Abstract
Mexico harbors great cultural and ethnic diversity, yet fine-scale patterns of human genome-wide variation from this region remain largely uncharacterized. We studied genomic variation within Mexico from over 1,000 individuals representing 20 indigenous and 11 mestizo populations. We found striking genetic stratification among indigenous populations within Mexico at varying degrees of geographic isolation. Some groups were as differentiated as Europeans are from East Asians. Pre-Columbian genetic substructure is recapitulated in the indigenous ancestry of admixed mestizo individuals across the country. Furthermore, two independently phenotyped cohorts of Mexicans and Mexican Americans showed a significant association between sub-continental ancestry and lung function. Thus, accounting for fine-scale ancestry patterns is critical for medical and population genetic studies within Mexico, in Mexican-descent populations, and likely in many other populations worldwide.
Understanding patterns of human population structure, where regional surveys are key for delineating geographically restricted variation, is important for the design and interpretation of medical genetic studies. In particular, we expect rare genetic variants, including functionally relevant sites, to exhibit little sharing among diverged populations (1). Native Americans display the lowest genetic diversity of any continental group, but there is high divergence among subpopulations (2). As a result, present-day American indigenous populations (and individuals with indigenous ancestry) may harbor local private alleles rare or absent elsewhere, including functional and medically relevant variants (3, 4). Mexico serves as an important focal point for such analyses because it harbors one of the largest sources of pre-Columbian diversity and has a long history of complex civilizations with varying contributions to the present-day population.
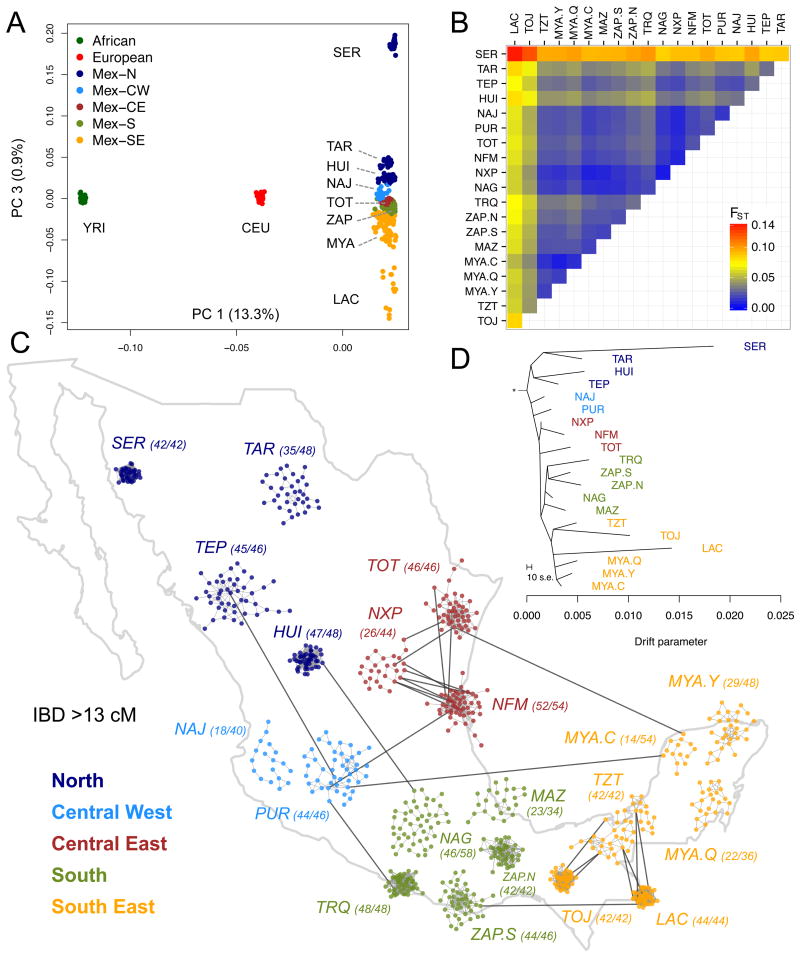
Previous estimates of Native Mexican genetic diversity examined single loci or were limited to a reduced number of populations or small sample sizes (5–8). We examined local patterns of variation from nearly one million genome-wide autosomal SNPs for 511 Native Mexican individuals from 20 indigenous groups, covering most geographic regions across Mexico (Table S1). Standard principal component analysis (PCA) summarizes the major axes of genetic variation in the dataset (see (9)). While PC1 and PC2 separate Africans and Europeans from Native Mexicans, PC3 differentiates indigenous populations within Mexico, following a clear northwest-southeast cline (Fig. 1A). A total of 0.89% of the variation is explained by PC3, nearly three times as much as the variation accounted for by the north-south axis of differentiation within Europe (0.30%, in (10)). The northernmost (Seri) and southernmost (Lacandon) populations define the extremes of the distribution, with very clear clustering of individuals by population, indicating high levels of divergence among groups (fig. S1). Seri and Lacandon show the highest level of population differentiation as measured with FST (0.136, Fig. 1B, Table S4), higher than the FST between Europeans and Chinese populations in HapMap3 (0.11) (11). Other populations within Mexico also show extreme FST values; for example, the Huichol and Tojolabal have a pairwise FST of 0.068, similar to that observed between the Gujarati Indians and the Chinese in HapMap3 (0.076).
