Abstract
RNA silencing suppressors from different plant viruses are structurally diverse. In addition to inhibiting the antiviral silencing response to condition susceptibility, many suppressors are pathogenicity factors that cause disease or developmental abnormalities. Here, unrelated suppressors from multiple viruses were shown to inhibit microRNA (miRNA) activities and trigger an overlapping series of severe developmental defects in transgenic Arabidopsis thaliana. This suggests that interference with miRNA-directed processes may be a general feature contributing to pathogenicity of many viruses. A normally labile intermediate in the miRNA biogenesis/RNA-induced silencing complex (RISC) assembly pathway, miRNA*, accumulated specifically in the presence of suppressors (P1/HC-Pro, p21, or p19) that inhibited miRNA-guided cleavage of target mRNAs. Both p21 and p19, but not P1/HC-Pro, interacted with miRNA/miRNA* complexes and hairpin RNA-derived short interfering RNAs (siRNAs) in vivo. In addition, p21 bound to synthetic miRNA/miRNA* and siRNA duplexes in vitro. We propose that several different suppressors act by distinct mechanisms to inhibit the incorporation of small RNAs into active RISCs.
Keywords: MicroRNA, RNA silencing, RNAi, virus-encoded suppressors, RISC
Systemic infection by plant viruses frequently results in disease symptoms that resemble developmental defects, including loss of leaf polarity, loss of proper control of cell division, and loss of reproductive functions (Hull 2001). These and other phenotypes are frequently associated with virus-encoded pathogenicity factors, many of which are suppressors of RNA silencing (Voinnet et al. 1999). RNA silencing functions as an adaptive immune response that restricts accumulation or spread of inducing viruses (Waterhouse et al. 2001). Suppressor proteins encoded by members of different virus families are distinct, suggesting that plant viruses evolved this counter-defensive mechanism independently on many occasions (Vaucheret et al. 2001; Tijsterman et al. 2002).
RNA silencing during virus infection is triggered by double-stranded RNA (dsRNA) generated during the course of virus replication or by the activity of a cellular RNA-dependent RNA polymerase (Ahlquist 2002). Processing of dsRNA by DICER or DICER-LIKE enzymes results in heterogeneous short interfering RNAs (siRNAs) of 21–25 nucleotides (Finnegan and Matzke 2003). siRNAs incorporate into RNA-induced silencing complexes (RISCs; Zamore et al. 2000; Elbashir et al. 2001b) and provide guide functions for sequence-specific ribonucleolytic activity. Protein components of RISC include ARGONAUTE family members, nucleases, and other factors (Hannon 2002).
microRNAs (miRNAs, ∼21 nucleotides) are chemically similar to siRNAs, but they arise from processing of imperfect hairpin-forming RNA precursors transcribed from miRNA genes (Ambros et al. 2003). miRNA processing occurs by a multistep mechanism involving DICER (DICER-LIKE1 [DCL1] in Arabidopsis) activity to excise the miRNA from the hairpin stem (Bartel 2004). miRNAs function as negative regulators of target mRNAs through directing either site-specific cleavage by RISCs or translational repression (Bartel 2004). In plants, miRNAs target a wide range of mRNAs encoding transcription factors required for development (Park et al. 2002; Rhoades et al. 2002; Palatnik et al. 2003). These include factors required for meristem identity and maintenance, patterning, cell division, hormone signal ing, and developmental timing. In addition, plant miRNAs also target mRNAs encoding miRNA metabolic factors and factors of unknown function (Rhoades et al. 2002; Xie et al. 2003). Loss of miRNA biogenesis or activity in Arabidopsis results in pleiotropic defects during embryonic, vegetative, and reproductive development (Park et al. 2002; Schauer et al. 2002; Kasschau et al. 2003).
Despite differences in precursor structure for siRNAs (perfectly paired dsRNA) and miRNAs (imperfect hairpins), functional siRNA and miRNA molecules are incorporated into RISC through an asymmetric strand selection process. Precursor processing yields a duplex intermediate containing perfectly (siRNA) or imperfectly (miRNA) paired strands with two unpaired bases at each 3′ end (Elbashir et al. 2001a). This duplex intermediate is unwound prior to, or during, incorporation of one strand into RISCs. Strand asymmetry depends on the strength of base-pair interactions at each end of the duplex, with the molecule containing the 5′ end participating in the weakest interaction preferentially used (Khvorova et al. 2003; Schwarz et al. 2003). The nonselected strand (siRNA* or miRNA*) is rapidly degraded (Bartel 2004).
