-K9 levels at Hoxb1 and its regulatory elements to the same extent as that seen when untreated cells are differentiated with RA (data not shown). However, as well as no induction of Hoxb expression, there was no visual decondensation of the locus in TSA-treated cells (Fig. 3A). Therefore, we conclude that there is an approximate ninefold transient decondensation of the Hoxb complex upon induction with RA, and that this decondensation is not just a simple consequence of increased levels of histone acetylation.
Extrusion from the CT accompanies Hoxb gene expression
Although we have detected changes in histone modification and chromatin decondensation upon induction of Hoxb, the timing of these events early in the induction program (by day 4) does not account for the temporal program of gene expression—in particular, why is Hoxb9 expression not seen until day 10? Domains of high transcriptional activity have been correlated with a distinctive chromatin structure at the cellular level, a looping out from CTs (Mahy et al. 2002a). Moreover, for some domains of coordinately regulated genes, the extrusion of these loops correlates with the up-regulation of gene expression (Volpi et al. 2000; Williams et al. 2002).
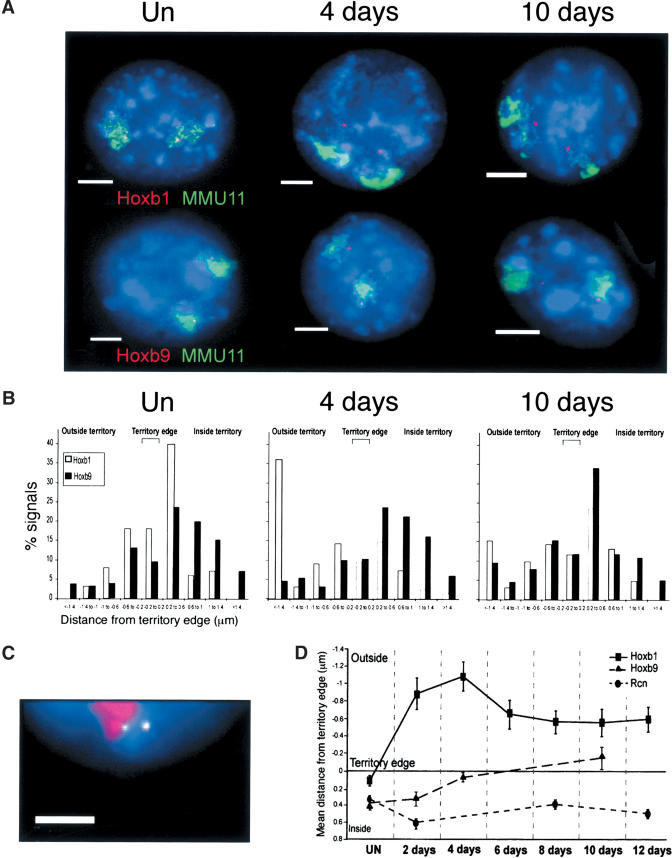
We therefore used FISH to examine how HoxB is organized relative to its CT, before and after induction. Probes encompassing Hoxb1 or Hoxb9 (Fig. 1A) and a paint for mouse chromosome 11 (MMU11) were cohybridized to nuclei from undifferentiated cells and from cells at day 4 or 10 of differentiation (Fig. 4A). We calculated the distance (in micrometers) between the Hoxb gene signals and the nearest CT edge as described previously (Fig. 4B; Table 1; Mahy et al. 2002a). In undifferentiated ES cell nuclei, most hybridization signals for both Hoxb1 and Hoxb9 were within the CT. By day 4 of differentiation, when Hoxb1 is expressed, most of its hybridization signals are now located well (>0.2 μm) outside of the CT, but Hoxb9 remains inside (Table 1; Fig. 4B,D). This was also confirmed by simultaneous detection of Hoxb1, Hoxb9, and MMU11 CT (Fig. 4C). A two-tailed distribution Student's t test showed that the relocalization of Hoxb1 outside of the CT upon differentiation is statistically significant (P < 0.000). By day 10, Hoxb1 expression is switched off and the gene relocalizes back toward the CT (P = 0.02;Fig. 4B,D). However, its location is still more exterior to the CT than in undifferentiated cells (P < 0.000). At day 10, Hoxb9 moves to the CT surface (Fig. 4D), and a significantly (P = 0.001) increased frequency of Hoxb9 signals are now found outside of the CT, coincident with the expression of this gene (Fig. 4B; Table 1).
Figure 4.