Fig. 1.
Genetic differentiation of Native Mexican populations. (A) Principal component analysis of Native Mexicans with HapMap YRI and CEU samples. Population labels as in Table S1. (B) Pairwise FST values among Native Mexican populations ordered geographically (see also Table S4). (C) Relatedness graph of individuals sharing more than 13 cM of the genome as measured by the total of segments identical-by-descent (IBD). Each node represents a haploid genome and edges within clusters attract nodes proportionally to shared IBD. The spread of each cluster is thus indicative of the level of relatedness in each population as determined by a force-directed algorithm. Only the layout of nodes within each cluster is the result of the algorithm, as populations are localized to their approximate sampling locations to ease interpretation. Parentheses indicate the number of individuals represented out of the total sample size (2N). The full range of IBD thresholds are shown in fig. S8. (D) TreeMix graph representing population splitting patterns of the 20 Native Mexican groups studied. The length of the branch is proportional to the drift of each population. African, European, and Asian samples were used as outgroups to root the tree (fig. S9).
The high degree of differentiation between populations measured by FST argues that these populations have experienced high degrees of isolation. Indeed, when autozygosity using runs of homozygosity (ROH) are inferred, all populations on average have long homozygous tracts, with the Huichol, Lacandon, and Seri all having on average over 10% of the genome in ROH (figs. S2, S3; (9)). These populations are especially small, increasing the effects of genetic drift and driving some of the high FST values. In contrast, the Mayan and Nahuan populations have much smaller proportions of the genome in ROH, consistent with ROH levels found in Near Eastern populations in HGDP (12). These populations are the descendants of large Mesoamerican civilizations, and concordant with large historical populations, have relatively low proportions of ROH. The high degree of variance in ROH between populations is an additional indicator of substructure between populations and suggests a large variance in historical population sizes. Comparing the observed ROH patterns to those derived from coalescent simulations, we find that Native American groups within Mexico are characterized by small effective population sizes under a model with a strong bottleneck, in agreement with other studies of Native American populations (13). The degree of population size recovery to the current day is consistent with the degree of isolation of the extant populations, ranging from 1196 chromosomes (95% CI 317–1548) for the Seri in the Sonora desert, to 3669 (95% CI 2588–5522) for the Mayans from Quintana Roo (figs. S4–S6; (9)).
Isolation also correlates with the degree of relatedness within and between ethnic groups, ultimately shaping the pattern of genetic relationships among populations. We built a relatedness graph (Fig. 1C) of individuals sharing >13 cM of the genome identically-by-descent (IBD) (corresponding to 3rd/4th cousins or closer relatives). Almost all the connections are within- vs. among-population, consistent with the populations being discrete rather than exhibiting large-scale gene flow (see figs. S7, S8, (9)). As seen with the ROH calculations, the Mayan and Nahuan groups have fewer internal connections. The few between-population connections appear in populations close to each coastline, such as the connections between the Campeche Mayans and populations to the west along the Gulf of Mexico.
The long-tract ROH and IBD analyses are especially relevant to the recent history of isolation of Native American populations. To model the branching order and gene flow among the Native American populations, we ran TreeMix (14) to generate a probabilistic model of divergence and migration among the Native American populations (Fig. 1D). The inferred tree with no migration paths recapitulates the north/south and east/west gradients of differentiation from the PCA and IBD analyses, with populations with high ROH values also exhibiting longer tip branches. The primary branches divide populations by geography. All northern populations (dark blue) branch from the same initial split at the root. We also find two additional major clades: a grouping of populations from the southern states of Guerrero and Oaxaca (green labels) and a “Mayan clade” composed of Mayan-speaking populations from Chiapas and the Yucatan peninsula in the southeast (orange labels). Introducing migratory edges to the model connects the Maya in Yucatan to a branch leading to the Totonac, whose ancestors occupied the large pre-Columbian city of El Tajin in Veracruz (15). This result points to an Atlantic coastal corridor of gene flow between the Yucatan Peninsula and Central/Northern Mexico (fig. S9), consistent with our IBD analysis. Indeed, the only Mayan language outside the Mayan territory is spoken by the Huastec, nearby in northern Veracruz, supporting a shared history (16).
