Abstract
To gain insight into the function and organization of proteins assembled on the DNA in response to genotoxic insult we investigated the phosphorylation of the Schizosaccharomyces pombe PCNA-like checkpoint protein Rad9. C-terminal T412/S423 phosphorylation of Rad9 by Rad3ATR occurs in S phase without replication stress. Rad3ATR and Tel1ATM phosphorylate these same residues, plus additional ones, in response to DNA damage. In S phase and after damage, only Rad9 phosphorylated on T412/S423, but not unphosphorylated Rad9, associates with a two-BRCT-domain region of the essential Rad4TOPBP1 protein. Rad9–Rad4TOPBP1 interaction is required to activate the Chk1 damage checkpoint but not the Cds1 replication checkpoint. When the Rad9-T412/S423 are phosphorylated, Rad4TOPBP1 coprecipitates with Rad3ATR, suggesting that phosphorylation coordinates formation of an active checkpoint complex.
Keywords: Checkpoint, DNA replication, fission yeast, DNA damage, ATR
In response to DNA damage, eukaryotic cells orchestrate a complex network of DNA damage responses (DDRs). These result in transient cell cycle arrest and repair of the damaged DNA (Zhou and Elledge 2000; Carr 2002). In multicellular eukaryotes, DDRs also interface with the apoptotic and senescence pathways to ensure that specific cell types that receive high levels of damage are removed from the cycling population (Wahl and Carr 2001). Failure of DDRs underlies many cancer-prone human genetic diseases, and mutations in DDR proteins are common events in the etiology of sporadic cancers.
Many of the DDRs require the activation of the ATR- and ATM-dependent DNA structure checkpoint pathways. Activation of the ATR and ATM kinases promotes a cascade of phosphorylation events. Among these are phosphorylation and activation of two downstream kinases Chk1 and Chk2 (Shiloh 2003). The phosphorylation of target proteins by ATM, ATR, Chk1, and Chk2 results in the regulation of transcription, changes in the profiles of protein stability, and changes to the subcellular localization of certain proteins (Liu et al. 2003; Shiloh 2003; Yao et al. 2003). Several target proteins have been identified, and in some instances, individual phosphorylation events have been ascribed specific functions. For example, phosphorylation of p53 and its E3 ubiquitin ligase Mdm2 influences the stability and function of p53, an important effector of checkpoint signals (Chene 2003). Because checkpoint pathways are vital for the maintenance of genomic stability and the suppression of carcinogenesis, many studies have aimed to understand the molecular mechanisms underpinning checkpoint pathway activation. Two distinct DNA-structure-responsive checkpoint pathways have been characterized.
The ATM-dependent checkpoint responds directly to DNA double-strand breaks (DSBs). Activation of ATM requires the presence of the Mre11–Rad50–Xrs2NBS1 complex (MRX), and studies in budding yeast demonstrate that the MRX complex binds to DNA double-strand ends at the site of damage and recruits the ATM homolog Tel1 (Nakada et al. 2003). In human cells, Nbs1 is required for ATM-dependent phosphorylation events in response to DNA damage (Girard et al. 2002; Uziel et al. 2003). ATM phosphorylates MRX in response to DNA damage and also targets a histone H2A variant, several checkpoint mediator proteins (including BRCA1, TOPBP1, P53BP1, and MDC1) and activates Chk2 (Shiloh 2003).
The ATR-dependent checkpoint responds to a variety of genotoxic insults, including UV-induced dimers and agents that stall DNA replication. ATR forms a stable protein complex with ATRIP, and this subunit is required to bind ATR–ATRIP to single-stranded DNA (ssDNA; Zou and Elledge 2003). In fact, ATR–ATRIP only associates with ssDNA when it is complexed with the single-strand-binding protein RPA. The association of ATR–ATRIP with regions of ssDNA-RPA apparently allows cells to respond to a wide variety of DNA lesions. However, these must first be processed by DNA repair enzymes to produce ssDNA (Carr 2003). Similarly, it is thought that stalled DNA replication results in increased quantities of ssDNA coated with RPA, and that this activates ATR (Sogo et al. 2002).
To activate the ATR-dependent checkpoint outside of DNA replication, sites of DNA damage must also recruit other checkpoint proteins. Rad17 is a checkpoint protein that shares homology to the five subunits of replication factor C (RFC). A discrete RFC-like checkpoint complex has been identified (Green et al. 2000) in which Rad17 replaces the large subunit of RFC. In much the same way that replicative RFC acts as a clamp loader for PCNA during DNA replication, the Rad17-Rfc[2–5] checkpoint clamp loader is required for the association of the heterotrimeric PCNA-like Rad9–Rad1–Hus1 (9–1–1) complex onto damaged chromatin in cells not undergoing DNA replication (Zou et al. 2003). Interestingly, the association of the PCNA-like 9–1–1 complex with DNA damage occurs independently of ATR–ATRIP association. In order for signals to be propagated from ATR to the downstream checkpoint kinase Chk1, mediator proteins such as BRCA1 are also required. In fission yeast, Crb2 is a BRCT-domain DNA damage checkpoint mediator protein that shares homology with Saccharomyces cerevisiae Rad9 and is thought to be a paralog of the human BRCT-domain checkpoint mediator proteins P53BP1 and BRCA1 (Saka et al. 1997). In fission yeast, Crb2 is recruited to sites of damage independently of Rad3ATR and Rad17 (Du et al. 2003). Because all of these components are required for Chk1 activation in response to DNA damage, checkpoint activation apparently requires the concomitant independent recruitment of ATR–ATRIP, the PCNA-like 9–1–1 complex, and the BRCT-domain mediator protein(s).
Less is known about how the ATR-dependent checkpoint is activated by stalled DNA replication. It is likely that ATR–ATRIP associates with the extended regions of ssDNA-RPA formed when replication stalls. However, the recruitment of the PCNA-like 9–1–1 complex is only implied by the fact that these proteins are required for intra-S-phase checkpoint signaling in fission yeast and mammalian cells. In extracts of Xenopus oocytes that can support DNA replication, it has been demonstrated that ATR and Rad9 associate with the replication complex during its assembly and with chromatin during normal DNA replication (You et al. 2002). This suggests that ATR and the 9–1–1 complex travel with the unperturbed fork. However, in S. cerevisiae, the ATR and Rad9 homologs have not been detected at sites of ongoing DNA replication (Katou et al. 2003), suggesting that they do not travel with the replication fork. Following replication fork arrest, however, these proteins are seen associated with the stalled replication fork.
