Abstract
A hallmark of Bartonella henselae is persistent bacteremia in cats despite the presence of a vigorous host immune response. To understand better the long-term survival of B. henselae in cats, we examined the feline humoral immune response to B. henselae outer membrane (OM) proteins in naturally and experimentally infected cats. Initially, a panel of sera (n = 42) collected throughout North America from naturally infected cats was used to probe B. henselae total membranes to detect commonly recognized antigens. Twelve antigens reacted with sera from at least 85% of cats, and five were recognized by sera from all cats. To localize these antigens further, OMs were purified on discontinuous sucrose density step gradients. Each membrane fraction (OM, hybrid or inner membrane [IM]) contained less than 1% of the total malate dehydrogenase activity (soluble marker), indicating very little contamination by cytoplasmic proteins. FtsI, an integral IM cell division protein, was used to identify the low-density fraction (ρ = 1.13 g/cm3) as putative IM (<5% of the total FtsI localized to the high-density fraction) while lipopolysaccharide (LPS) and Pap31, a homolog of the Bartonella quintana heme-binding protein A (HbpA), defined the high-density fraction (ρ = 1.20 g/cm3) as putative OM. Additionally, little evidence of cross-contamination between the IM and OM was evident by two-dimensional gel electrophoresis. When purified OMs were probed with feline sera, antigenic proteins profiles were very similar to those observed with total membranes, indicating that many, but not all, of the immunoreactive proteins detected in the initial immunoblots were OM components. Interestingly, two-dimensional immunoblots indicated that B. henselae LPS and members of the Hbp family of proteins did not appear to stimulate an humoral response in any infected cats. Seven proteins were recognized by at least 70% of sera tested, but only three were recognized by all sera. Nanospray-tandem mass spectrometry was used to identify OM components, including the immunodominant OM proteins. Recognition of the nonimmunogenic nature of the major OM components, such as LPS, and identification of the predominant immunogens should elucidate the mechanisms by which B. henselae establishes persistent bacteremic infections within cats. Additionally, the common antigens may serve as potential feline vaccine candidates to eliminate the pathogen from its animal reservoir.
Bartonella henselae, the causative agent of cat scratch disease (CSD) and bacillary angiomatosis, is an important emerging health concern in the United States, with an estimated 24,000 new CSD cases per year, 2,000 of which require hospitalization (5). B. henselae infections result in different manifestations depending on the immune status of the patient. CSD occurs in immunocompetent patients and is generally characterized by a self-limiting lymphadenopathy that usually resolves in 2 to 4 months but has been shown to persist for up to 2 years in some patients (22). In persons with a compromised immune system, such as human immunodeficiency virus patients, alcoholics, or organ transplant recipients, B. henselae infections are much more severe. The diseases caused by B. henselae in these patients include bacillary angiomatosis, peliosis hepatis, and endocarditis (24).
The virulence mechanisms by which B. henselae causes these diverse diseases are not understood, and in vivo investigations of human pathogenesis are difficult. However, B. henselae infections are zoonotic in origin, and studies using a natural animal host are less problematic. Cats are the major reservoir for B. henselae, with ∼40% of domestic cats harboring active infections and ∼80% testing seropositive from previous exposure (5). In the absence of ectoparasites, there is no horizontal transmission of B. henselae between cats (6, 42), so spread of the infection is thought to depend on the arthropod vector Ctenocephalides felis (6, 13). After transmission, B. henselae grows to high levels (104 to 106 CFU/ml) in the bloodstream of its feline host, establishing long-term infections within the red blood cells (RBC) (26). Other tissues that may be involved in bacterial persistence within the animal include the liver, brain, kidneys, heart, and lymph nodes (18). Infected cats generally display few clinical abnormalities, but when present, they include fever and lesions on internal organs (10).
B. henselae persists in the blood for several months, and occasionally a bacteremic state is maintained for years (19, 40). In some cases, bacteria appear to clear completely from the bloodstream, but random bacteremic relapses in these animals indicate that infection persists at other sites in the body (1, 16). However, if the infection is completely cleared, cats become immune to subsequent challenge with the same and other B. henselae strains (1, 39, 40). Importantly, cats develop a humoral response to B. henselae during infection, suggesting that antibodies directed against B. henselae components may contribute to acquired immunity. Studies utilizing B-cell- deficient mice revealed that the antibody response is crucial for elimination of bacteremia caused by Bartonella grahamii, a mouse pathogen closely related to B. henselae (34). Therefore, it seems likely that the feline humoral response is necessary for clearance of B. henselae from the bloodstream. Several bacterial antigens have been described that are commonly recognized by antibodies from infected felines (9, 18). Unfortunately, these antigenic proteins have not been identified, and their role in protective immune responses is unclear.
After infection of the feline host, B. henselae must evade the immune system to establish a persistent bacteremia. Concomitantly, the host immune system must recognize and eliminate the pathogen. The outer membrane (OM) of gram-negative bacteria acts as an interface between the pathogen and its host. The same components of the OM that promote bacterial survival in the host are prime targets of the humoral response. To understand better the interaction of B. henselae and the feline immune system, we identified B. henselae OM proteins that are immunoreactive in cats. Additionally, we determined that several major OM components, including lipopolysaccharide (LPS) and members of the Hbp family of proteins, are not recognized by the host humoral response. Determining why these major OM structures are not recognized by the immune system and the possible role of the “shared” OM antigens in protective immune responses should lead to a better understanding of the pathogenesis of B. henselae and its interaction with the host.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Virulent Houston-1 B. henselae (32) was grown in BBH-H medium as described previously (4). Cell densities of liquid cultures were determined by plate counts on BBH-H agar. Plates were incubated at 34°C under an atmosphere of 5% CO2, 18% O2, and 77% N2.
Isolation of membrane fractions.
