Abstract
The Rab11 GTPases and Rab11 family-interacting proteins (Rab11-FIPs) define integrated yet distinct compartments within the slow recycling pathway. The lipid content of these compartments is less well understood, although past studies have indicated phosphatidylserine (PS) is an integral component of recycling membranes. We sought to identify key differences in the presence of PS within Rab and Rab11-FIP containing membranes. We used live cell fluorescence microscopy and structured illumination microscopy to determine whether the previously published LactC2 probe for PS displays differential patterns of overlap with various Rab GTPases and Rab11-FIPs. Selective overlap was observed between the LactC2 probe and Rab GTPases when co-expressed in HeLa cells. Rab11-FIP1 proteins consistently overlapped with LactC2 along peripheral and pericentriolar compartments. The specificity of Rab11-FIP1 association with LactC2 was further confirmed by demonstrating that additional Rab11-FIPs (FIP2, FIP3, and FIP5) exhibited selective association with LactC2 containing compartments. Live cell dual expression studies of Rab11-FIPs with LactC2 indicated that PS is enriched along tubular compartments of the Rab11a-dependent recycling system. Additionally, we found that the removal of C2 domains from the Rab11-FIPs induced an accumulation of LactC2 probe in the pericentriolar region, suggesting that inhibition of trafficking through the recycling system can influence the distribution of PS within cells. Finally, we confirmed these findings using structured illumination microscopy suggesting that the overlapping fluorescent signals were on the same membranes. These results suggest distinct associations of Rab GTPases and Rab11-FIPs with PS-containing recycling system membrane domains.
Keywords: Rab11-FIP, Rab11a, Rab5, Rab7, Rab8a, live cell microscopy, phophatidylserine, structured illumination
Introduction
Cell membranes form a permeable barrier to extracellular elements as well as a dynamic network of internal compartments whose function is determined by the combination of proteins and lipids that associate with these organelles. In mammalian cells, endosomes are commonly characterized by the presence of one or more Rab GTPases that regulate the passage of membranes and proteins throughout the cell.1,2 Furthermore, visualization of cargo passage through endosomal compartments has added insight to the organization of these intracellular organelles.3,4 Recent efforts have focused on the characterization of the lipids within endosomes to contextualize the growing network of proteins that regulate intracellular traffic.5
Membrane trafficking involves transport of proteins and lipids between intracellular organelles and the cell surface. This process is regulated by small Rab GTPases that switch between GTP “active” and GDP “inactive” states.6 Rab11 proteins organize recycling of membranes and proteins to and from a pericentriolar, recycling compartment.7 Furthermore the presence of effector proteins known as the Rab11-family interacting proteins (Rab11-FIPs) coordinate transitions through the Rab11 containing compartments of the recycling pathway.8-12 Rab11-FIP1B, Rab11-FIP1C, Rab11-FIP2, and Rab11-FIP5 all contain N-terminal C2-domains.13 The C2 domain was originally characterized as one of the four conserved regions of Protein Kinase C (PKC), which was responsible for PKC-lipid interactions.14-16 Previous work has demonstrated an affinity of Rab11-FIP C2 domains for phospholipids, most notably phosphatidic acid, PI(3,4,5)P3, and phosphatidylserine (PS).17 Additionally, induction of phosphatidic acid production leads to translocation of FIP1C, FIP2, and FIP5 to the periphery of A431 cells,17 and diacylgylcerol kinase activity is required for translocation of FIP1C to pseudopods of A2780 cells.18 All these data point to an inherent role for phospholipid regulation of Rab11 and Rab11-FIP-dependent membrane recycling.
Phospholipids make up approximately 70% of the lipid composition within cells with the majority being phosphatidylcholine and phosphatidylethanolamine whereas phosphatidylserine (PS) is as much as 10% of the lipid composition within cells.19,20 Commonly known to serve as a negatively charged interface with which positively charged amino acids interact, PS is found widely throughout cells in a variety of compartments and organelles.21 In recent years, multiple groups have developed fluorescently labeled phospholipid binding domains as markers for specific compartments and lipid domains.22 Of particular interest is a (PS) binding probe made from the C2 domain of lactadherin (LactC2) that serves as a marker for discrete intracellular compartments.23 Previous studies have shown that the distribution of this probe overlaps with transferrin receptor, as well as a group of Rab small GTPases including Rab7, Rab23, and Rab35.23
Since Rab11-FIP proteins can potentially associate with not only phosphatidic acid, but also, phosphatidylserine, we have utilized LactC2 to examine the distribution of PS with Rab11 and Rab11-FIP containing compartments. Overall, we have found that Rabs and Rab11-FIPs exhibit differential patterns of overlap with the expressed LactC2 as an indicator of PS distribution. The inhibition of trafficking through the Rab11-containing recycling system with expression of carboxyl terminal regions of Rab11-FIP1C or Rab11-FIP2 or the tail of Myosin Vb caused accumulation of the LactC2 probe. However, the presence of a C2 domain in Rab11-FIPs did not necessarily correlate with overlap between Rab11-FIPs and LactC2. The data suggest that domains within the recycling system defined by Rab11-FIPs show distinct patterns for phosphatidylserine distribution.