Progressive looping of the HoxB cluster from 3′ to 5′. (A) FISH with MMU11 chromosome paint (green) and gene-specific probes (red) for Hoxb1 (top) or Hoxb9 (bottom) on MAA-fixed nuclei from undifferentiated (UN) cells, and from cells at 4 or 10 d of differentiation. Nuclei were counterstained with DAPI (blue). Bars, 5 μm. (B) Histograms showing the distribution of hybridization signals for Hoxb1 (open bars) or Hoxb9 (filled bars) relative to the MMU11 territory edge (μm). Negative distances indicate signals located beyond the visible limits of the detectable CT. n = 100 territories. (C) FISH with MMU11 chromosome paint (red) and gene-specific probes for Hoxb1 (green/red, 1:1) or Hoxb9 (green) on nuclei from cells at 10 d of differentiation. Nuclei were counter-stained with DAPI (blue). Bar, 5 μm. (D) Mean position (μm, ± S.E.M.) of Hoxb1 and Hoxb9 relative to the MMU11 territory edge during 12 d of differentiation. The position of Rcn in the MMU2 territory is shown as control (Mahy et al. 2002a). n = 100 territories. Normalization of the data to the size of the nucleus showed that the Hoxb movements are not due to the increased size of the nucleus and CT upon differentiation (data not shown).
Table 1.
Localization of Hoxb genes outside of CTs
| % signals outside of MMU11 CT
|
|||||
|---|---|---|---|---|---|
| Gene | Undiff. | Day 4 | Day 10 | TSA | TSA + 2 d |
| Hoxb1 (MAA) | 36 | 73 | 54 | 18 | 67 |
| Hoxb1 (pFa) | 15 | 65 | N/D | N/D | N/D |
| Hoxb9 (MAA) | 24 | 24 | 39 | N/D | N/D |
The percentage of Hoxb1 or Hoxb9 hybridization signals manually scored as being outside of the MMU11 CT by FISH to either MAA- or pFa-fixed nuclei from undifferentiated ES cells, and from cells after 4 and 10 d of differentiation. Undifferentiated cells that had been treated with TSA, and TSA-treated cells induced to differentiate with RA for 2 d, were also examined. N/D indicates not determined.
Relocalization outside the CT is not a general phenomena occurring upon differentiation, because the ubiquitously expressed Rcn gene locates inside of its MMU2 CT in both undifferentiated ES cells at all stages of differentiation (Fig. 4D; Mahy et al. 2002b). Consistent with the failure of TSA to induce chromatin decondensation at HoxB, we detected no movement of Hoxb1 outside of the CT of TSA-treated cells. However, TSA treatment does not compromise subsequent induction because 2 d after RA induction of these cells, Hoxb1 is expressed and loops out from the CT (Table 1).
We conclude that in undifferentiated ES cells Hoxb1 resides within, but close to, the surface of its CT. Upon induction with RA, it loops out away from the CT with kinetics that parallel those of its transcription, so that when Hoxb1 expression is silenced after day 4, the frequency and extent of its looping decreases. However, it does not return to the same location as it was in before induction. Compared with Hoxb1, Hoxb9 is located further inside of the CT in undifferentiated cells and remains there immediately after induction. Looping out of Hoxb9 is not detected until day 10, and it never reaches the same extent as that seen for Hoxb1 at day 4. Therefore, unlike the coordinate looping of the MHC (Volpi et al. 2000), the induction of expression from the Hoxb complex results in a progressive remodeling and a looping out of the region from the CT, which parallels the temporal activation of genes in the cluster.
Hoxb1 moves toward the center of the nucleus upon induction
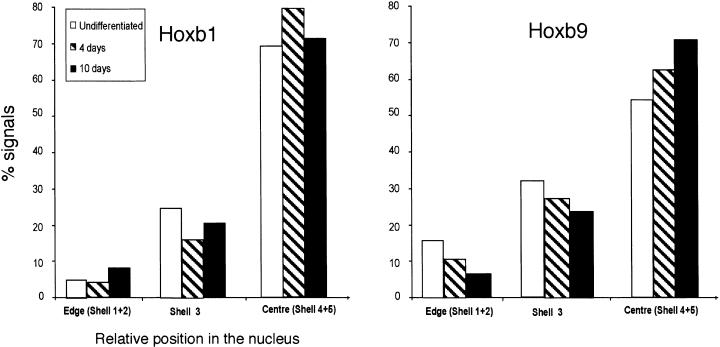
Looping could just be a passive consequence of chromatin decondensation, or it could represent the recruitment of the locus to a particular region of the nucleus or nuclear compartment. In the former case, there need be no directionality to the movement relative to the center or the periphery of the nucleus. In the latter case, there may be directional movement. To test this, we determined the distribution of Hoxb1 and Hoxb9 hybridization signals in images of nuclei eroded into concentric shells, from the edge (shells 1 and 2) to the center (shells 4 and 5) of the nucleus (Croft et al. 1999). A significant (P = 0.001) movement of Hoxb1 toward the nuclear center occurs at day 4 of differentiation, and by day 10, it has returned to its original nuclear location (Fig. 5). In contrast, there is a progressive movement of Hoxb9 toward the nuclear center between day 4 and 10 (P = 0.008; Fig. 5). There was no accompanying movement of the MMU11 CT in the same direction, suggesting that when HoxB loops out from the CT, it is specifically drawn toward the center of the nucleus.
