Abstract
The voltage-dependent anion channels (VDACs), prominently localized in the outer mitochondrial membrane, play important roles in the metabolite exchange, energy metabolism and mitochondria-mediated apoptosis process in mammalian cells. However, relatively little is known about the functions of VDACs in plants. To further investigate the function of AtVDAC1 in Arabidopsis, we analyzed a T-DNA insertion line for the AtVDAC1 gene. The knock-out mutant atvdac1 showed reduced seed set due to a large number of undeveloped ovules in siliques. Genetic analyses indicated that the mutation of AtVDAC1 affected female fertility and belonged to a sporophytic mutation. Abnormal ovules in the process of female gametogenesis were observed using a confocal laser scanning microscope. Interestingly, both mitochondrial transmembrane potential (ΔΨ) and ATP synthesis rate were obviously reduced in the mitochondria isolated from atvdac1 plants.
Introduction
Voltage-dependent anion channels (VDACs) are known as the most abundant proteins in the outer mitochondrial membrane [1]–[3]. As eukaryotic porins across the outer mitochondrial membrane, VDACs mediate the exchange of metabolites such as ATP, NADH, and ions [4]–[7] between the mitochondria and the cytoplasm, which is very significant for mitochondrial functions and cellular energy transactions [8]. VDAC, known as a component of the permeability transition pore in mammalian cells, is involved in mitochondria-mediated apoptosis together with the adenine nucleotide translocator and other molecules [9]–[12].
VDACs are widely present in fungi, plants and animals. Two forms of VDAC genes were found in yeasts [13], and three isoforms (VDAC1, VDAC2 and VDAC3) in mammalian cells [14]. Mammalian VDAC members function in male reproduction, the synaptic plasticity, and normal glucose homeostasis [15]–[17]. In plants, VDAC isoforms were identified in many species, such as Oryza sativa, Nicotiana tabacum, Lotus japonicus, Triticum aestivum, Solanum tuberosum, Arabidopsis thaliana [7], [18]–[26] and other species (see review [27]). The information about structure, subcellular location, and physiological functions of plant VDACs has been explored gradually, although our knowledge of VDACs in plants is much less than that in animals. VDACs in plants display similar electrophysiological and topological properties as other eukaryotic VDACs [27]; however, apart from roles in the growth and development, plant VDACs have their own specificities such as functions in biotic and abiotic stress responses [28]. The Arabidopsis thaliana genome encodes five VDAC isoforms, and four of them have been cloned [22]. In the past several years, T-DNA knockout mutants were used to analyze the functions of Arabidopsis VDAC members. In 2011, Tateda and coworkers first identified T-DNA insertion mutants of AtVDAC1 (At3g01280), AtVDAC2 (At5g67500), AtVDAC3 (At5g15090), AtVDAC4 (At5g57490), and reported that AtVDAC1, AtVDAC2 and AtVDAC4 played important roles in plant growth, leaf and pollen development, as well as in maintaining the steady state of the mitochondrial membrane potential; in particular, vdac1 plants produced slender and shorter siliques [29]. The number of pollen grains, the pollen germination rate, and the tube length of germinated pollen were dramatically reduced in the vdac1 mutant [28]. In 2012, Robert and coworkers demonstrated that three AtVDACs were dual-localized in both plasma membrane and mitochondrial membrane, and some mutant phenotypes were related to mitochondrial functions. They suggested that the decreased seed set in the atvdac1 mutant was not related to gametophytic sterility but rather followed from zygotic or early embryo lethality [30].
In the process of aerobic respiration, mitochondria are the pivotal organelles for ATP synthesis, through oxidative phosphorylation. Electron flow past the electron transport chain located in the inner mitochondrial membrane is coupled to proton translocation out of the matrix. The proton gradient across inner mitochondrial membrane drives ATP synthesis through oxidative phosphorylation, and the charge imbalance resulted from the generation of an electrochemical gradient across the inner mitochondrial membrane forms the basis of the inner mitochondrial transmembrane potential [31], [32]. Then what is the function of the abundant protein, VDAC, across the outer mitochondrial membrane? In mammalian cells, VDAC is involved in the permeability transition pore which mediates mitochondrial permeability transition leading to the loss of mitochondrial transmembrane potential, and participates in mitochondria-mediated apoptosis [33]. Do VDACs have underlying roles in regulation of mitochondrial transmembrane potential and/or ATP synthesis in plants? In Arabidopsis atvdac1 plants, what is responsible for reduced seed set? Is AtVDAC1 involved in male or female development? Furthermore, is AtVDAC1 involved in the processes of sporogenesis and gametogenesis before fertilization or the processes of zygogenesis and embryogenesis? Exploring these issues will help us to find out more information about functions of AtVDAC1.
In this study, we investigated the function of AtVDAC1 using the T-DNA insertion mutant atvdac1 in reproductive development. The phenotype in atvdac1 plants, reduced seed set, results from plenty of undeveloped ovules in siliques. Results indicate that AtVDAC1 is required for female development and involved in maintaining the steady state of mitochondrial transmembrane potential and ATP synthesis. This result is significant for further understanding of functions of AtVDAC1 in the reproductive process.
Results
Mutation of AtVDAC1 Causes Reduced Seed Set
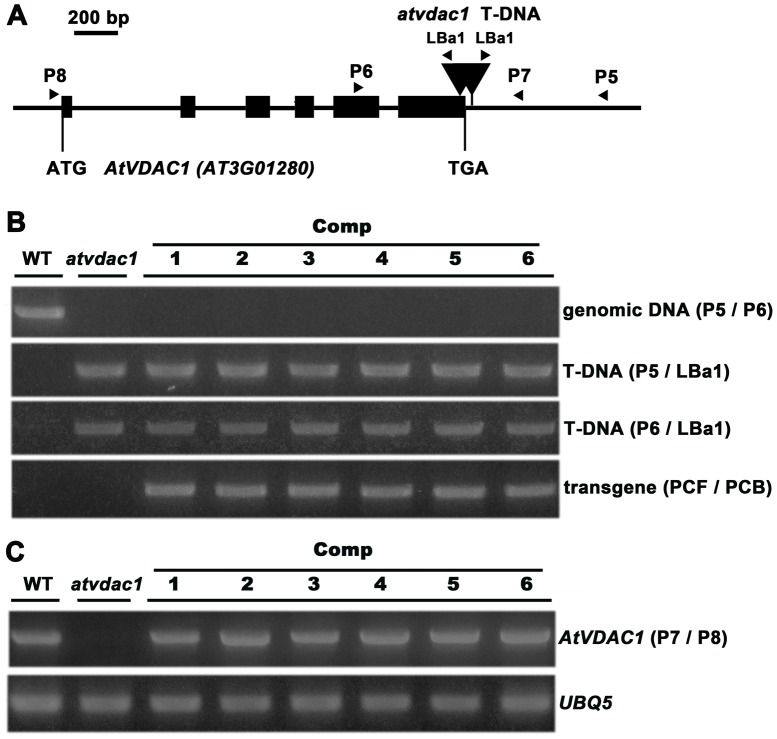
We identified a T-DNA insertion mutant (atvdac1) for the AtVDAC1 gene (At3g01280) from the Arabidopsis Biological Resource Center (ABRC), which had two T-DNAs inserted in the sixth exon and the 3′UTR of the AtVDAC1 gene (Figure 1A). The homozygous knockout mutant was isolated and confirmed by PCR analysis (Figure 1B, the left two lines), and the expression levels of AtVDAC1 gene were analyzed using RT-PCR (Figure 1C, the left two lines).