Results
Rab GTPases overlap with Lactadherin C2 (LactC2) domain in live HeLa cells
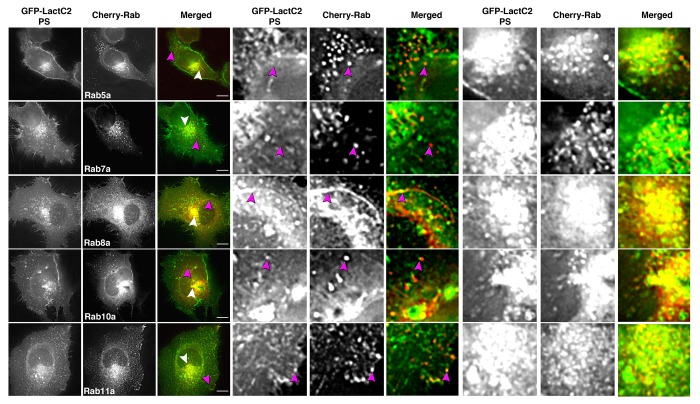
A combination of studies have suggested specificity in the phospholipid content of various endosomal compartments and other intracellular organelles.24 To elucidate differences in phosphatidylserine (PS) distribution within Rab compartments, we used live cell fluorescence microscopy to visualize the overlap between the PS probe, EGFP-Lactadherin C2 domain (EGFP-LactC2) and mCherry-chimeras of five Rab GTPases associated with endosomal compartments (Fig. 1). We found distinct separation between Rab5a and LactC2 in HeLa cells, especially in the periphery of cells. Of note we observed overlap between LactC2 and Rab7a in the pericentriolar region, as previously reported.23 However we also observed distinct separation of these two proteins in peripheral regions of the cell, which may be a result of cell-specific differences in lipid composition of endosomal compartments. Overlap was more readily visible between LactC2 and Rab8a or Rab10a in both the pericentriolar recycling compartment and vesicles in the periphery of cells. Rab11a also displayed consistent overlap with LactC2, supporting previous results that LactC2 is an efficient marker of transferrin-containing compartments particularly in the pericentriolar recycling endosome.23 We observed coordinated movement of compartments labeled for Rab11a and LactC2 over time and evidence of Rab11a localization to distal tips of PS-containing tubes (Vid. SV1). The data demonstrate differential labeling of various Rab-containing endosomal compartments by LactC2 and suggest that PS is enriched in the Rab11a-containing recycling system.
Figure 1. EGFP-LactC2 (PS) and mCherry-Rab GTPases in live HeLa cells show distinct patterns of overlap. HeLa cells expressing EGFP-LactC2 and mCherry-Rab proteins were imaged with live cell deconvolution microscopy for at least 1 min every 2 s. Rab5a and Rab7a showed limited overlap with LactC2 in the periphery of cells while Rab8a and Rab11a display consistent overlap with mCherry-LactC2 throughout the cell. Rab10 overlap with LactC2 was present but not consistent over multiple experiments. Data represent at least 3 independent experiments. Bars, 10 μm