We previously showed that the Turnip mosaic virus (TuMV) silencing suppressor, P1/HC-Pro, interferes with miRNA-guided regulation of at least 10 target mRNAs in infected or transgenic Arabidopsis plants. The suppressor also caused multiple developmental defects, including some resembling those associated with dcl1 mutants (Kasschau et al. 2003). Here, we show that interference with miRNA-guided target cleavage/degradation and development in Arabidopsis is a general property of several, unrelated silencing suppressors encoded by evolutionarily distinct viruses. Through analysis of miRNA processing intermediates and suppressor-containing complexes in vivo, three of these suppressors were shown to inhibit the RISC assembly pathway (by two distinct mechanisms) after DCL1-catalyzed formation of miRNA/miRNA* duplexes.
Results
Interference with miRNA-guided target cleavage and development in Arabidopsis by P1/HC-Pro was shown previously by using virus-infected and transgenic plants. To determine if miRNA interference is a general property of RNA silencing suppressors, the Beet yellows virus p21 (Reed et al. 2003), Tomato bushy stunt virus p19 (Voinnet et al. 1999; Silhavy et al. 2002), Turnip crinkle virus coat protein (CP; Qu et al. 2003; Thomas et al. 2003), and Cucumber mosaic virus 2b (Brigneti et al. 1998; Guo and Ding 2002) silencing suppressors were analyzed and compared with P1/HC-Pro (Anandalakshmi et al. 1998; Brigneti et al. 1998; Kasschau and Carrington 1998). These suppressors belong to evolutionarily and structurally unrelated protein families (Dolja and Koonin 1991; Koonin et al. 1991; Reed et al. 2003; Vargason et al. 2003). To facilitate detection and experimental manipulation, constructs were expressed by using Cauliflower mosaic virus 35S promoter and terminator sequences and engineered such that an influenza hemagglutinin (HA) epitope tag was added to the C terminus of each protein (Fig. 1A). The HA-tagged proteins were tested for functionality as RNA silencing suppressors using Agrobacterium-mediated transient assays in Nicotiana benthamiana leaves (Johansen and Carrington 2001). This assay measures the ability of a suppressor to inhibit silencing of the GFP sequence triggered by an inverted-repeat hairpin construct. All five suppressors inhibited silencing in transient assays, although 2b was relatively weak (Supplementary Fig. S1A–C). Only CP inhibited siRNA accumulation (Supplementary Fig. S1D). Functionality of the HA-tagged proteins was also tested in side-by-side comparisons to nontagged forms of each suppressor. Inhibition of silencing by each HA-tagged protein was comparable to inhibition by the corresponding nontagged form (data not shown).
Figure 1.
Developmental defects induced by silencing suppressors from five viruses. (A) Diagram of constructs used for expression of HA epitope-tagged viral suppressor proteins. Each construct contained the gene for one of the suppressors shown. (P35S) 35S promoter from Cauliflower mosaic virus; (TL) translational leader element from Tobacco etch virus; (T35S) 35S terminator from Cauliflower mosaic virus. (B) Expression of epitope-tagged silencing suppressors in transgenic Arabidopsis plants. Immunoblot analysis was done by using total protein samples (amounts shown) from plants transformed with empty vector (V) or constructs encoding P1/HC-Pro (HC), p21, 2b, CP, or p19. Mobility positions of 16–62-kDa protein standards are shown. Note that HC-Pro migrates as ∼50-kDa protein, but that breakdown fragments migrate at ∼25-kDa and ∼35-kDa positions. (C) Effects of silencing suppressors on leaf and rosette morphogenesis. Bars, 10 mm. (D) Effects of silencing suppressors on flowers (stage 11–12). Bars, 1 mm.
Developmental abnormalities in Arabidopsis expressing silencing suppressors
To determine if induction of developmental phenotypes is a general property of silencing suppressors, the five HA-tagged constructs, as well as empty vector, were introduced as transgenes into Arabidopsis Col-0 plants (Fig. 1B). Growth and development parameters were analyzed by using a minimum of 58 primary transformants expressing each suppressor.
Plants expressing P1/HC-Pro, p21, p19, and CP exhibited moderate to severe defects in leaf and rosette development (Fig. 1C). Rosette leaves (growth stage 5.1 according to the scale described by Boyes et al. [2001]) were narrow, lobed or serrated, or curled (Fig. 1C; Supplementary Table S1). Rosette diameter and leaf area were reduced, as were the weight of total aerial tissue and the length of the primary bolt (Supplementary Fig. S2). Plants expressing CP displayed leaf and rosette phenotypes that were generally mild compared with those in plants expressing P1/HC-Pro, p21, and p19. Plants expressing 2b were indistinguishable from vector-transformed plants with respect to leaf morphology (Fig. 1C), although modest reductions in aerial tissue weight, leaf area, and rosette diameter were detected (Supplementary Fig. S2A).
