Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are related intestinal pathogens that harbor highly similar pathogenicity islands known as the locus of enterocyte effacement (LEE). Despite their genetic similarity, these two pathogens disrupt epithelial tight junction barrier function with distinct kinetics. EHEC-induced reduction in transepithelial electrical resistance (TER), a measure of barrier function disruption, is significantly slower and more modest in comparison to that induced by EPEC. The variation in bacterial adherence only partially accounted for these differences. The LEE-encoded effector protein EspF has been shown to be critical for EPEC-induced alterations in TER. EspF from both EPEC and EHEC is expressed and secreted upon growth in tissue culture medium. The mutation of EHEC cesF suggested that the optimal expression and secretion of EHEC EspF required its chaperone CesF, as has been shown for EPEC. In contrast to EPEC espF and cesF, mutation of the corresponding EHEC homologs did not dramatically alter the decrease in TER. These differences could possibly be explained by the presence of additional espF-like sequences (designated U- and M-espF, where the letter designations refer to the specific cryptic prophage sequences on the EHEC chromosome closest to the respective genes) in EHEC. Reverse transcription-PCR analyses revealed coordinate regulation of EHEC U-espF and the LEE-encoded espF, with enhanced expression in bacteria grown in Dulbecco-Vogt modified Eagle’s medium compared to bacteria grown in Luria broth. Both EHEC espF and U-espF complemented an EPEC espF deletion strain for barrier function alteration. The overexpression of U-espF, but not espF, in wild-type EHEC potentiated the TER response. These studies reveal further similarities and differences in the pathogenesis of EPEC and EHEC.
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are evolutionarily related intestinal pathogens that infect at a low dose and cause diarrhea by unknown mechanisms (7). Both pathogens induce the formation of characteristic attaching and effacing (A/E) lesions in host cells and alter epithelial barrier function (17, 29, 31, 32). These noninvasive pathogens inject effector proteins directly into the host cytosol via a type III secretion system (TTSS). One of the secreted proteins, the translocated intimin receptor (Tir), inserts into the host membrane and interacts with intimin on the bacterial surface, leading to intimate attachment, actin polymerization within host cells, and the formation of a pedestal-like structure (38). The homologous loci of enterocyte effacement (LEE) in these pathogens share the same overall organization and encode the TTSS as well as various effector proteins required to form A/E lesions and mediate other host effects (11, 27).
Despite the similarities, there is mounting evidence that these bacteria differ significantly in their pathogenic mechanisms. The Shiga-like toxin (Stx) of EHEC is responsible for the bloody diarrhea that may progress to hemorrhagic colitis, and in some instances, hemolytic uremic syndrome (18). Stx-negative strains of EHEC retain the ability to induce A/E lesions and cause nonbloody diarrhea. In contrast, only a subset of EPEC strains produces a toxin, known as the EspC enterotoxin (26). This protein is therefore thought to play only an accessory role in pathogenesis. While the homologous LEE regions encoding the protein translocation complex are over 98% similar at the amino acid level, a divergence of up to 34% was observed in the genes encoding the secreted effector proteins (12). It is also increasingly apparent that the molecular mechanisms of host effects induced by these two pathogens differ in many respects (7). Transfer of the cloned EPEC LEE into E. coli K-12 confers the ability to induce A/E lesions and inject effector proteins into host cells (22). This strain was also able to displace occludin from the tight junctions (TJ) and disrupt the barrier function of host cells (34). In contrast, E. coli K-12 transformed with a clone containing the EHEC LEE was unable to induce A/E lesions or inject effector proteins into host cells, even when cotransformed with fragments from EPEC LEE (12). This suggests that there is a functional dissimilarity between one or more of the LEE-encoded effector proteins in the two pathogens or that determinants outside the LEE are required for EHEC pathogenesis.
Both EPEC and EHEC alter intestinal epithelial barrier function (17, 30). We have demonstrated previously that the EPEC-induced alteration of barrier function requires type III secretion and the type III secreted protein EspF. The studies reported in this paper were initiated to address our repeated observation that EPEC is more efficient at barrier function alteration than EHEC. Data presented here demonstrate several points of similarity, as well as differences, between EPEC- and EHEC-mediated alteration of the TJ barrier. A comparative analysis of the ΔespF and ΔcesF derivatives of EPEC and EHEC is also presented. These studies suggest the presence of an EHEC protein encoded outside the LEE that is coordinately regulated with genes in the pathogenicity island and is involved in pathogenesis.
MATERIALS AND METHODS
Cell culture.
The human intestinal epithelial T84 colon carcinoma-derived cell line was used in these experiments (6). Cells of the Caco-2 colon carcinoma cell line were grown in high glucose Dulbecco-Vogt modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. T84 cells were grown in a 1:1 (vol/vol) mixture of DMEM and Ham's F-12 medium supplemented with 6% newborn calf serum, passaged, and plated on either collagen-coated Transwell inserts (Costar, Cambridge, Mass.) or collagen-coated glass coverslips, as described previously (40).
Bacterial strains and infection of host cells.
E. coli strains E2348/69 (O127:H6; EPEC), 85-170 (O157:H7; EHEC), and their derivatives were used for all studies. The espF deletion (strain GH145) and cesF disruption (strain GH143) derivatives of strain 85-170 were generated by allelic exchange of the corresponding kanamycin-disrupted genes into the chromosome with the aid of SacB counter-selection as described earlier (21). The construction of the plasmids is described below. Confluent T84 or Caco-2 monolayers were infected with one of the above strains at the indicated multiplicity of infection (MOI) and assessed at the specified times postinfection.
DNA manipulations.