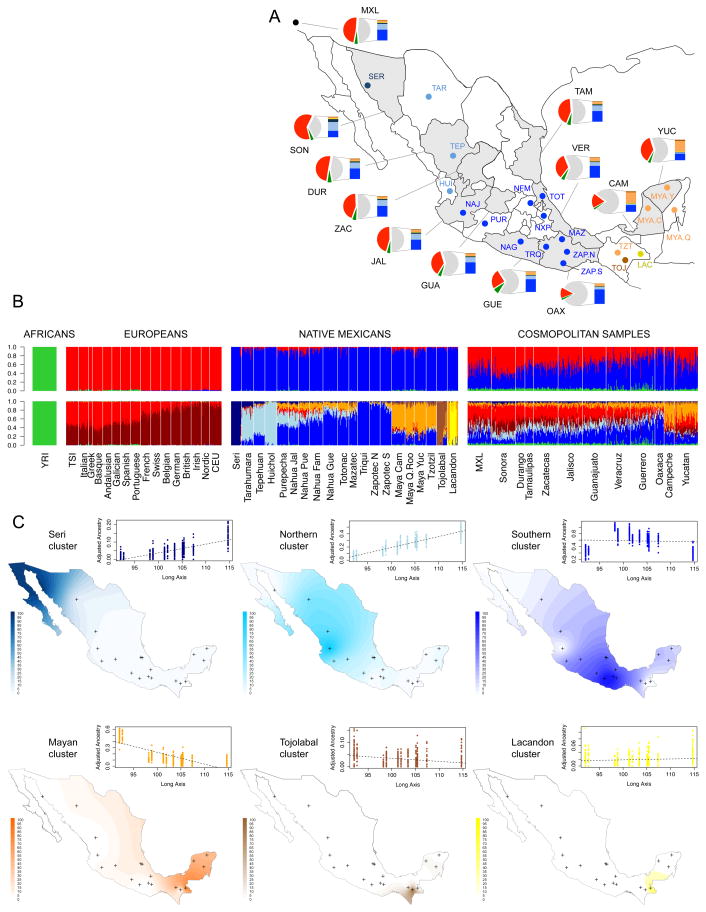
These signals remain today as a legacy of the pre-Columbian diversity of Mexican populations. Over the past 500 years, population dynamics have changed drastically. Today, the majority of Mexicans are admixed and can trace their ancestry back not only to indigenous groups but also to Europe and Africa. To investigate patterns of admixture, we combined data from continental source populations (including the 20 native Mexican groups, 16 European populations, and 50 West African Yorubas) with 500 admixed mestizo individuals from 10 Mexican states recruited by the National Institute of Genomic Medicine (INMEGEN) for this study, Mexicans from Guadalajara in the POPRES collection (17) and individuals of Mexican descent from Los Angeles in the HapMap Phase 3 project (Table S1). We ran the unsupervised mixture model algorithm ADMIXTURE (18) to estimate ancestry proportions for individuals in our combined dataset (Figs. 2, S10, Table S5). At the continental level of K = 3 ancestral clusters, we find that most individuals have a large amount of Native and European ancestry, with a small (typically <5%) amount of African ancestry. At the best-fit model for K = 9, the Native American cluster breaks down into six separate components (Fig. 2B). Three of these are mostly restricted to isolated populations (Seri: navy blue, Lacandon: yellow, and Tojolabal: brown). The other three show a wider but geographically well-defined distribution: a northern component (light blue) represented by Tarahumara, Tepehuano, and Huichol, gradually decreases southwards. Correspondingly, a southern component (blue), which includes Triqui, Zapotec, and Mazatec, gradually decreases northwards. In the Yucatan peninsula and the neighboring state of Chiapas, we found what we termed the “Mayan component” (orange in Fig. 2B, bottom panel), found primarily in Mayan-speaking groups. Interestingly, this Mayan component is also present at ~10–20% in central Mexican natives, consistent with the IBD and migration edges connecting the regions. This relationship between the Yucatan peninsula and central Mexico, seen in both recent shared IBD and genetic drift-based models of allele frequencies (TreeMix, ADMIXTURE), suggests that gene flow between the two regions has been ongoing for a long time. In contrast, Mayan admixture is not found at appreciable levels in highlanders of the southern state of Oaxaca (Triqui and Zapotec), where mountain ranges may have acted as geographic barriers to gene flow.