Plants expressing P1/HC-Pro, p21, p19, and CP also had obvious flower phenotypes and were generally infertile (Fig. 1D; Supplementary Fig. S2B). They failed to release pollen and, in the case of P1/HC-Pro-expressing plants, had split or nonfused carpels (Supplementary Table S1; Supplementary Fig. S2B). P1/HC-Pro, p21, and CP expressing plants had narrow and unusually long sepals, whereas plants expressing p19 had short sepals (Fig. 1D; Supplementary Fig. S2B; Supplementary Table S1). In all four cases, however, organs in the internal whorls were exposed prior to opening (Fig. 1D). At a low frequency, plants expressing P1/HC-Pro, p19, or 2b contained additional trichomes on abaxial or adaxial sepals (Supplementary Table S1). Plants expressing 2b were fertile, although they contained fewer flowers per plant compared with control plants (Supplementary Fig. S2B).
Therefore, the correlation between strong RNA silencing suppressor activity (as measured in the hairpin dsRNA silencing assay) and strong developmental phenotypes in Arabidopsis is relatively high. The protein with weak silencing suppressor activity in the transient assay (2b) caused only mild developmental abnormalities in transgenic plants. The data also support the hypothesis that pathogenicity associated with these proteins involves, at least partly, interference with growth and development during virus infection.
Interference with miRNA-guided mRNA cleavage by three silencing suppressors
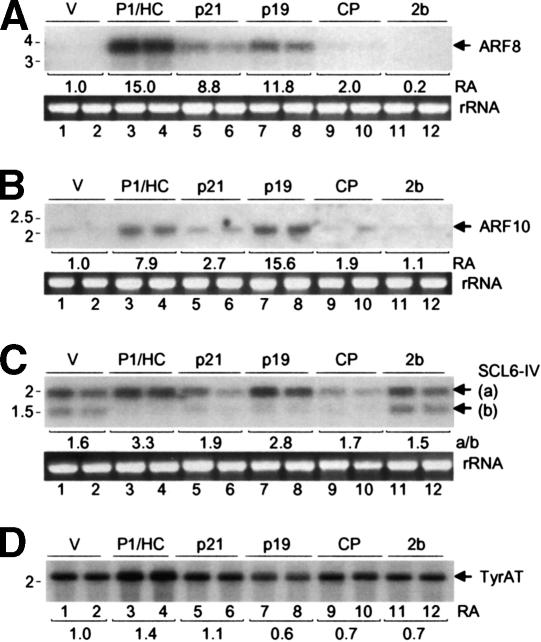
Developmental abnormalities in Arabidopsis plants expressing P1/HC-Pro correlate with inhibition of miRNA-guided target regulation (Kasschau et al. 2003). To determine if this is a general property of strong RNA silencing suppressors, the levels of three mRNAs (ARF8, ARF10, and SCL6-IV) that are normally under negative regulation by miRNAs (miR167, miR160, and miR171, respectively) were measured in transgenic plants expressing each of the suppressors and compared relative to the levels in vector-transformed plants. The ARF8 and ARF10 mRNAs accumulated to higher levels in plants expressing P1/HC-Pro, p21, and p19 (Fig. 2A,B, lanes 3–8). Also, the ratio of SCL6-IV full-length to 3′ cleavage product detected by blot assay was higher in plants expressing P1/HC-Pro, p21, and p19 (Fig. 2C, lanes 3–8). In plants expressing CP, only slight increases in ARF8 and ARF10 mRNA levels, and no change in the ratio of SCL6-IV RNAs, were measured (Fig. 2, lanes 9,10). Plants expressing 2b showed no increases in the levels or ratios of miRNA targets (Fig. 2, lanes 11,12).
Figure 2.
Blot analysis of miRNA targets in transgenic Arabidopsis plants. RNA samples from vector-transformed (lanes 1,2) and suppressor-expressing transgenic Arabidopsis plants (lanes 3–12) were analyzed in duplicate by hybridization with DNA probes. Ethidium bromide-stained 28S rRNA is shown for each blot. Mobility positions of relevant RNA size standards (kb) are shown. (A) Accumulation of ARF8 (At 5 g 37020) mRNA. Mean relative accumulation (RA) of ARF8 mRNA in suppressor expressing plants was calculated relative to that in vector-transformed plants, normalized against the accumulation of the control TyrAT (At 2 g 20610) mRNA. (B) Accumulation of ARF10 (At 2 g 28350) mRNA. Mean RA of ARF10 mRNA was calculated as in A.(C) Expression of full-length SCL6-IV (At 4 g 00150) mRNA. The mean ratio of full-length SCL6-IV mRNA (a) to the cleavage product (b) is shown. (D) Expression of TyrAT (At 2 g 20610) mRNA. This blot was first used to analyze ARF10 expression, then stripped and reprobed.