Figure 1. Molecular confirmation of isolated atvdac1 mutant and its gene complementation.
(A) Schematic diagram of the AtVDAC1 gene structure and the T-DNA insertion sites in the atvdac1 mutant. Closed boxes indicate exons, and arrowheads indicate the positions of primers used for genotyping. The start and stop codons are labeled. (B) Homozygous atvdac1 plants were complemented with the transgene (AtVDAC1 promoter:AtVDAC1 genomic DNA). Six independent transgenic lines, wild type (WT) and the atvdac1 mutant were analyzed by genomic PCR amplification. (C) Expression of the AtVDAC1 gene was analyzed by RT-PCR in the six independent complemented lines, WT and the atvdac1 mutant. The UBQ5 transcript was amplified as an internal control.
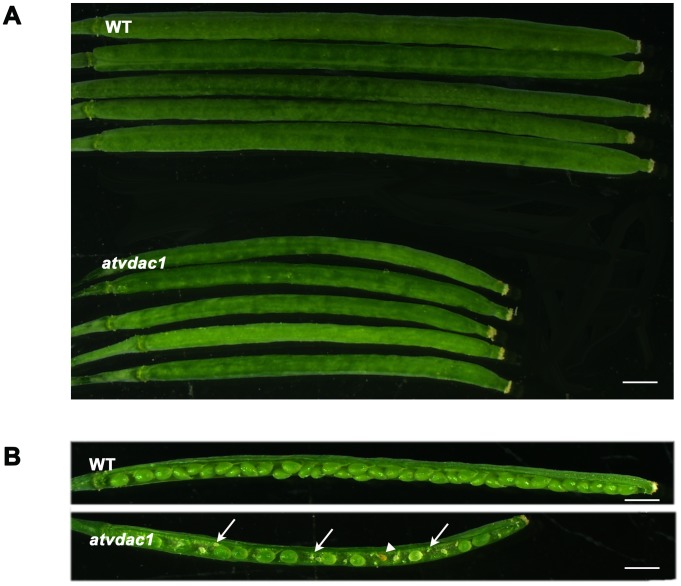
The homozygous atvdac1 mutant had apparently shorter siliques compared with the wild type (Figure 2A, Table S1), consistent with the observations by Tateda et al. (2011) [29]. Further microscopic observation revealed that the mutant atvdac1 had reduced seed set, that was, about 62.3% (Figure 2B, Table 1). There were two types of abnormalities in siliques from atvdac1: undeveloped ovules (33.4%) and aborted seeds (4.3%). As shown in Figure 2B, a plenty of undeveloped ovules were small, shrunken, and finally aborted. In contrast, full seed set was observed (Figure 2B), and the proportions of abnormalities were only about 0.8% and 0.3% respectively in wild type at the same growth condition (Table 1).
Figure 2. The atvdac1 mutation caused shorter siliques and reduced seed set.
(A) Representative siliques from wild-type (WT) and atvdac1 mutant plants. (B) Seed development in the representative siliques from WT and atvdac1 mutant plants. The arrows indicate undeveloped ovules and the arrowhead indicates the aborted seed. Bars = 1 mm.
Table 1. Analysis of seed development in siliques from wild type (WT), atvdac1 and reciprocal crosses between WT and atvdac1, showing that female fertility is affected.
| Crosses (female × male) | Normal seeds (%) | Aborted seeds (%) | Undeveloped ovules (%) | Total No. |
| WT selfeda | 98.9±0.2 | 0.3±0.1 | 0.8±0.2 | 2532 |
| atvdac1 selfeda | 62.3±6.0 | 4.3±1.3 | 33.4±4.7 | 2583 |
| WT × atvdac1 b | 98.0±0.5 | 0.6±0.6 | 1.4±0.3 | 1116 |
| atvdac1 × WTb | 54.5±5.9 | 5.5±1.6 | 39.9±4.4 | 1057 |
The statistical analysis was performed in siliques from 50-day-old plants after transplantion into the soil.
40 siliques were examined.
20 siliques were examined.
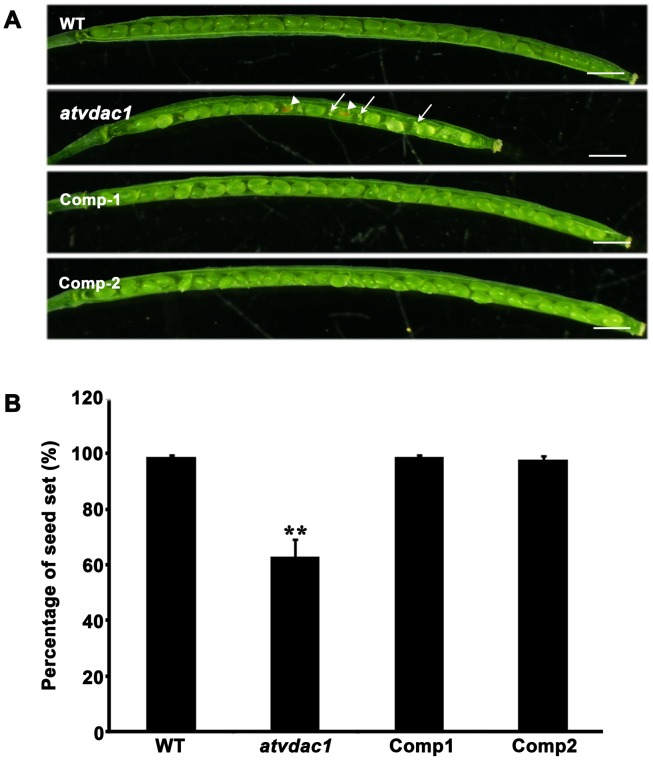
In order to confirm that the mutation of AtVDAC1 is responsible for the atvdac1 phenotype, we performed a complementation experiment. The complementation construction AtVDAC1 promoter:AtVDAC1 genomic DNA was introduced into atvdac1 plants. Six homozygous transformants with one copy of T-DNA insertion were obtained from the transgenic atvdac1 progeny. All these transformants were confirmed by PCR analysis using the pCAMBIA1300-specific primer PCF and AtVDAC1 gene-specific primer PCB. The genotype analysis of wild type and the atvdac1 mutant was also performed using primer pairs P5/LBa1, P6/LBa1 and P5/P6 as controls (Figure 1A, B). RT-PCR analysis showed that the expression of AtVDAC1 gene could be detected in the six independent complemented lines using primer pairs P7/P8 which corresponded to the 3′UTR/5′UTR of this gene respectively (Figure 1A, C). Phenotypic analysis of two complemented lines showed that the introduced AtVDAC1 gene rescued the defective phenotype observed in atvdac1 plants (Figure 3A, Table S2). The percentages of seed set from the complemented plants were recovered (Figure 3B). Above results demonstrate that the defective phenotype observed in the atvdac1 mutant results from the mutation of the AtVDAC1 gene.