Fig. 2.
Mexican population structure. (A) Map of sampled populations (detailed in Table S1) and admixture average proportions (Table S5). Dots correspond to Native Mexican populations color-coded according to K=9 clusters identified in B (bottom), and shaded areas denote states in which cosmopolitan populations were sampled. Pie charts summarize per-state average proportions of cosmopolitan samples at K=3 (European in red, West African in green, and Native American in gray). Bars show the total Native American ancestry decomposed into average proportions of the native subcomponents identified at K=9. (B) Global ancestry proportions at K=3 (top) and K=9 (bottom) estimated with ADMIXTURE including African, European, Native Mexican, and cosmopolitan Mexican samples (Tables S1–S2). From left to right Mexican populations are displayed North-to-South. (C) Interpolation maps showing the spatial distribution of the six native components identified at K=9. Contour intensities are proportional to ADMIXTURE values observed in Native Mexican samples, with crosses indicating sampling locations. Scatter plots with linear fits show ADMIXTURE values observed in cosmopolitan samples versus a distance metric summarizing latitude and longitude (long axis) for the sampled states. From left to right: Yucatan, Campeche, Oaxaca, Veracruz, Guerrero, Tamaulipas, Guanajuato, Zacatecas, Jalisco, Durango, and Sonora. Values are adjusted relative to the total Native American ancestry of each individual (9).
Patterns of Native American population substructure are recapitulated in the genomes of Mexican mestizos from cosmopolitan populations throughout Mexico. Sonora and neighboring northern states show the highest average proportions of the northern native component (15%, light blue in Fig 2B, bottom), while only traces are detected in Oaxaca and the Yucatan peninsula. Conversely, the southern native component is the most prevalent across states, reaching maximum values in Oaxaca and decreasing northwards. Cosmopolitan samples from the Yucatan peninsula have Native American fractions of the genome dominated by the Mayan component, which diminishes in northward populations. Likewise, Mayan-related local components, Tojolabal and Lacandon, are detected above 1% exclusively among individuals from the states neighboring the Yucatan peninsula. In contrast, Mexican-Americans sampled in Los Angeles (MXL) do not show a homogeneous pattern, consistent with their diverse origins within Mexico. Overall, the continuous geographic distribution of each Native American component across Mexico (fig. S12) demonstrates a high correlation of individual admixture proportions with geography, even in individuals of mixed ancestry (Fig 2C, NW-SE axis F-test for all Native clusters, p <10−16).
To further test if ancestral population structure is recapitulated in the genomes of mestizos, we used an Ancestry Specific PCA (ASPCA) approach (see fig. S13, (9, 19)). We estimated local ancestry using PCAdmix (20) to identify segments of the genome belonging to Native American, European, or African ancestries. This analysis is possible with any component of ancestry; here, we focused on only the European and Native American components of ancestry given the low proportions of African ancestry overall. We would expect the history of Spanish occupation and colonization in Mexico to be reflected in the European segments of Mexican mestizos, as has been seen previously (21). ASPCA of the European haplotypes in present-day Mexicans confirms this, as individuals cluster tightly with present-day Iberians even with a dense set of European populations (17, 22) (fig. S14).
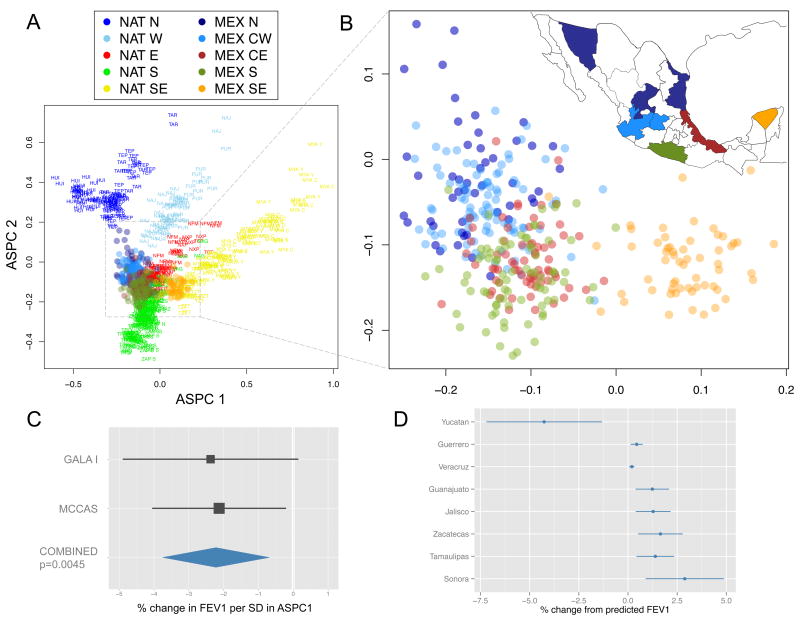
In contrast, given the complex demographic history of Native Americans, high isolation, and limited characterization of regional ancestry patterns (23, 24), it remains unknown if the correlation between genes and geography observed in Europe (10) can be similarly recapitulated within Mexico. We used ASPCA to uncover hidden population structure within Native American ancestry beyond that found solely in extant indigenous groups (Fig 3A). Consistent with the previous PCA analyses, we observe the most diverged indigenous populations defining the extremes of the top PCs due to high levels of genetic drift and isolation. However, including all the indigenous groups in the