Figure 5.
Movement of the Hoxb1 toward the center of the nucleus. The localization of Hoxb1 and Hoxb9 hybridization signals within erosion shells from the edge (shells 1 and 2) toward the center (shells 4 and 5) of the nucleus. Analysis was performed on MAA-fixed nuclei from undifferentiated ES cells (open bars) and from cells after 4 and 10 d of differentiation (hatched and filled bars, respectively). n = 50 cells.
Discussion
Changes in high-order chromatin structure and levels of gene expression
We have correlated the temporal pattern of gene expression from the Hoxb cluster with changes in nucleosome modification (Fig. 2), chromatin decondensation (Fig. 3), and an extrusion of HoxB genes out of the CT (Fig 4). We detect an increase (by day 4) of histone H3 acetylation (at K9), and methylation of H3 at K4, upon induction by RA. This is consistent with the known associations of these histone marks with states of gene activation (Lachner et al. 2003). However, the modifications are gained at both the transcriptionally activated Hoxb1 and at Hoxb9, which will remain silent for a further 6 d before its activation (Fig. 2). Therefore, these histone marks seem to be associated with poising of the genes for expression, rather than expression per se. We detect little met2H3-K9 at Hoxb1 or Hoxb9, no loss of met2H3-K9 upon induction, and no acquisition of this histone modification when Hoxb1 expression ceases after day 4. This suggests that this mark of gene repression is not involved in the early regulation of HoxB.
Although transcriptionally active chromatin is often considered to have an “open” chromatin structure, implying a decondensed chromatin structure, direct evidence for this has been lacking. Here we have shown for the first time that activation of an endogenous mammalian gene locus is accompanied by a visible approximately ninefold chromatin decondensation (Fig. 3). RA mediates its affects on Hoxb transcription via RA receptors that bind to RAREs such as the 3′DR2 (Marshall et al. 1994). Targeting of other nuclear hormone receptors to artificial arrays has also been shown to induce transient chromatin decondensation (Muller et al. 2001; Nye et al. 2002). However, compared with these studies, HoxB is several fold more decondensed, and the compaction is approximately consistent with formation of a 30-nm chromatin fiber
We have also shown that the induction of Hoxb expression is accompanied by a choreographed looping of the locus outside of CTs and toward the center of the nucleus, in a temporal pattern that parallels that of gene expression from the locus (Figs. 4, 5). Volpi et al. (2000) suggested that localization outside of CTs may be a visual manifestation of chromatin decondensation, and our data are consistent with this idea. Previously, we suggested that localization outside of CTs correlates with high levels of transcription (Mahy et al. 2002a). However, is it has also been suggested that looping outside of CTs can represent a poised state, prior to transcriptional activation (Ragoczy et al. 2003). In the very early embryo, prior to primitive streak formation, the 3′ end of HoxB is poised to respond to exogenous RA (Roelen et al. 2002). The rapid response to RA in ES cells (Fig. 1B; Papalopulu et al. 1991), and the absence of met2H3-K9 prior to induction, suggests that Hoxb1 is similarly poised for transcription in these cells. Because we have found that Hoxb1 is inside of its CT in undifferentiated ES cells (Fig. 4), we suggest that the looping out of decondensed chromatin from the territory represents the transcriptionally active state and not a poised state (Fig. 6).
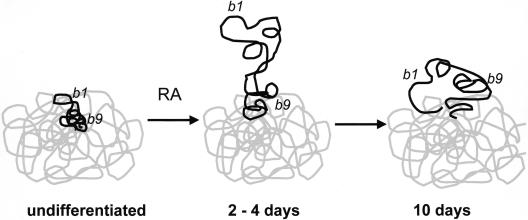
Figure 6.
Changes in chromatin structure at HoxB. In undifferentiated ES cells, the entire Hoxb locus (black) is condensed with the MMU11 CT (gray). However, the 3′ Hoxb1 gene is at the territory surface, poised to respond to retinoic acid (RA). The more 5′ Hoxb9 is further inside the territory. After 2 d of induction with RA, the Hoxb chromatin fiber decondenses, and the 3′ end of the locus, including Hoxb1, is extruded from the CT. By 10 d of differentiation, Hoxb1 is reeled in toward the CT, and Hoxb9 has now left the CT.