Figure 3. Complementation of the atvdac1 mutant rescued the phenotype of atvdac1 plants.
(A) Seed development in the representative siliques from wild-type (WT), atvdac1 mutant and complemented atvdac1 plants. The arrows indicate undeveloped ovules and arrowheads indicate aborted seeds. Bars = 1 mm. (B) Statistics of the seed set of complemented plants by comparison with that of atvdac1 mutant and WT plants. The statistical analysis was performed in siliques from 50-day-old plants after transplantion into the soil. Error bars represent SD, 40 siliques were examined for each genotype. Asterisks indicate a statistically significant difference between atvdac1 and WT (Student's t-test), **P<0.01.
AtVDAC1 Is Expressed throughout Reproductive Organs
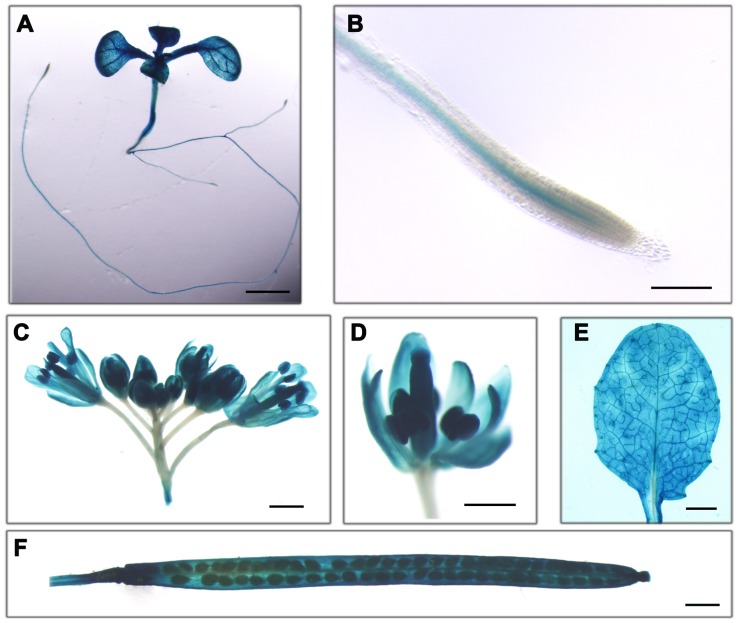
RT-PCR and quantitative RT-PCR assays showed that AtVDAC1 had a ubiquitous expression pattern with high transcription levels in roots, stems, leaves, inflorescences, siliques, and seedlings (data not shown). In order to investigate the AtVDAC1 expression pattern in tissues and organs in details, AtVDAC1 promoter:GUS construction was introduced into wild-type plants, and histochemical GUS staining assays were performed. GUS signals were detected in the cotyledons, true leaves and roots of seedlings and rosette leaves of adult plants, basically confirming the previous results by Tateda et al. (2011) [29] and Robert et al. (2012) [30]. Especially in reproductive organs, high level of GUS activity was detected in pistils, stamens, sepals and petals including siliques where the phenotype appeared in the atvdac1 mutant (Figure 4). This result is consistent with the analysis of microarray expression data which shows that AtVDAC1 displays a peak of expression in reproductive organs [27].
Figure 4. Expression pattern of AtVDAC1 in Arabidopsis, showing that GUS stains were observed in seedling (A), root tip (B), inflorescence (C), flower (D), rosette leaf (E), and silique (F) in transgenic plants carrying the AtVDAC1 promoter:GUS construction.
Bars = 2 mm (A), 200 µm (B), 1 mm (C), 500 µm (D), 2 mm (E), and 1 mm (F).
Mutation of AtVDAC1 Affects Female Development
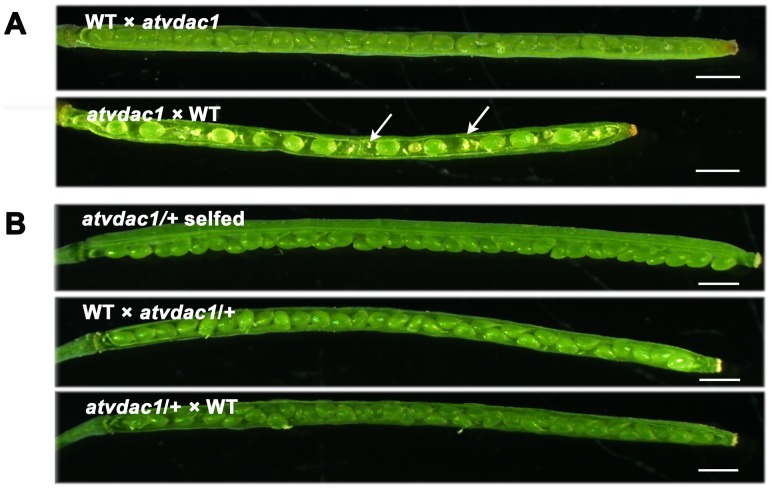
To determine whether the mutation of AtVDAC1 affects male or female fertility, reciprocal crosses were performed between atvdac1 and wild type. The crossed plants with atvdac1 as the female parent and wild type as the male parent, had a similar phenotype to atvdac1 plants. They harbored shorter siliques and showed a percentage of seed set close to that of atvdac1 selfed plants; however, when wild type was used as the female parent in crosses with atvdac1 plants, about 98.0% ovules produced normal seeds similar to wild-type plants (Figure 5A, Table 1, Table S1). These results indicate that the mutation of AtVDAC1 affects female fertility.
Figure 5. Reciprocal crosses show that female fertility was affected.
(A) Seed development in representative siliques of reciprocal crosses using pollen from atvdac1 plants to pollinate wild-type (WT) pistils and using pollen from WT plants to pollinate atvdac1 pistils. The arrows indicate undeveloped ovules. Bars = 1 mm. (B) Seed development in the representative atvdac1/+ selfed silique and in representative siliques from reciprocal crosses between WT and atvdac1/+. Bars = 1 mm.
All F1 plants from reciprocal crosses above were atvdac1 heterozygous mutants (atvdac1/+) confirmed by PCR assay (data not shown), and had normal siliques and seed set (Table S3, Figure 5B). The F2 progeny of atvdac1/+ plants segregated approximately 3∶1 (200 wild-type phenotype plants: 66 defective phenotype plants) as a single recessive mutation. Genotype analysis using PCR assay showed that all the 66 defective phenotype plants harbored the atvdac1 homozygous mutation (atvdac1/atvdac1), and there were 136 atvdac1 heterozygous (atvdac1/+) plants and 64 wild-type (+/+) plants in the 200 wild-type phenotype plants (Table 2). This result indicates the linkage between the phenotype and T-DNA insertion in the AtVDAC1 gene. Because the progeny of atvdac1/+ exhibited the Mendelian 1∶2∶1 (66∶136∶64) segregation pattern, the atvdac1 is a sporophytic mutant. In addition, reciprocal crosses between atvdac1/+ plants and wild-type (+/+) plants were also performed. As shown in figure 5B and Table S3, the crossed siliques had normal length and seed set. F1 plants from both crosses showed the same segregation ratio of nearly 1∶1 (with T-DNA insertion to without T-DNA insertion = 176∶192 and 239∶221, respectively) (Table 2), which means female and male transmissions of the atvdac1 allele from heterozygotes were not reduced. Therefore, the mutation of AtVDAC1 does not affect either male or female gametophytic development.