Cells were grown in 2 liters of BBH-H to a cell density of ∼1 × 108 to 2 × 108 CFU/ml and harvested by centrifugation (5,000 × g at 4°C for 20 min). The pellet was suspended in 300 ml of HEPES-NaCl buffer (20 mM HEPES, 25 mM NaCl [pH 7.6]) and collected as described above. The wash step was repeated twice, and the pellet was suspended in 2 ml of lysis buffer (20 mM HEPES, 5% sucrose [pH 7.6]). Cells were lysed by two passages though a chilled French pressure cell (14,000 lb/in2), and cell debris was removed by centrifugation (15,000 × g at 4°C for 15 min). To isolate total B. henselae membranes, the cell lysate was diluted to 50 ml with HEPES buffer (20 mM HEPES [pH 7.6]) and membranes were harvested by centrifugation (100,000 × g at 4°C for 2 h) in a type 45Ti rotor (Beckman Instruments, Inc., Palo Alto, Calif.). The resulting membrane pellet was suspended in 50 ml of HEPES buffer and harvested by centrifugation. The total membrane pellet was suspended in 1 ml of HEPES buffer, aliquoted, and stored at −80°C.
OM were isolated using a membrane isolation technique developed for Bartonella quintana (3) with the following modifications. Briefly, 2 ml of cell lysate was prepared as above, and a 0.3-ml aliquot of the lysate was saved prior to gradient centrifugation for the determination of total B. henselae protein composition. The remaining 1.7 ml was layered onto a discontinuous sucrose gradient (1.7 ml of sample, 6 ml of 25% sucrose, 6 ml of 35% sucrose, 8 ml of 45% sucrose, and 10 ml of 55% sucrose). All sucrose solutions were prepared in HEPES buffer. The sucrose gradient was centrifuged (100,000 × g at 4°C for 20 h) in a type SW28 rotor (Beckman Instruments, Inc.), 1.0-ml fractions were collected, and the refractive index of each fraction was measured using a densitometer (Bausch & Lomb, Rochester, N.Y.). Absorbances at 280 nm were determined for the fractions, and protein peaks were combined, diluted to 50 ml with HEPES buffer, and harvested by centrifugation (100,000 × g at 4°C for 5 h) in a type 45Ti rotor. The membrane pellets were diluted to 50 ml and harvested by centrifugation. The resulting pellets were suspended in 100 to 400 μl HEPES buffer depending on the size of the membrane pellet and were identified with the following markers. Malate dehydrogenase was used as a cytoplasmic marker, FtsI was used to identify inner membranes (IM), and LPS and HbpA were used to identify OM (the methods are described below). The protein concentrations were determined by the bicinchoninic acid assay (35), and the subcellular fractions containing purified OM, IM, and membrane hybrids were stored at −80°C.
SDS-PAGE, two-dimensional electrophoresis, and Western immunoblotting.
Proteins from subcellular fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 12.5% acrylamide gel (20) and visualized by Coomassie R-250 staining. For two-dimensional analysis, membrane samples containing 50 μg of protein were precipitated (at −20°C for 2 h) in 8 volumes of cold acetone, solubilized in immobilized pH gradient (IPG) rehydration buffer (8 M urea, 4% Triton X-100, 10 mM dithiothreitol, 1% biolytes), and used to rehydrate IPG strips (pH 3 to 10) as described by the manufacturer (Amersham Biosciences, Piscataway, N.J.). Isoelectric focusing was conducted for 18,000 Vh using the IPGphor system (Amersham Biosciences), followed by second-dimension electrophoresis on 1.5-mm SDS-PAGE slab gels of 12.5% polyacrylamide. Proteins were visualized by silver staining with the Plus One kit (Amersham Biosciences) or with the SilverQuest kit (Invitrogen, Carlsbad, Calif.) for mass spectrometry (MS)-based protein identification (the methods are described below) as specified by the respective manufacturers.
Immunoblot analyses were performed on B. henselae subcellular fractions to identify antigenic proteins and to establish membrane markers. Proteins from membrane fractions were separated by one- or two-dimensional SDS-PAGE and transferred to nitrocellulose (36). Immunoblots were probed with antibodies raised against the B. quintana protein HbpA (diluted 1:5,000; OM marker) and the Bartonella bacilliformis protein IalB (diluted 1:2,000), graciously provided by M. Minnick, Division of Biological Sciences, University of Montana, Missoula, Mont. Antibodies raised against the Escherichia coli protein FtsI (diluted 1:2,000, IM marker) were the kind gift of D. Weiss, Department of Microbiology, University of Iowa, Iowa City, Iowa. Sera were obtained from cats that were naturally and experimentally infected with B. henselae and have been characterized previously (15, 25). The sera were diluted according to determined titers prior to use (e.g., a serum with a titer of 1,024 was diluted 1:1,000 for use in immunoblotting) (15). Immunoreactive proteins were visualized by using horseradish peroxidase-conjugated goat anti-cat (Jackson Immunoresearch, West Grove, Pa.) or goat anti-rabbit (Sigma, St. Louis, Mo.) secondary antibodies and chemiluminescence (Enhanced Chemiluminescence; Amersham Biosciences). Image analysis was performed with EagleSight software, v. 3.2 (Stratagene, La Jolla, Calif.).
LPS and enzyme assays.
To determine which membrane fractions contained LPS, protein from each fraction (10 μg/lane) was separated by SDS-PAGE in 15% acrylamide gels and LPS was visualized with a modified silver stain (37). To determine the levels of cytoplasmic proteins present in the membrane fractions, malate dehydrogenase, a soluble enzyme, was assayed at 37°C as described previously (4).
MS protein identification.
Two-dimensional-gel-separated proteins were identified on reduced and alkylated trypsin-digested samples prepared as previously described (38). MS and protein identification were performed by the Mass Spectrometry Facility Research Technology Branch at the National Institute of Allergy and Infectious Diseases (Bethesda, Md.). Tandem MS (nano-LC-MS/MS) was performed on a QSTARXL (Applied Biosystems-Sciex, Ontario, Canada) hybrid quadropole-time-of-flight mass spectrometer. Mascot software (Matrix Sciences Inc., Beachwood, Ohio) was used to identify proteins from the NCBInr protein database from The National Center for Biotechnology Information at the National Institutes of Health.
