Abstract
In the Amazon Region, there is a virtual absence of severe malaria and few fatal cases of naturally occurring Plasmodium falciparum infections; this presents an intriguing and underexplored area of research. In addition to the rapid access of infected persons to effective treatment, one cause of this phenomenon might be the recognition of cytoadherent variant proteins on the infected red blood cell (IRBC) surface, including the var gene encoded P. falciparum erythrocyte membrane protein 1. In order to establish a link between cytoadherence, IRBC surface antibody recognition and the presence or absence of malaria symptoms, we phenotype-selected four Amazonian P. falciparum isolates and the laboratory strain 3D7 for their cytoadherence to CD36 and ICAM1 expressed on CHO cells. We then mapped the dominantly expressed var transcripts and tested whether antibodies from symptomatic or asymptomatic infections showed a differential recognition of the IRBC surface. As controls, the 3D7 lineages expressing severe disease-associated phenotypes were used. We showed that there was no profound difference between the frequency and intensity of antibody recognition of the IRBC-exposed P. falciparum proteins in symptomatic vs. asymptomatic infections. The 3D7 lineages, which expressed severe malaria-associated phenotypes, were strongly recognised by most, but not all plasmas, meaning that the recognition of these phenotypes is frequent in asymptomatic carriers, but is not necessarily a prerequisite to staying free of symptoms.
Keywords: Plasmodium falciparum, PfEMP1, variant proteins, cytoadherence
Plasmodium falciparum is the causative agent of the most dangerous form of malaria, a disease that affects approximately 200 million people every year in tropical areas of the world (WHO 2013). Immunity against malaria is only acquired after successive malaria episodes and is strongly dependent on the generation of specific B-cells, antibodies and a functional T-cell memory response (Marsh & Kinyanjui 2006, Crompton et al. 2010, Weiss et al. 2010, Portugal et al. 2013). Multiple targets of antibodies have been identified on the merozoite surface or localised to organelles within it and many of them are being considered as potential vaccine components (Vaughan & Kappe 2012). The infected red blood cells (IRBCs) and the variant parasite proteins directed to its surface are also important targets for antibodies and it has been shown that cumulative exposure increases the breadth of the recognition of antigenic variants that are displayed (Hviid 2005). A major portion of antibodies seems to be directed against variants of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) (Leech et al. 1984, Bull et al. 1998, Chan et al. 2012), which are important virulence factors because they mediate the cytoadhesion of IRBCs to a variety of receptors found in endothelial tissues [reviewed in Pasternak and Dzikowski (2009)], thus allowing the IRBCs to avoid spleen clearance. PfEMP1 variants are encoded by the var gene family and consist of 50-60 alleles per haploid genome (Baruch et al. 1995, Su et al. 1995); they are expressed in such a way so that only one or a few var gene loci (Joergensen et al. 2010) are active per infected RBC, leading to the exposure of only one (or a few) species of PfEMP1. In a timely fashion, the var gene locus activation is altered by chromatin modification so that PfEMP1 expression switches [reviewed by Guizetti and Scherf (2013)]. This leads to a constant immune evasion of the circulating IRBCs, which is termed antigenic variation. Due to the accelerated genetic recombination of var genes (Freitas-Júnior et al. 2000), the diversity of var genes in field isolates is very high [e.g., data described in Warimwe et al. (2009)] turning the task of developing antibodies against most of the variants into a long-lasting process. Importantly, most of the circulating variants show cytoadherence to CD36 and the presence of this phenotype was associated with non-severe malaria episodes (Ochola et al. 2011). Despite the great diversity of var genes and PfEMP1 variants, a number of alleles are fairly conserved between different isolates. An example of this are the var genes, which encode PfEMP1 versions expressed in pregnancy-associated malaria (PAM) (var2csa) (Lavstsen et al. 2003, Salanti et al. 2003); the recognition of these variants is associated with the decrease of PAM symptoms (Tutterrow et al. 2012). Another conserved group of var domains was recently revealed by the comparison of seven P. falciparum genomes (Rask et al. 2010). Three independent groups showed that up-regulation of the var genes expressing domain cassettes 8 (DC8) or DC 13 domains occurred in cases of severe childhood malaria (Avril et al. 2012, Claessens et al. 2012, Lavstsen et al. 2012). The receptor for these domains appears to be the endothelial protein C receptor (EPCR) (Turner et al. 2013). Another conserved phenotype of PfEMP1 encoded by DC4 domains (Oleinikov et al. 2009, Bengtsson et al. 2013) seems to be associated with ICAM1-binding, which itself was correlated with severe (cerebral) malaria (Turner et al. 1994). Very little is known about the adhesion binding properties and antigenic variation related to the var genes and PfEMP-1 proteins in Amazonian isolates. Previous reports by our group based on the analysis of PfEMP1-DBLα encoding tags showed that the var gene repertoires seem much smaller in Amazonian P. falciparum isolates compared to other endemic settings (Albrecht et al. 2006, 2010). It is also known that the absolute number of malaria-infected people in the Amazon may be underestimated due to a high incidence of asymptomatic infections in riverside settlements (Alves et al. 2002). Additionally, the occurrence of severe malaria is a rare event and seems to happen due to a late diagnosis outside the transmission area; consequently it is as lethal as in African settings. In absolute numbers, the mortality rate of P. falciparum malaria in Brazil is much lower than in Africa. Given a total of 265,000 cases and 76 deaths from malaria in Brazil in 2011 the mortality rate of malaria in Brazil was 0.029% compared to 0.3% worldwide in the same year [655,000 deaths out of 216 million cases worldwide in 2011 (WHO 2011)]. Based on this information, we set out to explore whether the recognition of possibly less circulating antigenic variants (due to redundant circulating var gene repertoires) by antibodies in patient sera were correlated to asymptomatic outcomes of malaria and whether the phenotype associated with severe malaria - a parasite line most likely expressing a DC8 domain encoding var gene - was recognised even in the virtual absence of severe malaria. For this, we phenotype-selected four Amazonian P. falciparum isolates plus the non-related 3D7 strain based on the receptors CD36 and ICAM1 expressed on CHO cells. We then quantified IRBC recognition by plasma antibodies using flow cytometry.