plot masks the signal contained in the indigenous segments of the Mexican mestizos. When plotting the ASPCA values for the admixed individuals only, we discover a strong correlation between Native American ancestry and geography within Mexico (Fig. 3B), with ASPC1 representing a west-to-east dimension and ASPC2 one from north to south. Both of these correlations are highly significant and linearly predictive of geographic location (Pearson’s r2 of 72% and 38% for ASPC1 and 2, respectively, both p-values < 10−5). The correlation is strong enough that the overall distribution of mestizo-derived indigenous haplotypes in ASPCA space resembles a geographic map of Mexico (Figs. 3B, S15). This finding suggests that the genetic composition of present-day Mexicans recapitulates ancient Native American substructure despite the potential homogenizing effect of post-colonial admixture. Fine-scale population structure going back centuries is not merely a property of isolated or rural indigenous communities. Cosmopolitan populations still reflect the underlying genetic ancestry of local native populations, arguing for a strong relationship between the indigenous and the Mexican mestizo population, albeit without the extreme drift exhibited in some current indigenous groups.
Fig. 3.
Sub-continental ancestry of admixed Mexican genomes and biomedical implications. (A) Ancestry-specific PCA (ASPCA) of Native American segments from Mexican cosmopolitan samples (colored circles) together with 20 indigenous Mexican populations (population labels). Samples with >10% of non-native admixture were excluded from the reference panel as well as population outliers such as Seri, Lacandon, and Tojolabal. (B) Zoomed detail of the distribution of the Native American fraction of cosmopolitan samples throughout Mexico. Native ancestral populations were used to define PCA space (prefixed by NAT) but removed from the background to highlight the sub-continental origin of admixed genomes (prefixed by MEX). Each circle represents the combined set of haplotypes called as Native American along the haploid genome of each sample with >25% of Native American ancestry. Inset map shows the geographic origin of cosmopolitan samples per state color-coded by region (9). (C) Coefficients and 95% confidence intervals for associations between ASPC1 and lung function (FEV1) from Mexican participants of the Genetics of Asthma in Latino Americans (GALA I) study, and the Mexico City Childhood Asthma Study (MCCAS), as well as both studies combined (Table S6, Fig. S17) (9). (D) Means and confidence intervals of predicted change in FEV1 by state extrapolated from the model in 3C.
Having found these hidden patterns of ancestry in the Native component of Mexican mestizos, we investigated whether this structure could have potential biomedical applications. Over the past decade, genetic ancestry has been associated with numerous clinical endpoints and disease risks in admixed populations, including neutrophil counts (25), creatinine levels (26), and breast cancer susceptibility (27). Similarly, ancestral background is especially important in pulmonary medicine, where different reference equations are used for different ethnicities defining normative predicted volumes and identifying thresholds for disease diagnosis in standard clinical practice (28). That is, depending on one’s ethnic background, the same value of Forced Expiratory Volume in 1 second (FEV1, a standard measure of lung function) could be either within the normal range or indicative of pulmonary disease. Previous work has shown that the proportion of African and European ancestry was associated with FEV1 in African Americans (29) and Mexicans (30), respectively, establishing the importance of genomic ancestry in lung function prediction equations.
To investigate possible associations between ancestral structure in Mexicans and FEV1, we applied our ASPCA approach to two studies measuring lung function in Mexican or Mexican-American children: the Mexico City Childhood Asthma Study (MCCAS) (31) and the Genetics of Asthma in Latinos Americans (GALA I) Study (32). Due to differences in protocols and genotyping platforms, we calculated ASPCA values for the two studies independently (fig. S17) using the same reference populations described above, then used fixed effects meta-analysis to combine the results (9).