RESULTS
B. henselae total membrane antigens.
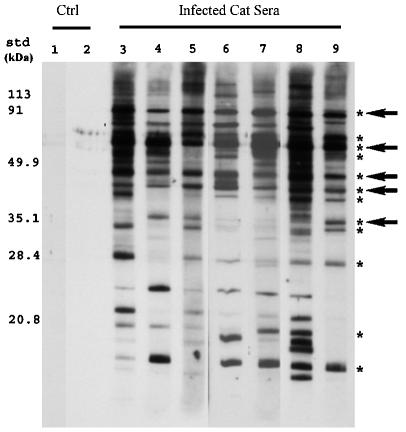
Sera from infected cats were previously shown to react with several proteins in the B. henselae cell lysate from naturally infected cats (9). Our initial localization of these immunoreactive proteins indicated that >90% localized to the total membrane fraction rather than the soluble cytoplasmic fraction (data not shown). To narrow the number of antigenic proteins to those which were most commonly associated with B. henselae infections, total membranes isolated from infectious strain Houston-1 were probed with a panel of sera collected from naturally infected cats from across North America (15). Sera from 42 cats seropositive (antibody titer, ≥256) for B. henselae exposure were used in immunoblot analyses with B. henselae total membranes (a representative blot is shown in Fig. 1). While multiple antigens were recognized by each of the serum samples, 12 membrane antigens reacted with sera from at least 85% of the samples screened (Fig. 1, asterisks). Of these 12 antigens, 5 (90, 67, 48, 44, and 35 kDa) were recognized by sera from 100% of cats (Fig. 1, arrows), and four of these antigens appeared to elicit a very strong humoral response. Previously, Freeland et al. and Kordick et al. detected antigens with similar molecular masses in studies that utilized whole-cell B. henselae antigen preparations (9, 18), and these new data suggest that these antigens localize to the membranes of B. henselae.
FIG. 1.
Western immunoblot demonstrating recognition of B. henselae membrane antigens by sera from naturally infected cats. Total membrane proteins were separated by SDS-PAGE, transferred to nitrocellulose, and screened with sera. Lane 1 was incubated with only secondary goat anti-cat antibodies, and lane 2 was incubated with serum from a specific-pathogen-free cat. Lanes 3 to 9 were incubated with sera from infected cats. Molecular mass standards (std) are indicated to the left. Antigens recognized by >85% of sera screened are marked with asterisks, and antigens recognized by 100% of sera are marked with arrows. Ctrl, control.
Membrane isolation and analysis.
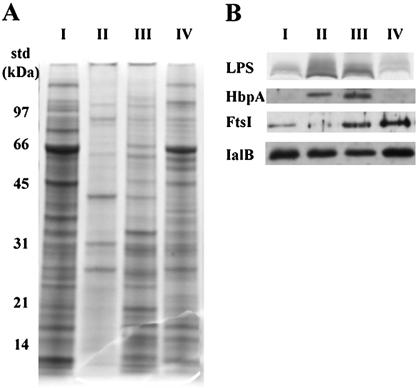
We were particularly interested in the commonly recognized OM antigens because of their potential direct interaction with the host immune system. To localize these antigens, purified OM with little contamination from other subcellular components was required. Initially, subcellular fractions were isolated by isopycnic centrifugation using a technique developed for blood agar-grown B. quintana (3). However, the densities of the B. henselae membrane fractions differed from those of B. quintana. Therefore, the sucrose concentrations of the gradient were optimized for the isolation of B. henselae OM. Another modification was to layer cell lysate on the sucrose gradient rather than total membranes isolated from the cell lysate by centrifugation. These steps decreased the formation of membrane hybrids and increased the yield of purified OM and IM. Centrifugation of the sucrose gradient yielded three membrane bands with densities of 1.20, 1.15, and 1.13 g/cm3. Initially, the isolated membrane fractions were analyzed by SDS-PAGE, and the high- and low-density fractions had distinct protein profiles (Fig. 2A). The fraction of intermediate density had a protein profile with similarities to high- and low-density fractions and probably contained both (Fig. 2A). The protein profiles suggest that the modified separation technique produced membrane fractions with protein compositions specific for distinct subcellular fraction.
FIG. 2.
Protein profiles of B. henselae subcellular fractions and localization of membrane marker proteins. Lanes: I, cell lysate; II, high-density membrane fraction; III, intermediate-density membrane fraction; IV, low-density membrane fraction. (A) Membrane fractions were isolated by sucrose gradient centrifugation, separated by SDS-PAGE, and visualized by Coomassie blue staining. Molecular mass standards (std) are indicated to the left. (B) Following separation by SDS-PAGE, membrane markers were visualized in membrane fractions by either modified silver staining for LPS or immunoblotting with antibodies raised against HbpA, FtsI, and IalB.
Protein markers were used to confirm the identity and assess the purity of membrane fractions with different densities. To determine the soluble protein contamination of different membrane fractions, malate dehydrogenase (a cytoplasmic enzyme) was used as a marker protein. Preliminary experiments with total membrane preparations localized malate dehydrogenase activity to the soluble fraction (data not shown). Malate dehydrogenase enzymatic assays on subcellular fractions established that less than 1% of the total malate dehydrogenase activity localized to the membrane fractions (data not shown), indicating very little contamination of the membrane fractions with cytoplasmic protein. To confirm the identity of the OM, LPS, a major component of the OM of gram-negative bacteria, was used as a marker. SDS-PAGE and a modified silver stain indicated that the majority of the LPS was localized to the high-density fraction (Fig. 2B, designated lane II, LPS). Additionally, antibodies raised against HbpA, an OM protein from the closely related species B. quintana (3), reacted strongly with a 31-kDa protein in the high-density fraction (Fig. 2B, lane II, HbpA). Taken together, these data identified the high-density fraction as OM. Lower levels of LPS and HbpA were also found in the intermediate-density fraction, consistent with its identification as hybrid membranes (Fig. 2B, lane III). Very little LPS or HbpA localized to the low-density membrane fraction (Fig. 2B, lane IV), suggesting that this membrane fraction was relatively free of OM contamination.