SUBJECTS, MATERIALS AND METHODS
Human plasma samples from symptomatic and asymptomatic individuals - Human plasma samples were obtained during an epidemiological survey from persons living in the surroundings of Porto Velho, the capital of the state of Rondônia (RO) (Brazil). Ethical clearance was obtained from the Ethical Board of the Institute for Biomedical Sciences at São Paulo University and the Institute of Research on Tropical Pathologies before the start of the trial (protocol 741/2006). Each individual expressed his/her written consent to participate in the study. Participants were considered symptomatic when they presented with at least one clinical symptom (fever > 37.8ºC, prostration, headache, chills, tremors, myalgia, nausea and/or vomiting) and positive parasitaemia was tested by a thick blood smear and/or polymerase chain reaction (PCR). Individuals who did not present with any clinical symptoms for up to 30 days after the blood withdrawal, but had tested positive for P. falciparum by PCR, were considered to be asymptomatic. All patients were treated with the adequate drugs (Coartem) directly after the molecular diagnosis. In total, 89 plasma samples from symptomatic individuals and asymptomatic individuals (the latter were residents of Santo Antônio, Bate-Estaca and Vila Candelária, communities next to the Madeira River) were tested.
Parasites and static cytoadherence assay - P. falciparum field isolates S20, 99, 106 and 134 were collected in RO (Albrecht et al. 2006). While the S20 strain was originally collected and culture-adapted by Segurado et al. (1997), the 99, 106 and 134 isolates were collected and culture-adapted during an effort to map the genetic diversity of P. falciparum in the western Amazon Region, but without the aim of monitoring cytoadherent phenotypes of the initial patient isolate [Research Project São Paulo State Research Foundation (01/09401-0)]. In consequence, the initial phenotypes were lost and not available for this work. The 3D7 strain was initially received by Walliker et al. (1987). All parasites were cultured in candle jars as described (Trager & Jensen 1976) using RPMI-1640 medium supplemented with 10% human plasma (type A or AB) and O- erythrocytes. The parasites were selected for the expression of the determined PfEMP1 variants using static cytoadherence assays as described by Golnitz et al. (2008). Briefly, 108 trophozoite infected RBCs were incubated over CHO-ICAM or CHO-CD36 cells (a generous gift from Dr Artur Scherf, Pasteur Institute, Paris) and were cultured to confluence in 25 cm2 culture bottles. Unbound parasites were then removed by three subsequent washings with RPMI at pH 6.8 and the still-bound parasites were detached by incubation with RPMI at pH 7.4 and cultivated with fresh RBCs. This procedure was repeated five times, after which a homogenous binding phenotype was achieved. To measure the achieved cytoadherence properties, 5 x 105 CHO cells expressing either CD36 or ICAM1 were seeded in labtec slides and were grown for 48 h. Mature trophozoite/early schizont stage parasites were prepared from mixed cultures by plasmagel flotation (Lelievre et al. 2005). Cytoadherence was then measured by the incubation of 5 x 106 trophozoite stage parasites over the cells for 1 h. After this, the supernatants were aspirated and the cells were washed three times with incomplete RPMI medium at pH 6.8. Next, the slides were stained and five fields were counted under a 1:1,000 magnification. The parasite lines were judged clonal after testing 11 microsatellites (Anderson et al. 1999) during previous research (L Albrecht, unpublished observations). The 3D7 lines, which dominantly transcribed var PF11_0521 and PFD0020c, were identified after clonal selection using limiting dilution on previously CHO-CD36 and CHO-ICAM panned parasites, respectively.
Reverse transcription (RT)-PCR and sequencing analysis - For determining the dominant var transcript in field isolates and 3D7 parasites, the RNAs of 108 ring stage parasites were recovered from each parasite line using the Trizol® method (Invitrogen). All of the RNAs were submitted to RT using random hexamer primers and RevertAid Reverse Transcriptase (Fermentas) following the manufacturer’s instructions. Then, the DBLα tag sequences were amplified using the oligonucleotides also employed by Warimwe et al. (2009). The resulting ~400 bp fragments were recovered from TAE-agarose gels and cloned into pGEM T-easy (Promega) vectors and 50 clones per lineage were sequenced in an ABI 3100 (Applied Biosystems) automated DNA sequencer. The sequences were analysed using CLUSTALX 1.83 and similarity to known DBLα tags was searched using the National Center for Biotechnology Information’s and PlasmoDBs BLAST (Altschul et al. 1997) website. The resulting DBLα tag sequences were then analysed for their distinct sequence identifiers (DSID) using the Perl script published by Bull et al. (2007). The DSID classification is based on the analysis of four semi-conserved tetrapeptides (positions of limited variability) and the number of cysteines found in the DBLα region flanked by the oligonucleotides used herein, resulting in six groups of DSIDs. By comparison with the previously established classification of the 5’ untranslated regions (UTR) of the var genes [upstream sequences (ups) (Lavstsen et al. 2003)], DBLα sequences of DSID groups 1-3 were mostly associated with upsA type 5’-UTR sequences, while DSID groups 4-6 were associated with upsB and upsC 5’-UTR sequences (Bull et al. 2007).
Immunofluorescence - Live parasite immunofluorescence assays were conducted with enriched P. falciparum trophozoites and human plasmas from asymptomatic and symptomatic carriers. For this, trophozoite/schizont stage parasites were incubated with 40 µg/mL of 4’-6-diamidino-2-phenylindole and HCS CellMask in 100 µL RPMI/1% bovine serum albumin for 60 min at 37ºC, followed by incubation with plasma (dilution 1:25) and an Alexa Fluor labelled antihuman-IgG antibody (Molecular Probes, 1:200 dilution). Between each incubation step, the material was washed three times with RPMI pH 7.2 and was visualised in fluorescence microscopy. As a control, we employed the antiglycophorin antibody (1:1,000 dilution) (Molecular Probes) and a secondary antibody, anti-mouse-IgG 1:500 (Alexa Fluor 594, Molecular Probes).
Flow cytometry - For the flow cytometry assays, the trophozoites were resuspended in RPMI pH 7.2 and 2% of bovine foetal serum (RPMI-S). Briefly, 5 x 106 parasites in blood at a parasitaemia of < 0.5% were incubated for 30 min with donor plasma, including as a control a plasma pool from persons who were never infected with malaria (1:10 dilution) in RPMI-S. This was immediately followed by antigoat IgG conjugated with Alexa Fluor 488 (1:200 dilution) for 30 min. After that, the parasites were incubated with ethidium bromide [100 µg/mL in phosphate buffered saline (PBS) 1X] for 20 min to perform the nuclear labelling of the parasitised erythrocytes. After washing, the RBC solutions were resuspended in PBS to an average concentration of 500 cells per µL and were analysed in a Guava EasyCyte Mini flow cytometer. For each plasma sample, 5,000 events were monitored and analysed using the Guava EasySoft software. The surface labelling of the IRBCs was expressed as a reactivity index (RI). For this, we established a cut-off value that measured baseline surface labelling using the non-reactive pool of plasma samples against each parasite lineage. Then, the average immunofluorescence values of each plasma sample measured in duplicate were divided by the average signal plus three standard deviations obtained from the negative plasma pool. All plasma samples with RI above 1 were considered reactive.