First, in GALA I we looked for significant ancestry-specific differences between Mexico City and the San Francisco Bay Area, the two recruitment sites. ASPCA values were associated with recruitment location, with the Receiver-Operator Characteristic curve from the Native American ancestry dimensions resulting in an Area Under the Curve (AUC) of 80% (fig S17). After we adjusted for overall ancestry proportions (here both African and Native American), both ASPCs were significant in a logistic regression: ASPC1 OR per SD: 0.44 (95% CI 0.22–0.68), p=3.8×10−4, ASPC2 OR per SD: 1.68 (95% CI 1.03–2.76), p=0.039. The ASPCs defined similar east-west and north-south axes as in the previous analysis (fig. S17) and show that Mexican-Americans in the San Francisco Bay Area tend to have increased Native American ancestry from Northwest Mexico compared to individuals from Mexico City (joint logistic regression likelihood ratio test p=6.4×10−5).
We then used the ASPCA values for both studies to test for an association with FEV1 as transformed to percentile of predicted “normal” function via the standard set of reference equations (28) for individuals of Mexican descent. These equations use population-specific demographic characteristics to account for age, sex, and height in estimates of lung function. Adjusting for overall ancestry proportions in linear regressions, we observed a significant association between FEV1 and the East-West component (ASPC1) in both studies with a meta-analysis p-value of 0.0045 (2.2% decrease in FEV1 per 1 SD, 95% CI 0.69–3.74). The effect sizes were homogeneous (Fig. 3C, Table S6) despite differences in recruitment strategy, geography, and genotyping platform (9). In contrast, ASPC2 showed no association with FEV1. Remarkably, while FEV1 has been previously associated with overall ancestry in several populations, the effect seen here is not correlated with overall admixture proportions, as we adjusted for those in the regression model. The combined results here indicate that sub-continental ancestry as measured by ASPCA is important for characterizing clinical measurements.
To estimate how variation in genetic ancestry within Mexico may impact FEV1, we used the results from GALA I and MCCAS to predict trait values by state (Fig. 3D) for the INMEGEN mestizo samples. We found that difference in sub-continental Native American ancestry as measured by ASPC1 results in an expected 7.3% change in FEV1 moving from the state of Sonora in the west to the state of Yucatan in the east. These results suggest that fine-scale patterns of Native ancestry alone could have significant impacts on clinical measurements of lung function in admixed individuals within Mexico.
This finding indicates that diagnoses of diseases like asthma and chronic obstructive pulmonary disease (COPD) relying on specific lung function thresholds may benefit from taking finer-scale ancestry into consideration. These changes due to ancestry are comparable to other factors affecting lung function. Comparing the expected effect of ancestry across Mexico with the known effects of age in the standard Mexican-American reference equations (28), the inferred 7.3% change in FEV1 associated with sub-continental ancestry is similar to the decline in FEV1 that a 30 year old Mexican-American individual of average height would experience by aging 10.3 years if male and 11.8 years if female. Similarly, comparing our results from the Mexican data with the model incorporating ancestry in African Americans, a difference of 7.3% in FEV1 would correspond to a 33% difference in African ancestry (29). Importantly, the association between FEV1 and ASPC1 is not an indicator of impaired lung function on its own – rather, it contributes to the distribution of FEV1 values and would modify clinical thresholds.
An important implication of our work is that multi- and trans-ethnic mapping efforts will benefit from including individuals of Mexican ancestry since the Mexican population harbors rich amounts of genetic variation that may underlie important biomedical phenotypes. A key question in this regard is whether existing catalogs of human genome variation capture genetic variation present in the samples analyzed here. We performed targeted SNP tagging and genome-wide haplotype sharing analysis within 100-Kb sliding windows to assess the degree to which haplotype diversity in the Mexican mestizo samples could be captured by existing reference panels (see figs. S18–20, (9)). Although Mexican-American samples (MXL) were included in both the HapMap and 1000 Genomes catalogs, average haplotype sharing for the INMEGEN mestizo samples is limited to 81.2% and to 90.5% when combined with all continental HapMap populations. It is only after including the Native American samples genotyped here that nearly 100% of haplotypes are shared, maximizing the chances of capturing most of the variation in Mexico.
Much effort has been invested in detecting common genetic variants associated with complex disease and replicating associations across populations. However, functional and medically relevant variation may be rare or population-specific, requiring studies of diverse human populations to identify new risk factors (4). Without detailed knowledge of the geographic stratification of genetic variation, negative results and lack of replication are likely to dominate the outcome of genetic studies in uncharacterized populations. Here, we demonstrate a high degree of fine-scale genomic structure across Mexico, shaped by pre-Columbian population dynamics and impacting the present-day genomes of Mexican mestizos, which is of both anthropological and biomedical relevance. Studies such as this one are crucial for enabling precision medicine, providing novel data resources, empowering the next generation of genetic studies, and demonstrating the importance of understanding and measuring fine-scale population structure and its associations with biomedical traits.