The mechanisms that mediated chromatin decondensation and extrusion from CTs have yet to be determined. It has been suggested that histone modifications may directly influence higher-order chromatin structures, for example, by altering nucleosome–DNA or nucleosome–nucleosome interactions, and by neutralizing charge in the histone N-terminal tails (Tse et al. 1998; Wolffe and Hayes 1999; Carruthers and Hansen 2000; Wang et al. 2001). However, increasing the levels of histone acetylation at HoxB experimentally with TSA does not lead to the changes in high-order chromatin structure that normally occur at the locus upon differentiation. Therefore, we suggest that chromatin compaction and nuclear organization represent a level of chromatin structure that is not simply a reflection of underlying histone acetylation.
Chromatin structure and the regulation of Hox gene expression
In vivo, Hoxb1 expression is initiated in primitive streak mesoderm early in gastrulation (E7.2); however, Hoxb1 and Hoxb2, but not more 5′ genes in the cluster, are poised to respond to exogenous RA before primitive streak formation (by E6.0; Roelen et al. 2002). In this regard, it is interesting to note that even in the nuclei of undifferentiated ES cells, the intrachromosomal organizations of Hoxb1 and Hoxb9 are different from each other. Whereas Hoxb1 is located at the CT surface, Hoxb9 is further inside the territory (Fig. 4D). The proximity of Hoxb1 to the territory surface may contribute to its poised transcriptional state and may underlie its ability to respond rapidly and robustly to RA (Fig. 6). It will therefore be interesting to analyze the nuclear organization of Hoxb1 when placed in inappropriate genomic locations (Kmita et al. 2000).
We propose a two-step model through which chromatin structure regulates Hoxb expression (Fig. 6). In response to RA, there appears to be a widespread alteration in histone modification and chromatin condensation that is not restricted to the most 3′ gene (Hoxb1). We suggest that this establishes a transcriptionally competent chromatin state. The progressive looping out of genes from the CT may then contribute to the temporal pattern of gene expression and be necessary to support high rates of transcription. This later step, but not the first one, has similarities to models that propose that a (3′ to 5′) opening of chromatin structure that leads to the sequential accessibility of Hox genes for transcription (Van der Hoeven et al. 1996, Kondo and Duboule 1999; Roelen et al. 2002).
Materials and methods
ES cells culture, differentiation, and TSA treatment
Undifferentiated OS25 ES cells were maintained on 0.1% gelatin-coated dishes in Glasgows' minimal Eagle medium containing 10% fetal calf serum, nonessential amino acids, 1 mM sodium pyruvate, 0.3 mg/ml L-glutamine, 0.1 mM 2-mercaptoethanol, 1000 U/mL human recombinant LIF, and 100 μg/mL G418 (Billon et al. 2002).
To induce differentiation, 5 × 105 cells were plated in 25-cm2 tissue culture flasks (Costar) without LIF or G418 for 1 d; 5 × 10–6 M RA was then added for 4 d. After 2 d (day 2 of differentiation) the cells were replated in 75-cm2 flasks, and the RA-containing medium was supplemented with 2.5 μM Ganciclovir to select against undifferentiated cells for another 2 d (day 4). Subsequently, media was supplemented only with Ganciclovir (no RA) for eight more days until day 12. The medium was changed every 2 d during the differentiation protocol (Billon et al. 2002).
The 5 × 106 undifferentiated ES cells, maintained in medium supplemented with G418 and LIF, were treated with 160 nM TSA for 24 h.
Real-time RT–PCR
Twenty micrograms of total RNA, prepared by using Bio/RNA-X-cell (Bio/Gene Limited), were treated with 20 U Rnase-free DNase I (Promega). After phenol/chloroform extraction, the RNA was ethanol precipitated in 1 v of 5 M ammonium acetate and dissolved in H2O. Two micrograms of RNA were reverse-transcribed by using Superscript II RNase H (Invitrogen) and 4 μL of Random Hexamers (50 μM; Amersham Pharmacia), and 1 μL was used as temple for real-time PCR, using the DNA-intercalating SYBR Green reagent and Master mix (Bio-Gene), on a lightcycler (Roche Molecular Biochemicals). Each reaction was performed in duplicate. PCR primers and conditions are described in Table 2. The 1:10, 1:100, and 1:1000 dilutions of β-actin PCR product of known concentration were used to produce a standard curve. The results were analyzed by Lightcycler software, version 3.5.
Table 2.