Rab11-FIP1 proteins consistently overlap with LactC2-labeled PS in live HeLa cells
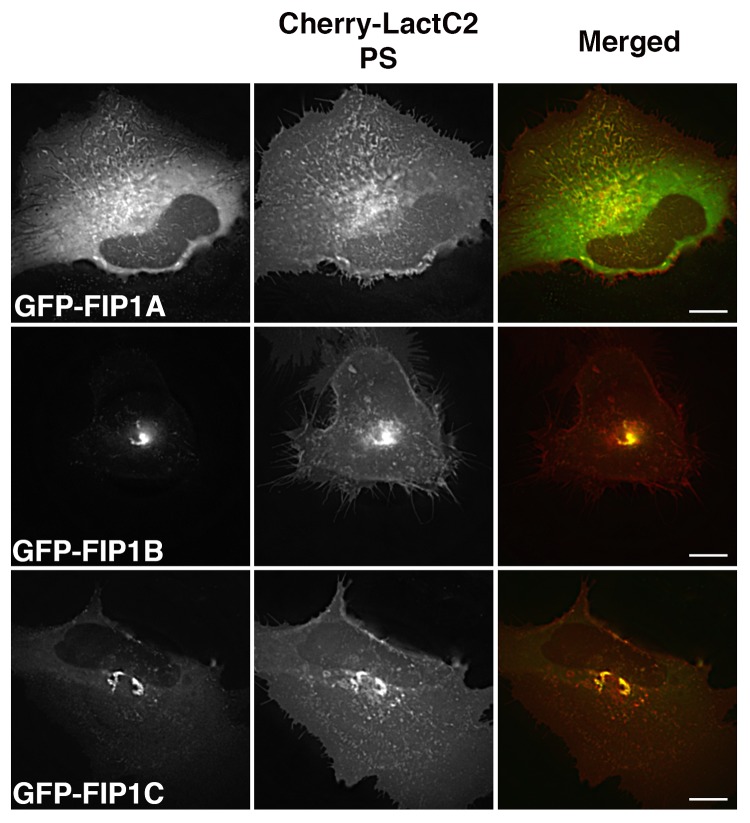
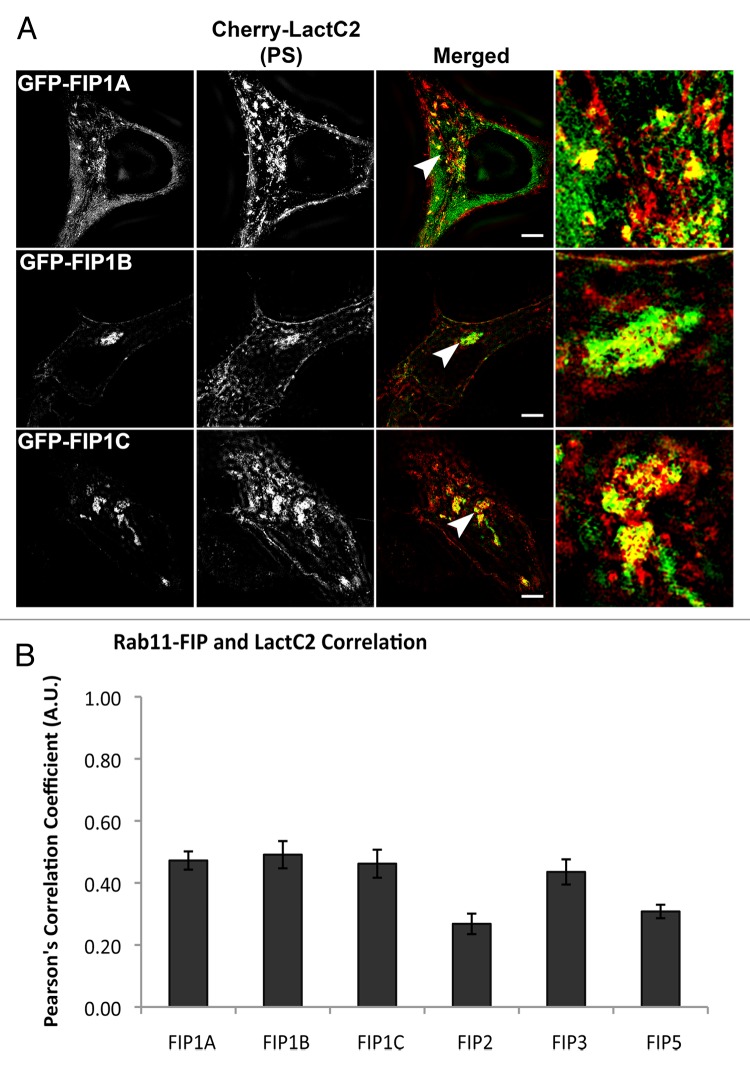
Based on the specific differences we observed in the overlap between the LactC2 PS probe and a select group of endosomal Rab GTPases, we next examined overlap between the PS probe and Rab11-FIPs using live cell deconvolution microscopy to visualize the overlap between Rab11-FIP1 proteins and LactC2 (Fig. 2). Rab11-FIP1A consistently overlapped with the LactC2 probe along endosomal tubules in live HeLa cells and both probes moved concurrently within cells, indicating occupation of the same compartment (Vid. SV2). Additionally, we found that Rab11-FIP1B and Rab11-FIP1C also overlapped with LactC2 in the pericentriolar region of HeLa cells and that LactC2 appeared to be accumulated in this area in the presence of overexpression of either of these two Rab11-FIP1 proteins. Of interest, Rab11-FIP1B and Rab11-FIP1C have C2 domains, while Rab11-FIP1A does not, suggesting that Rab11-FIP1A associates with PS-containing membranes through a mechanism other than a C2 domain binding to phospholipid. We further confirmed our findings with structured illumination microscopy (Fig. 3a) and quantitative analysis of the overlap between LactC2 and Rab11-FIP1 proteins revealed similar correlation coefficients (Rab11-FIP1A = 0.472 ± 0.029, Rab11-FIP1B = 0.491 ± 0.044, Rab11-FIP1C = 0.462 ± 0.045) that were not statistically different (P > 0.05) indicating Rab11-FIP1 proteins exhibit similar levels of association with LactC2 positive membranes (Fig. 3b). Of note, Rab11-FIP1A overlapped with LactC2 in pericentriolar and peripheral endosomes, while Rab11-FIP1B and Rab11-FIP1C overlapped primarily with LactC2 in the pericentriolar region only.
Figure 2. EGFP-Rab11-FIP1 proteins consistently overlap with mCherry-LactC2 (PS) in live HeLa cells. EGFP-Rab11-FIP1 proteins and mCherry-LactC2 overlapped in peripheral and pericentriolar compartments during imaging of live HeLa cells. FIP1B and FIP1C induced a partial accumulation of LactC2 in the pericentriolar compartments. Cells were imaged for at least one minute every 2 s. Data represent at least 3 independent experiments. Bars, 10 μm.