We have been intrigued by the fact that most of the checkpoint proteins are phosphorylated by ATR during the establishment of the checkpoint. In the fission yeast Schizosaccharomyces pombe and in other organisms, some of these, such as Rad26ATRIP, are differentially phosphorylated in response to either DNA damage of G2 cells or replication fork stalling by hydroxyurea (HU) treatment (Edwards et al. 1999). One protein that is phosphorylated in response to DNA damage in all organisms where it has been studied is Rad9, one of the PCNA-like checkpoint proteins. Initially, to detect S. pombe Rad9, we modified the genomic rad9+ locus to encode three HA epitopes (Caspari et al. 2000a). The rad9-HA strain was equivalent to wild-type cells in response to UV and ionizing irradiation and resistant to HU treatment. However, during later genetic analysis, we noted that cds1-d mutant cells containing the rad9-HA allele displayed increased sensitivity to HU when compared with cds1-d rad9+ cells (data not shown). This sensitivity correlated with an increase in aberrant mitosis, suggesting that the C terminus of Rad9 may play a specific role in checkpoint activation. In this study, we describe the analysis of Rad9 phosphorylation and demonstrate that Rad9 C-terminal phosphorylation allows Rad9 to interact with the TOPBP1 homolog Rad4 (also known as Cut5). Detailed examination of Rad9 phosphorylation and the phenotypes of rad9 phosphorylation-site mutants indicates that cells use differential phosphorylation events to distinguish the context in which ssDNA-RPA complexes are detected, presumably allowing the selection of the appropriate repair mechanisms.
Results
Rad9, in common with many checkpoint proteins, is phosphorylated in response to DNA damage in a rad3ATR-dependent manner (Caspari et al. 2000b). Rad3 is a member of the PI3 kinase-like protein kinase family (PIKK) and these unusual protein kinases favor the phosphorylation at TQ and SQ motifs (Kim et al. 1999). S. pombe Rad9 contains five T/SQ sites (Fig. 1A).
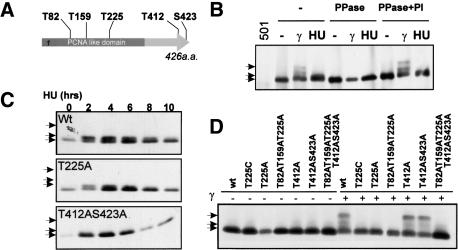
Figure 1.
Rad9 is phosphorylated differently after HU or γ-irradiation. (A) Schematic of the five S/TQ sites in Rad9. (B) Rad9-3HA was immunoprecipitated (α-HA) from extracts of HU (20 mM for 3 h) treated or γ-irradiated (500 Gy) cells. IPs were untreated (–), treated with phosphatase (PPase), or with phosphatase plus inhibitor (PPase + PI). (501) Untagged strain. (C) Extracts of HU (20 mM) treated wild-type or mutant rad9-HA cells probed with α-HA. HU-induced bandshift is dependent on T412 and S423 but not on T225. (D) Extracts of γ-irradiated rad9-HA cells (500 Gy) probed with α-HA. Hyperband-shift is dependent on T225 but not on T412 or S423. (B–D) Arrows (top) 225-dependent hypershift; (middle) 412/423-dependent shift; (bottom) basal. The T225C as well as the T225A mutant was used because the T225C mutant has a less severe phenotype than the T225A mutant, which might be because of structural abnormality in T225A.
Rad9 is differentially phosphorylated in response to DNA damage and S-phase arrest
In response to DNA damage by ionizing radiation (IR), Rad9 shows a significant mobility shift upon SDS-PAGE (Fig. 1B; Caspari et al. 2000b). However, in response to S-phase arrest by HU, Rad9 displays a distinct and less dramatic mobility shift (Fig. 1B). Both events are reversed by phosphatase (Fig. 1B) and are dependent on Rad3ATR (see Fig. 2C; Caspari et al. 2000b). We thus mutated the five PI3-like protein kinase consensus sites (S/TQ) to alanine and monitored the shifts in response to both treatments. Following incubation of cells for 2 h in HU, the small Rad9 mobility shift was seen to be dependent on both T412 and S423, both residing at the C terminus of Rad9 (Fig. 1C). The more pronounced mobility shift in response to IR was found only to be dependent on T225, which resides within the PCNA-like domain (Fig. 1D).
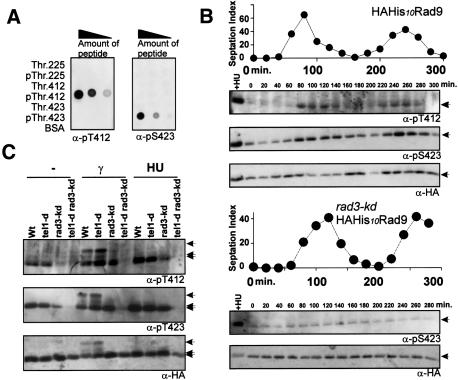
Figure 2.
The Rad9 C terminus (T412, S423) is phosphorylated during unperturbed S phase by Rad3(A) Antiphospho-T412 (α-pS412) or phospho-S423 (α-pS423) dot-blotted against dilutions of both unphosphorylated peptide and phosphopeptide (2.5-fold serial dilutions from 100 ng of peptides). (B) HAH10-rad9 (top) and HAH10-rad9 rad3-kd (bottom) cells synchronized in G2 and sampled every 20 min. Graph: septation index. Blots: Antibodies indicated. HU-treated extract (+HU) loaded as positive control. Phospho-T412 and phospho-S423 accumulated from 80 to 120 min and from 220 to 260 min, which corresponds to S phase. Note that S. pombe G1 is short; thus, septation index peak corresponds to S phase. (Arrow) Rad9p. (C) rad+ (wt), tel1 deleted (tel1-d), rad3 kinase dead (rad3-kd), and tel1-d rad3-kd cells treated with 20 mM HU for 3 h or 500 Gy of γ-irradiation. (B,C) HAH10-Rad9 pulled down by TALON beads from denatured extracts and probed for Rad9 phosphorylation. (Arrows, top) 225-dependent hypershift; (middle) 412/423-dependent shift; (bottom) basal.
To determine the phosphorylation state of these residues directly, we raised antibodies to phospho-peptides corresponding to each of the three sites (Fig. 2A). We were successful in raising phospho-specific antibodies for T412 and S423, but not for T225. Using the T412 and S423 phospho-specific antibodies as probes, we analyzed Rad9 C-terminal phosphorylation through the cell cycle (Fig. 2B). This revealed that Rad3ATR promotes phosphorylation of both sites during unperturbed S phase and that, in the absence of Rad3ATR activity, a basal level of phosphorylation was present throughout the cell cycle. Concomitant loss of both Rad3ATR and Tel1ATM resulted in the complete loss of signal detected by either antibody (Fig. 2C), suggesting that the basal level of phosphorylation observed was dependent on Tel1ATM. We next examined the C-terminal phosphorylation of Rad9 in response to HU treatment (Fig. 2C). We observed that C-terminally phosphorylated Rad9 accumulated as cells arrested in S phase. We cannot discern if this is indirect, caused by the accumulation of cells in S phase, a direct consequence of checkpoint activation, or a combination of both possibilities. Finally, we observed that, in response to IR, Rad3ATR and Tel1ATM both promoted C-terminal Rad9 phosphorylation (Fig. 2C). In addition, IR treatment also caused the Rad3- and T225-dependent mobility shift. It is clear that this shifted material contains Rad9 phosphorylated on the T412 and S423 C-terminal residues (Fig. 2C).