The ARF8, ARF10, or SCL6-IV mRNAs are targeted by three different miRNAs (Llave et al. 2002b; Rhoades et al. 2002; Kasschau et al. 2003). Increased accumulation of these target mRNAs in the presence of silencing suppressors could conceivably be the result of lower levels of miRNAs, although this would be inconsistent with previous studies (Mallory et al. 2002; Kasschau et al. 2003). Indeed, there was no consistent decrease among the three miRNAs caused by any of the suppressors (Fig. 3).
Figure 3.
miRNA and miRNA* accumulation in transgenic Arabidopsis plants. Small RNA samples from vector-transformed (lanes 1,2) and suppressor expressing transgenic Arabidopsis plants (lanes 3–12) were analyzed in duplicate by hybridization with oligonucleotide probes. Ethidium bromide-stained tRNA and 5S rRNA are shown below each blot, and mobility positions of 24- and 21-nucleotide RNA size standards are shown. Mean relative accumulation (RA) of miRNA or miRNA* signal relative to that in vector-transformed plants (lanes 1,2) is shown. (A) Accumulation of miR167 and miR167* from the miR167b locus. (B) Accumulation of miR160 and miR160* from the miR160c locus. (C) Accumulation of miR171 and miR171*.
Strong RNA silencing suppressors stabilize an intermediate in the miRNA pathway
The P1/HC-Pro, p21, and p19 silencing suppressors interfered with siRNA-guided and miRNA-guided target cleavage but not with siRNA or miRNA formation, suggesting that inhibition is likely associated with RISC assembly or RISC activity (Figs. 2, 3; Supplementary Fig. S1B–D). Assembly of RISCs containing miRNA requires unwinding of the miRNA/miRNA* duplex intermediate, followed by (or concurrent with) incorporation of miRNA into RISC and degradation of miRNA* (Khvorova et al. 2003; Schwarz et al. 2003). If the strong suppressors inhibit RISC assembly at the point of unwinding miRNA/miRNA*, we predicted that miRNA* species would accumulate specifically in plants expressing these proteins. The miR167b*, miR160c*, and miR171* levels were below the detection limit in vector-transformed plants (Fig. 3, lanes 1,2). However, they were each detected in plants expressing P1/HC-Pro, p21, and p19 (Fig. 3, lanes 3–8), with the highest levels of each accumulating in P1/HC-Pro-expressing plants. In contrast, each miRNA* accumulated to relatively low or nondetectable levels in plants expressing CP or 2b proteins (Fig. 3, lanes 9–12). Therefore, strong suppression of miRNA-guided target cleavage correlated with accumulation of normally labile miRNA* species, suggesting that the strong suppressors interfered with unwinding of miRNA/miRNA* duplexes.
Interaction of silencing suppressors and miRNA/miRNA* in vivo and in vitro
Tombusvirus p19 dimers bind duplex siRNAs (Silhavy et al. 2002; Vargason et al. 2003; Ye et al. 2003). This led to a competitive inhibition model in which p19 interferes with RISC assembly by sequestering siRNA intermediates (Lakatos et al. 2004). We tested the hypothesis that each of the three strong suppressors interacted with miRNA/miRNA* complexes in vivo by coimmunoprecipitation (co-IP) assays using anti-HA monoclonal antibody. Precipitated complexes from inflorescence (P1/HC-Pro and p21) and total aerial (P1/HC-Pro, p21, and p19) tissues were analyzed for suppressor protein, miRNAs, and miRNAs*. As controls, IP assays were done by using extracts from vector-transformed plants and by using a heterologous monoclonal antibody mixture specific for NIa and NIb proteins of Tobacco etch virus.
Each suppressor protein was detected in total extracts (IP inputs) and in immunoprecipitated material using HA antibody, but not in IP fractions using NIa/NIb antibody (Fig. 4A,B, top). The input extracts from the suppressor-expressing plants contained each of the miRNAs and miRNAs* (miR167, miR171, miR167b*, miR171*, and miR160c*) analyzed in the respective tissues (Fig. 4A [lanes 4,7], B [lanes 4,7,10]). No miRNAs or miRNAs* were detected in any IP fractions from plants expressing P1/HC-Pro (Fig. 4A,B, lanes 5,6). In contrast, each miRNA and miRNA*, but no 5S rRNA, coimmunoprecipitated with p21 in both tissue types (Fig. 4A,B, lane 9). Similarly, miR167, miR167b*, miR171, and miR160c* each specifically coimmunoprecipitated with p19 in aerial tissue extracts (Fig. 4B, lane 12), although the proportion of small RNA in the IP fractions relative to the input extract was less using p19 expressing plants compared to p21 expressing plants. In vector-transformed plant extracts, miR167, miR171, and 5S rRNA were detected (Fig. 4A,B, lane 1), but none were present in the IP fractions (Fig. 4, lanes 2,3). Faster migrating forms of some miRNAs and miRNAs* were detected in total extracts or IP fractions in the presence of P1/HC-Pro or p19 (Fig. 4, lanes 4,9,12). Truncated miRNAs in the presence of p19 were also observed previously by others (Papp et al. 2003).