To target a disruption in EHEC espF, the flanking regions of this gene were amplified and cloned on either side of the kanamycin-resistance-conferring aph gene in plasmid pVK4. pVK4 is similar to plasmid pVK3 except that the aph gene is in the opposite orientation (39). Specifically, the amplified upstream (with primers PF25 and PF26) and downstream (with primers PF27 and PF28) (Table 1) regions were sequentially cloned into the NotI-SpeI and EcoRI-SalI sites, respectively, of pVK4 to generate plasmid pMP2. The NotI-SalI fragment from pMP2 was transferred to the sacB-containing vector pKO3 (21), and the resulting plasmid pMP5 was used to target a deletion or disruption of espF in EHEC strain 85-170. To target a kanamycin cassette insertion in the EHEC cesF homolog, the corresponding region was first amplified (by primers SF5 and SF6; Table 1) and cloned into pGEMT-Easy (Promega, Madison, Wis.). The cesF region was then transferred as a NotI fragment to pKO-3 to generate plasmid pMP3. Subsequently, the kanamycin resistance cassette from pVK4 was cloned into the PstI site of pMP3, and the resulting plasmid pMP4 was used to target a disruption in the cesF region of EHEC strain 85-170. For complementation experiments, full-length EPEC espF and EHEC espF were amplified from the respective strains by colony PCR with the primers PF1 and PF3 (Table 1). U-espF was amplified by using the primers PF10 and PF11 (Table 1). In each case, the EcoRI-PstI-digested PCR fragments were cloned into pUC18 to generate the various complementing plasmids that were subsequently transformed to the EPEC espF deletion strain, UMD874 (a gift from Michael Donnenberg, University of Maryland), by electroporation. To generate His-tagged EPEC EspF, primers PF6 and PF7 were used to amplify espF directly from the EPEC chromosome by colony PCR, and the product was cloned into pDEST17 by using Gateway technology (Invitrogen, Carlsbad, Calif.). The resulting plasmid pIS4 encodes an N-terminal His-tagged EspF. When necessary, the clones were verified by sequence analysis.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, description, or sequence | Reference or source |
|---|---|---|
| EPEC strains | ||
| E2348/69 | Wild-type 0127:H7 | |
| UMD874 | espF deletion mutant | 23 |
| E2348/69 cesF | cesF disruption mutant | 9 |
| EHEC strains | ||
| EDL933 | Wild-type O157:H7 Stx+ | |
| 85-170 | O157:H7 ΔStx | 37 |
| GH143 | cesF::Kan derivative of 85-170 | This study |
| GH145 | espF deletion derivative of 85-170 | This study |
| Plasmids | ||
| pUC 18 | Cloning vector | |
| pDEST17 | Cloning vector | Invitrogen |
| pKO3 | Counterselectable vector | 21 |
| pVK4 | Source of Kan cassette | 39 |
| pMP2 | Intermediate for pMP5 construction | This study |
| pMP3 | Intermediate for pMP4 construction | This study |
| pMP4 | Plasmid used to generate GH143 (EHEC ΔcesF) | This study |
| pMP5 | Plasmid used to generate GH145 (EHEC ΔespF) | This study |
| pIS4 | EPEC espF in pDEST17 to produce His-tag protein | This study |
| pVK166 | Full-length EHEC espF in pUC18 | This study |
| pSL1 | Full-length EHEC espF in pUC18 | This study |
| pVK167 | Full-length EHEC U-espF in pUC18 | This study |
| Primers | ||
| PF1 | GGCAGAATTCATGCTTAATGGAATTAGTAA | |
| PF3 | AATTCTGCAGTTACCCTTTCTTCGATTGCT | |
| PF6 | AAAAGCAGGCTTTATGCTTAATGGAATTAG | |
| PF7 | AGAAAGCTGGGTTTACCCTTTCTTCGATTGC | |
| PF10 | GGTAGAATTCATGATTAACAATGTTTCTTC | |
| PF11 | TATACTGCAGTCACGAGCGCTTAGATGTAT | |
| PF18 | NNNNCTGCAGTTAAATGCCATGCTCTGCAAGATG | |
| PF25 | GGGGCGGCCGCCTGGGTAATTGATCATGGTC | |
| PF26 | TCGACTAGTGATACCTACAAGCTGCCGCC | |
| PF27 | CATGAATTCGCCTATGAGCAATCGAAGAA | |
| PF28 | CACGTCGACCTGAATAACACATCCCCGTC | |
| PF29 | TGGCCTGCAGCATTAAAAATCGTCCTCGCA | |
| SF5 | ACAGGGATCCGGAGTTGGTTAGTCAGTTTG | |
| SF6 | ATGAGGATCCAGAGAGAAGGTGCTGGATTG | |
| 16S-F | ACTCAAATGAATTGACGGGGGC | |
| 16S-R | AGGCCCGGGAACGTATTCAC |
RT-PCR analysis.
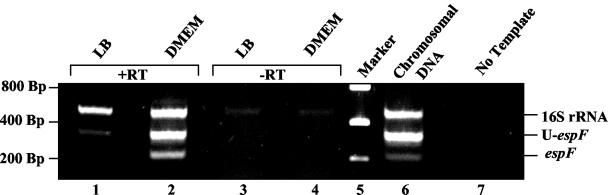
The expression of espF and U-espF was monitored by reverse transcription (RT)-PCR by using the primers PF1 and PF29 specific for the espF gene (product, 210 bp) and the primers PF10 and PF18 specific for the U-espF gene (product, 290 bp) (Table 1). Despite the presence of multiple, near-perfect binding sites for primer PF18, the most prominent product was the 290-bp fragment. Overnight cultures of EPEC and EHEC strain 85-170 grown in Luria-Bertani (LB) broth were subcultured in either LB or DMEM to mid-logarithmic phase. RNA was isolated from 1 ml of the mid-log cultures by using Trizol reagent (Invitrogen), treated with DNase I (Promega), and purified with the RNeasy kit (QIAGEN, Valencia, Calif.). Equal amounts of RNA were used in the RT-PCR experiments (Invitrogen), and the resulting fragments were analyzed on a 2% agarose gel. Primers specific to E. coli 16S rRNA (16S-F and 16S-R; product, 480 bp) (Table 1) were used as controls to ensure that variations observed in the levels of expression were not due to differences in initial RNA amounts or variability in RT reactions. Similar reactions were also carried out in the absence of reverse transcriptase to confirm the absence of DNA in the RNA samples.
Protein purification and antibody production.