Supplementary Material
Acknowledgments
We thank all volunteers for generously donating DNA samples and participating in the study. This project was possible with the joint support from multiple Institutions in Mexico and the United States. Stanford University supported CDB with funding from the Department of Genetics. The National Institute of Genomic Medicine (INMEGEN) received support from the Federal Government of Mexico, particularly the Ministry of Health, the Mexican Health Foundation (FUNSALUD) and the Gonzalo Río Arronte Foundation. State governments and universities of Durango, Campeche, Guanajuato, Guerrero, Oaxaca, Sonora, Tamaulipas, Veracruz, Yucatan, and Zacatecas contributed significantly to this work. This research was also supported by the George Rosenkranz Prize for Health Care Research in Developing Countries awarded to AM-E; UCSF Chancellor’s Research Fellowship, Dissertation Year Fellowship, and NIH Training Grant T32 GM007175 (to CRG); the RWJF Amos Medical Faculty Development Award; the Sandler Foundation; the American Asthma Foundation (to EGB); CONACYT Grant 129693 (to HR-V); BBSRC Grant BB/I021213/1 (to AR-L); and the National Institutes of Health (5R01GM090087, 2R01HG003229, ES015794, GM007546, GM061390, HL004464, HL078885, HL088133, RR000083, P60MD006902, ZIA ES49019). This work was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (to SJL). Some computations were performed using the UCSF Biostatistics High Performance Computing System. We also thank B. Henn, S. Gravel, and J. Byrnes for helpful discussions; C. Gunter and M. Carpenter for editing the manuscript; and M. Morales for informatics and programming support. CDB is on the advisory board of a project at 23andMe; and on the scientific advisory boards of Personalis, Inc.; InVitae; Etalon, Inc.; and Ancestry.com. The collections and methods for the Population Reference Sample (POPRES) are described by Nelson et al. (2008). The POPRES datasets used for the analyses described here were obtained from dbGaP through accession number phs000145.v1.p1. Access to the MCCAS dataset may be obtained under the terms of a data transfer agreement with NIEHS; the contact is SJL. Individual level genotypes for new data presented in this study are available, through a data access agreement to respect the privacy of the participants for transfer of genetic data, by contacting CDB, AM-E, and INMEGEN (http://www.inmegen.gob.mx/).
Footnotes
Materials and Methods
References and Notes
- 1.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proceedings of the National Academy of Sciences of the United States of America. 2011 Jul 19;108:11983. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007 Nov;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acuna-Alonzo V, et al. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in Native Americans. Hum Mol Genet. 2010 Jul 15;19:2877. doi: 10.1093/hmg/ddq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams AL, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2013 Dec 25; doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lisker R, Ramirez E, Babinsky V. Genetic structure of autochthonous populations of Meso-America: Mexico. Hum Biol. 1996 Jun;68:395. [PubMed] [Google Scholar]
- 6.Sandoval K, et al. Y-chromosome diversity in Native Mexicans reveals continental transition of genetic structure in the Americas. American journal of physical anthropology. 2012 Jul;148:395. doi: 10.1002/ajpa.22062. [DOI] [PubMed] [Google Scholar]
- 7.Gorostiza A, et al. Reconstructing the history of Mesoamerican populations through the study of the mitochondrial DNA control region. PLoS ONE. 2012;7:e44666. doi: 10.1371/journal.pone.0044666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reich D, et al. Reconstructing Native American population history. Nature. 2012 Jul 11;488:370. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See supplementary materials on Science Online.