St atistical analyses - All of the statistical analyses were performed using Prism GraphPad5 software. For the comparison of the ratio of responders/non-responders against phenotype-expressing parasite lines, Fisher’s exact test was applied. For the comparison of the RI values between different groups of plasma samples and/or parasite lines, Mann-Whitney’s U test was applied.
RESULTS
Cytoadherent-selected isolates show in part identical var DBLα sequences, but no selection of determined var type subclasses - In order to identify an association between var gene transcription and cytoadherent phenotypes in the Amazonian field isolates tested in this study, we used RT-PCR followed by cloning/sequencing of the amplified products. This is currently the only applicable non-high throughput method for var gene analysis with unknown genome sequences. Fifty clones were sequenced per isolate and adhesive phenotype (CHO-CD36 or CHO-ICAM), resulting in six-16 different sequences per isolate. All isolates showed one dominant DBLα sequence (Fig. 1). Severe malaria cases were recently related to the expression of upsA-related DBLα sequences (Warimwe et al. 2009) and these fall into DSID groups 1-3 [defined by Bull et al. (2005, 2007)]. Accordingly, we classified the sequences according to their DSID group using the criteria developed by Bull et al. (2007) described above. The abundance of sequences with specific DSID types varied between parasite lines. For example, the dominant sequence (46%) in the 3D7PFD0020c line was DSID type 1, while 38% were of DSID types 4-6 in this parasite line. However, panning of the parasites on CHO-ICAM, which should ideally result in DBLα DSID type 3 (PF11_0521) expressing parasites, failed to result in such parasites and all parasites panned over this receptor showed only low frequencies of DBLα DSID 1-3 expressing parasites that were previously associated with severe malaria (Bull et al. 2005) (Fig. 1). Alignment of all the sequences also showed that highly similar var DBLα sequences (> 95% sequence identity) were identified in 106ICAM, S20ICAM and 99ICAM, indicating that at least the DBLα domains in the expressed PfEMP1 proteins seem similar, reinforcing the somehow redundant var repertoires in these parasites. Unexpectedly, the selection over CHO-CD36 in two cases led to the expression of dominant DSID type 1-3 sequences in parasite lines S20 and 99. The selection of the 3D7 strain for CHO-ICAM adherence showed that the dominantly present var gene transcripts PFL1970w and PFD1000c were not exactly the ones identified previously by our group (Golnitz et al. 2008), nor were they PF11_0521, which contains the unique DBLβ/C2 domain that strongly interacts with ICAM1 in vitro (Oleinikov et al. 2009) or PFD1235w, as identified by Bengtsson et al. (2013). In fact, PFL1970w and PFD1000c do not even contain a DBLβ/C2 sequence at all (Rask et al. 2010), making their participation in ICAM1-binding doubtful. Selection of the 3D7 line on CHO-CD36 led to the expression of PFD0615c as previously published (Golnitz et al. 2008). We also tested two 3D7 lines that were previously encountered in cloning experiments and either expressed the var gene PFD0020c (3D7PFD0020c), which is associated with severe malaria (Lavstsen et al. 2012) or PF11_0521 (3D7PF11_0521), which contains the ICAM1 binding domain (Oleinikov et al. 2009). In a second approach, we tried to identify whether any of the identified DBLα sequences were similar to DBLα domains that were connected to the severe var-associated domains DC8 and DC13 and the ICAM1 binding phenotype DC4 in the seven P. falciparum genomes that were analysed by Rask et al. (2010). For this, we aligned the DBLα DNA sequences with sequences retrieved from their dataset. The sequence expressed in the CHO-CD36 selected S20 line (S20CD36) expressed a DBLα tag similar to IT4var35, a sequence that is closely related to var1csa, while the dominant sequence in the CHO-CD36 selected line 99 was identical to PFD1000c, which is an upsC1 type var gene (Rask et al. 2010). Because all of the remaining sequences from the 99 isolate were not similar to the 3D7 counterparts, we concluded that no cross-contamination had occurred during the selection process, which was also true for all other tested isolates (Fig. 1). No other DBLα sequences showed significant similarity to DBLα domains, which were connected to DC8, DC13 or DC4 domains from the published dataset from seven genomes (Rask et al. 2010), suggesting that they are most likely not part of the var genes encoding PfEMP1 containing this type of domain. We concluded that the produced phenotypes, despite their apparent adherence to given CHO lines expressing ICAM1 or CD36, do not necessarily express PfEMP1, which can be related to any specific disease-associated phenotype. At a minimum, the selection of 3D7 must have resulted in at least the marginal expression of either the PFD1235w or PF11_0521 var genes, which was not the case (Fig. 1, Supplementary data Fig. 4). Therefore, it can only be assumed that the selected parasite phenotypes express distinct PfEMP1s, with the exception of the lines 106ICAM, S20ICAM and 99ICAM.
Fig. 1. : frequency of clones with var transcripts from different parasite lineages suggests the expression of few dominantly expressed var genes in each lineage. Shown are the frequencies of occurrence of determined var DBLα sequences which were obtained by reverse transcription-polymerase chain reaction fragment cloning and sequencing of 50 clones per lineage. A: the frequency of sequence types in 3D7 parasite clones 3D7PFD0020c and 3D7Pf11_0521; B: the frequencies of sequence types detected in 3D7 parasites panned over CHO-CD36 or CHO-ICAM1; C-F: the results from panned parasite isolates S20, 99, 106 and 134. The most frequently found transcript sequences per isolate are indicated together with their distinct sequence identifiers classification (bold) and the identity of the most similar DBLα tag DNA sequence from the 3D7 genome are shown in parenthesis. The most similar 3D7 DBLα tags were S20ICAM (PFL1950w), S20CD36 (PFI1820w), 99ICAM (PFL1950w), 99CD36 (PFD1000c), 106ICAM (PFL1950w), 106CD36 (Mal6P1.316), 134ICAM (PFD0630c) and 134CD36 (PFD0995c). The x-axis numbers in each graph depict the sequences and the corresponding accessions can be retrieved in Supplementary data Fig. 4.