EPEC EspF protein fused to His6 was expressed in BL21(DE3) cells containing the expression clone pIS4. One liter of fresh LB medium was inoculated with the overnight culture, and the cells were grown to an optical density at 600 nm of 0.4 to 0.6. EspF expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and cells were grown for another 4 h. The culture was then centrifuged, and the pellet was resuspended in buffer (50 mM Tris-Hcl [pH 8.00], 50 mM NaCl, 1 mM EDTA, lysozyme and protease inhibitor cocktail; Sigma-Aldrich, St. Louis, Mo.) and sonicated. Inclusion bodies were separated by centrifugation and denatured in 50 mM Tris-HCl (pH 8.00) and 8 M guanidine-HCl. Insoluble material was removed by an additional centrifugation step. The protein was then refolded, dialyzed, filtered through a 0.22-mm-pore-size filter (Millipore, Bedford, Mass.) and applied to a Ni2+ nitrilotriacetic acid column (QIAGEN,). Pure protein was eluted with 350 mM imidazole, dialyzed, and used to immunize NZW rabbits (Covance, Princeton, N.J.), and the resulting polyclonal antiserum was used for EspF detection. The antibody specifically detected EspF from wild-type EPEC as well as infected T84 cells. The band corresponding to EspF was absent in the espF deletion strain UMD874 (data not shown). This antibody also detected the EHEC EspF but not the EspF-like sequences of EHEC.
Western blot analyses.
A Western blot analysis was performed to verify the absence of EspF in the deletion strain GH145. To determine the role of CesF on EspF expression, mid-log phase DMEM cultures of EHEC strains 85-170 and GH143 were centrifuged, and the pellets were resuspended in loading buffer and assessed by Western blot analysis. Supernatants from the corresponding cultures were precipitated with 24% trichloroacetic acid in the presence of 0.02% deoxycholate and washed with acetone, and the dry pellet was resuspended in loading buffer. Half of each sample was then run on a 12% polyacrylamide gel, transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, Calif.), and probed for EspF. For immunoblot analysis of occludin, proteins were extracted from cells by using RIPA buffer, and protein concentration was determined by the Bradford protocol (Bio-Rad). Equal amounts of protein (50 mg) were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and electrophoretically transferred onto nitrocellulose membranes with a Trans-blot cell apparatus (Bio-Rad). Membranes were blocked by using Zymed blocking solution for 1 h at room temperature (RT). Following three 5-minute washes with Tris-buffered saline-Tween, the membranes were incubated with occludin primary antibody (1:1,000 dilution) for 1 h at RT, followed by incubation with alkaline phosphatase-conjugated secondary antibodies for 1 h at RT. Color development was attained via a nitro blue tetrazolium-5-bromo-4-cloro-indolylphosphate premixed solution (Zymed, San Francisco, Calif.).
Measurement of TER and determination of bacterial adherence.
T84 monolayers on Transwell inserts (Costar) were infected with mid-log growth phase bacteria, and the transepithelial electrical resistance (TER) was measured at 2, 4, and 6 h postinfection as previously described (34). Monolayers infected with EPEC or EHEC at an MOI of 100 (unless otherwise indicated) for 1 h were washed to remove nonadherent bacteria and incubated for additional time (1, 3, or 5 h) in the presence of fresh DMEM. The initial inoculum was monitored by measuring the optical density at 600 nm and confirmed by plating serially diluted samples on LB agar plates. The number of attached bacteria was determined immediately following the wash at 1 h postinfection or following the TER reading at 6 h postinfection. Cells and attached bacteria were removed from filters by scraping, serially diluted, and plated on LB, and the number of CFU was determined.
Immunofluorescence microscopy.
T84 or Caco-2 cells were grown to confluence on glass coverslips and infected with EPEC and EHEC strains and their derivatives for 6 h. Monolayers were washed after infection with cold phosphate-buffered saline; they were then fixed and permeabilized by using 3.7% paraformaldehyde and 0.2% Triton X-100, respectively, at RT for 10 min. Cells were blocked by using 1% bovine serum albumin for 20 min, incubated for 1 h with occludin antibody (Zymed) at a concentration of 4 μg/ml and then with fluorescein isothiocyanate-conjugated secondary antibody (1:100 dilution). Coverslips were mounted on slides with ProLong Antifade medium (Molecular Probes, Eugene, Oreg.) and visualized on a Nikon Opti-Phot inverted microscope equipped with the Spot-RT digital imaging system (Diagnostic Instruments, Sterling Heights, Mich.).
Statistical analysis.
Data were analyzed by using Student's t test for independent samples. Differences were considered significant if P was ≤0.05.
RESULTS
Differences between EPEC and EHEC attachment rates do not account for the diminished effect of EHEC on barrier function.
Although EPEC and EHEC have been previously reported to disrupt barrier function (17, 20, 29, 31), the present study was undertaken to directly compare the effect of the two pathogens on this key physiological function. There was no significant difference in the ability of Stx+ (EDL933) and Stx− (85- 170) EHEC 0157:H7 strains to decrease TER (at 6 h postinfection, the percent change in TER with Stx+ was −38% ± 4% and with Stx− was −37% ± 5% [mean ± standard error]; n = 12), similar to earlier reports for a related EHEC strain (29). Also, purified Stx did not alter epithelial barrier function (data not shown). Therefore, the stx mutant EHEC 0157:H7 strain 85-170 was used in all subsequent experiments. This strain is referred to as EHEC throughout this paper. While results with the T84 cell line are reported, all observations were independently confirmed in the Caco-2 colon carcinoma cell line (data not shown).