Figure 4.
Interactions between suppressor proteins and small RNAs. (A) Co-IP of suppressor proteins, miRNAs, and miRNAs* from transgenic Arabidopsis inflorescence tissue. Immunoblot analysis was done using IP input (in) samples and immunoprecipitated fractions using HA or NIa/NIb monoclonal antibodies. Mobility positions of 16–62-kDa protein size standards are shown. Blot hybridization assays were done by using RNA recovered from immunoprecipitates. Mobility positions of 21- and 24-nucleotide RNA size standards are shown. (B) Co-IP of suppressor proteins, miRNAs, and miRNAs* from transgenic Arabidopsis total aerial tissue. (C) Co-IP of suppressor proteins and GFP-specific siRNAs from Agrobacterium-infiltrated N. benthamiana leaf tissue. (D) Electrophoretic mobility shift assays using purified p21 and siRNA duplex, miR171, miR171*, and miR171/miR171* duplex. Complexes were analyzed by native PAGE. The positions of p21:RNA complexes and free probes are shown.
To confirm that co-IP of p21 and p19 with miRNA and miRNA* was not a peculiar artefact of the transgenic system, IP assays were also done by using extracts from N. benthamiana leaves expressing the GFP hairpin RNA construct and each suppressor. siRNAs related to the GFP sequence ranged in size between 21 and 24 nucleotides, as shown previously (Fig. 4C, lanes 1,4,7). siRNAs specifically coimmunoprecipitated with both p21 and p19 (Fig. 4C, lanes 6,9). Only the small size-class (∼21 nucleotides) coimmunoprecipitated with p19, as expected from the well-characterized binding properties (Fig. 4C, lane 9; Silhavy et al. 2002; Vargason et al. 2003; Ye et al. 2003). No siRNA was detected in IP fractions from tissue expressing GFP siRNA and P1/HC-Pro, although the efficiency of IP of P1/HC-Pro after transient expression was relatively low (data not shown).
We tested the hypothesis that p21 binds small RNA duplexes directly by electrophoretic mobility shift assays by using purified recombinant p21 and synthetic miR171, miR171*, miR171/miR171* duplex, or an siRNA duplex. The miR171/miR171* duplex contained two mismatched positions and two G:U base-pairs. In the absence of p21 protein, each single-stranded and duplex RNA migrated to near the bottom of the gel (Fig. 4D, lanes 1,3,5,7). In the presence of p21, slower-migrating complexes were detected by using the siRNA duplex and miR171/miR171* duplex (Fig. 4D, lanes 2,8). No p21 complexes were detected using single-stranded miR171 or miR171* (Fig. 4D, lanes 4,6). These data indicate that p21 interacts directly with small RNA duplexes, regardless of whether or not the duplex contains perfectly complementary (siRNA) or mismatched (miRNA) strands. By using DNA oligonucleotides corresponding to a duplex siRNA, low levels of p21 complex were detected (<10% of the level detected using the RNA duplexes; data not shown).
Discussion
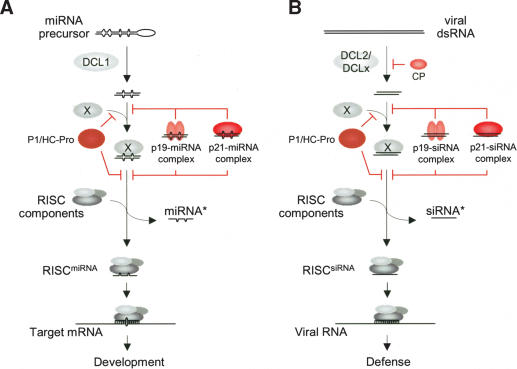
Among the five silencing suppressors analyzed, three (P1/HC-Pro, p21, and p19) were characterized as strong due to their effects on siRNA-guided cleavage, miRNA-guided cleavage of target mRNAs, and development. However, we propose that these three suppressors function by at least two distinct mechanisms to arrest the RISC assembly pathway. Although each suppressor inhibited turnover of miRNA* species, a feature we interpret is due to lack of unwinding of miRNA/miRNA* duplexes, only two (p21 and p19) could be detected in a complex with miRNAs and miRNAs* in vivo. For both p21 and p19, a direct binding model seems quite likely given the clear duplex siRNA- and miRNA-binding properties of p21 and p19. Sequestration of miRNA/miRNA* duplexes may occur in the cytoplasm after processing by DCL1 in the nucleus and subsequent nucleocytoplasmic transport (Zamore 2004). Cytoplasmic localization of p21 is in accord with this hypothesis (Reed et al. 2003).