The phenotypes associated with phosphorylation-site mutants
To determine the function of Rad9-412/423 (T412/S423) phosphorylation, we examined the phenotype of C-terminal phosphorylation-site mutants. rad9-T412A cells showed IR sensitivity equivalent to chk1-d cells (Fig. 3A), and the double mutant showed very little increase in sensitivity compared with the two single mutants. We have also extensively characterized rad9-S423A (which showed milder sensitivity than rad9-T412A) plus rad9-T412C and rad9-T412AS423A mutant cells (Fig. 3; data not shown). The latter both consistently showed characteristics equivalent to rad9-T412 cells. This indicates that the two C-terminal phosphorylation sites affect the same process and are both required. To examine the DNA damage checkpoint response, we irradiated wild-type, rad9-T412A, and chk1-d cells synchronized in G2. Like chk1 mutants, rad9-T412A cells were G2 DNA-damage-checkpoint defective (Fig. 3B). Consistent with this, we were unable to detect phosphorylation of Chk1 (as judged by Chk1 mobility shift assays; Walworth and Bernards 1996) when rad9-T412A cells were irradiated with IR (Fig. 3C). In contrast, rad9-T412A cells were only mildly sensitive to HU (Fig. 3A), arrested mitosis in response to HU treatment for a considerable period of time (Fig. 3B), and were able to activate Cds1 kinase to an extent similar to that seen in rad9+ cells (Fig. 3C). Taken together, these data suggest that Rad9 C-terminal phosphorylation is specifically required for the activation of Chk1. This requirement does not arise from disruption of the 9–1–1 complex itself because we can coimmunoprecipitate Rad9-T412A-HA protein with α-Rad1 antibody as effectively as we coimmunoprecipitate Rad9-HA (data not shown).
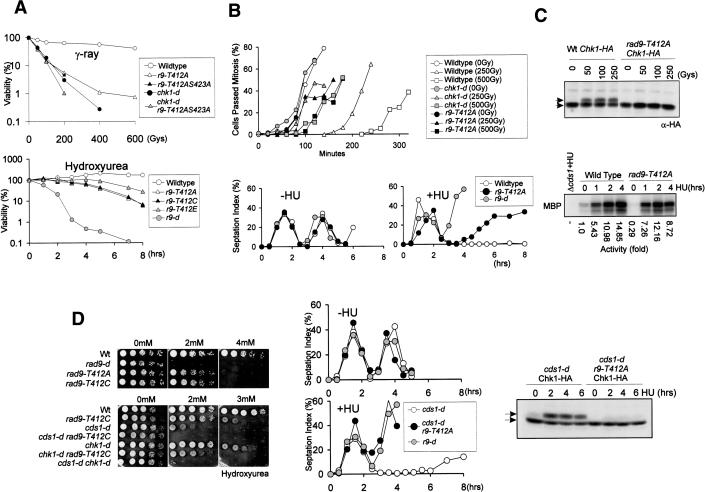
Figure 3.
Thr 412 and Ser 423 are required for checkpoint arrest after γ-irradiation and replication fork collapse but not after replication fork stall. (A, top) γ-Ray sensitivity. Cells are plated after irradiation of indicated dose of γ-ray. rad9-T412A (r9-T412A), rad9-T412AS423A (r9-T412AS423A), and chk1 deleted (chk1-d) cells show similar sensitivities and rad9-T412A chk1-d double-mutant cells showed little increase in sensitivity. (Bottom) HU sensitivity of specified mutants. Cells are plated after incubation with 20 mM HU for the hours shown. rad9-T412A and rad9-T412C cells lose their viability more slowly than rad9 deleted (r9-d) cells. Phosphomimic mutant rad9-T412E (r9-T412E) cells died more slowly than rad9-T412A cells. (B) Checkpoint analysis for G2 synchronized rad9+ (Wildtype), rad9-d, rad9-T412A, and chk1-d cells. Cells are synchronized in G2 by lactose gradients and γ-irradiated (top) or HU treated (bottom). (C) rad9-T412A cells are deficient in Chk1 phosphorylation (top) after γ-irradiation (extracts probed with α-HA) but proficient in activation of Cds1 (bottom) after HU exposure (Cds1 immunoprecipitated from extracts and in vitro activity measured using MBP; Lindsay et al. 1998). (D) Activation of Chk1 after replication fork collapse is defective in rad9-T412A cells. (Left panel) Sensitivity of rad9+ (wt) rad9-d, rad9-T412A, and rad9-T412C cells to continuous exposure to low dose HU. The rad9-T412C mutant shows the same HU sensitivity as rad9-T412A and is used for epistasis analysis with chk1-d and cds1-d. (Middle panel) Checkpoint analysis of cds1-d, rad9-d, and cds1-d rad9-T412A cells upon HU exposure, details as in B. Whereas cds1-d is arrested in S/G2 phase, the cds1-d r9-T412A cells prematurely entered into mitosis as did the checkpoint-deficient rad9-d cells. “Cut” phenotypes are seen in these mutants (data not shown). (Right panel) Extracts from cds1-d chk1-HA and cds1-d chk1-HA rad9-T412A cells after HU treatment for the indicated times were probed for Chk1 (α-HA). (C,D, top arrow) Phospo-Chk1-HA.
In addition to activation of Chk1 in response to DNA damage in G2, we and others have also found that Chk1 is activated in response to replication problems induced by HU in the absence of functional Cds1 kinase (Lindsay et al. 1998). The accepted explanation for this is that Cds1 activation is required to stabilize replication forks that are arrested by HU or by DNA damage. In the absence of Cds1 activity, stalled replication proteins dissociate from the sites of DNA replication (often referred to as “fork collapse”) and that this results in the accumulation of potentially lethal DNA structures that activate the Rad3ATR–Chk1 checkpoint. This phenomenon has been observed by two-dimensional DNA electrophoresis (Sogo et al. 2002; data not shown). To establish if the rad9 C-terminal phosphorylation site mutants were also defective in activating Chk1 in response to fork collapse, we created double mutants between cds1-d and either rad9-T412A or rad9-T412C and assayed sensitivity to HU by monitoring the ability to form colonies in the presence of low concentrations of HU. As previously seen for chk1 mutants (Lindsay et al. 1998), rad9 C-terminal phosphorylation-site mutants showed synergistic sensitivity with cds1 deletion (Fig. 3D; data not shown). This correlated with an inability to arrest mitosis (Fig. 3D) and loss of the Chk1 mobility shift (Fig. 3D), which is evident in cds1-d rad9+ cells after treatment with HU (Lindsay et al. 1998).