F low cytometry analysis revealed a consistent presence of antibodies against the 3D7 PFD0020c and 3D7 Pf11_0521 phenotypes - We tested the IRBC surface recognition of antibodies contained in plasma samples from 44 symptomatic and 45 asymptomatic individuals. Immunofluorescence assays revealed that IRBCs were recognised in a specific way and showed labelling only on the surface of IRBC, but not on normal RBCs. Additionally, no staining of parasites inside the IRBCs was observed (Supplementary data Fig. 1). We then measured the recognition of field isolates/phenotypes by antibodies contained in the plasma of symptomatic and asymptomatic individuals by flow cytometry and calculated the RI for each plasma and parasite line tested. When we compared the percentage of positive plasma samples for parasite lines (RI higher than 1), we observed that the 3D7 isolates selected for CHO-ICAM1 and CHO-CD36 were recognised by more plasma samples from asymptomatic individuals than those from symptomatic individuals (Fig. 2) (Fisher’s exact test, p = 0.0184 and p = 0.0079, respectively). However, recognition of the 3D7Pf11_0521 phenotype was more frequent in symptomatic individuals (p = 0.0148). No significant differences in the number of plasma samples recognising all other parasite lines were detected (p > 0.05) when comparing the recognition by plasma samples from symptomatic and asymptomatic individuals. When comparing the recognition of the different 3D7 phenotypes by plasma samples from asymptomatic individuals, the 3D7Pf11_0521, 3D7ICAM and 3D7CD36 phenotypes were less frequently recognised than 3D7PFD0020c (p < 0.0001). Most of the phenotypes, excluding the phenotype-selected 3D7 parasites, were recognised by 40-80% of the plasma samples (Fig. 2). The 3D7PFD0020c lineage was recognised by the most plasma samples and it also had the highest RI (Fig. 2, Table I). In order to estimate the previous exposure of all of the individuals to malaria attacks, we measured antibodies against a recombinant version of the highly conserved C-terminus from merozoite protein 1 as a surrogate marker, given that the number of informed, previous malaria attacks was not predictive for a stronger or weaker recognition (Supplementary data, Table). As shown in Supplementary data (Table), the majority of individuals presented detectable to high titres of antibodies against PfMSP119. On the other hand, low titres were not paralleled by a weak or absent reaction against the RBC surface, suggesting diversity in the response to malarial antigens. The same was true when comparing the number of informed infections with either titre level in the MSP119 ELISAs or IRBC recognition. We then individually analysed whether any of the phenotypes were recognised more strongly by plasma samples from either asymptomatic or symptomatic individuals. Significant differences and a stronger recognition (higher RI values) by plasma samples from asymptomatic individuals were indeed observed for the parasite lines S20CD36, 3D7PFD0020c, 3D7ICAM and 3D7CD36, while 134CD36 and 3D7Pf11_0521 had higher RI with plasma samples from symptomatic individuals. The remaining phenotypes were not differentially recognised by symptomatic or asymptomatic donor plasma samples (Table II, Supplementary data Fig. 2). Additionally, we asked whether plasma samples recognised specifically more of the CHO-ICAM or the CHO-CD36 selected phenotype of each isolate. We also compared the recognition intensities of the specific 3D7 phenotypes 3D7PFD0020cand 3D7Pf11_0521 with their panned counterparts 3D7CD36 and 3D7ICAM. Eventual differences here may inform about the relative participation of antiPfEMP1 antibodies in recognition when we assume that all other possibly exposed variant antigens, such as RIFINs, STEVORs and others, have either a smaller participation in recognition or are not differently expressed after the phenotype selection process. Interestingly, in all cases, the ICAM1 phenotype was more strongly recognised than the CD36 phenotype, independently of the origin of the plasma. In the majority of cases, the difference in recognition was significantly different (Table I, Supplementary data Fig. 2). As was true for the percentage of plasma samples that reacted against this phenotype, the recognition of 3D7PFD0020c was significantly higher in all plasma samples compared to the recognition of any other 3D7 phenotype. The phenotype 3D7Pf11_0521 was also very strongly recognised. Both phenotypes were significantly more likely to be recognised than the other 3D7 phenotypes 3D7CD36 and 3D7IC AM (Table I, Supplementary data Fig. 2), although plasma samples from asymptomatic carriers did not react differently against the 3D7Pf11_0521 phenotype compared to the 3D7ICAM or 3D7CD36 phenotypes.
Fig. 2. : most plasma recognises the 3D7PFD0020c phenotype. The percentage of responders of each plasma donor group was plotted against the recognised parasite lineage. Asterisks mean significant differences between the percentages of recognising plasmas from the asymptomatic and symptomatic group (Fisher’s exact test).
TABLE I. Many plasma recognise more the CHO-ICAM selected lineage of parasites than the CHO-CD36 selected lineage.
| p (plasmas symptomatic individuals) | p (plasmas asymptomatic individuals) | |
|---|---|---|
| S20ICAM↑ vs. S20CD36 | 0.0085 | 0.9706 |
| 99ICAM↑ vs. 99CD36 | 0.0319 | 0.0137 |
| 106ICAM vs. 106CD36 | 0.0623 | 0.9005 |
| 134ICAM↑ vs. 134CD36 | 0.0359 | 0.8116 |
| 3D7PFD0020c vs. 3D7Pf11_0521↑ | 0.3682 | < 0.0001 |
| 3D7ICAM↑ vs. 3D7CD36 | 0.0446 | 0.0366 |
| 3D7PFD0020c↑ vs. 3D7ICAM | < 0.0001 | < 0.0001 |
| 3D7PFD0020c↑ vs. 3D7CD36 | < 0.0001 | < 0.0001 |
| 3D7Pf11_0521↑ vs. 3D7ICAM | < 0.0001 | 0.3508 |
| 3D7Pf11_0521↑ vs. 3D7CD36 | < 0.0001 | 0.2850 |
the significance of differences in reactivity indices of plasmas either from asymptomatic or symptomatic individuals reacting either against the CHO-ICAM or the CHO-CD36-selected parasite lineage was evaluated by Mann-Whitney’s U test without normalisation of the data. Shown are the resulting p values. The arrows indicate which lineage was statistically more recognised.
TABLE II. Comparison of reactivity indices (RI) from plasmas originating from asymptomatic and symptomatic individuals shows differing patterns of recognition.
| Tested parasite lineage | S20ICAM | S20CD36 | 99ICAM | 99CD36 | 106ICAM | 106CD36 | 3D7PFD0020c | 3D7Pf11_0521 | 134ICAM | 134CD36 | 3D7ICAM | 3D7CD36 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p | 0.8417 | 0.0458 | 0.8310 | 0.8432 | 0.2331 | 0.5736 | 0.0100 | 0.0252 | 0.0707 | 0.0113 | 0.0023 | 0.0098 |
the significance of difference of the RI from asymptomatic and symptomatic patient’s plasmas reacting against the shown parasite lineages was evaluated using Mann-Whitney’s U test without normalisation of the data. Note that 134CD36 and 3D7Pf11_0521 were recognised stronger by plasmas from symptomatic infections.
The huge range of reactivity observed against the different 3D7 phenotypes expressing different alleles of PfEMP1, seen as the presence of dominant transcripts of the different var genes in the corresponding parasites, appears to highlight the importance of PfEMP1 recognition in the overall response against IRBC.