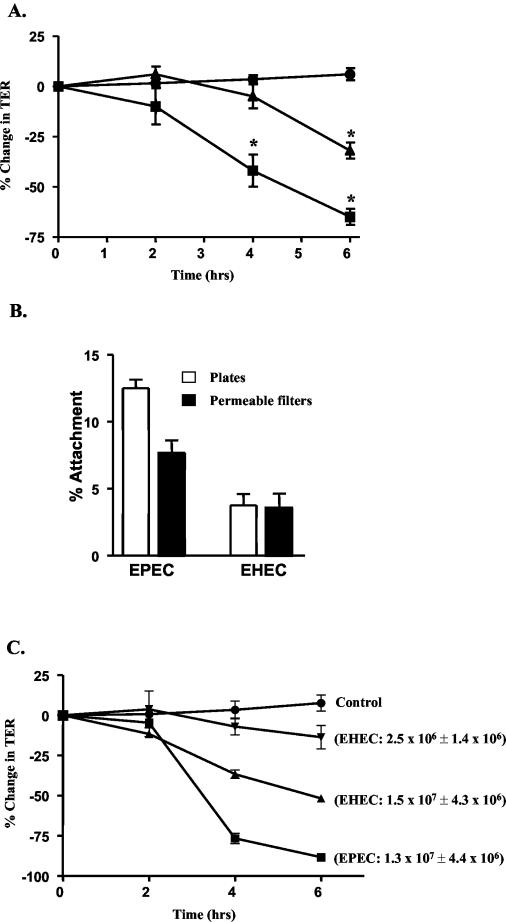
A time course of infection revealed that EPEC and EHEC infection of intestinal epithelial cells at an MOI of 100 resulted in distinct kinetics for TER alterations. EPEC-induced TER reduction was evident as early as 2 h, progressing to a 60% decrease at 6 h postinfection (Fig. 1A). In contrast, the EHEC-associated decrease in TER was not significant until 6 h postinfection and only dropped 30% by this time (Fig. 1A).
FIG. 1.
EPEC and EHEC alter barrier function with distinct kinetics. (A) T84 monolayers on permeable filters were treated with serum-free, antibiotic-free DMEM buffer (•) or infected with EPEC (▪) or EHEC (▴), and TER was measured at 2, 4, and 6 h postinfection. Values represent the percent change in TER compared to baseline measurements at time zero (n ≥ 5; asterisks correspond to values significantly different than controls where P is ≤0.005). (B) To estimate bacterial attachment to T84 cells, monolayers on plates or permeable filters were infected at an MOI of 100 for 1 h at 37°C. The infected monolayers were washed, scraped, serially diluted, and plated on LB to determine the number of CFU. The percentage of attached bacteria relative to the original inoculum is indicated (n = 4; P < 0.003 for EPEC versus EHEC on plates). (C) To determine the effect of increased attachment of EHEC on barrier function, monolayers were infected with various MOIs of EHEC (results of two are shown) and TER was followed. The TER of control uninfected (control) and EPEC-infected monolayers was also assessed for comparison. The number of adherent bacteria (± standard error) at 6 h postinfection is indicated in parenthesis.
The more modest effect of EHEC on intestinal epithelial barrier function could reflect a difference in the attachment rates of EHEC and EPEC. Bundle forming pili (BFP) are responsible for promoting the initial attachment of EPEC to epithelial cells (35). EHEC does not express BFP proteins. Therefore, the attachment of EPEC and EHEC to polarized intestinal epithelial cells grown on plates and permeable filters was determined. Epithelial cells grown on plates or Transwell filters were infected with EPEC or EHEC at an equivalent MOI, and the number of attached bacteria was determined as described in Materials and Methods. While 8 to 12% of the original EPEC inoculum attached to host cells at 1 h, less than 5% of EHEC attached at this time (Fig. 1B). To determine if a difference in attachment rates was responsible for the more modest effect of EHEC on TER, the number of attached EHEC cells was equalized to that of EPEC by increasing the MOI of EHEC. A 10-fold increase in the MOI of EHEC yielded comparable numbers of attached EPEC and EHEC cells at 6 h postinfection (Fig. 1C). Despite equivalent numbers of attached bacteria, the disruption of barrier function by EPEC remained greater than that induced by EHEC (Fig. 1C).
EPEC and EHEC similarly disrupt occludin from TJ.
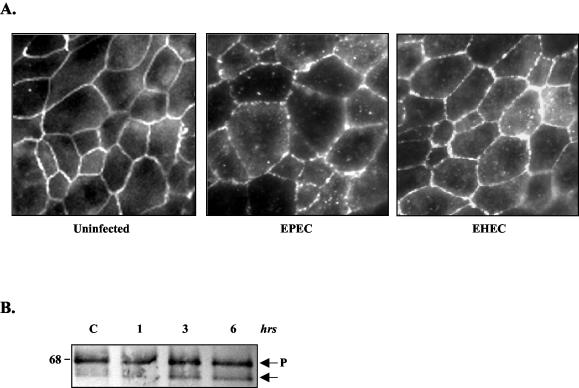
EPEC has been shown to redistribute the transmembrane TJ protein occludin, an event that correlates with both occludin dephosphorylation and a decrease in TER (34). Occludin contributes to formation of the TJ barrier, consistent with its uniform and peripheral distribution in uninfected cells, as seen in Fig. 2A. In view of the significant disparity in the ability of EHEC to reduce TER, we examined the effect of EHEC on occludin. Like EPEC, EHEC also redistributed occludin, as shown by a loss of protein from the membrane and increased intracellular localization (Fig. 2A) (34). When analyzed by Western blotting, occludin resolves according to its phosphorylation status, with phosphorylated forms migrating as a higher-molecular-weight band than the lesser or nonphosphorylated forms. As was previously reported with EPEC, EHEC caused the progressive dephosphorylation of occludin (Fig. 2B).
FIG. 2.
EHEC redistributes occludin in T84 cells. (A) Occludin immunofluorescence studies were performed on uninfected control monolayers and EPEC- or EHEC-infected monolayers for 6 h. Both pathogens induced a punctuate pattern of occludin distribution in the membrane, as well as enhanced staining in the cytoplasm. (B) Protein extracts from control (C) and EHEC-infected monolayers (1, 3, and 6 h) were separated on a denaturing polyacrylamide gel, transferred to nitrocellulose membranes, and immunoblotted with antioccludin antibody. Occludin migrates as two or more bands, based on its phosphorylation status; P refers to phosphorylated occludin (33). The amount of lesser-phosphorylated occludin increases as EHEC infection progresses, as previously reported for EPEC (33, 34).
Deletion of espF or its chaperone cesF has no significant impact on the decrease in TER by EHEC.