Table 2. The segregation ratio of progeny from atvdac1/+ selfed and reciprocal crosses between wild type (WT) and atvdac1/+.
| Crosses (female × male) | atvdac1/atvdac1 with T-DNA insertion (W1) | atvdac1/+ with T-DNA insertion (W2) | +/+ without T-DNA insertion (WO) | Ratio (W1+W2):WO | TEF | TEM |
| atvdac1/+ selfeda | 66 | 136 | 64 | 3.16∶1 | NA | NA |
| WT × atvdac1/+ b | 0 | 176 | 192 | 0.92∶1 | NA | 92% |
| atvdac1/+ × WTb | 0 | 239 | 221 | 1.08∶1 | 100% | NA |
TE, transmission efficiency = (W1+W2)/WO ×100%; TEF, female transmission efficiency.
TEM, male transmission efficiency; NA, not applicable.
The ratio was calculated using seeds from ten atvdac1/+ plants.
The ratio was calculated using seeds from eight plants.
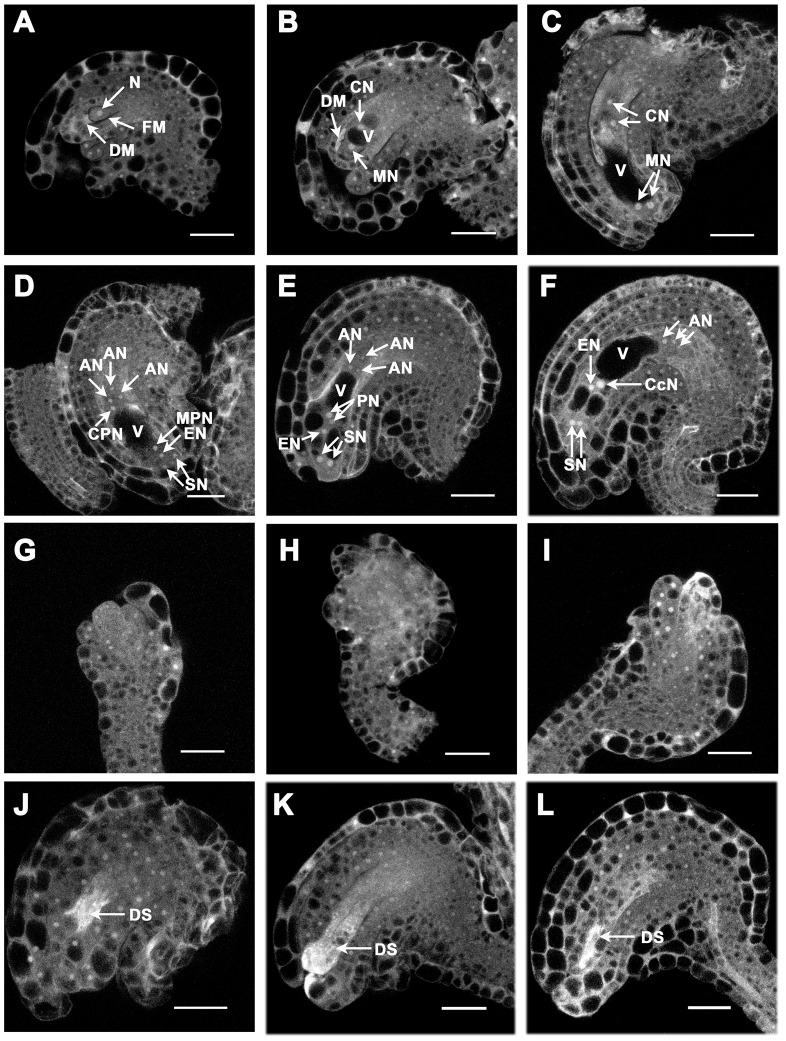
To further investigate the phenotype of undeveloped ovules observed in atvdac1 siliques, ovule development in wild-type and mutant plants was examined via confocal laser scanning microscopy (CLSM) [34]. In wild-type plants, a single hypodermal archesporial cell directly differentiates into a megaspore mother cell that undergoes meiosis to generate four haploid megaspores (stage FG0). As shown in Figure 6A, the chalazal-most megaspore survived, enlarged and became the functional megaspore (FM); and the micropylar megaspores degenerated (DM), as shown by their strong autofluorescence in wild type (stage FG1). The haploid functional megaspore underwent three rounds of mitosis with cellularization and fusion of polar nuclei (stages FG3–FG6, Figure 6B-F). But in atvdac1 pistils, we observed smaller ovules that lacked the embryo sac structure with one functional megaspore when other normal ones finished sporogenesis and reached the FG1 stage (Figure 6G). This result suggests that the process of sporogenesis was affected so the uninucleate female gametophyte could not form in abnormal ovules. During FG3-FG6 stages, abnormal ovules without nuclei and embryo sac structures or with degraded structures (DS) shown as strong autofluorescence, were also observed (Figure 6H-L).
Figure 6. Ovule development in wild-type (WT) and atvdac1 siliques revealed by confocal laser scanning microscopy (CLSM).
(A–F) Female gametogenesis in WT siliques at stage FG1 (A), FG3 (B), FG4 (C), early FG5 (D), late FG5 (E), FG6 (F). (G–L) Abnormal ovules in atvdac1 siliques at stage FG1 (G), FG3 (H), FG4 (I), FG5 (J, K), FG6 (L). Abbreviations: AN, antipodal nucleus (nuclei); CN, chalazal nucleus (nuclei); CcN, central cell nucleus; CPN, chalazal polar nucleus; DM, degenerated megaspores; DS, degenerated structure; EN, egg cell nucleus; FM, functional megaspore; MN, micropylar nucleus (nuclei); MPN, micropylar polar nucleus; N, nucleus; PN, polar nuclei; SN, synergid nuclei; V, vacuole. Bars = 20 µm.
Mutation of AtVDAC1 Affects ΔΨ and ATP Synthesis
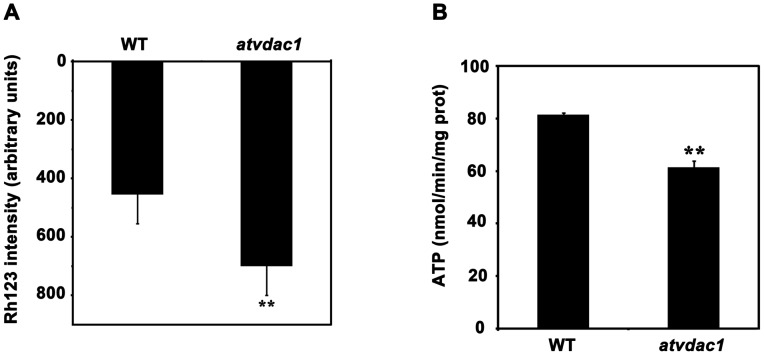
In order to find out whether AtVDAC1 is involved in regulation of the mitochondrial transmembrane potential (ΔΨ) in Arabidopsis, we isolated mitochondria from wild-type and atvdac1 plants respectively, and measured the ΔΨ using Rhodamine123. As shown in Figure 7A, the ΔΨ in atvdac1 was obviously lower than that in wild type. Mitochondria are the energy powerhouse in eukaryotic cells, so oxidative phosphorylation efficiency may be affected as a result of abnormal functional state of mitochondria. ATP synthesis rate was then measured using isolated mitochondria. The mitochondria from atvdac1 plants produced approximately 25% less ATP than that from wild-type plants (Figure 7B). These results indicate that the mutation of AtVDAC1 affects ΔΨ and ATP synthesis of mitochondria.