PCR primers and amplification conditions
| Gene/locus | Primers | Product size (bp) | PCR conditions | |
|---|---|---|---|---|
| RT—PCR | Oct4 | GGCGTTCTCTTTGGAAAGGTGTTC CTCGAACCACATCCTTCTCT | 312 | 15 min 95°C (30 sec 95°C, 30 sec 55°C; 30 sec 72°C) × 30 |
| Gapdh | CTCAAGATTGTCAGCAATGCA CCTTCCACAATGCCAAAGTT | 70 | 15 min 95°C (30 sec 95°C, 30 sec 61°C, 25 sec 72°C) × 25 | |
| Hoxb1 | CCATATCCTCCGCCGCAG CGGACTGGTCAGAGGCATC | 450 | 15 min 95°C (30 sec 95°C, 30 sec 62°C, 40 sec 72°C) × 35 | |
| Hoxb9 | CAGGGAGGCTGTCCTGTCTAATC CTTCTCTAGCTCCAGCGTCTGG | 176 | 15 min 95°C (30 sec 95°C, 30 sec 68°C, 20 sec 72°C) × 30 | |
| Hoxb13 | CTGGAACAGCCAGATGTGTT CCTGCTAAAGGTGTCATCTC | 445 | 15 min 95°C (30 sec 95°C, 30 sec 57°C, 40 sec 72°C) × 35 | |
| β-actin | GGTCAGAAGGACTCCTATGTGG TCTCAGCTGTGTGGTGAAG | 475 | ||
| ChIP | Hob1 r4 + promoter + initiation site | TTCCATGTCGCTCTCAGATG TAGGAAGGGGCTAGGGAGTG | 172 | 15 min 95°C (30 sec 95°C, 30 sec 64°C, 20 sec 72°C) × 25 |
| Hoxb1 3′DR2 | GACAGTGGGTCCTGGGGGT GGTGCTCAACAGGCTTGTTTGGA | 110 | 15 min 95°C (30 sec 95°C, 30 sec 61°C, 20 sec 72°C) × 25 | |
| Hoxb1 exon 1 | CCATATCCTCCGCCGCAG TCTGACGAGCAAGGCGATTCCTTG | 104 | ||
| Hoxb1 promoter | CCGCTTAGCCCATTGGCCT TAGGAAGGGGCTAGGGAGTG | 80 | ||
| Hoxb9 promoter | TCCGAAAGCCCTCTGCAC GACATTCTCAGACATTATCCGCG | 110 | ||
| Hoxb9 exon 1 | GCACGCCCGAGTACAGTTTG CCTCTCTTTGTCCTCGCTTCCT | 112 |
Chromatin immunoprecipitation
The ChIP protocol was modified from that of Christenson et al. (2001). ES cells were replated onto 10-cm tissue culture dishes. One day later, formaldehyde (Sigma), to a final concentration of 1%, was added to the medium for 10 min at room temperature. Cells were washed once in cold phosphate-buffered saline (PBS) before scraping in buffer A (85 mM KCl, 5.5% sucrose, 10 mM Tris at pH 7.5, 0.2 mM EDTA, 0.5 mM spermidine, 250 μM PMSF, and 1:100 protease inhibitor cocktail; Roche), containing 0.5% NP40. The dish was rinsed with buffer A, and the cell suspension incubated for 5 min on ice and then centrifuged at 2000g for 3 min. The pellet was resuspended in 1% SDS, 10 mM EDTA, 50 mM Tris at pH8, and 250 μM PMSF and sonicated three times for 15 sec each at 10 μm of amplitude by using a Soniprep 150. Chromatin fragments were centrifuged for 3 min, 4°C, at 16000g, and the OD260nm was measured. Ten micrograms of sonicated salmon sperm DNA (sssDNA) and 100 μL of protein G–agarose (Amersham Biosciences), prewashed two times in buffer A, were added to the supernatant and incubated for 1 h at 4°C on a rotating wheel. After centrifugation (2 min, 4°C, 2000g) to remove aggregated protein G, the supernatant was divided into aliquots of 150 μg DNA. One aliquot was kept aside (input), and one was incubated with no antibody as a negative control (mock IP).
Each test aliquot was diluted in 250 μL IP dilution buffer (100 mM Tris at pH 8, 150 mM NaCl, and 0.5% Triton X-100) and rotated overnight at 4°C with the following antibodies: 3 μL of antibody that recognizes the C terminus of H3 (Verreault et al. 1996), 5 μL anti-met2H3-K4 (Upstate Biotechnology, 07-212), and anti-acH3-K9 (Upstate Biotechnology, 06-942) and anti-met2H3-K9 (Abcam, ab7312). After antibody binding, 100 μg sssDNA and 100 μL 50% (v/v) protein G–agarose were added and incubated for 2 h at 4°C and then centrifuged for 2 min, 4°C, at 2000g. The chromatin-antibody/protein G–agarose complexes were washed for 5 min with 500 μL of each of the following buffers: IP dilution buffer, IP dilution buffer and 500 mM NaCl, wash buffer (10 mM Tris at pH 8.0, 500 mM LiCl, 1% NP40, and 1% deoxycholate), and TE (pH 8.0). One hundred microliters of TE (pH 8.0) with 500 μg/mL of RNaseA were added to the beads for 30 min at 37°C. Samples were eluted in 1% SDS and 0.1 M NaHCO3. NaCl was added to 200 mM and the mixture heated for 6 h at 65°C to reverse the formaldehyde cross-links. Samples were digested with 20 μg/mL proteinase K for 1 h at 55°C and the DNA purified by phenol/chloroform extraction and ethanol precipitation. The DNA pellet was resuspended in 20 μL of sterile H2O, and 1 μL aliquots were used as templates in real-time PCR.