Although P1/HC-Pro was the most effective miRNA pathway suppressor tested, we obtained no evidence that it interacts with miRNAs or miRNAs* in vivo. However, it clearly inhibited miRNA* turnover, again suggesting that miRNA/miRNA* unwinding and RISC assembly was suppressed. One possibility is that the interaction between P1/HC-Pro and miRNA/miRNA* is too weak to be detected by using an IP assay. However, one would not expect the suppressor causing the strongest effects on inhibition of miRNA-guided target cleavage, promotion of miRNA* accumulation, and development to interact most weakly with small RNAs if direct binding was the mode of action. We suggest an alternative model in which P1/HC-Pro interferes with a protein (Fig. 5, protein X) or complex associated with miRNA/miRNA* duplexes (as well as siRNA duplexes). P1/HC-Pro might also inhibit the miRNA/miRNA* complex indirectly by suppressing a factor required for production of one or more components associated with the complex. In either case, interaction between P1/HC-Pro and a pathway component would prevent unwinding and assembly of active RISC. Among the known RNAi factors from animals, the dsRNA-binding protein R2D2 plays a role in transfer of siRNAs between DICERs and RISCs (Liu et al. 2003). A functionally equivalent protein in plants, however, has yet to be identified.
Figure 5.
Mechanisms of suppression of miRNA and siRNA pathways by silencing suppressors. (A) microRNA pathway. (B) Antiviral siRNA pathway. p19 and p21 sequester miRNA/miRNA* and siRNA duplexes, stabilizing both strands. The point in the pathway at which p19 and p21 sequester small RNA duplexes is not known. P1/HC-Pro inhibits duplex unwinding and therefore stabilizes both strands, but by a mechanism that does not involve direct binding to small RNA duplexes. Protein X is a hypothetical protein with a role comparable to animal R2D2 (Liu et al. 2003). In both pathways, interference by HC-Pro, p19, and p21 prevents RISC assembly and subsequent target RNA degradation. The TCV CP is proposed to interfere with DCL2 or other DICER-LIKE activities required for TCV-derived siRNA formation (Xie et al. 2004), but not DCL1.
Among the other two silencing suppressors analyzed in this study, TCV CP functioned effectively as an inhibitor of siRNA formation but only weakly as a miRNA pathway suppressor. This suppressor may primarily inhibit DCL2 or other DCL activities required for production of TCV-derived siRNA (Xie et al. 2004), but not DCL1 required for miRNA processing. Nevertheless, CP had small but measurable effects on miRNA-directed target cleavage, which may partly explain the developmental consequences of CP expression in Arabidopsis. The CMV 2b protein had very little effect on miRNA-guided functions and development in Arabidopsis, and may function by mechanisms that are quite distinct from the others analyzed (Brigneti et al. 1998; Guo and Ding 2002).
Finally, is there a physiologic advantage to viruses that interfere with miRNA-guided gene regulation? If miRNAs are required for defense responses or for expression of genes required for susceptibility to a broad range of viruses, then evolution of virus-encoded miRNA inhibition functions can be easily rationalized. However, in view of the functions of known miRNA target genes in plants, and the lack of effects of dcl1 (miRNA-deficient) mutations on virus susceptibility (Z. Xie and J.C. Carrington, unpubl.), no evidence for such requirements is available. It seems more likely that interference with the miRNA pathway by suppressors such as P1/HC-Pro, p21, and p19 is a consequence of inhibition of shared steps in the silencing pathways involving siRNAs for antiviral defense and miRNAs for development.
Materials and methods
Genes and constructs
Suppressors were derived from the following viruses: P1/HC-Pro, TuMV; p21, Beet yellows virus; p19, Tomato bushy stunt virus; CP, Turnip crinkle virus; and 2b, Cucumber mosaic virus. Coding sequences for each suppressor were amplified by PCR using a 3′ primer that added a C-terminal HA epitope (primer sequences available upon request). Resulting DNA fragments were cloned into a modified pCB-302 plant transformation vector (Peng and Dolja 2000) using NcoI and XbaI sites. Suppressor constructs contained a Cauliflower mosaic virus 35S promoter anda5′ nontranslated leader sequence from Tobacco etch virus (Carrington and Freed 1990). Resulting constructs were introduced into Agrobacterium tumefaciens strains GV2260 and GV3101.
Transgenic plants
Arabidopsis thaliana Col-0 plants were transformed by the vacuum-infiltration method (Clough and Bent 1998) by using A. tumefaciens GV3101 carrying constructs for expression of epitope-tagged P1/HC-Pro, p21, p19, CP, 2b, or the empty expression vector. Seed from primary transformants was grown under selection for phosphoinothricin resistance in a standard greenhouse.