Figure 7. Mitochondrial transmembrane potential (ΔΨ) and ATP synthesis rate were lower in atvdac1 than those in wild type (WT).
(A) Isolated mitochondria (1 mg/mL) prepared from WT and atvdac1 mutant plants were treated with Rhodamine123 (Rh123) for 20 min, and ΔΨ was measured by Rh123 uptake. (B) ATP synthesis rate was measured in mitochondria prepared from WT and atvdac1 mutant plants. Error bars represent SD (n≥3). Asterisks indicate statistically significant differences between atvdac1 and WT (Student's t-test), **P<0.01.
Discussion
VDACs play crucial roles in regulating metabolic and energetic functions of mitochondria [4], [5], [8], [35] and are involved in mitochondria-mediated apoptosis in mammalian cells [10], [36]. But the knowledge on plant VDACs is still limited. As to the function of AtVDAC1 on reproductive development, one group reported that the pollen development was affected in vdac1 plants [29]. The other group suggested that the decreased seed production resulted from zygotic or early embryo lethality [30]. In this work, we found that there were plenty of undeveloped ovules in atvdac1 siliques which mainly resulted from the defective female development. Results indicated that mitochondrial transmembrane potential (ΔΨ) and ATP synthesis rate were reduced in the mitochondria isolated from atvdac1 plants.
We analyzed the T-DNA insertion mutant atvdac1 (SALK_034653) for the AtVDAC1 gene (At3g01280). In atvdac1 siliques, approximately 37.7% of ovules did not develop into mature seeds (Table 1). Only the homozygous atvdac1 mutant (atvdac1/atvdac1) had the defective phenotype (Figure 2) and the atvdac1 heterozygous mutant (atvdac1/+) had full seed set (Figure 5B), so the reduced seed set did not result from zygotic or embryo lethality. Genetic analyses were then used to investigate the reason of the reduced seed set. Firstly, reciprocal crosses between homozygous atvdac1 and wild type were performed according to the approach described by Motamayor et al. (2000) [37] and Wang et al. (2012) [38]. When wild-type pistils were pollinated by mutant pollen, the crossed siliques had normal seed set; however, atvdac1 siliques showed the same defective phenotype regardless of the pollen was from wild-type or mutant plants (Figure 2B, 5A; Table 1), indicating that female fertility in atvdac1 plants was affected.
Sporophytic mutations affect the diploid sporophyte generation of the life cycle of plants and exhibit Mendelian 1∶2∶1 segregation patterns; on the contrary, gametophytic mutations affect the gametophyte generation and exhibit altered segregation patterns [39], [40]. The progeny of atvdac1/+ exhibited the Mendelian 1∶2∶1 segregation pattern (Table 2). The results from reciprocal crosses between atvdac1/+ plants and wild-type plants indicated that female and male transmissions of the atvdac1 allele from heterozygotes were not affected (Table 2). Accordingly, the mutation of AtVDAC1 does not affect either male or female gametophytic development and is a sporophytic mutation. Interestingly abnormal ovules were observed using a confocal laser scanning microscope in stage FG1 (Figure 6G), pointing to the earlier defect in the process of megasporogenesis. As ATP synthesis rate was reduced in the mitochondria isolated from atvdac1 plants (Figure 7B), we hypothesize that the process of meiosis might be affected during the stage of megasporogenesis. Many motor proteins driven by ATP hydrolysis are involved in meiosis; therefore, enough amount of ATP is needed in this process [41], [42]. In Arabidopsis, many sporophytic mutants affecting sporogenesis have been reported [37], [43]–[46]. Moreover, abnormal ovules without nuclei and embryo sac structures or with degraded structures (DS) were also observed at stages FG3-FG6 (Figure 6H-L). We suggest that the defects might directly result from the abnormal megasporogenesis or from the affected sporophytic tissues. Previous studies demonstrated that integuments in sporophytic tissues played important roles on the control of megagametogenesis [47]–[49].
Several lines of evidence have been shown that mitochondria are deeply involved in male gametophytic development and many mutations of mitochondrial genes affect male gametophytic development, even resulting in cytoplasmic male sterility (CMS) [50], [51]. In our study, only female development was affected by genetic analyses (Table 1, 2) in atvdac1 plants, which may be result from the sporophytic mutation of atvdac1. The female gametocyte is included in the diploid sporophytic tissues; therefore, female development may be affected by the defect of sporophytic tissues. Till now, we cannot exclude that the microsporogenesis is affected in the atvdac1 mutant only according to the results from genetic analyses. Future investigations on the microsporogenesis will provide further information for this issue.
Interestingly, in the study of Tateda and coworkers, neither vdac1-3 (SALK_039833) nor vdac1-5 (SALK_058473C) homozygous mutant was identified in the progeny of self-crossed heterozygous plants [29]. The positions of T-DNA insertions within the 5′ region of the AtVDAC1 gene may result in complete disruption of this gene in vdac1-3 and vdac1-5 plants, implying that AtVDAC1 plays important roles in the plant growth and development. In our study, the atvdac1 (SALK_034653) mutant harbored two T-DNAs inserted in the 3′ end (the last exon and 3′UTR) of the AtVDAC1 gene (Figure 1A). The AtVDAC1 transcripts including first five exons were detected in homozygous atvdac1 plants by RT-PCR analysis (Figure S1), implying that the atvdac1 plants might accumulate truncated AtVDAC1 polypeptides retaining partial activity. Therefore, the homozygous atvdac1 plants could be obtained but the female development was affected, different from the vdac1-6 (SALK_011520C) mutant with a T-DNA in the fifth intron, exhibiting defective pollen development [29]. Different phenotypes might result from the different positions of T-DNA insertions. This phenomenon that different mutations in the same gene exhibit different phenotypes, was also observed in other genes such as SWI1 [52].