A distinction between the DNA-damage checkpoint and the response to fork collapse
In response to replication fork collapse (HU treatment of cds1-d cells), we observed that Rad9 displays a mobility shift indistinguishable to that seen when G2 cells are irradiated with IR (Fig. 4A). Like the IR-induced shift, the fork collapse-induced shift is dependent on the Rad9 T225 residue, because it is not seen in mutants where this site is mutated to a nonphosphorylatable residue (Fig. 4A; data not shown). However, unlike the IR-induced shift (Fig. 1D), the fork collapse-induced shift is dependent on the C-terminal phosphorylation sites T412 (Fig. 4A) and S423 (data not shown). One interpretation of this is that, in the context of replication fork collapse, the Rad9 C terminus must have previously been phosphorylated when cells enter S phase before further phosphorylation (of T225) and the subsequent mobility shift can occur. A similar observation was made when cells were treated with camptothecin (Fig. 4E), which is also proposed to induce replication fork collapse. This contrasts with G2 cells irradiated with IR, where Rad9 C-terminal phosphorylation is not a pre-requisite for further Rad9 T225-dependent mobility shift (Fig. 1D). We have shown that, in response both to IR and to fork collapse, C-terminal Rad9 phosphorylation is required for the phosphorylation of Chk1 and for the subsequent cell cycle arrest. We thus sought to ascertain if there were further distinct requirements for the fork collapse-induced T225-dependent mobility shift when compared with the shift induced by IR. We tested cells containing the crb2-d, chk1-d, and tel1-d deletion mutants and cells containing the temperature-sensitive rad4 allele cut5-T401 (Fig. 4B,C; data not shown). Only the cells harboring the cut5-T401 allele (at 30°C, a semipermissive temperature) failed to show the Rad9 T225-dependent shift when treated with HU in the absence of cds1. Previously, we have demonstrated that rad4ts mutants at the restrictive temperature of 36°C still exhibit the T225-dependent mobility shift of Rad9 after IR treatment but that Chk1 is not phosphorylated (Harris et al. 2003).
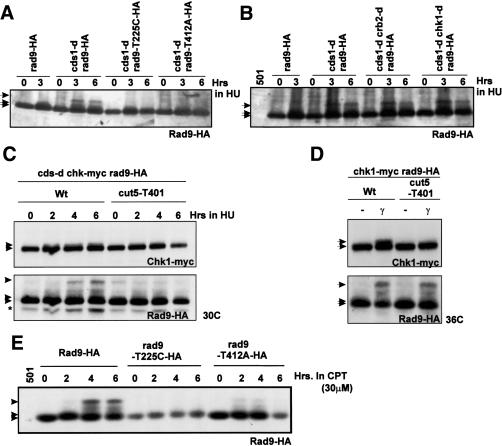
Figure 4.
A distinct damage response after replication fork collapse compared with G2. (A–C) Protein extracts from rad9-3HA cells or cells harboring a mutant version of rad9-3HA probed with α-HA. The strains shown were incubated with 20 mM HU for the indicated times at 30°C and compared for the T225-dependent hyper-shift that occurs in the absence of cds1. (501) Untagged strain. (D) rad4+ (cut5+) (wt) and rad4 (cut5-T401) mutant cells carrying the rad9-3HA allele were γ-irradiated (500 Gy) at 36°C. Extracts were prepared immediately and probed for Rad9-HA. Chk1-myc is also genomically expressed in C and D. (E) Cells carrying rad9-3HA or mutant versions of rad9-3HA were incubated with 30 μM of camptothecin (CPT) for the indicated times at 30°C. Extracts were probed for Rad9-HA. (Asterisk) Nonspecific band. (A–E) Rad9 arrows indicate 225-dependent hypershift (top); 412/423-dependent bandshift (middle); basal (bottom). (C,D) Chk1 arrows indicate phosphorylated (top) and basal (bottom).
Because 30°C represents a semipermissive temperature for cut5-T401 (Harris et al. 2003) and because we had noticed that the fork collapse-induced Chk1 phosphorylation was reduced at this temperature (Fig. 4C), we confirmed that, for the rad4 allele cut5-T401, Rad9 phosphorylation after IR was present but Chk1 phosphorylation was absent when treated at the restrictive temperature of 36°C (Fig. 4D) as previously published. The presence of T225 supershift in crb2-d and chk1-d cells after replication fork collapse demonstrates that the absence of T225 supershift in the rad4 mutant (cut5-T401) is not due to the cell cycle arrest defect evident at this temperature (Harris et al. 2003).
Rad9 C-terminal phosphorylation promotes association with Rad4TOPBP1
Because Rad4TOPBP1 (Cut5) interacts with Crb2 and Chk1, our data lead us to predict that C-terminal phosphorylation of Rad9 serves to recruit Rad4TOPBP1, bringing Rad4, Crb2, and Chk1 into proximity with Rad3ATR, and thus allowing propagation of the checkpoint signal. This would also be consistent with previous data that identified a potential interaction between the S. cerevisiae Rad9 homolog Ddc1 and the Rad4TOPBP1 homolog Dpb11 and the interaction between the human homologs (Makiniemi et al. 2001; Wang and Elledge 2002).
To test this, we created an N-terminal TAP-tagged rad4 allele (TAP-rad4) and used the protein A domains to precipitate Rad4TOPBP1 from soluble extract. Coprecipitation of Rad9 was monitored using the α-HA antibody. We found that we could coprecipitate Rad9 only in circumstances in which we had previously shown that Rad9 was C-terminally phosphorylated, such as after treatment with IR or following HU arrest and during unperturbed S phase. In addition, we could not coprecipitate Rad9 protein from extracts made from rad9-T412A mutant cells or when rad3 and tel1 were mutated (Fig. 5A). Consistent with a phosphorylation-dependent interaction between Rad4TOPBP1 and Rad9, we were able to disrupt the interaction by phosphatase (PPase) treatment (Fig. 5B). The modest release of Rad9 from Rad4 precipitates after PPase treatment most likely reflects the short incubation time and low temperature that were necessary to avoid protein degradation. Alternatively, it may reflect lack of accessibility of PPase to the phosphorylated residue, or a mixture of both. To further explore the interaction, we followed the profiles of Rad4TOPBP1 and Rad9 upon size exclusion chromatography following preparation from logarithmically growing cells or cells arrested in S phase with HU (Fig. 5C). A small proportion of Rad9 prepared from rad9+ cells displayed a shift in size after checkpoint activation, cofractionating with Rad4TOPBP1. This effect was largely lacking in extracts from rad9-T412A mutant cells, consistent with the formation of a complex between Rad4TOPBP1 and Rad9 that is dependent on C-terminal phosphorylation of Rad9.