DISCUSSION
Antibodies directed to the surface of P. falciparum-IRBC are important players in combating the symptoms and virulence of malaria and the recognition of PfEMP1 seems to be of major importance (Bull et al. 1998, Chan et al. 2012). Many studies from epidemic sites in Africa and Southeast Asia have shown that the cumulative acquisition of antibodies against a great number of antigenic phenotypes provides protection from malaria. The degree of acquired immunity ranges from protection initially from death, then from severe outcomes and finally from symptomatic malaria (Hviid 2005, Marsh & Kinyanjui 2006). As a consequence, it is believed that continuously exposed individuals do not necessarily suffer from malaria, but become asymptomatic carriers. Additionally, it is hypothesised that during the first infections (at least in childhood malaria in Africa), a few conserved PfEMP1 phenotypes are expressed and lead to an increased severity of disease, which potentially occurs via the mechanism postulated by Turner et al. (2013). The authors proposed that by blocking the interaction of soluble protein C through the binding of a DC8/DC13 PfEMP1 domain, endothelial activation is induced, which then leads to the up-regulation of inflammatory signals, including the increased expression of endothelial ICAM1 and other receptors. As a consequence, the DC8/DC13 containing PfEMP1 phenotypes would be the first to be recognised by persons who did suffer severe malaria episodes. P. falciparum infection in the Brazilian Amazon is currently decreasing and transmission is unstable in many places. Symptomatic cases seem to accumulate in areas with a strong influx of non-immune populations, such as rural settlements, wood logging or mining activities (Oliveira-Ferreira et al. 2010). Perhaps as a consequence of low transmission and quick treatment, the overall diversity of variant protein encoding genes, such as the var genes, in the Amazon seems much smaller than in any other endemic area so far tested (Albrecht et al. 2006); development to severe malaria is also likely prevented by quick treatment. The herein used isolates possess var genes, which represent part of the repertoire of circulating strains in the Amazon (Albrecht et al. 2010). In order to elucidate the differential presence of antibodies against the IRBC surface in asymptomatic (“protected”) and symptomatic (“susceptible”) individuals, we tested the recognition of P. falciparum isolates from the western Brazilian Amazon and compared them to four lineages of 3D7 that expressed different var genes/PfEMP1. These included the virulence-associated phenotypes, which expressed either the var gene PFD0020c (Lavstsen et al. 2012, Turner et al. 2013) or Pf11_0521, one of the ICAM1-binding PfEMP1 variants of the 3D7 genome (Oleinikov et al. 2009).
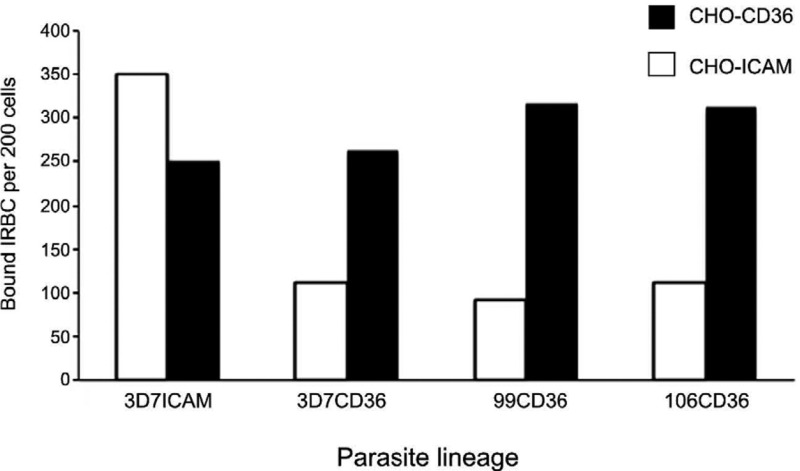
For the preparation of relevant phenotypes in natural isolates, we tried to focus on two main cytoadherent phenotypes that are supposedly expressed either in non-severe malaria (CD36 adherence) or severe malaria (ICAM1 adherence) (Ochola et al. 2011), although the latter possibly occurs in consequence to previous EPCR-adherence (Turner et al. 2013). We tried to prepare these phenotypes by panning the isolates over receptor-transfected CHO cells (Hasler et al. 1993). Although our group had previously shown the usefulness of this quite economic cytoadherence selection approach using the 3D7 strain (Golnitz et al. 2008), this method cannot warrant that the selected parasites indeed use the most affine PfEMP1 for a defined receptor. One example is the 3D7 line selected on CHO-ICAM cells. Although the parasite line also showed the PFD1000c transcript, the dominant transcript was now PFL1970w (Fig. 1) and not PFF0845c, as described previously by us (Golnitz et al. 2008). On the other hand, PfEMP1, which contains the most domains that bind strongly to ICAM1, is present in PF11_0521 (Oleinikov et al. 2009), although the upsA type PFD1235w also contains ICAM1 binding domains (Bengtsson et al. 2013). This means that other receptors - and not ICAM1 - seem to be recognised. Several issues must be taken into account when using the assay employed in this study: first, there are additional receptors on CHO cells, which can be recognised by PfEMP1 domains (Andrews et al. 2005). Potentially the binding affinity to ICAM1 on the CHO-ICAM cells is not superior to the other unknown receptors present on the CHO cells, leading to varying dominant transcripts in different experiments that use the CHO-ICAM cells. Given the var transcript sequence results from each parasite line panned over CHO-ICAM, we deduced that the method was not suitable for the directed selection of truly ICAM binding IRBCs. Second, the expression of the desired receptor molecules varied on the CHO-cell lines and frequent checks are necessary to assure that the receptors are indeed still expressed. During our experiments, we checked whether the cells sufficiently expressed the respective receptor using specific antibodies against human CD36 and ICAM1 in immunofluorescence assays (data not shown). We also checked whether some isolates showed the most cytoadhesion to the CHO-cell line they were selected for. As depicted in Supplementary data Fig. 3, all isolates did adhere more to the cell line they were selected for, but they also showed cytoadherence to other CHO lines. The observation of simultaneous CD36 and ICAM1-binding phenotypes has been documented before [for example, Scherf et al. (1998)]. In favour of the system used in this study, cytoadherence to receptors was partially inhibited by co-incubation with specific antibodies to CD36 and ICAM1 (data not shown). One alternative to cellular systems for cytoadherence selection would be the use of purified receptors, which has been done successfully by other groups, at least for CSA or ICAM1-binding parasite lines [see protocols published by J Reeder in Moll et al. (2008)]. We tried using commercially available purified human ICAM1 spotted on plastic dishes, but the yields per selection round were too poor and this approach was abandoned (BC Carlos, unpublished observations). An additional concern regarding this method is the fact that naturally occurring receptors are embedded in or associated to membrane bilayers and are surrounded by vast arrays of glycosyl rests that are present on each cell surface. This is not the case when purified and recombinant receptors are used, which may consequently influence the cytoadherence strength.