Figure 3. The Rab11-FIP1 proteins are within 100–200nm of LactC2 by SIM. (A) HeLa cells transfected with EGFP-Rab11-FIP1 proteins and mCherry-LactC2 were imaged on coverslips using structured illumination microscopy. Each Rab11-FIP1 protein displayed overlap with LactC2. Images were collected over a 1 μm stack of individual HeLa cells. Bars, 10 μm. (B) Pearson’s correlation coefficients were analyzed for each condition. Rab11-FIP1A (0.472 ± 0.029, n = 14 cells), Rab11-FIP1B (0.491 ± 0.044, n = 9 cells), and Rab11-FIP1C (0.462 ± 0.045, n = 8 cells) had statistically similar overlap with LactC2 (P > 0.05) Results were analyzed using an unpaired, two-tailed, Student’s t test and presented as Mean ± SEM.
In addition to fluorescence microscopy, we isolated GFP-Rab11-FIPs from HEK293 cell lysates in the presence and absence of detergent and analyzed recovered mCherry-LactC2 by SDS-PAGE and western blot. We found that mCherry-LactC2 was not recovered with any GFP-Rab11-FIPs when detergent was included in the preparation. However, mCherry-LactC2 was recovered with Rab11-FIP1A, Rab11-FIP1C, and Rab11-FIP2 when the detergent was omitted (Fig. S1). The lower band present in the detergent-free preparation is likely a degraded form of mCherry-LactC2 that retains the ability to bind Rab11-FIP containing membranes. These data suggest that mCherry-LactC2 is specifically associated with Rab11-FIPs in the presence of membranes, but that these associations do not result from GFP-Rab11-FIPs binding directly to the LactC2 probe. The combined data demonstrate that Rab11-FIP1 proteins associate with LactC2 containing membranes and that these associations may be mediated by domains other than the N-terminal C2 domains.
Distinct patterns of LactC2 association with compartments containing Rab11-FIP2, Rab11-FIP3, or Rab11-FIP5
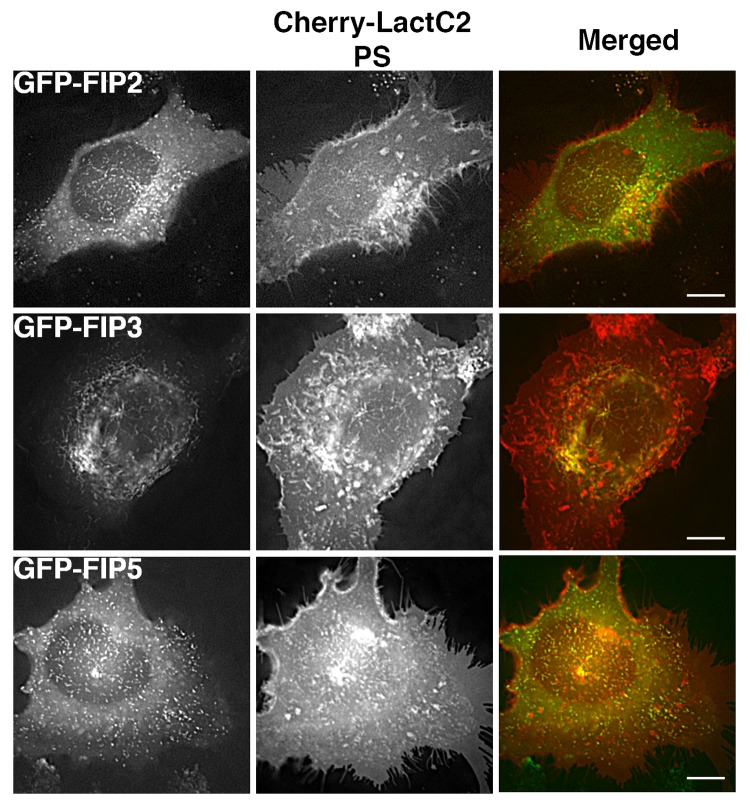
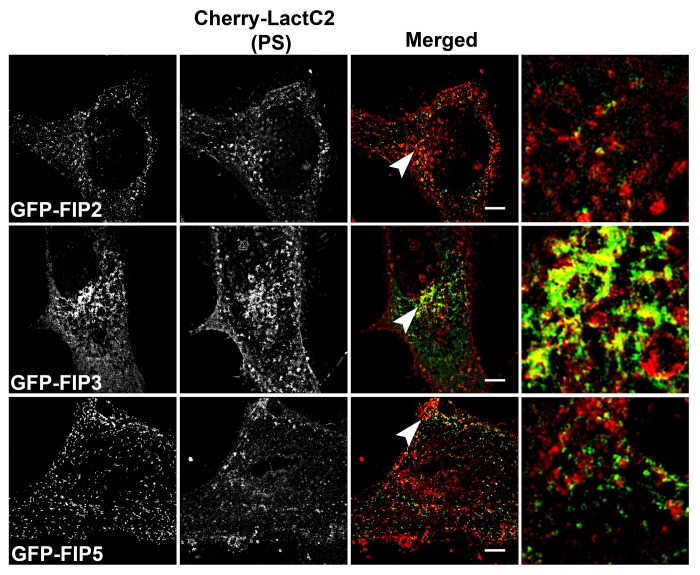
We next examined the overlap of mCherry-LactC2 co-expressed with other GFP-Rab11-FIPs (FIP2, FIP3, and FIP5) in HeLa cells (Fig. 4). We observed distinct separation between mCherry-LactC2 and either GFP-Rab11-FIP2 or GFP-Rab11-FIP5 in the periphery of cells. Similarly, we observed points of separation between mCherry-LactC2 and GFP-Rab11-FIP3. However, we did note considerable overlap between mCherry-LactC2 and GFP-Rab11-FIP3 in the pericentriolar area along elaborate branching tubules, suggesting that the GFP-Rab11-FIP3 and mCherry-LactC2 were present along coincident membranes. Additionally, we further imaged these conditions using structured illumination microscopy and found that the patterns of overlap and separation observed with deconvolution microscopy were consistent with that of structured illumination microscopy (Fig. 5a). Analysis of the correlation between LactC2 and the Rab11-FIPs studied in this experiment showed that Rab11-FIP2 (0.268 ± 0.033) and Rab11-FIP5 (0.308 ± 0.022), while having similar levels of overlap with LactC2 (P > 0.05), each had significantly lower correlation coefficients (P < 0.05) than either Rab11-FIP1 proteins or Rab11-FIP3 with LactC2 (0.435 ± 0.041) (Fig. 3b). Rab11-FIP3 overlap with LactC2 was not statistically different from the overlap between LactC2 and the Rab11-FIP1 proteins (P > 0.05) (Fig. 3b). These combined data indicate that additional Rab11-FIPs outside the Rab11-FIP1 group also occupy PS containing membrane domains, but not with the same uniformity as the Rab11-FIP1 proteins. Furthermore these data confirm that the association of Rab11-FIPs with PS containing membranes is not solely dependent upon the presence of the C2 domain found in Rab11-FIP2 and Rab11-FIP5, yet absent in Rab11-FIP3.