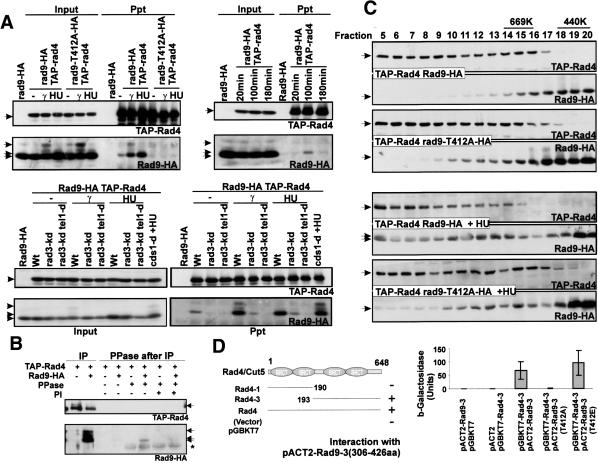
Figure 5.
Rad9 interacts with Rad4(Cut5) in a phosphorylation-dependent manner. (A) TAP-Rad4(Cut5) was immunoprecipitated from extracts of rad9-HA TAP-rad4 cells. Coprecipitation of Rad9-HA is detected using α-HA. (Top left) Rad9-HA but not Rad9-T412A-HA coprecipitates with TAP-Rad4(Cut5) after HU or γ-ray treatment. Cells were incubated with or without 20 mM HU for 4 h or treated with 500 Gy of γ-radiation. (Top right) Rad9-HA coprecipitates with TAP-Rad4(Cut5) in unperturbed S phase. Cells were synchronized in G2 and allowed to progress into S phase. Extracts were prepared from G2 (20 min), S phase (100 min), and the following G2 (180 min). (Bottom) The Rad9–Rad4 interaction is dependent on Rad3 and Tel1. TAP-rad4 rad9-HA strains containing the rad3 kinase dead allele (rad3-kd), the rad3-kd and the tel1 deletion (rad3-kd tel1-d), or the cds1-d allele were treated with γ-radiation (500 Gy) or 20 mM HU (4 h). (B) Rad9-HA association with TAP-Rad4 in phosphorylation-dependent manner. Immunoprecipitated Rad4–Rad9 complexes from the extract of γ-treated cells are either untreated, treated with CIP (PPase), or treated with PPase in presence of phosphatase inhibitor (PI) at 30°C for 15 min. Arrows represent Rad9-isoforms, and the asterisk represents α-HA cross-reaction with phosphatase. (C) Rad4(Cut5) changes slightly, but Rad9 drastically shifts to a high-molecular-weight complex after HU treatment (20 mM, 3 h) in a T412-dependent manner during gel filtration on a Superose 6 column. Rad9-HA is also observed in fractions 5–8, and this is much reduced in rad9-T412A mutant extracts. (D) Schematic of Rad4(Cut5) fragments used for two-hybrid (numbers are amino acids). (+) Strong blue color on plate assay. (–) Negative result. The bar chart quantifies a liquid β-gal ONPG assay. The Rad9 C terminus (amino acids 306–426) interacted with the Rad4 C terminus. The interaction was disrupted by the rad9-T412A mutation and enhanced in a phosphomimic rad9-T412E mutant.
To identify the region of Rad4TOPBP1 that interacted with Rad9 C terminus in a phospho-specific manner, we cloned a fragment encoding the C terminus of Rad9, fulllength rad4, and several subfragments of rad4 into two hybrid vectors and examined the interactions of the encoded products by the β-galactosidase activity induced when these were cotransfected into S. cerevisiae. In this way, we identified an interaction between the Rad9 C terminus and a fragment containing the two C-terminal BRCT domains of Rad4TOPBP1. Mutation of the codon encoding Rad9-T412 to encode alanine resulted in the interaction being lost. Mutation of the same codon to encode glutamic acid, which introduces a charged residue thought to mimic phosphorylation, resulted in an increase in β-galactosidase activity, indicating that the interaction was more robust (Fig. 5D).
An interaction between Rad4TOPBP1 and Rad3
Because Rad4TOPBP1 appeared to associate with Rad9 in a phosphorylation-specific way, and because this interaction correlated with Rad3ATR-dependent phosphorylation events, we examined whether Rad4TOPBP1 could coprecipitate Rad3ATR. We observed an increase in the TAP-Rad4-dependent coprecipitation of Myc-Rad3 in response to DNA damage or S-phase arrest in extracts from rad9+ cells that was not observed from extracts prepared from cells harboring the rad9-T412A mutation (Fig. 6A). This suggests that an interaction between Rad4TOPBP1 and Rad3ATR occurs that is dependent on the C-terminal phosphorylation of Rad9. One interpretation of these data is that Rad3ATR-dependent phosphorylation of Rad9 T412 and S423 residues promotes the formation of a Rad3ATR–Rad4TOPBP1–Rad9 protein complex, thus linking active Rad3 to downstream substrates. Consistent with this, we observe that phosphorylation of Rad4TOPBP1 and of Crb2 are dependent on the C-terminal phosphorylation sites of Rad9 (Fig. 6B).
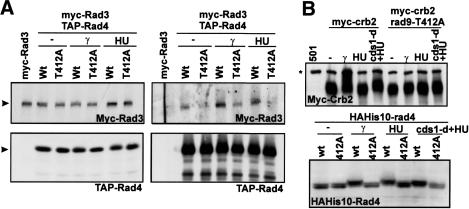
Figure 6.
Rad3 associates with Rad4 after DNA damage in rad9-T412 dependent manner. (A) Rad3 co-IPs with Rad4(Cut5) in a rad9-T412-dependent manner. myc6-rad3 TAP-rad4 strains with rad9+ or rad9-T412A backgrounds were untreated, incubated with 20 mM HU for 4 h, or treated with 500 Gy of γ-radiation. TAP-Rad4 precipitated from extracts with IgG beads, Myc6-Rad3 was detected with α-Myc monoclonal (9E10). (B) Crb2 (top) and Rad4 (bottom) bandshifts were dependent on rad9-T412 after HU, cds1-d + HU, or γ-irradiation (500 Gy) treatment. Extracts from myc8-crb2, myc8-crb2 rad9-T412A, HAH10-rad4, or HAH10-rad4 rad9-T412A are blotted by either α-myc (9E10) or α-HA (16b12) antibody. (Asterisk on Crb2 blot) α-myc cross-reacted band.
Discussion
It has previously been reported that ATR–ATRIP complexes in human and yeast cells associate with DNA damage sites in a manner independent of the 9–1–1 complex and that the 9–1–1 complex similarly associates with sites of DNA damage independently of ATR–ATRIP (Melo et al. 2001; Zou et al. 2003). A recent report in fission yeast further suggests that the mediator protein Crb2 (potentially an analog of P53BP1 and BRCA1) can also associate with sites of DNA damage, and that this is independent of both Rad3ATR and of the 9–1–1 complex (Du et al. 2003). Thus, several distinct protein complexes may be able to independently bind to damaged chromatin. Previously, we and others have demonstrated that successful transmission of the checkpoint signal to Chk1 requires the presence of all three of these protein complexes (Caspari and Carr 1999). From studies of the individual phosphorylation events, it is also known that Rad3ATR can phosphorylate its partner Rad26ATRIP independently of other checkpoint components (Edwards et al. 1999) and that the Rad3ATR–Rad26ATRIP complex can phosphorylate 9–1–1 components, but not Crb2 or Chk1, in the absence of Rad4TOPBP1 function (Harris et al. 2003). These data suggested a hierarchical organization of the checkpoint.