After the panning selection using the field isolates, we observed dominant var transcripts in all parasite lines, indicating that we were potentially analysing phenotypically homogenous cultures. The DBLα-specific oligonucleotides used for the var transcript amplification were previously used in a number of studies by us and others [e.g., Warimwe et al. (2009)] and permitted estimation of the corresponding sequences were associated to specific ups (upsA, B and C) using the DSID analysis approach (Bull et al. 2007). Importantly, the var genes preceded by the upsA type sequences were found to be expressed in cases of severe malaria (Bull et al. 2005, Warimwe et al. 2009), while the upsB and upsC sequences were not. The classification was greatly refined with the analysis of the variant genes from seven P. falciparum genomes (Rask et al. 2010) and a novel quantitative PCR based approach now permits a rather detailed classification of var transcripts (Lavstsen et al. 2012). When trying to classify the obtained field isolate transcripts, we observed that most of these fell into upsB and upsC types of transcripts (DSID types 4-6) and unexpectedly, only the dominant sequences from S20CD36 and 106CD36 presented the upsA type sequence. The identified sequence of S20CD36, however, was quite similar to the var1csa sequence [most similar to ITvar35, upon alignment to the dataset that was published in Rask et al. (2010)], which is a transcript consistently transcribed by many parasites, but apparently not encoding a functional PfEMP1 protein (Kyes et al. 2003). It is difficult to interpret why CHO-CD36 panning induced a parasite line to express an upsA sequence type. Considering the procedure by which the isolates and the panned 3D7 strains were phenotypically selected for and given that the detected var gene transcripts, with two exceptions, did not fall in the DSID groups that were associated with severe malaria cases, we suggest that the phenotypes we looked at will most likely not be found in severe cases in a field situation, but in non-severe cases. When we monitored the antibody recognition of the different parasite lines, we observed a strong recognition of severe malaria-associated phenotypes, such as the 3D7PFD0020c and 3D7Pf11_0521 parasite lines. This indicates that the severe malaria associated DC8/DC13 phenotype is present and circulating in Amazonian isolates. This strong recognition occurred in a similar form with plasmas from symptomatic and asymptomatic individuals, although we did not detect antibodies in all samples. Following the hypothesis that the first malaria infections lead to more severe disease in non-immune carriers, which is characterised by the expression of var genes from the DC8 and DC13 groups, including PFD0020c (Avril et al. 2012, Claessens et al. 2012, Lavstsen et al. 2012) and perhaps the DC4 group var genes (Bengtsson et al. 2013), one would expect that at least asymptomatic plasmas would contain specific antibodies for this phenotype. Potentially not all Amazonian P. falciparum isolates contain severe malaria associated var genes or there may be antigenically very different, not yet described variants that we did not test in this study. The existence of a selection system that permits purifying and enriching EPCR-binding phenotypes would be very useful to discover Amazonian severe var variants. However, it is possible that the severe var phenotype is only induced after a certain duration of infection and quick treatment (less than 48 h after onset) as is common in the Amazon would prevent the quantitative outgrowth of this phenotype. In consequence, these individuals would not necessarily react to the severe var phenotype. In one study, the controlled natural infection of non-immune individuals with the 3D7 strain led to the expression of severe var PFD0020c or PFD1235w in only two out of six volunteers only on days 3-4 after the onset of infection (Lavstsen et al. 2005).
Based on the analysis of the DBLα motifs, we have previously found that almost no sequences are shared between Amazonian isolates and the 3D7 strain (Albrecht et al. 2010). This finding may explain why the panned 3D7 lines that adhere to CD36 and ICAM1 and express non-PoLV1-3 (non upsA) type var genes were only weakly recognised by the plasma samples and significantly less by the plasma samples from symptomatic patients. The recognition of the field isolates occurred in approximately 40-70% of each lineage by the plasma samples from each group, while the panned 3D7 variants were recognised much less frequently. As expected, the recognition of phenotypes was often stronger by the asymptomatic plasma samples. In previous studies, the reaction of antibodies against the IRBC surface was estimated using parasite lines that specifically expressed no PfEMP1 (PfEMP1 knockdown) or had impaired transport of PfEMP1 and other IRBC surface localised proteins (SBP-1 knockout). The knockdown of PfEMP1 only decreased the IRBC surface recognition by antibodies from a serum pool of malaria-experienced persons by approximately 70% compared to the normal PfEMP1-displaying IRBCs (Voss et al. 2006). In the absence of other proteins besides PfEMP1 in the SBP-1 knockouts, there was a 90% decrease in IRBC surface recognition by immune sera (Maier et al. 2007). This indicates that a major portion of the antibodies against the IRBC surface is directed against the PfEMP1 proteins.
One of the goals of this study was to reveal whether the occurrence of asymptomatic malaria was paralleled by a significantly stronger presence of anti-IRBCs. While this was the case in recognition frequency and the antibody RI against many of the parasite lineages tested in this study, there were still a number of plasma samples in the asymptomatic group that reacted poorly against many phenotypes. This indicates that protection in these specific individuals is potentially quite fragile and is directed against a very limited repertoire of variant antigens, most of which were not present in this study. Another explanation is that the recognition of the IRBC surface is not crucial for the establishment of an asymptomatic status, at least not under the transmission conditions where the plasma donors lived. We did not test the IgG subclasses that reacted to the surface of the parasites and this may also be an important factor for achieving humoral immunity against the IRBC surface. A recent study using plasma samples from the same site tested the recognition of merozoite surface antigens in asymptomatic and symptomatic populations (Medeiros et al. 2013) and the authors observed similar patterns of recognition: while many of the asymptomatic patients recognised most of the antigens tested, a number of individuals did not recognise those antigens. Therefore, we propose that the recognition of many interchangeable targets may provide protection from symptoms under the transmission conditions of the western Amazon. The recent detection of EPCR as relevant receptor for cytoadherent IRBC (Turner et al. 2013) and the establishment of a system to quickly select for parasite phenotypes that express the severe var related PfEMP1 should provide a useful tool to focus attention on the question of whether P. falciparum with decreased virulence exist, which could potentially provide a biological explanation for the virtual absence of severe malaria in the Brazilian Amazon.
ACKNOWLEDGEMENTS
To Wolfgang Fischer and Márcio Yamamoto, for sequencing of DBLα tag sequences, and to Drs Mauro Shugiro Tada and Tony Katsuragawa, for help with data and sample collection at the sites.