Figure 4. Alternative EGFP-Rab11-FIPs exhibit selective overlap with mCherry-LactC2. HeLa cells coexpressing EGFP-Rab11-FIPs and mCherry-LactC2 were imaged using live cell deconvolution microscopy. Images were collected every 2 s for at least 1 min. EGFP-Rab11-FIP2 and EGFP-Rab11-FIP5 were separate from mCherry LactC2 particularly in the periphery of HeLa cells. EGFP-Rab11-FIP3 was overlapped with mCherry-LactC2 in the pericentriolar region of the cell on distinct tubular membranes. Data represent at least 3 independent experiments. Bars,10 μm.
Figure 5:
mCherry-LactC2 with alternative EGFP-Rab11-FIPs is limited to EGFP-FIP3 within the pericentriolar region by SIM. A. EGFP-Rab11-FIPs encoded by genes other than Rab11-FIP1 do not overlap as extensively with LactC2 in HeLa cells. HeLa cells expressing EGFP-Rab11-FIPs and mCherry-LactC2 were imaged on coverslips by structured illumination microscopy. EGFP-Rab11-FIP3 was observed along mCherry-LactC2 positive compartments within the perincentriolar region while EGFP-Rab11-FIP2 and EGFP-Rab11-FIP5 did not overlap with mCherry-LactC2. Bars,10 μm. B. Pearson’s correlation coefficients were analyzed for each condition. Rab11-FIP2 (0.268 ± 0.033, n = 8 cells) and Rab11-FIP5 (0.308 ± 0.022, n = 10 cells) had significantly lower correlation coefficients (P < 0.05) than other Rab11-FIPs. Rab11-FIP3 (0.435 ± 0.041, n = 17 cells; P > 0.05) was not significantly different than Rab11-FIP1 proteins. Results were analyzed using an unpaired, two-tailed, Student’s t test and presented as Mean ± SEM.
Phosphatidylserine is enriched within tubular compartments of Rab11-FIPs
We next examined the co-expression of multiple Rab11-FIPs in HeLa cells in the presence of LactC2 to discern whether PS is enriched along tubular endosomal compartments that contain multiple Rab11-FIPs. We used live cell deconvolution microscopy to image HeLa cells expressing Cerulean-Rab11-FIP3, Venus-Rab11-FIP1A, and mCherry-LactC2. Previous studies have shown that Rab11-FIP3 and Rab11-FIP1A form a series of subdomains along tubular endosomal compartments that move in concert throughout the pericentriolar region.25 Therefore, we used this same combination of proteins to determine whether LactC2, as an indicator of PS-containing membranes, is enriched along specific tubular recycling system compartments. We found that mCherry-LactC2 was visible along tubular endosomes co-labeled with both Rab11-FIP1A and Rab11-FIP3 (Fig. 6). The movement of these compartments together suggests that tubular endosomal compartments are triple labeled for Rab11-FIP3, Rab11-FIP1A, and PS (Vid. SV3). Neither Rab11-FIP1A nor Rab11-FIP3 has a C2 domain, yet they were associated with the membranes enriched in PS. Thus Rab11-FIP1A and Rab11-FIP3 both associate with PS-containing membranes through a mechanism independent of integral C2 domains.