Two BRCT domains in Rad4TOPBP1 define a phospho-specific protein interaction
In this report, we provide evidence that Rad3ATR (and to a lesser extent Tel1ATM) must phosphorylate the C terminus of Rad9 to promote association of Rad9 with Rad4TOPBP1. In two-hybrid experiments, this interaction that depends on the C-terminal pair of BRCT domains in Rad4TOPBP1, is lost when Rad9-T412 is mutated to alanine and becomes stronger when the residue is replaced with a charged amino acid. These data strongly indicate a direct interaction between Rad9 and a pair of Rad4TOPBP1 BRCT domains, although it remains formally possible that a heterologous S. cerevisiae protein bridges the interaction. During the preparation of this manuscript, it was reported that tandem C-terminal BRCT domains from human PTIP and BRCA1 directly associate with phospho-SQ peptides but not with unphosphorylated controls (Manke et al. 2003). A separate report demonstrated that the BRCA1 tandem C-terminal domain also associates specifically with a phospho-SP motif of the DEAH-box helicase BACH1 (Yu et al. 2003). Our data are consistent with at least a subset of tandem BRCT domains defining a phospho-specific protein interaction motif. We show here that such a phospho-specific protein interaction underlies the assembly of a functional checkpoint unit following the independent recruitment of checkpoint proteins to the sites of DNA damage. Because the BRCT domains of Rad4TOPBP1, the C-terminal phosphorylation of Rad9, and the interaction between these two proteins are all conserved through evolution (Makiniemi et al. 2001; Wang and Elledge 2002) and the C-terminal phosphorylation of hRad9 has been implicated in checkpoint signaling in mammalian cells using dominant negative constructs (Chen et al. 2001; Roos-Mattjus et al. 2003; St Onge et al. 2003), our data have significant implications for understanding the organization of checkpoint responses in human cells and thus the maintenance of genomic instability and cancer avoidance.
Rad9 C-terminal phosphorylation in response to DNA damage
Following DNA damage in G2 cells the phosphorylation of the C terminus of Rad9 to promote the association of Rad9 with Rad4TOPBP1 correlates with the subsequent ability of Rad3ATR to phosphorylate the Rad4TOPBP1-interacting proteins Crb2 and Chk1 and thus the activation of the DNA damage checkpoint in G2. One interpretation of this would be as follows: C-terminal Rad9 phosphorylation by either Rad3ATR or Tel1ATM upon DNA damage in G2 cells promotes the formation of a Rad9–Rad4TOPBP1 complex that acts as a signal initiation complex. Rad9–Rad4TOPBP1 complex then promotes formation of a stable complex between Rad4TOPBP1 and Rad3, organizing these three discrete protein complexes in such a manner to allow Rad3 to phosphorylate other substrates, including Crb2 and Chk1.
Consistent with such a model, we have shown the coprecipitation of C-terminally phosphorylated Rad9 with Rad4TOPBP1 and the Rad9 C-terminal phosphorylation-dependent coprecipitation of Rad4TOPBP1 with Rad3ATR in response to checkpoint activation. However, we are unable to directly demonstrate formation of a Rad9–Rad4TOPBP1–Rad3ATR complex. We suspect this problem is technical. We are unable to prepare sufficient material to reproducibly show serial coprecipitation. This may reflect the fact that these phosphorylation events occur in the insoluble fraction, associated with damaged chromatin. After cell lysis, a fraction of the proteins remain insoluble, and protein interactions may not remain stable following solubilization.
Rad9 C-terminal phosphorylation and DNA replication
Our data demonstrate that a Rad4TOPBP1–Rad9 complex is formed during normal S phase in S. pombe. The coprecipitation of Rad3ATR with Rad4TOPBP1 during S-phase arrest further suggests that this complex also contains Rad3ATR. By reference to work reported using Xenopus extracts, where both ATR and the PCNA-like checkpoint protein Hus1 were shown to associated with the replication apparatus during the initiation stage of replication (You et al. 2002) and to remain chromatin-associated until replication was completed, we speculate that Rad3 phosphorylates the Rad9 C terminus during the initiation step of replication, and that this results in the binding of Rad9 to Rad4TOPBP1. Such a scenario would predict that Rad9 and Rad3 travel with the replication fork, although the technology used in this study cannot address this point directly. Alternatively, Rad9–Rad4TopBP1 can be recruited transiently onto the stalled or collapsed fork. The recent study in budding yeast showed that the spRad9 homolog Ddc1 is recruited when the replication fork stalls (Katou et al. 2003). However, the ddc1-null mutant in budding yeast does not show a replication checkpoint deficiency, unlike in the fission yeast rad9-null mutant. We further speculate that each organism may have evolved differently, and that these apparent incompatibilities may reflect different aspects of the role of specific checkpoint proteins at replication fork.
What then would be the purpose of the Rad9–Rad4TOPBP1 interaction during unperturbed S phase? It is unlikely to reflect detection of endogenous DNA damage because we do not see phosphorylation of other proteins such as Chk1 or Cds1 or phosphorylation of Rad9-T225. Based on the different requirements for Rad4TOPBP1 for the T225-dependent Rad9 mobility shift seen after IR and after the collapse of replication forks, we propose that the preassembly of a Rad9–Rad4TOPBP1 complex at the initiation of DNA replication might allow cells to determine the context of ATR-activation by the ssDNA-RPA complex and thus determine the appropriate response to either DNA damage in G2 or DNA damage as a result of replication fork collapse.
In Figure 7, we present a model that would account for our data, and that may provide a basis for future experiments. In this model, the initial response to replication fork arrest is Rad3ATR- and Mrc1-dependent activation of Cds1, which stabilizes the fork (Alcasabas et al. 2001; Tanaka and Russell 2001). We have made the assumption that the presence of Mrc1 occludes the possible activation of Chk1 (one way in which this could be achieved would be if Mrc1 physically prevented the association of Crb2 with Rad4TOPBP1). When fork stabilization fails and the replication fork collapses, the presence of a preformed Rad4TOPBP1–Rad9 complex promotes phosphorylation events that include those resulting in the T225-dependent mobility shift of Rad9. In the absence of the preformed complex, Rad3 is unable to phosphorylate Rad9 in this way. From our data, this is the key distinction between DNA damage resulting from IR treatment of G2 cells and DNA damage caused by replication fork collapse. However, we do not know the molecular details underlying how this vital context information is obtained. One possibility is that the presence of the Rad4TOPBP1–Rad9 complex recruits DNA processing enzymes that reveal ssDNA in sufficient quantity to activate Rad3ATR, and that beyond this point the response is equivalent to the response to IR in G2. A second possibility is that Rad4TOPBP1–Rad9 interaction is necessary for recruitment of Rad4TOPBP1 to the damage caused by a collapsed fork, distinguishing this form of damage from that resulting from IR irradiation of G2 cells.