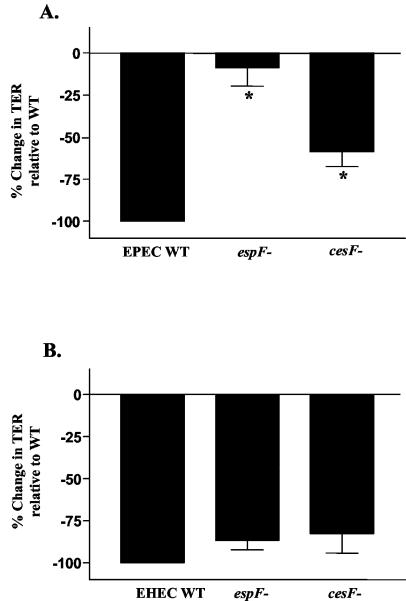
Some of the differences in the mechanisms of EPEC and EHEC pathogenesis have been attributed to variation in the sequences of type III secreted proteins (1). Previous studies have shown that the EPEC LEE-encoded protein EspF is largely responsible for the effects on the TJ barrier (24). EHEC also encodes a homolog of EspF within its LEE that displays over 75% identity to EPEC EspF. We therefore investigated the role of the EHEC EspF in barrier function alteration by creating an EHEC espF deletion strain, GH145, via allelic recombination. GH145, UMD874 (the EPEC espF deletion mutant), and the respective isogenic parent strains were assessed for their effects on TER. As in previous studies, the EPEC ΔespF strain was markedly impaired for altering host epithelial barrier function (Fig. 3A) (24). In sharp contrast, mutation of espF in EHEC had no significant impact on the drop in TER (Fig. 3B).
FIG. 3.
EHEC ΔespF and ΔcesF strains are not impaired for barrier function alterations. T84 monolayers were treated with DMEM or infected with EPEC (A) or EHEC (B) and their corresponding ΔespF and ΔcesF derivatives. The percent change in TER at 6 h postinfection relative to the wild-type (WT) baseline is indicated (n = 15; asterisks indicate P < 0.003 for EPEC ΔespF and ΔcesF strains relative to wild-type EPEC; P > 0.5 for EHEC ΔespF and ΔcesF strains relative to wild-type EHEC).
The EPEC EspF chaperone CesF has been shown to be important for the efficient folding and secretion of EPEC EspF (9). Strains in which cesF has been deleted express and secrete diminished quantities of EspF and display a marked decrease in their ability to reduce TER (9). The corresponding EHEC homolog of this protein is 78% identical and 84% similar to EPEC CesF. Database searches revealed the presence of a single homolog for CesF in EHEC. To evaluate the impact of CesF on EHEC-induced barrier function alterations, an EHEC cesF mutant strain was created as described in Materials and Methods. The EPEC ΔcesF strain was impaired for reducing TER, compared to wild-type EPEC, which is consistent with its reduced ability to secrete EspF (Fig. 3A) (9). In contrast, infection with the EHEC ΔcesF strain decreased TER to levels that were not significantly different from wild-type EHEC (Fig. 3B).
EHEC EspF expression is upregulated in DMEM and is CesF dependent.
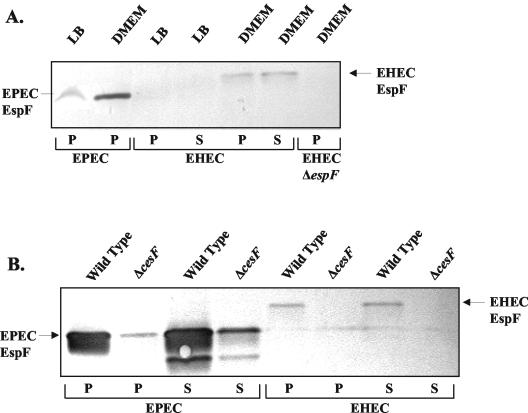
Having found that EHEC EspF and CesF play only modest roles in barrier function disruption compared to EPEC EspF and CesF, we considered whether this may be a function of differential regulation or secretion of these proteins by EPEC and EHEC. EPEC EspF has previously been shown to be expressed at low levels in LB and highly upregulated in DMEM in a CesF-dependent manner (9). To compare EspF expression by EPEC and EHEC, whole bacterial and secreted proteins of bacteria grown in LB and DMEM were resuspended in loading buffer and assessed by Western blot analysis. Antibody raised against EPEC EspF also detected EHEC EspF (Fig. 4A). As with EPEC EspF, EHEC EspF is up-regulated in bacteria grown in DMEM and is secreted into the medium (Fig. 4A). The lower intensity of the bands for EHEC compared to EPEC may reflect differences in expression or in the affinity of the antibody. The protein expression pattern correlates with RT-PCR analyses (Fig. 5), suggesting that the regulation is primarily at the transcriptional level. As expected, EHEC EspF was absent in extracts of the ΔespF strain (Fig. 4A).
FIG. 4.
EHEC EspF is expressed and secreted in a CesF-dependent process. (A) Proteins from whole bacterial pellets (P) or precipitated supernatants (S) from various strains were separated by polyacrylamide gel electrophoresis and immunoblotted by using antibody raised against EPEC EspF. The lanes are as follows: whole bacterial proteins from EPEC grown in DMEM (lane 1); EHEC whole bacterial proteins (lane 2) and secreted proteins (lane 3) grown in LB; and EHEC whole bacterial protein (lane 4), EHEC secreted proteins (lane 5), and EHEC ΔEspF whole bacterial proteins (lane 6) grown in DMEM. (B) The role of CesF in the expression and secretion of EspF was determined by immunoblotting proteins from bacterial pellets (P) and secreted (S) proteins from wild-type (lanes 1, 3, 5, and 7) and ΔcesF derivatives (lanes 2, 4, 6, and 8) of both EPEC and EHEC. Deletion of cesF decreased the amount of EspF expressed and secreted in both EPEC and EHEC.
FIG. 5.
EHEC espF and U-espF are coordinately regulated. Equivalent amounts of (DNase-treated) RNA from EHEC strain 85-170 grown to mid-logarithmic phase in LB or DMEM were subjected to RT-PCR analysis by using gene-specific primers to espF and U-espF. Lanes 3 and 4 represent controls in which the reverse transcriptase was omitted. Lanes 5, 6, and 7 correspond to molecular weight markers, positive (EHEC chromosomal DNA template), and negative (no template) controls, respectively.