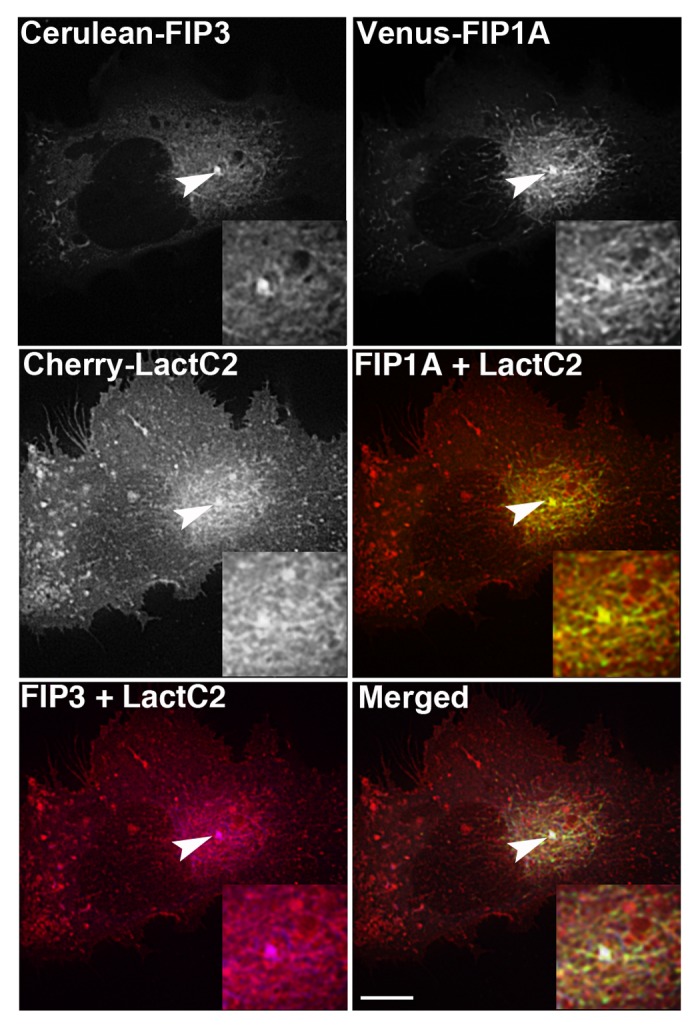
Figure 6. mCherry-LactC2 is enriched along tubular compartments of the Rab11-FIP network. Cerulean-Rab11-FIP3, Venus-Rab11-FIP1A, and mCherry-Rab11-LactC2 expressed in HeLa cells overlap along endosomal tubules in the pericentriolar region. Images were collected every 3 s for at least 1 min. Bars, 10μm.
Inhibition of trafficking through the recycling system induces an accumulation of PS
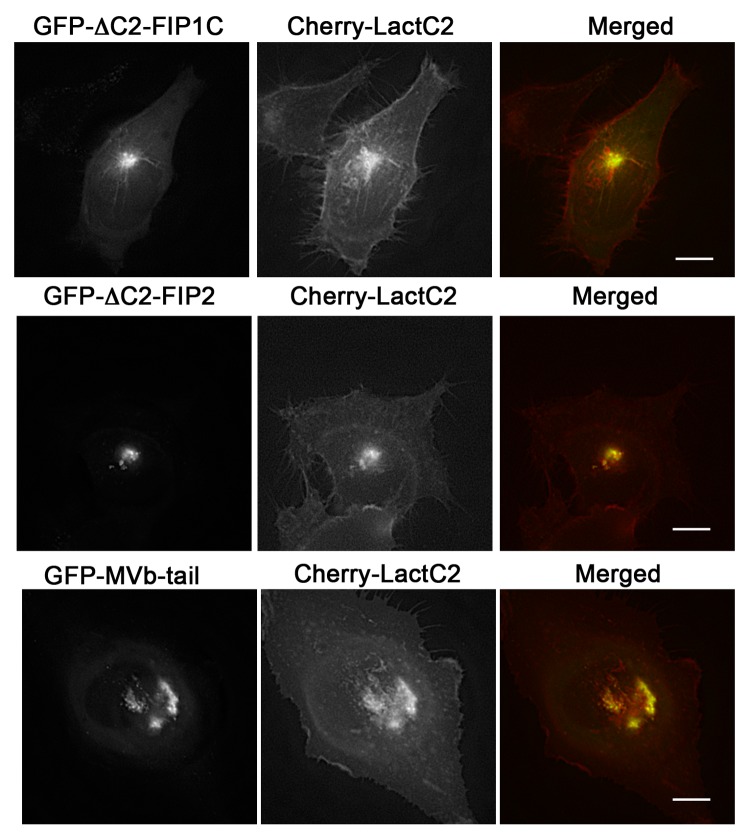
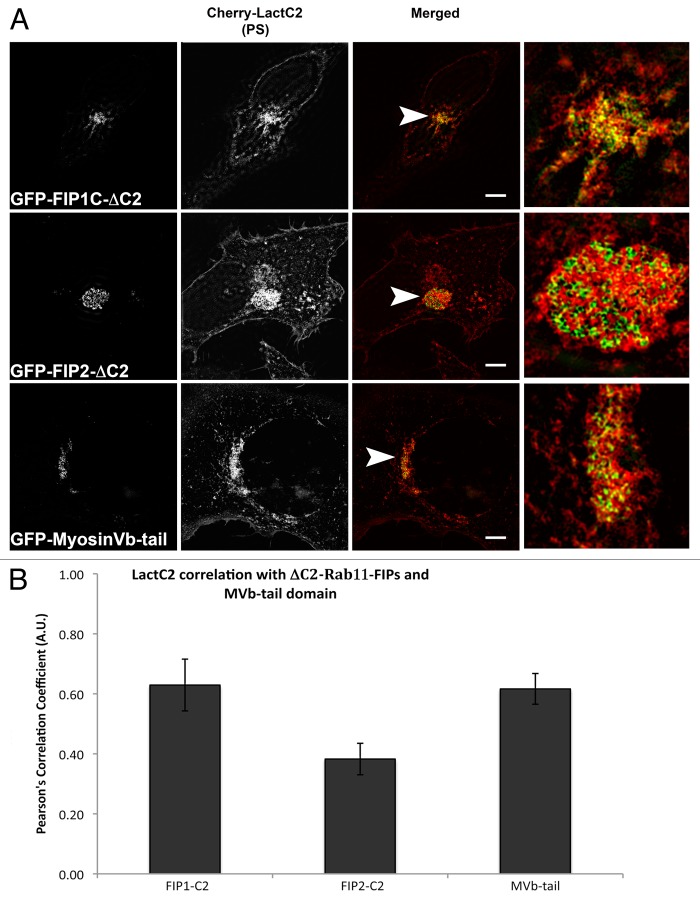
Previous studies have determined that N-terminal truncations of Rab11-FIP1C and Rab11-FIP2 that delete the C2-domains (ΔC2-Rab11-FIP1C or ΔC2-Rab11-FIP2, respectively) inhibit transferrin trafficking through the Rab11a-containing recycling compartment.12 Similarly, expression of the motorless tail of Myosin Vb also potently inhibits recycling of transferrin.26 We therefore evaluated whether these inhibitory constructs could influence the localization of LactC2 as a reflection of PS. ΔC2-Rab11-FIP1C or ΔC2-Rab11-FIP2 were expressed with mCherry-LactC2 and we used deconvolution microscopy to visualize the distribution of mCherry-LactC2 under these conditions (Fig. 7). Both ΔC2-Rab11-FIP1C and ΔC2-Rab11-FIP2 induced an accumulation of LactC2 in the pericentriolar recycling compartment. We also examined these transfected cells using structured illumination microscopy and found that, while we detected overlap between ΔC2-Rab11-FIP1C and mCherry-LactC2, we noticed distinct separation of ΔC2-Rab11-FIP2 and mCherry-LactC2 even within the tight tubular network in the pericentriolar recycling compartment (Fig. 8). When we expressed the Myosin Vb tail, we