To confirm the identity and assess the purity of the low-density membrane fraction, several antibodies to E. coli IM proteins were used as probes against purified membrane fractions in an immunoblot analysis. These included the c subunit of the F1F0ATPase, SecE, the magnesium transporter CorA, and the cell division protein FtsI. Antibodies against the c subunit, SecE, and CorA failed to react with B. henselae proteins (data not shown), indicating either that B. henselae does not possess homologs of these proteins or that there is low amino acid sequence conservation between homologs. FtsI antibodies reacted with a 67-kDa B. henselae protein present predominantly in the intermediate- and low-density membrane fractions (Fig. 2B). The size of this reactive protein was very similar to that of the FtsI homolog in the closely related bacterium, Brucella suis. Based on size and reactivity, this protein most probably represented the FtsI homolog of B. henselae; its presence confirmed the low-density fraction as IM. The relative absence of FtsI (<5% as determined by densitometry) in the OM revealed low levels of IM contamination of the purified OM. Taken together, these data confirmed the identity of the various membrane fractions and demonstrated low levels of cross contamination between subcellular fractions.
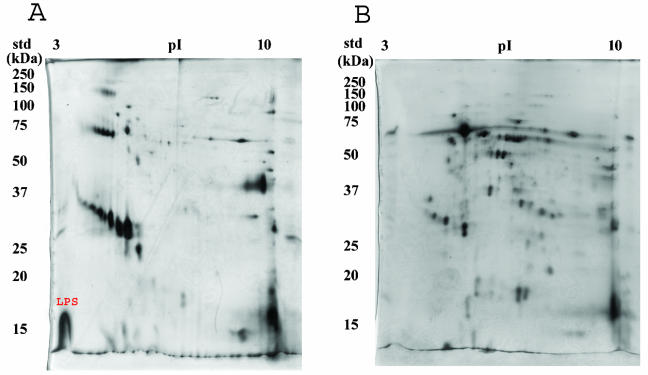
Two-dimensional protein profiles of the B. henselae OM and IM were clearly distinct, with a much more complex protein composition of the IM than of the OM (Fig. 3). A 70-kDa protein was the dominant protein observed in the IM fraction (Fig. 3B), while the OM had major proteins with estimated molecular masses of 41, 25, and 16.5 kDa and two closely grouped protein clusters. A large portion of the total OM protein content occurred in a cluster of at least seven proteins ranging is mass between 28 and 34 kDa. A second, smaller cluster of five proteins with masses ranging between 67 and 70 kDa was also present in the OM (Fig. 3A). The darkly stained, acidic, low-molecular-weight molecule present in the OM but not in the IM is probably LPS, which typically migrates in two-dimensional gels as a large, vertical streak at the dye front on the acidic edge of the gel (Fig. 3A, LPS). Taken together, these analyses of the membrane fractions further demonstrated that the OMs were relatively free of contamination with other subcellular components.
FIG. 3.
Two-dimensional SDS-PAGE profiles of B. henselae OM (A) and IM (B). Isolated OM and IM fractions were separated first by pI and then by molecular mass. Proteins were visualized by silver staining with the PlusOne kit. LPS is labeled in red in panel A. The pH gradient is indicated at the top, and molecular mass standards (std) are indicated to the left.
IalB localization.
IalB has been strongly implicated in RBC invasion by B. bacilliformis, and B. henselae contains a homolog (8, 30). Coleman and Minnick localized IalB to the IM of B. bacilliformis grown on blood agar (7); therefore, IalB was investigated as a possible IM marker for B. henselae by using immunoblots probed with specific IalB antiserum. Unlike IalB localization in B. bacilliformis, however, significant amounts of IalB were present in all B. henselae membrane fractions (Fig. 2B, IalB). In the previous publication, the authors reported that IalB contained a putative signal sequence and were surprised that IalB localized to the IM. Thus, the localization of IalB in the OM of B. henselae was more consistent with the protein harboring an N-terminal export signal. Additionally, B. henselae grown in liquid medium appeared to produce much larger quantities of the protein than did B. bacilliformis grown on blood agar (8). Taken together, the localization of IalB in B. henselae and the increase in the amount of protein detected in purified OM may more accurately reflect the cellular location of the protein, where it could interact with eukaryotic cells during RBC or endothelial cell invasion.
Antigenic OM proteins.
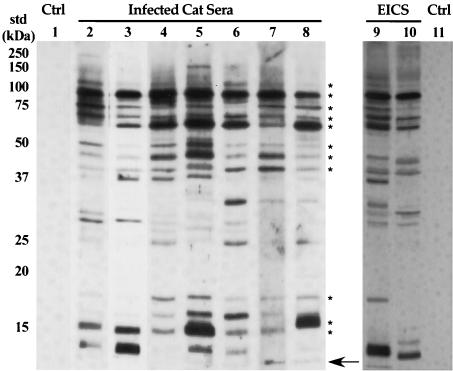
The purified B. henselae OM were also used to localize conserved antigens recognized by sera from naturally infected cats. OM proteins reacted strongly with feline sera, and 11 antigens were detected by all (n = 14) naturally infected cat sera (Fig. 4, asterisks). Additionally, sera from two experimentally infected cats were used to screen B. henselae OM. The experimentally infected cats were part of a prior study and demonstrated protection against homologous-strain challenge after the initial B. henselae infection was cleared by antibiotic treatment (25). Serum samples collected from these cats 1 week after secondary challenge were used to screen B. henselae OM. The pattern of OM antigen recognition in experimentally infected cats was similar, although not identical, to that in naturally infected cats (Fig. 4). The differences in B. henselae antigen recognition between naturally and experimentally infected animals were primarily in low-molecular-mass OM proteins. These data revealed that B. henselae OM proteins stimulate a strong humoral response in cats and that several of these OM antigens were recognized by all serum samples. Moreover, these antigens were recognized by sera from cats with naturally acquired B. henselae infections or experimentally induced with laboratory-grown B. henselae. Interestingly, no sera recognized antigens where LPS typically migrates (Fig. 4, arrow; Fig. 5A and B) (see below). Previous research by Liberto et al. and Matera et al. revealed that Bartonella LPS induces an atypical cell-mediated immune response (21, 23), and we were not able to detect a humoral response to B. henselae LPS. Taken together, these data reveal that there are multiple antigens present in the B. henselae OM that stimulate a humoral response but LPS does not.