PCR on immunoprecipitated DNA
One microliter of immunoprecipitated DNA was quantified by real-time PCR. For PCR in the 3′DR2 region and exon 1, DNA-intercalating SYBR Green reagent and The Master Mix (Bio-Gene) was used. The QuantiTect SYBR Green PCR kit (Qiagen) was used for the Hoxb1 and Hoxb9 promoter. Each ChIP and each real-time PCR was done at least twice. Input sonicated DNA (150 μg) from ES cell lysate was diluted 1:10, 1:100, and 1:1000, and 1 μL amplified in parallel with the immunoprecipitated DNA (IP) to produce a standard curve. Each 10-fold dilution increased the CT by about three cycles. For each amplification, the background corresponding to the mock IP was subtracted from the IP, and the specific enrichment was calculated from the standard curve.
FISH
Paint for mouse chromosome 2 (MMU2) was PCR-labeled with biotin-16-UTP as previously described (Mahy et al. 2002b). Biotin-16-UTP–labeled paint for MMU11 was purchased from Cambio. BACs 50j5 and 67L02 for Rcn and Pax6, respectively (Mahy et al. 2002b), were labeled with digoxigenin or biotin by nick-translation. The 50 kb BAC MMP-4 (T. Jinks, M.T. Martinez-Pastor, and R. Krumlauf, unpubl.) encompassing Hoxb1 (Fig. 1A) was labeled with digoxigenin-11-dUTP by nick translation. A plasmid containing Hoxb9 (P. Hunt and R. Krumlauf, unpubl.) was labeled with digoxigenin-11-dUTP or biotin-16-UTP. Approximately 200 ng of paint and 150 ng BAC/plasmid were used per slide, together with 15 μg mouse Cot1 DNA (GIBCO BRL) and 5 μg sssDNA.
Two-dimensional analysis was as described previously (Croft et al. 1999; Mahy et al. 2002b). To preserve three-dimensional nuclear structure, ES cells grown on slides were washed three times in PBS before permeabilization in CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES at pH 6.8, and 0.5% Triton X100) for 5 min on ice. After washing three times in PBS, slides were fixed with 4% pFa/PBS for 10 min before freeze-thaw in 20% glycerol/PBS and FISH as described previously (Kurz et al. 1996; Mahy et al. 2002a,b).
Image analysis
Two-dimensional samples were examined on a Zeiss Axioplan fluorescence microscope fitted with a triple band-pass filter (Chroma no. 83000). Gray-scale images were captured with a cooled CCD camera (Princeton Instruments Pentamax) and custom IPLab scripts. For three-dimensional analysis, a focus motor was used to collect image stacks at 0.5-μm intervals along the z-axis. Images were captured with a Xillig CCD camera.
Analysis of probe position relative to CTs in 2D was as previously described (Mahy et al. 2002a,b). Because the size of the nucleus and the CTs increased upon differentiation, the locus to territory edge distance was also normalized by dividing it by the radius of the circle of equal area to that of the territory (Mahy et al. 2002b).
To calculate the d between two probes in two dimensions the hybridization signals were manually segmented and the xy coordinates of the centroid of each were determined. d was then calculated from d = [(x1 – x2)2 + (y1 – y2)]2. Calculation of d in stacks of three-dimensional images was similar. Briefly, the xyz coordinates of the centroid of each signal were determined. The distance between the two probes (or space diagonal) was the distance between two spots at opposite corner of a cuboid. dabc = (a2 + b2 + c2); a, b, and c are the length of the side of the cuboid.
Probe position relative to the nuclear periphery and the nuclear center was determined by erosion of the nuclear area into five concentric shells (1 through 5) of equal area from the periphery of the nucleus to the center (Croft et al. 1999). The proportion of DAPI fluorescence and CT signal was calculated for each of the five shells, and the shell containing the gene signal was manually recorded. To do a t test for the gene probe, an arbitrary value was given to each shell (one at the periphery and five at the more center shells), and the number of probes located in each was multiplied by this value. To perform a t test for the CT position, the percentage of signal was multiplied by the mean radius for each shell. In a plane-projected sphere, the mean radius of random a point for each shell was calculated as 2/3[(rb3 – ra3)/(rb2 – ra2)].