Several studies have been performed to determine the localization of AtVDACs using various experimental methods. The mitochondrial localization of AtVDACs was observed in the protoplasts of AtVDACs-GFP transgenic plants and in onion epidermal cells using AtVDACs–GFP fusion constructs [22], [29]. However, AtVDAC3 was also identified in plasma membrane fractions of Arabidopsis cell suspensions [53]. Recently, biochemical evidence has revealed that AtVDAC1, AtVDAC2 and AtVDAC3 were targeted to both mitochondria and plasma membrane in differential abundance but their functions seemed to be mainly associated with mitochondria [30]. In plants, evidence regarding the role of VDACs in mitochondrial function is mainly based on their mitochondrial localization [28]. Several lines of evidence show that the Bcl-2 family proteins could interact with VDAC to regulate the mitochondrial transmembrane potential (ΔΨ) during mitochondria-mediated apoptosis in mammalian cells [10], [11], [54]. Several lines of evidence indicate cytological and biochemical similarities between plant and mammalian apoptosis [55]. In order to investigate the relationship between plant VDAC and the ΔΨ, we measured ΔΨ in isolated mitochondria prepared from wild-type and atvdac1 mutant plants. Our result indicated that the mutation of AtVDAC1 resulted in the decline of ΔΨ (Figure 7A), similar as that detected in protoplasts [29]. Therefore we suppose that AtVDAC1 plays an important role in maintaining the steady state of ΔΨ in Arabidopsis. That VDACs are involved in mitochondrial respiration in other species have been reported [15],[56],[57]. In human cells, diminished VDAC1 expression resulted in decreased ATP synthesis and cytosolic ADP and ATP levels [58]. In Arabidopsis, we previously reported that AtVDAC3 was involved in regulating respiration pathways and ATP levels during seed germination at low temperature [59]. In atvdac1 mutant, ATP synthesis rate was also obviously reduced in isolated mitochondria (Figure 7B). Genetic characterization of mutants indicates that mitochondria-associated proteins play important roles in female development in Arabidopsis [60]–[63]. For example, a mutation of SDH1-1 encoding the flavoprotein subunit of mitochondrial complex II (succinate dehydrogenase [SDH]) affects the embryo sac development [62]. The decline of ΔΨ and ATP synthesis means the abnormal function state of mitochondria and reduced energy supply. Shortage of energy supply could compel plants to finish the reproductive process as soon as possible. As to a silique, shortage ATP supply would not meet the need of the whole ovules development, so reduced seed set could be observed.
Materials and Methods
Plant Material and Growth Conditions
All Arabidopsis thaliana plants used in this study were derived from the Columbia ecotype background (Col-0). Seeds from atvdac1 (SALK_034653) were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/) [30]. The genotype of the T-DNA insertion line for AtVDAC1 (At3g01280) was confirmed by PCR analysis with primers: P5, P6 and LBa1. Seeds were surface-sterilized with chloride gas. After cold treatment at 4°C for 2 days, seeds were sown on the medium containing 0.5 × MS salts (MS, Sigma-Aldrich, St. Louis, MO, USA), 1% sucrose and 0.8% agar which was supplemented with 15 mg/L hygromycin as required. 7-day-old seedlings were transferred to soil and grown at 22°C under a 16-hr-light/8-hr-dark cycle.
AtVDAC1 Complementation Experiment
The genomic DNA fragment of AtVDAC1 (5′UTR, exons, introns and 3′UTR included) and the 1.8 kb promoter were amplified using Arabidopsis genomic DNA as templates with primer pairs as follows: for the promoter using V1PF and V1PBG, for the genomic DNA fragment using V1GF and V1GB. The two fragments were cloned into the pCAMBIA1300 vector (harboring the resistant gene to hygromycin) by the sites of SacI/BamHI and BamHI/XbaI to generate AtVDAC1 promoter:AtVDAC1 genomic DNA construction which then was introduced into atvdac1 plants according to the method of Clough and Bent (1998) [64]. The transformants were selected on 15 mg/L hygromycin and further verified by PCR assays using primer pairs PCF and PCB. UBQ5 (At3g62250) was amplified using primer pairs UBQF and UBQB in RT-PCR analyses.
Construction of AtVDAC1 Promoter:GUS and Histochemical GUS Staining Assays
The AtVDAC1 promoter (1953 bp upstream from the start codon) was amplified from Arabidopsis genomic DNA with the primer pairs, V1PFP and V1PBC. The AtVDAC1 promoter fragment was then cloned into pCAMBIA1300221-GUS vector (harboring the resistant gene to hygromycin) by the PstI/BamHI sites to generate AtVDAC1 promoter:GUS construction.
Plant tissue samples were prefixed with cold 90% acetone for 15 min, and then washed twice with sterilized water. Samples were incubated in the GUS staining solution (1 mg/mL 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 2 mM potassium ferricyanide and potassium ferrocyanide, 10 mM EDTA and 0.1% Triton X-100 in 50 mM sodium phosphate buffer, pH 7.0) at 37°C for 6–8 hr after vacuum-infiltrated for 10 min. Stained samples were cleared in 75% ethanol solution and 25% acetic acid solution, and afterward observed with a Olympus SZX16 microscope (Olympus, Tokyo, Japan).
Confocal Laser Scanning Microscopic (CLSM) Observation
The CLSM observation of ovules was performed as described by Christensen et al. (1997) [34] and Shi et al. (2005) [65] with slight modifications. The pistils were fixed in 4% glutaraldehyde (in 12.5 mM cacodylate, pH 6.9) and kept under a vacuum for 2 hr. After further fixation overnight at room temperature, tissues were dehydrated through an ethanol series. Then the tissues were cleared in 2∶1 (v/v) benzyl benzoate:benzyl alcohol for at least 1 hr. The pistils were dissected, sealed under cover slips and then observed with a Zeiss LSM510 META confocal laser scanning microscope (Zeiss, Jena, Germany).
Isolation of Mitochondria
The rosette leaves of plants grown in a greenhouse for 4 weeks were used as starting materials. Isolation of mitochondria was performed as described by Day and Hanson (1978) [66] with following modifications. The materials were ground in an ice-cold mortar with extraction buffer (0.3 M sucrose, 1 mM EDTA, 1 mg/mL bovine serum albumin (BSA), 0.6% [w/v] polyvinylpyrrolidone 40, 0.5 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and 20 mM This-HCl, pH 8.0). The homogenate was filtered through 4 layers of gauze and centrifuged at 3,000 g for 10 min. The supernatant was centrifuged at 11,000 g for 20 min to collect the pellet. The pellet was resuspended in extraction buffer and subjected to low-speed (1,500 g) and high-speed (11,000 g) centrifugations. The pellet was resuspended in a small volume of extraction buffer and loaded on top of a sucrose “cushion” (0.6 M sucrose, 1 mM EDTA, 1 mg/mL BSA, 0.5 mM DTT, 0.2 mM PMSF, and 20 mM This-HCl, pH 8.0). The cushion was centrifuged at 11,000 g for 20 min. The pellet was washed twice in resuspension buffer (0.3 M Sucrose, 1 mg/mL BSA, 20 mM This-HCl, pH 8.0).
Measurement of ΔΨ and ATP synthesis rate in isolated mitochondria
Measurement of mitochondrial transmembrane potential (ΔΨ) in isolated mitochondria was performed with the method described by Narita et al. (1998) [67]. ΔΨ was assessed by measuring the ΔΨ-dependent uptake of Rhodamine123 (Rh123) by using a spectrophotometer (Hitachi F-4500, Tokyo, Japan) with excitation at 505 nm and emission at 534 nm after addition of 10 µM Rh123 to a mitochondrial suspension. Measurement of ATP synthesis rate in isolated mitochondria was performed according to the method of Meyer et al. (2009) [68] using the ATP Bioluminescent Assay Kit (Sigma-Aldrich, St. Louis, MO, USA). Mitochondria were incubated in respiration buffer containing 10 mM Glu, 10 mM malate, 12 µM CoA, 0.2 mM TPP, 2 mM NAD+, and 1 mM ADP for exactly 10 min. Then, TCA was added to a final concentration of 2.3% (w/v). After centrifugation for 15 min at 20,000 g, the supernatant was diluted for 100 times using 25 mM Tricine buffer (pH 7.8) and then used for ATP production estimation.