FIG. 4.
Western immunoblot of B. henselae OM proteins screened with sera from infected cats. After SDS-PAGE separation, OM proteins were transferred to nitrocellulose and incubated with sera from naturally infected cats (lanes 2 to 8), sera from experimentally infected cats (designated EICS; lanes 9 and 10), and sera from naive cats (lanes 1 and 11). OM antigens recognized by all sera from naturally infected cats are marked with asterisks on the right. The arrow on the right indicates the dye front, where LPS migrates. Molecular mass standards (std) are indicated to the left. Ctrl, control.
FIG. 5.
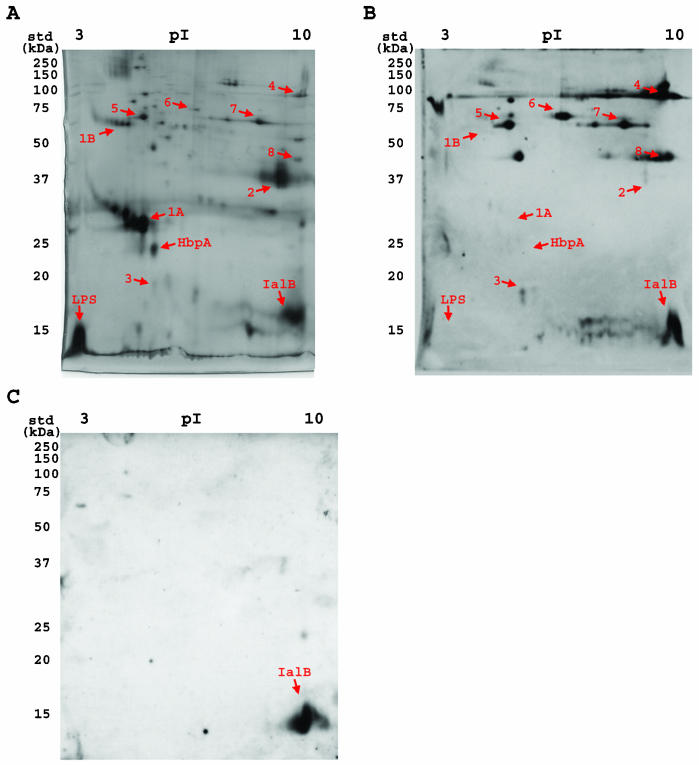
Two-dimensional characterization of B. henselae OM proteins and their recognition by feline immune serum. Following separation by two-dimensional SDS-PAGE, OM proteins were either visualized by silver staining (A) or transferred to nitrocelluose and probed with sera from infected cats (B) (representative immunoblot) or with specific antibodies raised against IalB (C). LPS and IalB are labeled on the panels. Proteins marked with arrows are discussed in the text. Molecular mass standards (std) are indicated to the left.
Identification of OM antigens.
Two-dimensional immunoblots of B. henselae OM proteins were probed with sera from eight naturally infected and two experimentally infected cats (a representative blot is displayed in Fig. 5B). Comparison of the silver-stained gels with the immunoblots revealed several interesting observations. First, the major cluster of OM proteins between 28 and 34 kDa was not recognized by immune cat sera (Fig. 5A and B, no. 1A). Using nanospray-MS/MS and confirmed with MALDI-ToF MS, the 25-kDa protein located near this cluster of proteins was identified as Pap31, a B. henselae homolog of HbpA (Fig. 5A and B, HbpA). Additionally, the amino acid sequence acquired by nanospray-MS/MS from several members of the major cluster of proteins identified them as Pap31, suggesting that they are a family of heme-binding proteins, similar to the Hbp proteins present in B. quintana. The second, smaller cluster of proteins evident in B. henselae OM (Fig. 5A and B, no. 1B) also had amino acid sequences homologous to Pap31, suggesting that B. henselae heme-binding proteins may exist either as high-molecular-mass variants, as is the case with B. quintana HbpB (56 kDa), or as multimers that are not fully dissociated by the denaturing conditions used during two-dimensional electrophoresis. Second, LPS was not recognized by any of the feline immune sera screened (Fig. 5A and B). Third, the 40-kDa major OM protein (Fig. 5B, no. 2) stimulated variable humoral responses in cats, ranging from weak (Fig. 5B) to intense (data not shown). This protein was identified as OMP43, a previously characterized B. henselae surface protein that has homology to the OMP2 family of porins from Brucella spp. OMP43 may also function as an adhesin during B. henselae interactions with eukaryotic cells (2). Fourth, IaIB was localized to both the IM and OM of B. henselae. The presence of IalB (pI ≈ 9.0) in the OM was confirmed by probing two-dimensional immunoblots with IalB-specific antiserum (Fig. 5C). While IaIB was recognized by ∼70% of the cat sera tested, the strength of the reaction was highly variable (as observed for OMP43) (Fig. 5A and B). Finally, the most immunogenic B. henselae OM proteins were relatively minor components of the OM.