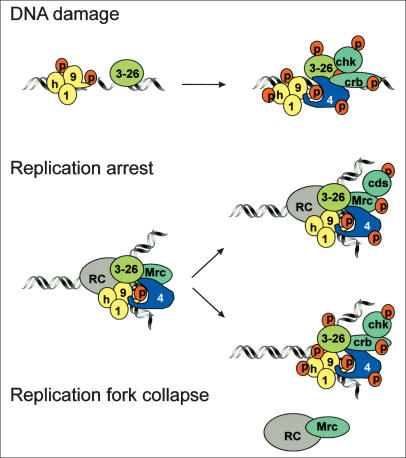
Figure 7.
Model: (Top) After DNA damage, Rad3ATR–Rad26ATRIP (3–26) and Rad9 (9) are independently recruited. Rad3ATR phosphorylates Rad9 on T225 and T412/S423. Rad9-C-terminal T412/S423 phosphorylation promotes association with Rad4TOPBP1 (4), which brings Crb2 and Chk1 into proximity with Rad3ATR. (Middle) Replication fork assembly recruits Rad3ATR–Rad26ATRIP (3–26) and Rad9 (9) to the replication complex (RC). Rad3-dependent phosphorylation of Rad9 C-terminal T412/S423 allows association with Rad4TOPBP1. In S phase, Rad4TOPBP1 may associate with Mrc1 (Mrc). Upon fork arrest by HU, Mrc1 recruits Cds1 into proximity with Rad3ATR allowing it to be phosphorylated and activated (Alcasabas et al. 2001; Tanaka and Russell 2001). At the same time, we propose that Mrc1 binding occludes Crb2 (note Crb2 is not phosphorylated in HU arrest; Fig. 6B). (Bottom) When a replication fork collapses (e.g., after treatment of cds1-d cells with HU or CPT), Rad3ATR phosphorylates T225 on Rad9, and also phosphorylates other proteins such as Rad26ATRIP (Edwards et al. 1999) and Hus1 (Caspari et al. 2000a). The loss of Mrc1 and the replication complex allows the Rad9–Rad4TOPBP1 complex to recruit Crb2 and Chk1 into proximity with Rad3ATR.
Conclusion
Most of the proteins involved in the DNA damage and DNA replication checkpoint have been identified, and the majority are highly conserved through evolution. Many features of the checkpoint pathways remain unexplained, including their apparent complexity and the fact that many of the same proteins participate in both the DNA damage and DNA replication responses. The data presented here begin to uncover the molecular organization of the checkpoint proteins following their recruitment to sites of DNA damage or collapsed DNA replication forks. We suggest that one of the reasons for the apparent complexity of the system is because it allows cells to distinguish between similar biochemical consequences of DNA damage (such as ssDNA-RPA complexes) that occurs in distinct circumstances (such as induced damage in G2 and collapsed replication forks). It is important to make these distinctions because different signaling responses to the cell cycle and the DNA repair apparatus will be appropriate in each case. Our data suggest that the Rad3ATR-dependent phosphorylation of Rad9 promotes association between Rad9 and Rad4TOPBP1 through phospho-specific BRCT-domain interactions during unperturbed S phase, and that this helps cells distinguish collapsed forks from DNA damage in G2 cells. It is intriguing that a phospho-specific BRCT-mediated interaction between BRCA1 and BACH1 in human cells (Yu et al. 2003) is promoted by cyclin-dependent kinase activity against BACH1 in G2 and also by the G2 checkpoint. Together, these observations suggest that a combination of phosphorylation events can orchestrate the organization of the checkpoint apparatus before it is activated and that, upon activation, the consequent phospho-specific protein interactions dictate the downstream consequences of this activation.
Materials and methods
Strain constructions
Rad9 phosphorylation sites mutants were constructed by transforming mutagenized DNA fragment into an h-rad9::ura4 ade6-704 leu1-32 ura4-D18 strain and selecting for 5-FOA resistance. Colonies were verified by PCR, sequencing, and Western blot analysis. rad9 phosphorylation-site mutant genes were C-terminally tagged (3xHA) as previously described (Bahler et al. 1998; Caspari et al. 2000a). An N-terminally His10-HA tag was introduced at the rad9 locus by transforming a PCR-amplified His10-HA encoding fragment into a strain in which ura4+ replaced the first ATG codon of rad9+. For biochemical analysis, cells were synchronized by elutriation. Otherwise, synchrony was achieved by lactose gradients (Carr and Murray 1996). Protocols for checkpoint measurements, cell scoring, irradiation, centrifugal elutriation, and lactose gradient centrifugation were as previously described (Edwards and Carr 1997).
Western blot, immunoprecipitation, and gel filtration
Phospho-specific Ab (antibody) against Ser 412 and Thr 423 were generated by BETHYL laboratories (Thr 412 peptide CDAEFGPT-(PO3)QAEQSY; Ser 423, peptide CSYHGIFS-(PO3)QED). For Western blotting of crude extract, protein was prepared by TCA (trichloro-acetic acid) extraction (Caspari et al. 2000a). Detection of by phospho-specific Abs was by the guanidine chloride method (Liu et al. 2003). Briefly, protein was extracted from 1 × 109 His10-HA-rad9 cells and His10-HA-Rad9 purified from cleared extracts using TALON beads (Clontech). After four washes with 8 M urea samples were resuspended in SDS sample buffer for Western blot. Anti-phosphopeptide antibodies were used in 1/20 (α-pT412) or 1/50 (α-pS423). Extracts for co-IP experiments were made using the liquid nitrogen method (Caspari et al. 2000a) except that TEG buffer (50 mM Tris-HCl at pH 7.5, 10 mM EDTA, 10% glycerol, 0.1% IGEPAL) supplemented with protease inhibitor cocktail (Complete; Roche), 10 mM N-ethylmaleimide (Sigma), and 1 mM phenylmethylsulfonyl fluoride (Sigma) was used as the extraction buffer. TAP-Rad4 was precipitated by Rabbit IgG-coated magnetic beads (Liu et al. 2003). For phosphatase treatment on IPed Rad4–Rad9 complex, IPed Rad4–Rad9 is washed by phosphatase buffer (50 mM Tris at pH 7.5, 1 mM MgCl2) three times. And 100 mM Na2HPO4 is used for protein phosphatase inhibitor. The fact that only a percentage of the material is released presumably reflects the occlusion of the phosphate by the protein–protein interaction. Gel filtration was performed as described (Edwards et al. 1999) except Superose 6 column was used. Extracts were prepared by the liquid nitrogen method. For extraction, TEG buffer was used, and for column equilibration and running, buffer B (50 mM NaH2PO4–Na2HPO4 at pH 7, 150 mM NaCl, 0.1% IGEPAL [Sigma], 10% glycerol, 0.5 mM DTT, 5 mM EGTA, 60 mM β-glycerophosphate, 0.1 mM NaF, 1 mM orthovanadate) was used.