The expression and secretion of EspF in ΔcesF strains of EPEC and EHEC were also examined. EPEC ΔcesF was impaired for EspF expression and secretion as previously reported (Fig. 4B) (9). The EHEC ΔcesF strain was similarly impaired for expression and secretion of EspF (Fig. 4B). The role of CesF for stabilization and/or secretion of EspF-like sequences (described below) could not be evaluated due to the lack of antibodies.
EHEC EspF and U-EspF are coordinately regulated.
We were intrigued to find that while EPEC EspF is indispensable for disrupting the barrier function of the intestinal epithelial monolayer, EHEC strains lacking EspF were not significantly impaired for inducing barrier function alterations. The finding that mutation of neither espF nor cesF altered the TER phenotype of EHEC led us to examine the potential roles of the other EspF-like protein encoded by EHEC in more detail. In addition to the LEE-encoded EspF homolog, EHEC encodes two additional EspF-like sequences elsewhere on the chromosome (16, 28). We have designated these genes U-espF and M-espF (the letter designations refer to the specific cryptic prophage sequences on the EHEC chromosome closest to the respective genes) (28). The sequence relationship between EspF and the EspF-like open reading frames is presented in Table 2. The predicted sequence of M-EspF corresponds to the C-terminal region of U-EspF, presumably lacking the N-terminal sequence required for translocation through the TTSS. Therefore, M-espF was not explored further. The protein encoded by U-espF could not be detected by the antibody raised against EPEC EspF. Therefore, the expression and secretion of these proteins could not be assessed directly. As a preliminary step in assessing the role of U-espF in EHEC pathogenesis, the expression of this gene was monitored by RT-PCR. These experiments revealed that both espF and U-espF are expressed poorly, if at all, in LB-grown cultures, while growth in DMEM activates the transcription of both these genes, a feature shared by many type III effectors (Fig. 5) (4). The expression and regulation of U-espF is thus highly suggestive of a role for this gene in pathogenesis.
TABLE 2.
Sequence relationships between EspF and the EspF-like open reading framesa
| EspF homolog | Sequence length (no. of amino acids) | No. of proline-rich repeats | % Identify between sequencesb
|
|||
|---|---|---|---|---|---|---|
| EPEC EspF | EHEC EspF | EHEC U-EspF | EHEC M-EspF | |||
| EPEC EspF | 206 | 3 | 100 | 87 | X | X |
| EHEC EspF | 248 | 4 | 75 | 100 | X | X |
| EHEC U-EspF | 337 | 7 | 24 | 28 | 100 | 97 |
| EHEC M-EspF | 189 | 5 | 28 | 27 | 55 | 100 |
Pairwise global alignment of the predicted amino acid sequences with the Blosum 62 scoring matrix is shown in normal type. Sequence similarity between EspF and U- and M-EspF occurs primarily in the proline-rich sequences. Pairwise local alignment (boldface type) without gaps reveals the closest sequence relationship between the LEE-encoded EspF of EPEC and EHEC and between the U- and M-EspF sequences. Alignments were performed by using the Align program (Scientific and Educational Software, Durham, N.C.)
X, absence of extended similarity between the respective sequences.
EHEC EspF and U-EspF are functionally equivalent to EPEC EspF.
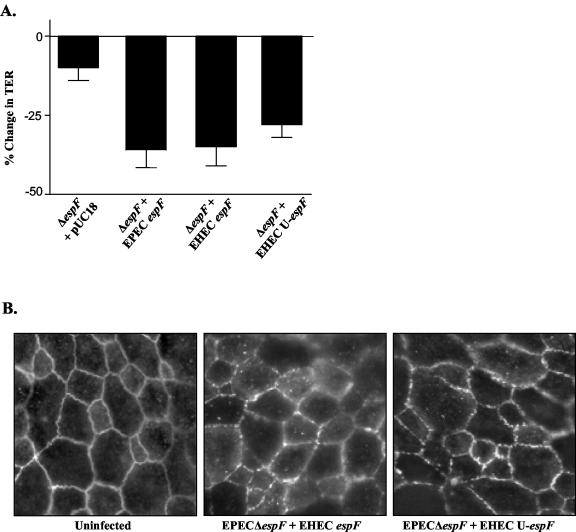
Having found that EspF and U-EspF of EHEC are similarly regulated by DMEM, we next compared the functional capacities of EHEC EspF, U-EspF, and EPEC EspF. If U-EspF could also influence epithelial TJ alterations, this functional redundancy would explain the lack of a phenotype for the EHEC espF-deletion strain. To determine if EHEC EspF and U-EspF are functionally equivalent to EPEC EspF, the ability of EHEC espF or U-espF to complement an EPEC ΔespF strain, UMD874, was examined. Unlike the EHEC ΔespF strain, the EPEC ΔespF strain displays an unambiguous phenotype with reference to TJ structure and barrier function alterations (Fig. 3A) (24). The effects of complementation would therefore be clearly evident in this strain. As shown in Fig. 6A, complementation of UMD874 with plasmids encoding either EHEC espF or U-espF restored the TER phenotype. Despite variability in the primary sequences of EPEC espF, EHEC espF, and EHEC U-espF, there was no significant difference in the time course or degree of barrier function alteration induced by these clones. In addition, immunofluorescence studies revealed that complementation of UMD874 with EHEC espF or U-espF similarly restored the ability to redistribute occludin (Fig. 6B). These data confirm the functional equivalence of EHEC EspF and U-EspF to EPEC EspF.
FIG. 6.
EHEC EspF and U-EspF can functionally substitute for EPEC EspF. The EPEC ΔespF strain UMD874 was complemented with either the pUC18 vector alone or plasmids encoding EPEC EspF, EHEC EspF, or EHEC U-EspF. (A) These strains were used for infection of T84 monolayers on permeable supports, and TER measurements were recorded at 6 h postinfection (n ≥ 5, P > 0.1 for EPEC espF versus EHEC espF or U-espF). (B) Additionally, immunofluorescence studies were performed to assess the impact of these strains on occludin distribution.
U-EspF contributes to the disruption of the TJ barrier by EHEC.