- 10.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008 Nov 06;456:98. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010 Sep 2;467:52. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henn B, Hon L, Macpherson J, Eriksson N. Cryptic Distant Relatives Are Common in Both Isolated and Cosmopolitan Genetic Samples. PLoS ONE. 2012 doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hey J. On the number of New World founders: a population genetic portrait of the peopling of the Americas. PLoS Biology. 2005 Jul 01;3:e193. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pascual Soto A. El Tajín. En busca de los orígenes de una civilización. UNAM-INAH; Mexico: 2006. [Google Scholar]
- 16.Campbell L, Kaufman T. Mayan Linguistics: Where Are We Now? Annual Review of Anthropology. 1985;14:187. [Google Scholar]
- 17.Nelson MR, et al. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008 Sep 01;83:347. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009 Sep 01;19:1655. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno-Estrada A, et al. Reconstructing the population genetic history of the Caribbean. PLoS genetics. 2013 Nov;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brisbin A, et al. PCAdmix: Principal Components-Based Assignment of Ancestry Along Each Chromosome in Individuals with Admixed Ancestry from Two or More Populations. Hum Biol. 2012 Aug;84:343. doi: 10.3378/027.084.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson NA, et al. Ancestral components of admixed genomes in a mexican cohort. PLoS Genet. 2011 Dec;7:e1002410. doi: 10.1371/journal.pgen.1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botigue LR, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci U S A. 2013 Jul 16;110:11791. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, et al. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008 Apr 01;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva-Zolezzi I, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci USA. 2009;21:8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nalls MA, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008 Jan;82:81. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peralta CA, et al. The Association of African Ancestry and elevated creatinine in the Coronary Artery Risk Development in Young Adults (CARDIA) Study. Am J Nephrol. 2010;31:202. doi: 10.1159/000268955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fejerman L, et al. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Research. 2008 Dec 01;68:9723. doi: 10.1158/0008-5472.CAN-08-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. American Journal of Respiratory and Critical Care Medicine. 1999 Jan;159:179. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 29.Kumar R, et al. Genetic ancestry in lung-function predictions. New England Journal of Medicine. 2010 Jul 22;363:321. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salari K, et al. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005 Jul;29:76. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- 31.Hancock DB, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS genetics. 2009 Aug 01;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torgerson DG, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012 May 12; doi: 10.1016/j.jaci.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao X, et al. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007 Jun;80:1171. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug 01;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 36.Weir BS, Hill WG. Estimating F-statistics. Annual Review of Genetics. 2002;36:721. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 37.Wickham H. Use R. Springer; New York: 2009. ggplot2: Elegant Graphics for Data Analysis; p. VIII.p. 213. [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep 01;81:559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nalls MA, et al. Measures of autozygosity in decline: globalization, urbanization and its implications for medical genetics. PLoS genetics. 2009 Apr 01;5:e1000415. doi: 10.1371/journal.pgen.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jobin M, Mountain J. REJECTOR: Software for Population History Inference from Genetic Data via a Rejection Algorithm. Bioinformatics. 2008;24:2936–2937. doi: 10.1093/bioinformatics/btn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auton A, et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009 Mar 13;:1. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proceedings of the National Academy of Sciences. 2011 Apr 29;108:5154. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen GK, Marjoram P, Wall JD. Fast and flexible simulation of DNA sequence data. Genome research. 2009 Jan;19:136. doi: 10.1101/gr.083634.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. American journal of human genetics. 2007 Nov 01;81:1084. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning SR. Missing data imputation and haplotype phase inference for genome-wide association studies. Human Genetics. 2008 Dec 01;124:439. doi: 10.1007/s00439-008-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gusev A, et al. Whole population, genome-wide mapping of hidden relatedness. Genome research. 2009 Mar 01;19:318. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henn BM, et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012 Jan;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kidd JM, et al. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet. 2012 Oct 5;91:660. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raiko T, Ilin A, Karhunen J. Principal component analysis for large scale problems with lots of missing values. Machine Learning: ECML. 2007;2007:691. [Google Scholar]
- 50.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genetic Epidemiology. 2005 Jun;28:289. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 51.Torgerson DG, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nature Genetics. 2011 Sep;43:887. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burchard EG, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004 Feb 1;169:386. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 53.Galanter JM, et al. Cosmopolitan and ethnic-specific replication of genetic risk factors for asthma in 2 Latino populations. The Journal of allergy and clinical immunology. 2011 Jul;128:37. doi: 10.1016/j.jaci.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bigham A, et al. Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data. PLoS genetics. 2010 Sep 09;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, et al. Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. J Allergy Clin Immunol. 2010 Feb;125:321. doi: 10.1016/j.jaci.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008 Mar 22;319:1100. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 57.McVean G. A genealogical interpretation of principal components analysis. PLoS genetics. 2009 Oct;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aguirre Beltran G. The Slave Trade in Mexico. The Hispanic American Historical Review. 1944;24:412. [Google Scholar]
- 59.Lao O, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008 Aug 26;18:1241. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 60.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. American journal of human genetics. 2005 Apr 01;76:449. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American journal of human genetics. 2001 May 01;68:978. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Villarreal-Molina MT, et al. Association of the ATP-binding cassette transporter A1 R230C variant with early-onset type 2 diabetes in a Mexican population. Diabetes. 2008 Feb;57:509. doi: 10.2337/db07-0484. [DOI] [PubMed] [Google Scholar]
- 63.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics. 2008 Dec;40:1461. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21:263. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.