also observed an accumulation of LactC2 (Figs. Seven and 8). Measurement of the Pearson’s correlation coefficient between LactC2 and the ΔC2-Rab11-FIPs or Myosin Vb tail indicated that ΔC2-Rab11-FIP2 (0.383 ± 0.052) had significantly lower correlation with LactC2 (P < 0.05) than ΔC2-Rab11-FIP1C (0.629 ± 0.086) and Myosin Vb tail (0.616 ± 0.051) (P > 0.05) (Fig. 8b). Together these results suggest that inhibition of trafficking through the recycling system leads to accumulation of PS in inhibited membrane cisternae.
Figure 7. EGFP-Rab11-FIPs lacking the N-terminal C2 domain induce accumulation of LactC2 in the pericentriolar region. EGFP-Rab11-FIP1CΔC2 or EGFP-Rab11-FIP2ΔC2 cause an accumulation of mCherry-LactC2 in the pericentriolar region of live HeLa cells. Expression of MyosinVb-tail induced a similar accumulation of mCherry-LactC2. Images were collected every 2 s for at least 1 min. Data are representative of 2 independent experiments. Bar, 10μm.
Figure 8:
The mCherry-LactC2 and Rab11-FIP-ΔC2 proteins exhibit selective overlap in the pericentriolar region. (A) EGFP-Rab11-FIPΔC2 constructs coexpressed with mCherry-LactC2 on coverslips and imaged using structured illumination microscopy demonstrated distinct associations between LactC2 and Rab11-FIP1C or Myosin Vb, but visible separation between LactC2 and Rab11-FIP2 . Bars, 10 μm. (B) ΔC2-Rab11-FIP2 (0.383 ± 0.052, n = 9 cells) and LactC2 (P < 0.05) had significantly lower correlation than ΔC2-Rab11-FIP1C (0.629 ± 0.086, n = 3 cells) or Myosin Vb tail (0.616 ± 0.051, n = 9 cells; P > 0.05). Results were analyzed using an unpaired, two-tailed, Student’s t test and presented as Mean ± SEM.