Acknowledgments
S.C. was supported by fellowships from La Ligue Contre le Cancer and the Wellcome Trust (GR071481). W.A.B. is a Centennial fellow of the James S. McDonnell foundation. We thank Austin Smith (Institute for Stem Cell Research, Edinburgh) for OS25 cells, and Robb Krumlauf (Stowers Institute, Kansas City) for Hoxb BACs and for stimulating our research in this area. The H3-C terminal antibody was a gift of Alain Verreault (Cancer Research UK, Clare Hall). We are grateful to Nick Hastie, Robin Allshire, and Richard Meehan for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.292104.
References
- Andersson K., Bjorkroth, B., and Daneholt, B. 1984. Packing of a specific gene into higher order structures following repression of RNA synthesis. J. Cell. Biol. 98: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon N., Jolicoeur, C., Ying, Q.L., Smith, A., and Raff, M. 2002. Normal timing of oligodendrocyte development from genetically engineered, lineage-selectable mouse ES cells. J. Cell. Sci. 115: 3657–3665. [DOI] [PubMed] [Google Scholar]
- Brown D.G., Warren, V.N., Pahlsson, P., and Kimber, S.J. 1993. Carbohydrate antigen expression in murine embryonic stem cells and embryos, I: Lacto and neo-lacto determinants. Histochem. J. 25: 452–463. [DOI] [PubMed] [Google Scholar]
- Bulger M., Schubeler, D., Bender, M.A., Hamilton, J., Farrell, C.M., Hardison, R.C., and Groudine, M. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 23: 5234–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers L.M. and Hansen, J.C. 2000. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275: 37285–37290. [DOI] [PubMed] [Google Scholar]
- Christenson L.K., Stouffer, R.L., and Strauss III, J.F. 2001. Quantitative analysis of the hormone-induced hyperacetylation of histone H3 associated with the steroidogenic acute regulatory protein gene promoter. J. Biol. Chem. 276: 27392–27399. [DOI] [PubMed] [Google Scholar]
- Croft J.A., Bridger, J.M., Perry, P., Boyle, S., Teague, P., and Bickmore, W.A. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell. Biol. 145: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S., Schiebel, K., Little, G., Edelmann, P., Rappold, G.A., Eils, R., Cremer, C., and Cremer, T. 1999. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp. Cell. Res. 252: 363–375. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang, Y., and Allis, C.D. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell. Biol. 15: 172–183. [DOI] [PubMed] [Google Scholar]
- Gavalas A. and Krumlauf, R. 2000. Retinoid signalling and hindbrain patterning. Curr. Opin. Genet. Dev. 10: 380–386. [DOI] [PubMed] [Google Scholar]
- Hunt P. and Krumlauf, R. 1992. Hox codes and positional specification in vertebrate embryonic axes. Annu. Rev. Cell Biol. 8: 227–256. [DOI] [PubMed] [Google Scholar]
- Kim A. and Dean, A. 2003. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 23: 8099–8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M. and Duboule, D. 2003. Organizing axes in time and space: 25 years of colinear tinkering. Science 301: 331–333. [DOI] [PubMed] [Google Scholar]
- Kmita M., van der Hoeven, F., Zakany, J., Krumlauf, R., and Duboule, D. 2000. Mechanisms of Hox gene colinearity: Transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes & Dev. 14: 198–211. [PMC free article] [PubMed] [Google Scholar]
- Kondo T. and Duboule, D. 1999. Breaking colinearity in the mouse HoxD complex. Cell 97: 407–417. [DOI] [PubMed] [Google Scholar]
- Kondo T., Zakany, J., and Duboule, D. 1998. Control of colinearity in AdbB genes of the mouse HoxD complex. Mol. Cell 1: 289–300. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. 1994. Hox genes in vertebrate development. Cell 78: 191–201. [DOI] [PubMed] [Google Scholar]
- Kurz A., Lampel, S., Nickolenko, J.E., Bradl, J., Benner, A., Zirbel, R.M., Cremer, T., and Lichter, P. 1996. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell. Biol. 135: 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O'Sullivan, R.J., and Jenuwein, T. 2003. An epigenetic road map for histone lysine methylation. J. Cell. Sci. 116: 2117–2124. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson, M., Gaszner, M., Allis, C.D., and Felsenfeld, G. 2001. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293: 2453–2455. [DOI] [PubMed] [Google Scholar]
- Maconochie M.K., Nonchev, S., Studer, M., Chan, S.K., Popperl, H., Sham, M.H., Mann, R.S., and Krumlauf, R. 1997. Cross-regulation in the mouse HoxB complex: The expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes & Dev. 11: 1885–1895. [DOI] [PubMed] [Google Scholar]
- Mahy N.L., Perry, P.E., and Bickmore, W.A. 2002a. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell. Biol. 159: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy N.L., Perry, P.E., Gilchrist, S., Baldock, R.A., and Bickmore, W.A. 2002b. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell. Biol 157: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H., Studer, M., Popperl, H., Aparicio, S., Kuroiwa, A., Brenner, S., and Krumlauf, R. 1994. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 370: 567–571. [DOI] [PubMed] [Google Scholar]