Supplementary data
TABLE. The reactivity index (RI) analysis reveals stronger recognition of a number of parasite lineages by asymptomatic patient plasmas
Fig. 1. : immunofluorescence of intact infected red blood cell shows labelling exclusively of the RBC surface. Immunofluorescence assays were done as described in Subjects, Materials and Methods. BF: bright field; DAPI (4’-6-diamidino-2-phenylindole): nuclei; merged: overlay of all fluorescent signals; αGlyA + Alexa 594: antiglycophorin antibody labelled with Alexa 594; αIgG + Alexa 488: human antibodies detected with anti-human IgG labelled with Alexa 488.
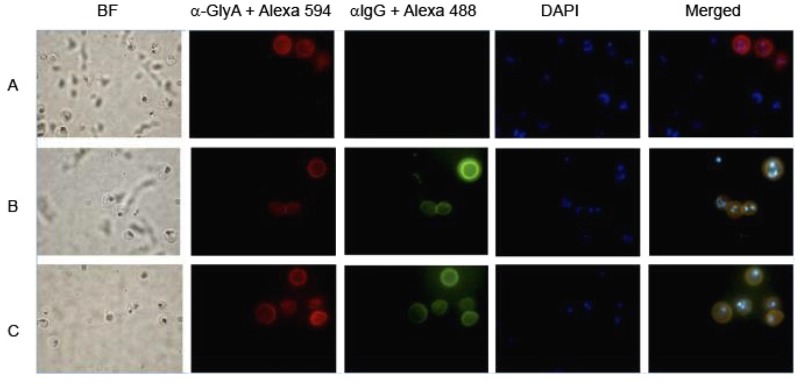
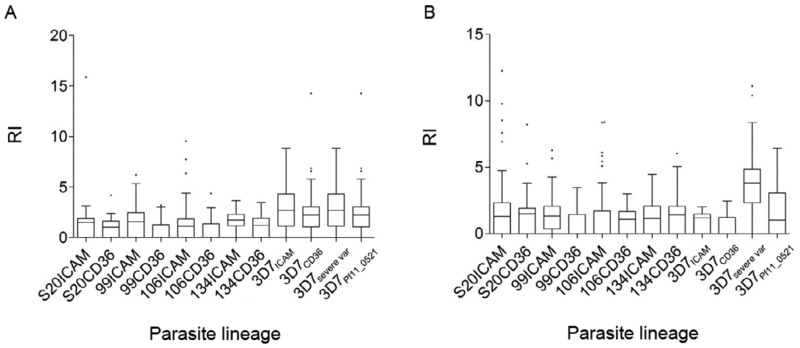
Fig. 2. : reactivity indices (RI) complementary to Fig. 3. Shown are median values, 75% percentiles and outliers depicted as whiskers. A: RI for symptomatic plasmas; B: values for asymptomatic plasmas.
Fig. 3. : cytoadherence-selected parasites adhere most to the CHO cell line they were selected for, but also adhere significantly to other cells. The indicated parasites were cytoadherence-selected as described. Trophozoite stage parasites were then tested over CHO-ICAM and CHO-CD36 cell lines and the cytoadherent parasites were counted as described. Shown are data from one of two independent experiments with similar results.
Fig. 4. : GenBank or PlasmoDB entries of the different DBLα sequences from the analysed parasite lineages after selection by panning (3D7, 99, 106, 134, S20) or cloning (3D7PFD0020c and 3D7Pf11_0521).
Footnotes
Financial support: FAPESP (2009/17114-3), CNPq (576128/2008-2)
FTMC and GW are CNPq fellows.
REFERENCES
- Albrecht L, Castiñeiras C, Carvalho BO, Ladeia-Andrade S, Silva NS, Hoffmann EHE, dalla Martha RC, Costa FTM, Wunderlich G. The South American Plasmodium falciparum var gene repertoire is limited, highly shared and possibly lacks several antigenic types. Gene. 2010;453:37–44. doi: 10.1016/j.gene.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Albrecht L, Merino EF, Hoffmann EHE, Ferreira MU, Ferreira RGM, Osakabe AL, dalla Martha RC, Ramharter M, Durham AM, Ferreira JE, del Portillo HA, Wunderlich G. Extense variant gene family repertoire overlap in western Amazon Plasmodium falciparum isolates. Mol Biochem Parasitol. 2006;150:157–165. doi: 10.1016/j.molbiopara.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves FP, Durlacher RR, Menezes MJ, Krieger H, Silva LH, Camargo EP. High prevalence of asymptomatic Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg. 2002;66:641–648. doi: 10.4269/ajtmh.2002.66.641. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- Andrews KT, Adams Y, Viebig NK, Lanzer M, Schwartz-Albiez R. Adherence of Plasmodium falciparum infected erythrocytes to CHO-745 cells and inhibition of binding by protein A in the presence of human serum. Int J Parasitol. 2005;35:1127–1134. doi: 10.1016/j.ijpara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Avril M, Tripathi AK, Brazier AJ, Andisi C, Janes JH, Soma VL, Sullivan DJ, Bull PC, Stins MF, Smith JD. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc Natl Acad Sci USA. 2012;109:1782–1790. doi: 10.1073/pnas.1120534109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Bengtsson A, Joergensen L, Rask TS, Olsen RW, Andersen MA, Turner L, Theander TG, Hviid L, Higgins MK, Craig A, Brown A, Jensen ATR. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J Immunol. 2013;190:240–249. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull PC, Berriman M, Kyes S, Quail MA, Hall N, Kortok MM, Marsh K, Newbold CI. Plasmodium falciparum variant surface antigen expression patterns during malaria. e26PLoS Pathog. 2005;1 doi: 10.1371/journal.ppat.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull PC, Kyes S, Buckee CO, Montgomery J, Kortok MM, Newbold CI, Marsh K. An approach to classifying sequence tags sampled from Plasmodium falciparum var genes. Mol Biochem Parasitol. 2007;154:98–102. doi: 10.1016/j.molbiopara.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J-A, Howell KB, Reiling L, Ataide R, Mackintosh CL, Fowkes FJI, Petter M, Chesson JM, Langer C, Warimwe GM, Duffy MF, Rogerson SJ, Bull PC, Cowman AF, Marsh K, Beeson JG. Targets of antibodies against Plasmodium falciparum-infected erythrocytes in malaria immunity. J Clin Invest. 2012;122:3227–3238. doi: 10.1172/JCI62182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens A, Adams Y, Ghumra A, Lindergard G, Buchan CC, Andisi C, Bull PC, Mok S, Gupta AP, Wang CW, Turner L, Arman M, Raza A, Bozdech Z, Rowe JA. A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci USA. 2012;109:1772–1781. doi: 10.1073/pnas.1120461109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, Tan X, Doumbo S, Doumtabe D, Kone Y, Narum DL, Liang X, Doumbo OK, Miller LH, Doolan DL, Baldi P, Felgner PL, Pierce SK. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas LH, Júnior, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Golnitz U, Albrecht L, Wunderlich G. Var transcription profiling of Plasmodium falciparum 3D7: assignment of cytoadherent phenotypes to dominant transcripts. 14Malar J. 2008;7 doi: 10.1186/1475-2875-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol. 2013;15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler T, Albrecht GR, van Schravendijk MR, Aguiar JC, Morehead KE, Pasloske BL, Ma C, Barnwell JW, Greenwood B, Howard RJ. An improved microassay for Plasmodium falciparum cytoadherence using stable transformants of Chinese hamster ovary cells expressing CD36 or intercellular adhesion molecule-1. Am J Trop Med Hyg. 1993;48:332–347. doi: 10.4269/ajtmh.1993.48.332. [DOI] [PubMed] [Google Scholar]