Protein and RNA blot analysis
Protein extracts were prepared and normalized for SDS-PAGE by using the Bradford assay (Bio-Rad). Immunoblot analysis of total protein samples (10 μg) was done using anti-HA-peroxidase conjugate (Roche). Total RNA was extracted from independent pools of leaf or inflorescence tissues by using Trizol reagent (Johansen and Carrington 2001). Low-molecular-weight RNA was isolated with RNA/DNA Midi Kits (Qiagen). Blot hybridization of normalized total or low-molecular-weight RNA (5 μg) was done as described (Llave et al. 2002a), and hybridization intensities were quantified by using a PhosphorImager or Scanning Densitometer (Molecular Dynamics). 32P-radiolabeled probes for mRNAs were synthesized by random-priming of cloned genomic sequences (Feinberg and Vogelstein 1983). 32P-Radiolabeled miRNA probes were produced by end-labeling of complementary oligonucleotides. Sequences for miRNA* probes were predicted from miRNA precursor structures (Llave et al. 2002a; Reinhart et al. 2002). Accumulation of miRNA-targeted mRNAs was normalized to levels of mRNA from the control gene TyrAT (At 2 g 20610). Accumulation of SCL6-IV full-length mRNA (“a” form) relative to the 3′ cleavage product (“b” form) was represented as a ratio as described (Llave et al. 2002b).
Immunoprecipitation
Aerial or inflorescence tissue from transgenic Arabidopsis plants (5 wk old), and leaf tissue from Agrobacterium-infiltrated N. benthamiana, were ground under liquid nitrogen and homogenized in 5 mL/g lysis buffer (50 mM Tris-HCl at pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 0.1% NP-40, and 2× complete protease inhibitor cocktail; Roche). Cell debris was pelleted by centrifugation for 15 min at 9500 × g. The clarified lysate was precleared for 20 min at 4°C with 10 μL bed volume protein A-agarose (30 μg protein A) per milliliter. Precleared lysates were reacted with 4 μg anti-HA (Roche) or anti-NIa/NIb (Slade et al. 1989) per milliliter for 1 h at 4°C, then with 50 μL bed volume protein A-agarose (150 μg protein A) per milliliter for 3 h at 4°C. Precipitates were washed three times in lysis buffer and divided for protein and RNA analysis. Nucleic acid was recovered by treatment with 3 v proteinase K solution (100 mM Tris-HCl at pH 7.4, 10 mM EDTA, 150 mM NaCl, 2% SDS, and 0.2 μg/μL proteinase K) for 15 min at 65°C, extraction with saturated phenol and phenol:chloroform, and ethanol precipitation. For miRNA and miRNA* blot assays, 5 μg of RNA recovered from the input extract, or RNA from IP fractions representing 150 mg tissue, was used. Five micrograms of RNA recovered from input extracts corresponded to the equivalent of ∼5 mg tissue. As a control, blots were stripped and rehybridized with an oligonucleotide probe specific to 5S rRNA.
For IP assays using Agrobacterium-infiltrated leaves, GFP silencing was induced as described (Johansen and Carrington 2001). A. tumefaciens cultures were injected at the following concentrations: 35S:dsGFP-FAD2, O.D.600 = 0.1; 35S:vector,35S:P1/HC-Pro, 35S:p21, or 35S:p19, O.D.600 = 0.9. Infiltrated tissue was harvested and processed 72 h postinjection. 32P-Radiolabeled probes for siRNAs were synthesized by random-priming of cloned smGFP sequence (Johansen and Carrington 2001).
Electrophoretic mobility shift assays
The p21 coding sequence was amplified by PCR using a 5′ primer that added an N-terminal hexahistidine tag (primer sequences available upon request). Resulting DNA fragments were cloned into pET16b (Novagen) by using NcoI and BamHI sites. The resulting construct was introduced into Escherichia coli strain BL21(DE3) (Novagen), and protein was expressed and purified under native conditions by using Ni-NTA resin (Qiagen) following the recommendations of the manufacturer.
Probes were synthesized by end-labeling RNA oligonucleotides miR171 (5′-UGAUUGAGCCGCGCCAAUAUC-3′), miR171* (5′-UAUUGGCCUGGUUCACUCAGA-3′), siRNA (5′-CGUAC GCGGAAUACUUCGAUU-3′), or siRNA* (5′-UCGAAGUAUUC CGCGUACGUG-3′; Dharmacon) using [32P]ATP. Duplexes were formed during annealing reactions similar to those described by others (Silhavy et al. 2002). Formation of duplexes was confirmed by electrophoresis mobility assays. p21 complex formation reactions contained 1 μM p21 and 0.1 μM oligonucleotide in 10 μL binding buffer (0.1 M KCL, 25 mM HEPES, and 10 mM DTT at pH 7.6; Ye et al. 2003) and were done for 15 min at 23°C.