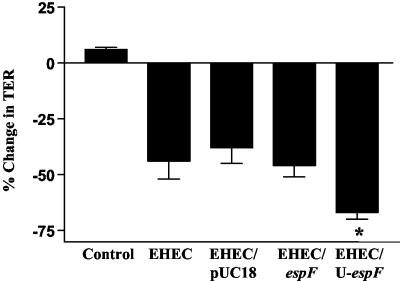
The ability of U-espF to complement the diminished phenotype of UMD874 is suggestive of a potential role for this protein in EHEC pathogenesis. To evaluate the potential relevance of EspF and U-EspF in EHEC pathogenesis, wild-type EHEC was transformed with multicopy plasmids encoding espF or U-espF. These strains were evaluated for their ability to enhance the barrier disruption mediated by EHEC. The presence of U-espF, but not espF, on a multicopy plasmid caused a significantly greater reduction in TER compared to EHEC transformed with pUC18 alone (Fig. 7). This finding suggests that U-EspF likely contributes to the barrier function alterations mediated by EHEC.
FIG. 7.
U-EspF can enhance EHEC-induced alteration of epithelial barrier function. The EHEC strain 85-170 was transformed with the pUC18 empty vector or vectors encoding full-length EHEC espf or U-espF. T84 cells were infected with these strains or treated with media alone (control), and TER measurements were recorded at 6 h postinfection (n > 10; asterisk denotes significant difference for U-espF compared to pUC18, where P is <0.0005; P > 0.1 for espF compared to pUC18).
DISCUSSION
The comparison of EHEC- and EPEC-derived strains in this study highlights a number of differences between these two pathogens in their ability to disrupt the intestinal epithelial barrier. While the two pathogens displayed distinct kinetics for the alteration of TER, a measure of epithelial barrier function, both appeared to mediate these effects through the redistribution of TJ proteins such as occludin. Although the regulation of EspF expression for EPEC and EHEC was similar in responsiveness to DMEM and dependence on CesF, the role of EspF in decreasing TER was distinctly different. EPEC ΔespF and ΔcesF strains are markedly impaired in inducing barrier function alterations, while a mutation of espF or cesF in EHEC had little to no impact.
One possible reason for the lack of a distinct phenotype for the EHEC ΔespF strain could be the presence of redundant EspF-like proteins in EHEC (M-EspF and U-EspF). M-EspF lacks the N-terminal signal sequence for secretion and therefore is likely not injected into host cells. However, a role for U-espF in EHEC pathogenesis is implicated both by its expression and coordinate regulation with the LEE encoded espF. Like many type III effector genes including EPEC and EHEC espF, U-espF transcription was activated upon growth in DMEM (10).
Wild-type EHEC containing plasmids encoding EHEC U-espF but not espF caused a greater drop in TER than did the parent strain, suggesting a dose-dependent relationship between U-EspF and EHEC-induced barrier function alterations. The inability of additional espF copies to induce a similar effect may suggest that this protein is produced at saturating levels in the parent strains. Alternatively, this inability may reflect variability in the expression of this protein from the transformed plasmid. Clones containing either EHEC espF or U-espF did, however, efficiently complement an EPEC ΔespF strain. The absence of a phenotype for the EHEC ΔespF strain could reflect the redundancy of U-EspF in this function. It is not clear if U-EspF requires CesF or is folded and secreted by an unrelated chaperone. Implicit in our EPEC complementation experiments is that EHEC EspF and U-EspF are efficiently folded in EPEC and, further, that these proteins are translocated into host cells via the EPEC type III secretion pore (9). This idea is particularly intriguing, given the differences between EPEC EspF and the EHEC EspF and EspF-like sequences and the inability of anti-EPEC EspF antibodies to detect U-EspF(s). It is possible, however, that U-EspF may be folded and secreted in a chaperone-independent manner.
The most dramatic effects of EHEC infection, bloody diarrhea and hemolytic uremic syndrome, are primarily due to the expression of the Shiga-like toxin. Strains lacking the Shiga-like toxins, however, cause a watery diarrhea akin to that caused by EPEC. Among other differences between the two pathogens is the presence of the BFP attachment factor present in EPEC but not in EHEC (8, 14). While both pathogens have homologous LEE regions, the regulation of the genes in the pathogenicity island may differ. The transcriptional regulator Per, encoded on the EPEC adherence factor plasmid but absent in EHEC, is a global regulator of transcription that controls the expression of BFP proteins and genes in the LEE (25). The consequent differences in attachment and virulence gene expression may explain the more modest barrier function alterations induced by EHEC compared to EPEC (Fig. 1).
It is increasingly evident that even the LEE homologs do not function in an identical fashion in the two pathogens (1, 3). For instance, while tyrosine phosphorylation of Tir (Y474) in the epithelial cell is critical for EPEC-induced changes, EHEC Tir mediates host effects in the absence of tyrosine phosphorylation (5). Furthermore, these two molecules are not functionally interchangeable (19). The consequence of this difference is that EPEC Tir recruits the host protein Nck to the pedestal, while EHEC Tir does not. The specificity of this interaction was recently narrowed to a 12-amino-acid region including Y474 (1, 15). Two other signaling molecules, Grb2 and CrkII, are recruited only to EPEC pedestals but not to EHEC pedestals (reviewed in reference 2). Differences in another LEE-encoded protein, intimin, in these two pathogens correlates with their distinct colonization niches in the intestine (13, 36). The present study identifies one more LEE-encoded locus, espF, that seems to be different in its relevance to the respective pathogens, despite the homology and apparent similarity in function of the EspF proteins.
These studies suggest that other determinants on the EHEC chromosome may be required to enable the LEE-encoded proteins to function. It was recently suggested that the EHEC genome encodes a second TTSS that could mediate the secretion of the LEE-encoded effector protein EspB in an E. coli K-12 background (16). This would imply that the inability of EHEC LEE to confer pathogenic properties to E. coli K-12 might be due to the inefficiency of the LEE-encoded secretion apparatus. This would be surprising, given the high degree of similarity between the two pathogens in the genes encoding the secretion apparatus (27). Given the abundance of EspF-like sequences and the critical role of EPEC EspF in disruption of the TJ barrier, it is curious that EHEC typically engenders only a modest alteration in barrier function compared to EPEC. The inefficient secretion or expression of effector molecules by EHEC could be one explanation for this difference. A detailed comparison of the expression and secretion of known EPEC and EHEC effector proteins should clarify this issue.