- Hviid L. Naturally acquired immunity to Plasmodium falcipa-rum malaria in Africa. Acta Trop. 2005;95:270–275. doi: 10.1016/j.actatropica.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Joergensen L, Bengtsson DC, Bengtsson A, Ronander E, Berger SS, Turner L, Dalgaard MB, Cham GKK, Victor ME, Lavstsen T, Theander TG, Arnot DE, Jensen ATR. Surface co-expression of two different PfEMP1 antigens on single Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. e1001083PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes SA, Christodoulou Z, Raza A, Horrocks P, Pinches R, Rowe JA, Newbold CI. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T, Magistrado P, Hermsen CC, Salanti A, Jensen ATR, Sauerwein R, Hviid L, Theander TG, Staalsoe T. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. 21Malar J. 2005;4 doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T, Salanti A, Jensen ATR, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. 27Malar J. 2003;2 doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, Seguin-Orlando A, Willerslev E, Gilbert MTP, Lusingu J, Theander TG. PNAS Plus: Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci USA. 2012;109:e1791–e1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech JH, Barnwell JW, Miller LH, Howard RJ. Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J Exp Med. 1984;159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre J, Berry A, Benoit-Vical F. An alternative method for Plasmodium culture synchronization. Exp Parasitol. 2005;109:195–197. doi: 10.1016/j.exppara.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Maier AG, Rug M, O’Neill MT, Beeson JG, Marti M, Reeder J, Cowman AF. Skeleton-binding protein 1 functions at the parasitophorous vacuole membrane to traffic PfEMP1 to the Plasmodium falciparum-infected erythrocyte surface. Blood. 2007;109:1289–1297. doi: 10.1182/blood-2006-08-043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh K, Kinyanjui S. Immune effector mechanisms in malaria. Parasite Immunol. 2006;28:51–60. doi: 10.1111/j.1365-3024.2006.00808.x. [DOI] [PubMed] [Google Scholar]
- Medeiros MM, Fotoran WL, dalla Martha RC, Katsuragawa TH, Silva LHP, Wunderlich G. Natural antibody response to Plasmodium falciparum merozoite antigens MSP5, MSP9 and EBA175 is associated to clinical protection in the Brazilian Amazon. 608BMC Infect Dis. 2013;13 doi: 10.1186/1471-2334-13-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll K, Ljungström I, Perlmann H, Scherf A, Wahlgren M. Methods in malaria research. 5th. MR4/ATCC; Manassas/BioMalPar; Paris: 2008. 330 [Google Scholar]
- Ochola LB, Siddondo BR, Ocholla H, Nkya S, Kimani EN, Williams TN, Makale JO, Liljander A, Urban BC, Bull PC, Szestak T, Marsh K, Craig AG. Specific receptor usage in Plasmodium falciparum cytoadherence is associated with disease outcome. e14741PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0014741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinikov AV, Amos E, Frye IT, Rossnagle E, Mutabingwa TK, Fried M, Duffy PE. High throughput functional assays of the variant antigen PfEMP1 reveal a single domain in the 3D7 Plasmodium falciparum genome that binds ICAM1 with high affinity and is targeted by naturally acquired neutralizing antibodies. e1000386PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Ferreira J, Lacerda MV, Brasil P, Ladislau JL, Tauil PL, Daniel-Ribeiro CT. Malaria in Brazil: an overview. 115Malar J. 2010;9 doi: 10.1186/1475-2875-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak ND, Dzikowski R. PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum. Int J Biochem Cell Biol. 2009;41:1463–1466. doi: 10.1016/j.biocel.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Portugal S, Pierce SK, Crompton PD. Young lives lost as B cells falter: what we are learning about antibody responses in malaria. J Immunol. 2013;190:3039–3046. doi: 10.4049/jimmunol.1203067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask TS, Hansen DA, Theander TG, Pedersen AG, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes - divide and conquer. e1000933PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, Hviid L, Theander TG. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado AA, di Santi SM, Shiroma M. In vivo and in vitro Plasmodium falciparum resistance to chloroquine, amodiaquine and quinine in the Brazilian Amazon. Rev Inst Med Trop Sao Paulo. 1997;39:85–90. doi: 10.1590/s0036-46651997000200004. [DOI] [PubMed] [Google Scholar]
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Turner GD, Morrison H, Jones M, Davis TM, Looareesuwan S, Buley ID, Gatter KC, Newbold CI, Pukritayakamee S, Nagachinta B. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JEV, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutterrow YL, Avril M, Singh K, Long CA, Leke RJ, Sama G, Salanti A, Smith JD, Leke RGF, Taylor DW. High levels of antibodies to multiple domains and strains of VAR2CSA correlate with the absence of placental malaria in Cameroonian women living in an area of high Plasmodium falciparum transmission. Infect Immun. 2012;80:1479–1490. doi: 10.1128/IAI.00071-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AM, Kappe SHI. Malaria vaccine development: persistent challenges. Curr Opin Immunol. 2012;24:324–331. doi: 10.1016/j.coi.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Warimwe GM, Keane TM, Fegan G, Musyoki JN, Newton CRJC, Pain A, Berriman M, Marsh K, Bull PC. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci USA. 2009;106:21801–21806. doi: 10.1073/pnas.0907590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GE, Traore B, Kayentao K, Ongoiba A, Doumbo S, Doumtabe D, Kone Y, Dia S, Guindo A, Traore A, Huang CY, Miura K, Mircetic M, Li S, Baughman A, Narum DL, Miller LH, Doumbo OK, Pierce SK, Crompton PD. The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. e1000912PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO - World Health Organization World malaria report 2011. 2011 who.int/malaria/world_malaria_report_2011/en/
- WHO - World Health Organization World Malaria Report 2013. 2013 who.int/malaria/publications/world_malaria_report_2013/en/