Acknowledgments
We thank Shou-Wei Ding, Herman Scholthof, and Jack Morris for providing us with cDNA clones encoding viral RNA silencing suppressors. We also thank Kristin D. Kasschau and Lisa K. Johansen for helpful discussions throughout the course of this study, and Taiowa Montgomery and Karen A. Kraxberger for assistance with data collection. This work was supported by grants from the National Science Foundation (MCB-0209836), the National Institutes of Health (AI43288), and the U.S. Department of Agriculture (NRI 2002-35319-11560) to J.C.C., and from the National Institutes of Health (GM053190) to V.V.D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1201204.
References
- Ahlquist P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. 2003. A uniform system for microRNA annotation. RNA 9: 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi R., Pruss, G.J., Ge, X., Marathe, R., Smith, T.H., and Vance, V.B. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. 95: 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed, A.M., Ascenzi, R., McCaskill, A.J., Hoffman, N.E., Davis, K.R., and Görlach, J. 2001. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G., Voinnet, O., Wan-Xiang, L., Ding, S.W., and Baulcombe, D.C. 1998. Viral pathogenicity determinants are suppressors of transgene silencing. EMBO J. 17: 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carrington J.C. and Freed, D.D. 1990. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 64: 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dolja V.V. and Koonin, E.V. 1991. Phylogeny of capsid proteins of small icosahedral RNA plant viruses. J. Gen. Virol. 72: 1481–1486. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P. and Vogelstein, B. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 257: 8569–8572. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J. and Matzke, M.A. 2003. The small RNA world. J. Cell. Sci. 116: 4689–4693. [DOI] [PubMed] [Google Scholar]
- Guo H.S. and Ding, S.W. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21: 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Hull R. 2001. Matthews' plant virology, 4th ed. Academic Press, San Diego, CA.
- Johansen L.K. and Carrington, J.C. 2001. Silencing on the spot: Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D. and Carrington, J.C. 1998. A counter-defensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95: 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Khvorova A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Choi, G.H., Nuss, D.L., Shapira, R., and Carrington, J.C. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. 88: 10647–10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L., Szittya, G., Silhavy, D., and Burgyán, J. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23: 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Rand, T., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. 2002a. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002b. Cleavage of Scarecrow-like mRNA targets is directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart, B.J., Bartel, D., Vance, V.B., and Bowman, L.H. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. 99: 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Papp I., Mette, M.F., Aufsatz, W., Daxinger, L., Schauer, S.E., Ray, A., van der Winden, J., Matzke, M., and Matzke, A.J. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C.W. and Dolja, V.V. 2000. Leader proteinase of the beet yellows closterovirus: Mutation analysis of the function in genome amplification. J. Virol. 74: 9766–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Ren, T., and Morris, T.J. 2003. The coat protein of Turnip crinkle virus suppresses posttranscriptional gene silencing at an early step. J. Virol. 77: 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.C., Kasschau, K.D., Prokhnevsky, A.I., Gopinath, K., Pogue, G.P., Carrington, J.C., and Dolja, V.V. 2003. Suppressor of RNA silencing encoded by Beet yellows virus. Virology 306: 203–209. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Schauer S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. 2002. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487–491. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvágner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Silhavy D., Molnár, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M., and Burgyán, J. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21: 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D.E., Johnston, R.E., and Dougherty, W.G. 1989. Generation and characterization of monoclonal antibodies reactive with the 49-kDa proteinase of tobacco etch virus. Virology 173: 499–508. [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Leh, V., Lederer, C., and Maule, A.J. 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306: 33–41. [DOI] [PubMed] [Google Scholar]
- Tijsterman M., Ketting, R.F., and Plasterk, R.H. 2002. The genetics of RNA silencing. Annu. Rev. Genet. 36: 489–519. [DOI] [PubMed] [Google Scholar]
- Vargason J.M., Szittya, G., Burgyán, J., and Tanaka Hall, T.M. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Béclin, C., and Fagard, M. 2001. Post-transcriptional gene silencing in plants. J. Cell. Sci. 114: 3083–3091. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto, V.M., and Baulcombe, D.C. 1999. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. 96: 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang, M.B., and Lough, T. 2001. Gene silencing as an adaptive defence against viruses. Nature 411: 834–842. [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau, K.D., and Carrington, J.C. 2003. Negative feedback regulation of Dicer-like1 (DCL1) in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13: 784–789. [DOI] [PubMed] [Google Scholar]
- Xie Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Malinina, L., and Patel, D.J. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426: 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D. 2004. Plant RNAi: How a viral silencing suppressor inactivates siRNA. Curr. Biol. 14: R198–R200. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]