Acknowledgments
We gratefully acknowledge James Kaper for providing us with strain 85-170 and Michael Donnenberg for strain UMD874.
These studies were supported by NIH KO-1 award DK63030 to V.K.V., grants DK50694 and DK58964 from the NIH, and the VA Merit Review and VA Research Enhancement Awards Program of the Department of Veterans Affairs to G.H.
Editor: J. T. Barbieri
REFERENCES
- 1.Campellone, K. G., A. Giese, D. J. Tipper, J. M. Leong. 2002. A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol. Microbiol. 5:1227-1241. [DOI] [PubMed] [Google Scholar]
- 2.Campellone, K. G., and J. M. Leong. 2003. Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 6:82-90. [DOI] [PubMed] [Google Scholar]
- 3.Ceponis, P. J., D. M. McKay, J. C. Ching, P. Pereira, and P. M. Sherman. 2003. Enterohemorrhagic Escherichia coli O157:H7 disrupts Stat1-mediated gamma interferon signal transduction in epithelial cells. Infect. Immun. 71:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 5.DeVinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowski, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dharmsathaphorn, K., J. A. McRoberts, K. G. Mandel, L. D. Tisdale, and H. Masui. 1984. A human colonic tumor cell line that maintains vectorial electrolyte transport. Amer. J. Physiol. 246:G204-G208. [DOI] [PubMed] [Google Scholar]
- 7.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Invest. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S., H. Z. Zhang, and K. D. Stone. 1997. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability. Gene 192:33-38. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, S. J., C. B. O'Connell, A. Koutsouris, C. Brinkley, M. S. Donnenberg, G. Hecht, and J. B. Kaper. 2002. A gene from the locus of enterocyte effacement that is required for enteropathogenic Escherichia coli to increase tight-junction permeability encodes a chaperone for EspF. Infect. Immun. 70:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, S. J., J. Yu, and J. B. Kaper. 1999. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67:4260-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzhenry, R. J., D. J. Pickard, E. L. Hartland, S. Reece, G. Dougan, A. D. Phillips, and G. Frankel. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giron, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 15.Gruenheid, S. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856-859. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 17.Hecht, G. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. VII. Enteropathogenic Escherichia coli: physiological alterations from an extracellular position. Amer. J. Physiol. Gastrointest. Liver Physiol. 281:G1-G7. [DOI] [PubMed] [Google Scholar]
- 18.Kaper, J. B. 1998. Enterohemorrhagic Escherichia coli. Curr. Opin. Microbiol. 1:103-108. [DOI] [PubMed] [Google Scholar]
- 19.Kenny, B. 2001. The enterohaemorrhagic Escherichia coli (serotype O157:H7) Tir molecule is not functionally interchangeable for its enteropathogenic E. coli (serotype O127:H6) homologue. Cell. Microbiol. 3:499-510. [DOI] [PubMed] [Google Scholar]
- 20.Li, Z., E. Elliott, J. Payne, J. Isaacs, P. Gunning, and V. E. O'Loughlin. 1999. Shiga toxin-producing Escherichia coli can impair T84 cell structure and function without inducing attaching/effacing lesions. Infect. Immun. 67:5938-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 23.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 24.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 26.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 29.Philpott, D. J., C. A. Ackerley, A. J. Kiliaan, M. A. Karmali, M. H. Perdue, and P. M. Sherman. 1997. Translocation of verotoxin-1 across T84 monolayers: mechanism of bacterial toxin penetration of epithelium. Amer. J. Physiol. 273:G1349-G1358. [DOI] [PubMed] [Google Scholar]
- 30.Philpott, D. J., D. M. McKay, W. Mak, M. H. Perdue, and P. M. Sherman. 1998. Signal transduction pathways involved in enterohemorrhagic Escherichia coli-induced alterations in T84 epithelial permeability. Infect. Immun. 66:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philpott, D. J., D. M. McKay, P. M. Sherman, and M. H. Perdue. 1996. Infection of T84 cells with enteropathogenic Escherichia coli alters barrier and transport functions. Amer. J. Physiol. 270:G634-G645. [DOI] [PubMed] [Google Scholar]
- 32.Roe, A., and D. Gally. 2000. Enteropathogenic and enterohaemorrhagic Escherichia coli and diarrhoea. Curr. Opin. Infect. Dis. 13:511-517. [DOI] [PubMed] [Google Scholar]
- 33.Sakakibara, A., M. Furuse, M. Saitou, Y. Ando-Akatsuka, and S. Tsukita. 1997. Possible involvement of phosphorylation of occludin in tight junction formation. J. Cell Biol. 137:1393-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonovic, I., J. Rosenberg, A. Koutsouris, and G. Hecht. 2000. Enteropathogenic Escherichia coli dephosphorylates and dissociates occludin from intestinal epithelial tight junctions. Cell. Microbiol. 2:305-315. [DOI] [PubMed] [Google Scholar]
- 35.Tobe, T., and C. Sasakawa. 2001. Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell. Microbiol. 3:579-585. [DOI] [PubMed] [Google Scholar]
- 36.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzipori, S., H. Karch, K. I. Wachsmuth, R. M. Robins-Browne, A. D. O'Brien, H. Lior, M. L. Cohen, J. Smithers, and M. M. Levine. 1987. Role of a 60-megadalton plasmid and Shiga-like toxins in the pathogenesis of infection caused by enterohemorrhagic Escherichia coli O157:H7 in gnotobiotic piglets. Infect. Immun. 55:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viswanathan, V. K., P. H. Edelstein, C. D. Pope, and N. P. Cianciotto. 2000. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 68:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuhan, R., A. Koutsouris, S. D. Savkovic, and G. Hecht. 1997. Enteropathogenic Escherichia coli-induced myosin light chain phosphorylation alters intestinal epithelial permeability. Gastroenterol. 113:1873-1882. [DOI] [PubMed] [Google Scholar]