Proteins that were recognized by all sera tested include proteins with approximate molecular masses of 90, 65, and 70 kDa (Fig. 5, no. 4, 5, and 6, respectively). The antigen at approximately 90 kDa was intensely reactive with all sera screened, despite the relatively low level of this protein evident in OM fractions (Fig. 5, no. 4). Nanospray-MS/MS identified this protein as OMP89, a possible surface protein containing a zinc metalloprotease domain (43). Protein 5 was identified as GroEL, a chaperonin that localizes to multiple subcellular fractions in Bartonella species (29). Amino acid sequence acquired for protein 6 did not match any protein sequence available in public databases. The other OM antigens, 3, 7, and 8, were recognized by ≥80% of sera screened. As with protein 6, amino acid sequence from protein 3 did not match any protein sequence available in public databases. Protein 7 was identified as HtrA, an envelope-associated protease often involved in bacterial stress responses. Unfortunately, no sequence data were obtained for protein 8 despite multiple attempts, probably because of the relatively small amounts of this protein and its migration at the extreme basic end of the first dimension. The identification and immunoreactivity of these proteins are summarized in Table 1. Once the completed B. henselae genome is released, identification of the novel B. henselae OM proteins 3 and 6 should be possible.
TABLE 1.
Identification and immunoreactivity of B. henselae OM components
| OM componenta | Identification | Antigen recognition (%)b |
|---|---|---|
| LPS | LPSc | 0 |
| IalB | IalBd | 70 |
| HbpA | Pap31e | 0 |
| 1 | Pap31e | 0 |
| 2 | OMP43e | 90 |
| 3 | Unknown proteine,f | 90 |
| AVTLVGAR | ||
| IALAGQRK | ||
| MVLEFGVSK | ||
| 4 | OMP89e | 100 |
| 5 | GroELe | 100 |
| 6 | Unknown proteine,f | 100 |
| GITLSVRP | ||
| LLDNYG | ||
| AQLVLRHR | ||
| 7 | HtrAe | 80 |
| 8 | No sequence | 80 |
Components as labeled in Fig. 5.
Recognition is presented as the percentage of sera reactive with the OM component through two-dimensional immunoblotting.
Identified through characteristic migration and staining properties.
Identified through two-dimensional immunoblotting with specific antibody.
Identified through nanospray-MS/MS.
Peptide sequences are listed for proteins with no match in the databases.
DISCUSSION
The interaction of B. henselae with the feline immune system has been the subject of intense investigation since the bacterium was first isolated from cats in 1992 (9, 18). This interest intensified when protective immunity in experimentally infected cats was described (10), with the goal of identifying the proteins responsible for eliciting this protective response. While several major antigens were detected by using sera from naturally or experimentally infected cats, these proteins were not localized or identified (9, 18). Of the antigens detected, those that localize to the cell surface would be the most important for understanding the protective response and the interaction of B. henselae with its host.
To identify cell surface antigens, we developed an OM isolation procedure for B. henselae that yielded distinct OM which were free of contamination with other subcellular components. Through two-dimensional immunoblot analyses, seven distinct B. henselae OM proteins were detected that elicited an humoral response in at least 70% of infected cats. In some cases, OM antigens that appeared as single bands in one-dimensional SDS-PAGE were resolved into multiple antigenic proteins with similar sizes but different pIs through two-dimensional immunoblot analyses, suggesting that multiple antigens are responsible for the humoral response previously attributed to single B. henselae proteins. Some proteins, such as IalB and OMP43, induced antibody responses that varied greatly between cats. Generally, the proteins that appeared to stimulate the strongest humoral response were relatively minor components of the OM, although it is possible that the expression levels of these proteins are significantly different in vitro and in vivo. The most immunogenic protein present in B. henselae OM was OMP89, a putative metalloprotease. Interestingly, several of the most antigenic proteins, GroEL, HtrA, and OMP89, are involved in either protein folding or degradation. This may be attributable to environmental stresses due to the intracellular persistence of B. henselae. Alternately, the proteases HtrA and OMP89 may function to provide B. henselae with amino acids, the organism's preferred carbon source (4). Based on their surface location and in vivo expression, these and the other commonly recognized OM proteins are probably involved in the ability of B. henselae to establish persistent infections within the feline host.
In the process of establishing chronic bacteremia, B. henselae appears to utilize several strategies to avoid clearance by the host immune system. Analysis of the OM composition revealed that relatively few proteins comprise the majority of the total protein present in the OM fraction, consistent with data presented in previous reports (2, 27). Other pathogenic bacteria, such as Treponema pallidum, express few OM proteins to avoid the humoral response and promote long-term survival in their respective hosts (31). It seems likely that the less complex protein profile in B. henselae reflects a similar adaptation for survival in host animals. Additionally, two-dimensional immunoblotting revealed that the major OM proteins of B. henselae did not stimulate an immune response in infected cats. The group of 28- to 34-kDa heme-binding proteins stimulated virtually no antibody production in any sera tested. This was surprising because these proteins were dominant in the OM. There are at least two possible explanations for these observations. The first is that these proteins are expressed only in vitro or in the arthropod vector and therefore are not presented to the feline immune system. However, this seems very unlikely because these proteins are involved in heme uptake and B. henselae appears to utilize heme as its sole iron source (4, 33). It is more plausible that these proteins are not immunogenic or are undergoing antigenic variation. Genes encoding multiple HbpA homologs in B. quintana have been identified (28). Therefore, it seems most likely that B. henselae also harbors multiple hbp genes, which provide a genetic reservoir for HbpA variability. Taken together, these data indicate that B. henselae is well adapted for immune evasion and long-term survival in the host.
The host response to endotoxin during infection is a common cause of symptoms, such as fever. LPS is a potent stimulator of cell-mediated immunity, and gram-negative bacteria present in the bloodstream induce the production of numerous cytokines, including tumor necrosis factor alpha. High levels of cytokine production can induce septic shock, a typical host response to bacteremia. Bartonella LPS, however, does not induce the production of tumor necrosis factor alpha (23) and has low endotoxic activity as measured by toll-like receptor 2 signaling (41). This atypical host response to Bartonella LPS is most probably due to an unusual composition of the lipid A and attached long-chain fatty acids (11, 14, 41). The altered LPS of Bartonella may be responsible for the pathogen's ability to establish persistent bacteremic infections without causing significant clinical pathology within the host. Consistent with this, LPS from B. henselae did not stimulate humoral response in cats, indicating that Bartonella endotoxin, unlike that of most gram-negative bacteria, is not immunogenic. Taken together, the immunoevasive characteristics of the LPS and HbpA suggest that B. henselae is well adapted for long-term survival within its natural host.