Supporting Information
RT-PCR analysis of AtVDAC1 gene transcripts in wild type (WT) and the atvdac1 mutant. (A) Schematic diagram of the AtVDAC1 gene structure and the T-DNA insertion sites in the atvdac1 mutant. Closed boxes indicate exons, and arrowheads indicate the positions of primers. The start and stop codons are labeled. (B) RT-PCR analysis of AtVDAC1 gene transcripts in WT and atvdac1 mutant. The UBQ5 transcript was amplified as an internal control.
(TIF)
Statistics for lengths of siliques from wild type (WT), atvdac1 and reciprocal crosses between wild type (WT) and atvdac1 .
(DOCX)
Statistics for lengths of siliques from wild type (WT), atvdac1 and complemented lines.
(DOCX)
Statistics for lengths of siliques from atvdac1 /+ selfed and reciprocal crosses between wild type (WT) and atvdac1 /+.
(DOCX)
Sequences of the primers used in this study.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (Project 31071259). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mannella CA, Bonner WD Jr (1975) X-ray diffraction from oriented outer mitochondrial membranes. Detection of in-plane subunit structure. Biochim Biophys Acta 413: 226–233. [DOI] [PubMed] [Google Scholar]
- 2. Zalman LS, Nikaido H, Kagawa Y (1980) Mitochondrial outer membrane contains a protein producing nonspecific diffusion channels. J Biol Chem 255: 1771–1774. [PubMed] [Google Scholar]
- 3. Hoogenboom BW, Suda K, Engel A, Fotiadis D (2007) The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol 370: 246–255. [DOI] [PubMed] [Google Scholar]
- 4. Zizi M, Forte M, Blachly-Dyson E, Colombini M (1994) NADH regulates the gating of VDAC, the mitochondrial outer membrane channel. J Biol Chem 269: 1614–1616. [PubMed] [Google Scholar]
- 5. Rostovtseva TK, Bezrukov SM (1998) ATP transport through a single mitochondrial channel, VDAC, studied by current fluctuation analysis. Biophys J 74: 2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smack DP, Colombini M (1985) Voltage-dependent channels found in the membrane fraction of corn mitochondria. Plant Physiol 79: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blumenthal A, Kahn K, Beja O, Galun E, Colombini M, et al. (1993) Purification and characterization of the voltage-dependent anion-selective channel protein from wheat mitochondrial membranes. Plant Physiol 101: 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blachly-Dyson E, Forte M (2001) VDAC channels. IUBMB Life 52: 113–118. [DOI] [PubMed] [Google Scholar]
- 9. Crompton M, Virji S, Ward JM (1998) Cyclophilin-D binds strongly to complexes of the voltage-dependent anion channel and the adenine nucleotide translocase to form the permeability transition pore. Eur J Biochem 258: 729–735. [DOI] [PubMed] [Google Scholar]
- 10. Shimizu S, Narita M, Tsujimoto Y (1999) Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 399: 483–487. [DOI] [PubMed] [Google Scholar]
- 11. Shimizu S, Matsuoka Y, Shinohara Y, Yoneda Y, Tsujimoto Y (2001) Essential role of voltage-dependent anion channel in various forms of apoptosis in mammalian cells. J Cell Biol 152: 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brenner C, Grimm S (2006) The permeability transition pore complex in cancer cell death. Oncogene 25: 4744–4756. [DOI] [PubMed] [Google Scholar]
- 13. Young MJ, Bay DC, Hausner G, Court DA (2007) The evolutionary history of mitochondrial porins. BMC Evol Biol 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saccone C, Caggese C, D'Erchia AM, Lanave C, Oliva M, et al. (2003) Molecular clock and gene function. J Mol Evol 57 Suppl 1 S277–S285. [DOI] [PubMed] [Google Scholar]
- 15. Sampson MJ, Decker WK, Beaudet AL, Ruitenbeek W, Armstrong D, et al. (2001) Immotile sperm and infertility in mice lacking mitochondrial voltage-dependent anion channel type 3. J Biol Chem 276: 39206–39212. [DOI] [PubMed] [Google Scholar]
- 16. Weeber EJ, Levy M, Sampson MJ, Anflous K, Armstrong DL, et al. (2002) The role of mitochondrial porins and the permeability transition pore in learning and synaptic plasticity. J Biol Chem 277: 18891–18897. [DOI] [PubMed] [Google Scholar]
- 17. Anflous-Pharayra K, Cai Z-J, Craigen WJ (2007) VDAC1 serves as a mitochondrial binding site for hexokinase in oxidative muscles. Biochim Biophys Acta 1767: 136–142. [DOI] [PubMed] [Google Scholar]
- 18. Roosens N, Al Bitar F, Jacobs M, Homble F (2000) Characterization of a cDNA encoding a rice mitochondrial voltage-dependent anion channel and its gene expression studied upon plant development and osmotic stress. Biochim Biophys Acta 1463: 470–476. [DOI] [PubMed] [Google Scholar]
- 19. Al Bitar F, Roosens N, Smeyers M, Vauterin M, Van Boxtel J, et al. (2003) Sequence analysis, transcriptional and posttranscriptional regulation of the rice vdac family. Biochim Biophys Acta 1625: 43–51. [DOI] [PubMed] [Google Scholar]
- 20. Al Bitar F, Roosens N, Boxtel JV, Dewaele E, Jacobs M, et al. (2002) Expression of the rice vdac isoform2: histochemical localization and expression level. Biochim Biophys Acta 1579: 133–141. [DOI] [PubMed] [Google Scholar]
- 21. Wandrey M, Trevaskis B, Brewin N, Udvardi MK (2004) Molecular and cell biology of a family of voltage-dependent anion channel porins in Lotus japonicus . Plant Physiol 134: 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee SM, Hoang MH, Han HJ, Kim HS, Lee K, et al. (2009) Pathogen inducible voltage-dependent anion channel (AtVDAC) isoforms are localized to mitochondria membrane in Arabidopsis . Mol Cells 27: 321–327. [DOI] [PubMed] [Google Scholar]
- 23. Elkeles A, Devos KM, Graur D, Zizi M, Breiman A (1995) Multiple cDNAs of wheat voltage-dependent anion channels (VDAC): isolation, differential expression, mapping and evolution. Plant Mol Biol 29: 109–124. [DOI] [PubMed] [Google Scholar]
- 24. Elkeles A, Breiman A, Zizi M (1997) Functional differences among wheat voltage-dependent anion channel (VDAC) isoforms expressed in yeast. Indication for the presence of a novel VDAC-modulating protein? J Biol Chem 272: 6252–6260. [DOI] [PubMed] [Google Scholar]
- 25. Heins L, Mentzel H, Schmid A, Benz R, Schmitz UK (1994) Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J Biol Chem 269: 26402–26410. [PubMed] [Google Scholar]
- 26. Tateda C, Yamashita K, Takahashi F, Kusano T, Takahashi Y (2009) Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Rep 28: 41–51. [DOI] [PubMed] [Google Scholar]
- 27. Homble F, Krammer EM, Prevost M (2012) Plant VDAC: Facts and speculations. Biochim Biophys Acta 1818: 1486–1501. [DOI] [PubMed] [Google Scholar]
- 28. Takahashi Y, Tateda C (2013) The functions of voltage-dependent anion channels in plants. Apoptosis 18: 917–924. [DOI] [PubMed] [Google Scholar]
- 29. Tateda C, Watanabe K, Kusano T, Takahashi Y (2011) Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis . J Exp Bot 62: 4773–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robert N, d'Erfurth I, Marmagne A, Erhardt M, Allot M, et al. (2012) Voltage-dependent-anion-channels (VDACs) in Arabidopsis have a dual localization in the cell but show a distinct role in mitochondria. Plant Mol Biol 78: 431–446. [DOI] [PubMed] [Google Scholar]