- Muller W.G., Walker, D., Hager, G.L., and McNally, J.G. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell. Biol. 154: 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye A.C., Rajendran, R.R., Stenoien, D.L., Mancini, M.A., Katzenellenbogen, B.S., and Belmont, A.S. 2002. Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 22: 3437–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveen T., Niederreither, K., Dolle, P., Chambon, P., Meijlink, F., and Deschamps, J. 2003. Retinoids regulate the anterior expression boundaries of 5′ Hoxb genes in posterior hindbrain. EMBO J. 22: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalopulu N., Lovell-Badge, R., and Krumlauf, R. 1991. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucl. Acids Res. 19: 5497–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popperl H., Bienz, M., Studer, M., Chan, S.K., Aparicio, S., Brenner, S., Mann, R.S., and Krumlauf, R. 1995. Segmental expression of Hoxb-1 is controlled by a highly conserved auto-regulatory loop dependent upon exd/pbx. Cell 81: 1031–1042. [DOI] [PubMed] [Google Scholar]
- Ragoczy T., Telling, A., Sawado, T., Groudine, M., and Kosak, S.T. 2003. A genetic analysis of chromosome territory looping: Diverse roles for distal regulatory elements. Chrom. Res. 11: 513–525. [DOI] [PubMed] [Google Scholar]
- Roelen B.A.J., de Graaff, W., Forlani, S., and Deschamps, J. 2002. Hox cluster polarity in early transcriptional availability: A high order regularity level of clustered Hox genes in the mouse. Mech. Develop. 119: 81–90. [DOI] [PubMed] [Google Scholar]
- Sachs R.K., Van den Engh, G., Trask, B., Yokota, H., and Hearst, J.E. 1995. A random-walk/giant-loop model for interphase chromosomes. Proc. Natl. Acad. Sci. 92: 2710–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Francastel, C., Cimbora, D.M., Reik, A., Martin, D.I., and Groudine, M. 2000. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes & Dev. 14: 940–950. [PMC free article] [PubMed] [Google Scholar]
- Simeone A., Acampora, D., Arcioni, L., Andrews, P.W., Boncinelli, E., and Mavilio, F. 1990. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346: 763–766. [DOI] [PubMed] [Google Scholar]
- Simeone A., Acampora, D., Nigro, V., Faiella, A., D'Esposito, M., Stornaiuolo, A., Mavilio, F., and Boncinelli, E. 1991. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech. Develop. 33: 215–227. [DOI] [PubMed] [Google Scholar]
- Studer M., Gavalas, A., Marshall, H., Ariza-McNaughton, L., Rijli, F.M., Chambon, P., and Krumlauf, R. 1998. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125: 1025–1036. [DOI] [PubMed] [Google Scholar]
- Tse C., Sera, T., Wolffe, A.P., and Hansen, J.C. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18: 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Hashiguchi, N., Janicki, S.M., Tumbar, T., Belmont, A.S., and Spector, D.L. 2000. Visualization of gene activity in living cells. Nat. Cell. Biol. 2: 871–878. [DOI] [PubMed] [Google Scholar]
- Tumbar T., Sudlow, G., and Belmont, A.S. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell. Biol. 145: 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Engh G., Sachs, R., and Trask, B.J. 1992. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk. Science 257: 1410–1412. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven F., Zakany, J., and Duboule, D. 1996. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell 85: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman, P.D., Kobayashi, R., and Stillman, B. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87: 95–104. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret, E., Jones, T., Vatcheva, R., Williamson, J., Beck, S., Campbell, R.D., Goldsworthy, M., Powis, S.H., Ragoussis, J., et al. 2000. Large-scale chromatin organisation of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell. Sci. 113: 1565–1576. [DOI] [PubMed] [Google Scholar]
- Wang X., He, C., Moore, S.C., and Ausio, J. 2001. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 276: 12764–12768. [DOI] [PubMed] [Google Scholar]
- Williams R.R.E., Broad, S., Sheer, D., and Ragoussis, J. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell. Res. 272: 163–175. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Hayes, J.J. 1999. Chromatin disruption and modification. Nucl. Acids Res. 27: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Hu, Y.F., Zhong, H., Nye, A.C., Belmont, A.S., and Li, R. 2001. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell. Biol. 155: 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota H., Singer, M.J., van den Engh, G.J., and Trask, B.J. 1997. Regional differences in the compaction of chromatin in human G0/G1 interphase nuclei. Chrom. Res. 5: 157–166. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Reinberg, D. 2001. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 15: 2343–2360. [DOI] [PubMed] [Google Scholar]