The RBC invasion protein IalB is a B. bacilliformis IM protein produced in small quantities, and the B. henselae genome encodes a homolog (30). However, B. henselae produced large amounts of the protein, and a significant portion of IalB localized to both the OM and IM of the bacterium. A possible explanation for these data is that the localization studies of IalB in B. bacilliformis were done on membrane fractions isolated from cells grown on blood agar plates, while the present study utilized broth grown bacteria. Bartonella cells grown on blood agar, compared to those grown in coculture with endothelial cells, have a slow replication rate (doubling time, ∼15 h) and contain significantly fewer ribosomes which could affect protein expression (17). However, the BBH-H-grown B. henselae used in this study has a relatively high growth rate (approximately five times faster than blood agar-grown bacteria) and probably also has a greater capacity for protein synthesis (4, 17). This may reflect more closely what occurs in vivo, since B. henselae cocultured with eukaryotic cells exhibits a growth rate closer to that of broth-grown cells than to that of blood agar-grown bacteria (17).
In this study we have identified seven proteins present in the OM (OMP89, GroEL, HtrA, IalB, OMP43, and two novel proteins) of B. henselae that are targeted in the feline humoral response. Ever since immunity to homologous rechallenge was first reported (10, 12, 39), vaccination of cats against B. henselae has been suggested as a means of reducing the public health risk (10, 12). Clearly, the OM immunogens described here are prime candidates for the development of an efficacious vaccine against B. henselae.
Acknowledgments
We thank Tim Hoover for valuable insight and suggestions and Carl Hammer and Mary Ann Robinson for nanospray-MS/MS protein identification.
This work was supported in part by a grant from the University of Georgia Research Foundation. M.R.C. was supported in part by NSF research training grant BIR-9413235.
Editor: J. B. Bliska
REFERENCES
- 1.Abbott, R. C., B. B. Chomel, R. W. Kasten, K. A. Floyd-Hawkins, Y. Kikuchi, J. E. Koehler, and N. C. Pedersen. 1997. Experimental and natural infection with Bartonella henselae in domestic cats. Comp. Immunol. Microbiol. Infect. Dis. 20:41-51. [DOI] [PubMed] [Google Scholar]
- 2.Burgess, A. W., and B. E. Anderson. 1998. Outer membrane proteins of Bartonella henselae and their interaction with human endothelial cells. Microb. Pathog. 25:157-164. [DOI] [PubMed] [Google Scholar]
- 3.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. F. Minnick. 2000. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chenoweth, M. R., G. A. Somerville, D. C. Krause, K. L. O'Reilly, and F. C. Gherardini. 2004. Growth characteristics of Bartonella henselae in a novel liquid medium: primary isolation, growth-phase-dependent phage induction, and metabolic studies. Appl. Environ. Microbiol. 70:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomel, B. B., R. C. Abbott, R. W. Kasten, K. A. Floyd-Hawkins, P. H. Kass, C. A. Glaser, N. C. Pedersen, and J. E. Koehler. 1995. Bartonella henselae prevalence in domestic cats in California: risk factors and association between bacteremia and antibody titers. J. Clin. Microbiol. 33:2445-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomel, B. B., R. W. Kasten, K. Floyd-Hawkins, B. Chi, K. Yamamoto, J. Roberts-Wilson, A. N. Gurfield, R. C. Abbott, N. C. Pedersen, and J. E. Koehler. 1996. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34:1952-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, S. A., and M. F. Minnick. 2003. Differential expression of the invasion-associated locus B (ialB) gene of Bartonella bacilliformis in response to environmental cues. Microb. Pathog. 34:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, S. A., and M. F. Minnick. 2001. Establishing a direct role for the Bartonella bacilliformis invasion-associated locus B (Ia1B) protein in human erythrocyte parasitism. Infect. Immun. 69:4373-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeland, R. L., D. T. Scholl, K. R. Rohde, L. J. Shelton, and K. L. O'Reilly. 1999. Identification of Bartonella-specific immunodominant antigens recognized by the feline humoral immune system. Clin. Diagn. Lab. Immunol. 6:558-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene, C. E., M. McDermott, P. H. Jameson, C. L. Atkins, and A. M. Marks. 1996. Bartonella henselae infection in cats: evaluation during primary infection, treatment, and rechallenge infection. J. Clin. Microbiol. 34:1682-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 12.Gurfield, A. N., H. J. Boulouis, B. B. Chomel, R. Heller, R. W. Kasten, K. Yamamoto, and Y. Piemont. 1997. Coinfection with Bartonella clarridgeiae and Bartonella henselae and with different Bartonella henselae strains in domestic cats. J. Clin. Microbiol. 35:2120-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins, J. A., S. Radulovic, D. C. Jaworski, and A. F. Azad. 1996. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae). J. Med. Entomol. 33:490-495. [DOI] [PubMed] [Google Scholar]
- 14.Hornef, M. W., M. J. Wick, M. Rhen, and S. Normark. 2002. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 3:1033-1040. [DOI] [PubMed] [Google Scholar]
- 15.Jameson, P., C. Greene, R. Regnery, M. Dryden, A. Marks, J. Brown, J. Cooper, B. Glaus, and R. Greene. 1995. Prevalence of Bartonella henselae antibodies in pet cats throughout regions of North America. J. Infect. Dis. 172:1145-1149. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya, H., S. Maruyama, M. Irei, R. Takahashi, M. Yamashita, and T. Mikami. 2002. Genomic variations among Bartonella henselae isolates derived from naturally infected cats. Vet. Microbiol. 89:211-221. [DOI] [PubMed] [Google Scholar]