- 31. Mitchell P, Moyle J (1965) Evidence discriminating between the chemical and the chemiosmotic mechanisms of electron transport phosphorylation. Nature 208: 1205–1206. [DOI] [PubMed] [Google Scholar]
- 32. Mitchell P, Moyle J (1965) Stoichiometry of proton translocation through the respiratory chain and adenosine triphosphatase systems of rat liver mitochondria. Nature 208: 147–151. [DOI] [PubMed] [Google Scholar]
- 33. Tsujimoto Y, Shimizu S (2007) Role of the mitochondrial membrane permeability transition in cell death. Apoptosis 12: 835–840. [DOI] [PubMed] [Google Scholar]
- 34. Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64. [Google Scholar]
- 35. Pedersen PL (2007) Warburg, me and Hexokinase 2: Multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the “Warburg Effect”. i.e., elevated glycolysis in the presence of oxygen. J Bioenerg Biomembr 39: 211–222. [DOI] [PubMed] [Google Scholar]
- 36. Tsujimoto Y, Shimizu S (2000) VDAC regulation by the Bcl-2 family of proteins. Cell Death Differ 7: 1174–1181. [DOI] [PubMed] [Google Scholar]
- 37. Motamayor JC, Vezon D, Bajon C, Sauvanet A, Grandjean O, et al. (2000) Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sex Plant Reprod 12: 209–218. [Google Scholar]
- 38. Wang Y, Hou Y, Gu H, Kang D, Chen Z, et al. (2012) The Arabidopsis APC4 subunit of the anaphase-promoting complex/cyclosome (APC/C) is critical for both female gametogenesis and embryogenesis. Plant J 69: 227–240. [DOI] [PubMed] [Google Scholar]
- 39. Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, et al. (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124. [DOI] [PubMed] [Google Scholar]
- 41. Jurgens G (2005) Cytokinesis in higher plants. Annu Rev Plant Biol 56: 281–299. [DOI] [PubMed] [Google Scholar]
- 42. Kull FJ, Endow SA (2013) Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci 126: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2 . Plant Cell 8: 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schneitz K, Hulskamp M, Kopczak SD, Pruitt RE (1997) Dissection of sexual organ ontogenesis: a genetic analysis of ovule development in Arabidopsis thaliana . Development 124: 1367–1376. [DOI] [PubMed] [Google Scholar]
- 45. Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, et al. (2003) TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson-Beers K, Pruitt RE, Gasser CS (1992) Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, et al. (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS (1997) Interactions among genes regulating ovule development in Arabidopsis thaliana . Genetics 145: 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Linke B, Borner T (2005) Mitochondrial effects on flower and pollen development. Mitochondrion 5: 389–402. [DOI] [PubMed] [Google Scholar]
- 51. Carlsson J, Leino M, Sohlberg J, Sundstroem JF, Glimelius K (2008) Mitochondrial regulation of flower development. Mitochondrion 8: 74–86. [DOI] [PubMed] [Google Scholar]
- 52. Boateng KA, Yang X, Dong F, Owen HA, Makaroff CA (2008) SWI1 is required for meiotic chromosome remodeling events. Mol Plant 1: 620–633. [DOI] [PubMed] [Google Scholar]
- 53. Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, et al. (2004) Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics 3: 675–691. [DOI] [PubMed] [Google Scholar]
- 54. Shimizu S, Shinohara Y, Tsujimoto Y (2000) Bax and Bcl-xL independently regulate apoptotic changes of yeast mitochondria that require VDAC but not adenine nucleotide translocator. Oncogene 19: 4309–4318. [DOI] [PubMed] [Google Scholar]
- 55. Vianello A, Zancani M, Peresson C, Petrussa E, Casolo V, et al. (2007) Plant mitochondrial pathway leading to programmed cell death. Physiol Plant 129: 242–252. [Google Scholar]
- 56. Anflous K, Armstrong DD, Craigen WJ (2001) Altered mitochondrial sensitivity for ADP and maintenance of creatine-stimulated respiration in oxidative striated muscles from VDAC1-deficient mice. J Biol Chem 276: 1954–1960. [DOI] [PubMed] [Google Scholar]
- 57. Graham BH, Li Z, Alesii EP, Versteken P, Lee C, et al. (2010) Neurologic dysfunction and male infertility in Drosophila porin mutants: a new model for mitochondrial dysfunction and disease. J Biol Chem 285: 11143–11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abu-Hamad S, Sivan S, Shoshan-Barmatz V (2006) The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc Natl Acad Sci USA 103: 5787–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang X-Y, Chen Z-W, Xu T, Qu Z, Pan X-D, et al. (2011) Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell 23: 1093–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Skinner DJ, Baker SC, Meister RJ, Broadhvest J, Schneitz K, et al. (2001) The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13: 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Portereiko MF, Sandaklie-Nikolova L, Lloyd A, Dever CA, Otsuga D, et al. (2006) NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant physiol 141: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant physiol 143: 1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan XY, Liu XL, Wang W, Jia DJ, Chen LQ, et al. (2010) Mutations in the Arabidopsis nuclear-encoded mitochondrial phage-type RNA polymerase gene RPOTm led to defects in pollen tube growth, female gametogenesis and embryogenesis. Plant Cell Physiol 51: 635–649. [DOI] [PubMed] [Google Scholar]
- 64. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 65. Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, et al. (2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Day DA, Hanson JB (1978) On methods for the isolation of mitochondria from etiolated corn shoots. Plant Sci Lett 11: 99–104. [Google Scholar]
- 67. Narita M, Shimizu S, Ito T, Chittenden T, Lutz RJ, et al. (1998) Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc Natl Acad Sci USA 95: 14681–14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, et al. (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR analysis of AtVDAC1 gene transcripts in wild type (WT) and the atvdac1 mutant. (A) Schematic diagram of the AtVDAC1 gene structure and the T-DNA insertion sites in the atvdac1 mutant. Closed boxes indicate exons, and arrowheads indicate the positions of primers. The start and stop codons are labeled. (B) RT-PCR analysis of AtVDAC1 gene transcripts in WT and atvdac1 mutant. The UBQ5 transcript was amplified as an internal control.
(TIF)
Statistics for lengths of siliques from wild type (WT), atvdac1 and reciprocal crosses between wild type (WT) and atvdac1 .
(DOCX)
Statistics for lengths of siliques from wild type (WT), atvdac1 and complemented lines.
(DOCX)
Statistics for lengths of siliques from atvdac1 /+ selfed and reciprocal crosses between wild type (WT) and atvdac1 /+.
(DOCX)
Sequences of the primers used in this study.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.