Two-hybrid assay
The protocols were as previously described in Caspari et al. (2000a). For in-plate X-gal assay, transformants were patched on X-gal (80 mg/L) and BU salts (70 g of Na2HPO4 and 30 g of NaH2PO4/L) containing plates. For the liquid ONPG assay, cells disrupted by repetitive freeze and thawed in liquid nitrogen were suspended in buffer and incubated with ONPG at 30°C. Here 1 unit of β-galactosidase activity is defined as the hydrolysis of 1 μmole of ONPG to o-nitrophenol (assayed as O.D.420) and D-galactose per minute per cell. O.D.420 was divided by incubated time and cell number.
Acknowledgments
K.F. acknowledges Wellcome Travelling Fellowship 062903/Z/00/Z. This work was supported by the MRC (G0001129).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.291104.
References
- Alcasabas A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Carr A.M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair 1: 983–994. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Beginning at the end. Science 300: 1512–1513. [DOI] [PubMed] [Google Scholar]
- Carr A.M. and Murray, J.M. 1996. DNA repair and checkpoint controls in fission yeast: A practical guide. In Microbial genome methods (ed. K.W. Adolph), pp. 133–147. CRC Press, Boca Raton, FL.
- Caspari T. and Carr, A.M. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81: 173–181. [DOI] [PubMed] [Google Scholar]
- Caspari T., Dahlen, M., Kanter-Smoler, G., Lindsay, H.D., Hofmann, K., Papadimitriou, K., Sunnerhagen, P., and Carr, A.M. 2000a. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20: 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T., Davies, C., and Carr, A.M. 2000b. Analysis of the fission yeast checkpoint rad proteins. Cold Spring Harbor Symposium on Quantitative Biology LXV: 451–456. [DOI] [PubMed] [Google Scholar]
- Chen M.J., Lin, Y.T., Lieberman, H.B., Chen, G., and Lee, E.Y. 2001. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J. Biol. Chem. 276: 16580–16586. [DOI] [PubMed] [Google Scholar]
- Chene P. 2003. Inhibiting the p53–MDM2 interaction: An important target for cancer therapy. Nat. Rev. Cancer 3: 102–109. [DOI] [PubMed] [Google Scholar]
- Du L.L., Nakamura, T.M., Moser, B.A., and Russell, P. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23: 6150–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.J. and Carr, A.M. 1997. Analysis of radiation-sensitive mutants of fission yeast. Methods Enzymol. 283: 471–494. [DOI] [PubMed] [Google Scholar]
- Edwards R.J., Bentley, N.J., and Carr, A.M. 1999. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1: 393–398. [DOI] [PubMed] [Google Scholar]
- Girard P.-M., Riballo, E., Begg, A., Waugh, A., and Jeggo, P.A. 2002. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene 21: 4191–4199. [DOI] [PubMed] [Google Scholar]
- Green C.M., Erdjument-Bromage, H., Tempst, P., and Lowndes, N.F. 2000. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10: 39–42. [DOI] [PubMed] [Google Scholar]
- Harris S., Kemplen, C., Caspari, T., Chan, C., Lindsay, H.D., Poitelea, M., Carr, A.M., and Price, C. 2003. Delineating the position of rad4+/cut5+ within the DNA-structure checkpoint pathways in Schizosaccharomyces pombe. J. Cell Sci. 116: 3519–3529. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kim S.T., Lim, D.S., Canman, C.E., and Kastan, M.B. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274: 37538–37543. [DOI] [PubMed] [Google Scholar]
- Lindsay H.D., Griffiths, D.J.F., Edwards, R.J., Christensen, P.U., Murray, J.M., Osman, F., Walworth, N., and Carr, A.M. 1998. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Dev. 12: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Powell, K.A., Mundt, K., Wu, L., Carr, A.M., and Caspari, T. 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes & Dev. 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiniemi M., Hillukkala, T., Tuusa, J., Reini, K., Vaara, M., Huang, D., Pospiech, H., Majuri, I., Westerling, T., Makela, T.P., et al. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276: 30399–30406. [DOI] [PubMed] [Google Scholar]
- Manke I.A., Lowery, D.M., Nguyen, A., and Yaffe, M.B. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639. [DOI] [PubMed] [Google Scholar]
- Melo J.A., Cohen, J., and Toczyski, D.P. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 15: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Matsumoto, K., and Sugimoto, K. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes & Dev. 17: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos-Mattjus P., Hopkins, K., Oestreich, A., Vroman, B., Johnson, K., Naylor, S., Lieberman, H., and Karnitz, L. 2003. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J. Biol. Chem. 278: 24428–24437. [DOI] [PubMed] [Google Scholar]
- Saka Y., Esashi, F., Matsusaka, T., Mochida, S., and Yanagida, M. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes & Dev. 11: 3387–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 3: 155–168. [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Lopes, M., and Foiani, M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 279: 599–602. [DOI] [PubMed] [Google Scholar]
- St Onge R., Besley, B., Pelley, J., and Davey, S. 2003. A role for the phosphorylation of hRad9 in checkpoint signaling. J. Biol. Chem. 278: 26620–26628. [DOI] [PubMed] [Google Scholar]
- Tanaka K. and Russell, P. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3: 966–972. [DOI] [PubMed] [Google Scholar]
- Uziel T., Lerenthal, Y., Moyal, L., Andegeko, Y., Mittelman, L., and Shiloh, Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22: 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G.M. and Carr, A.M. 2001. The evolution of diverse biological responses to DNA damage: Insights from yeast and p53. Nat. Cell Biol. 3: E277–E286. [DOI] [PubMed] [Google Scholar]
- Walworth N. and Bernards, R. 1996. rad-dependent responses of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271: 353–356. [DOI] [PubMed] [Google Scholar]
- Wang H. and Elledge, S.J. 2002. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 160: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Zhang, Z., An, X., Bucci, B., Perlstein, D.L., Stubbe, J., and Huang, M. 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. 100: 6628–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Kong, L., and Newport, J. 2002. The role of single-stranded DNA and polymerase α in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 30: 27088–27093. [DOI] [PubMed] [Google Scholar]
- Yu X., Chini, C.C., He, M., Mer, G., and Chen, J. 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642. [DOI] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge, S.J. 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]
- Zou L., Liu, D., and Elledge, S.J. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. 100: 13827–13832. [DOI] [PMC free article] [PubMed] [Google Scholar]