- 17.Kempf, V. A., M. Schaller, S. Behrendt, B. Volkmann, M. Aepfelbacher, I. Cakman, and I. B. Autenrieth. 2000. Interaction of Bartonella henselae with endothelial cells results in rapid bacterial rRNA synthesis and replication. Cell. Microbiol. 2:431-441. [DOI] [PubMed] [Google Scholar]
- 18.Kordick, D. L., T. T. Brown, K. Shin, and E. B. Breitschwerdt. 1999. Clinical and pathologic evaluation of chronic Bartonella henselae or Bartonella clarridgeiae infection in cats. J. Clin. Microbiol. 37:1536-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kordick, D. L., K. H. Wilson, D. J. Sexton, T. L. Hadfield, H. A. Berkhoff, and E. B. Breitschwerdt. 1995. Prolonged Bartonella bacteremia in cats associated with cat-scratch disease patients. J. Clin. Microbiol. 33:3245-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Liberto, M. C., G. Matera, A. G. Lamberti, G. S. Barreca, A. Quirino, and A. Foca. 2003. In vitro Bartonella quintana infection modulates the programmed cell death and inflammatory reaction of endothelial cells. Diagn. Microbiol. Infect. Dis. 45:107-115. [DOI] [PubMed] [Google Scholar]
- 22.Maguina, C., and E. Gotuzzo. 2000. Bartonellosis. New and old. Infect. Dis. Clin. North Am. 14:1-22, vii. [DOI] [PubMed] [Google Scholar]
- 23.Matera, G., M. C. Liberto, A. Quirino, G. S. Barreca, A. G. Lamberti, M. Iannone, E. Mancuso, E. Palma, F. A. Cufari, D. Rotiroti, and A. Foca. 2003. Bartonella quintana lipopolysaccharide effects on leukocytes, CXC chemokines and apoptosis: a study on the human whole blood and a rat model. Int. Immunopharmacol. 3:853-864. [DOI] [PubMed] [Google Scholar]
- 24.Maurin, M., R. Birtles, and D. Raoult. 1997. Current knowledge of Bartonella species. Eur. J. Clin. Microbiol. Infect. Dis. 16:487-506. [DOI] [PubMed] [Google Scholar]
- 25.McDermott, M. S. 1998. Bartonella henselae infection: diagnosis, treatment and immunological response. M.S. thesis. University of Georgia, Athens.
- 26.Mehock, J. R., C. E. Greene, F. C. Gherardini, T. W. Hahn, and D. C. Krause. 1998. Bartonella henselae invasion of feline erythrocytes in vitro. Infect. Immun. 66:3462-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minnick, M. F. 1994. Identification of outer membrane proteins of Bartonella bacilliformis. Infect. Immun. 62:2644-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minnick, M. F., K. N. Sappington, L. S. Smitherman, S. G. Andersson, O. Karlberg, and J. A. Carroll. 2003. Five-member gene family of Bartonella quintana. Infect. Immun. 71:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minnick, M. F., L. S. Smitherman, and D. S. Samuels. 2003. Mitogenic effect of Bartonella bacilliformis on human vascular endothelial cells and involvement of GroEL. Infect Immun 71:6933-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, S. J., and M. F. Minnick. 1995. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63:1552-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radolf, J. D., E. J. Robinson, K. W. Bourell, D. R. Akins, S. F. Porcella, L. M. Weigel, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect. Immun. 63:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regnery, R. L., B. E. Anderson, J. E. d. Clarridge, M. C. Rodriguez-Barradas, D. C. Jones, and J. H. Carr. 1992. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J. Clin. Microbiol. 30:265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sander, A., S. Kretzer, W. Bredt, K. Oberle, and S. Bereswill. 2000. Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol. Lett. 189:55-59. [DOI] [PubMed] [Google Scholar]
- 34.Seubert, A., R. Schulein, and C. Dehio. 2002. Bacterial persistence within erythrocytes: a unique pathogenic strategy of Bartonella spp. Int. J. Med. Microbiol. 291:555-560. [DOI] [PubMed] [Google Scholar]
- 35.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 36.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 38.Wolf, K., E. Fischer, D. Mead, G. Zhong, R. Peeling, B. Whitmire, and H. D. Caldwell. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69:3082-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, K., B. B. Chomel, R. W. Kasten, C. C. Chang, T. Tseggai, P. R. Decker, M. Mackowiak, K. A. Floyd-Hawkins, and N. C. Pedersen. 1998. Homologous protection but lack of heterologous-protection by various species and types of Bartonella in specific pathogen-free cats. Vet. Immunol. Immunopathol. 65:191-204. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto, K., B. B. Chomel, R. W. Kasten, C. M. Hew, D. K. Weber, W. I. Lee, J. E. Koehler, and N. C. Pedersen. 2003. Infection and re-infection of domestic cats with various Bartonella species or types: B. henselae type I is protective against heterologous challenge with B. henselae type II. Vet. Microbiol. 92:73-86. [DOI] [PubMed] [Google Scholar]
- 41.Zahringer, U., B. Lindner, Y. A. Knirel, W. M. R. van der Akker, R. Hiestand, H. Heine and C. Dehio. 2004. Structure and biological activity of the short-chain lipopolysaccharide from Bartonella henselae ATCC 49882T. J. Biol. Chem., in press. [DOI] [PubMed]
- 42.Zanutto, M. S., E. M. Mamizuka, R. Raiz-Junior, T. M. Lima, C. L. Diogo, T. S. Okay, and M. K. Hagiwara. 2001. Experimental infection and horizontal transmission of Bartonella henselae in domestic cats. Rev. Inst. Med. Trop. Sao Paulo 43:257-261. [DOI] [PubMed] [Google Scholar]
- 43.Zimmermann, R., V. A. Kempf, E. Schiltz, K. Oberle, and A. Sander. 2003. Hemin binding, functional expression, and complementation analysis of Pap 31 from Bartonella henselae. J. Bacteriol. 185:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]