Discussion
Our investigations have demonstrated that Rab GTPases and Rab11-FIPs associate with LactC2-labeled (PS-containing) membranes at select locations within HeLa cells as determined by live cell and structured illumination microscopy. Co-expression of Rab GTPases with LactC2 in HeLa cells highlighted distinct overlapping patterns between LactC2 and Rab8a, Rab10, and Rab11a, while Rab5a and Rab7a displayed distinct separation from the PS probe in the periphery of cells. Rab11-FIP1 proteins each appeared to overlap with LactC2 especially in the pericentriolar region of the cell despite the differences in the presence or absence of C2 domains. Similarly other members of the Rab11-FIPs displayed selective association with LactC2, but Rab11-FIPs with C2 domains did not necessarily overlap with PS-containing membranes consistently. We did find that LactC2 was enriched along tubular compartments containing multiple Rab11-FIPs that do not possess C2 domains, supporting our earlier findings and suggesting that C2 domains are not the sole mechanism for Rab11-FIPs to associate with PS-containing membranes. Finally we found that truncated Rab11-FIP chimeras induced an accumulation of PS containing membranes in the pericentriolar region, suggesting that perturbation of Rab11a dependent vesicle recycling influences the distribution and movement of PS-containing membranes within the recycling system. Overall, this study indicates that the plasma membrane recycling system contains multiple discrete domains, which are defined by Rab11-FIPs and selective associations with PS containing membranes.
The diverse associations between Rab11-FIPs and LactC2 positive membranes observed in the current study may result from multiple potential modes of interaction for these effector proteins with PS-containing membranes. Rab11-FIPs primarily associate with lipids and membranes via their interaction with Rab1113 or an N-terminal phospholipid-binding C2 domain.17 Purified C2 domains of Rab11-FIPs bind a variety of acidic lipids17 and C2 domains in general bind a variety of phospholipids often in a calcium dependent manner.15,16 Thus, the overlap we observed among Rab11-FIPs and LactC2 may be a result of binding between Rab11-FIPs and multiple phospholipids on the same membrane as LactC2. The removal of the C2 domains from Rab11-FIPs induced an accumulation of LactC2 in the pericentriolar compartment, yet we observed a clear separation of LactC2 from ΔC2-Rab11-FIP2 in the pericentiolar cisternae. The lack of co-localization between LactC2 and either Rab11-FIP2 or Rab11-FIP5 in the cell periphery also suggested that Rab11-FIPs might associate with phospholipids other than PS,17 different subpopulations of PS,27 and perhaps even compete for the same pool of PS as LactC2. Heterodimerization among Rab11-FIPs and interactions with Myosin Vb to form protein complexes that regulate membrane trafficking might also explain the diverse patterns of localization described in the current study.12,25,26,28,29 Furthermore, the assembly of Rab11-FIP1A and Rab11-FIP3 along LactC2 membrane domains indicated that additional binding motifs exist and that the formation of Rab11-FIP complexes may facilitate associations between Rab11-FIPs and phospholipids. Finally, interactions of Rab11-FIPs with other Rabs such as Rab430 or Rab14,31,32 and previously characterized interactions of Rab11-FIP3 (also known as Arphophilin-1) with Arf-GTPases may also promote associations with a variety of phosphoinositides.33,34 The current data confirm the previous reports of PS as a component and potential target lipid in the Rab11-containing recycling system and indicate a range of possibilities for these associations to occur.
The occupation of membranes by Rab11-FIPs and LactC2 highlights potential sorting domains that have important implications for membrane trafficking.35 Membranous compartments within cells are continuous yet distinct based on their composition36,37 and the development of reliable lipid probes has allowed for visualization of Rab11-FIPs in the context of a dynamic lipid environment.22 While lipids are commonly thought of as substrates for proteins to bind and associate with various compartments, the current technology permits visualization of the coordinated movement of Rab11-FIPs with PS-containing compartments.38 We observed coordinated movement among the Rab11-FIP1 proteins with LactC2 that was more apparent than we observed between LactC2 and Rab11-FIP2 or Rab11-FIP5, indicating differences in association with lipids and potential mechanisms for sorting cargoes based on the lipid content of membrane domains. The LactC2 probe permitted visualization of inhibited PS movement in the presence of truncated forms of the Rab11-FIPs. Similarly, the patchwork assembly of Rab11-FIP1A and Rab11-FIP3 along PS-containing domains coupled with the selective nature of Rab11-FIP associations with LactC2 membrane domains indicates that multiple pathways may be at work in the same recycling system.39 Previous reports have indicated that the formation of such complexes is required to construct defined domains.40 Furthermore, the assembly of these complexes promotes critical events such as fusion of vesicle membranes.41 Each of these findings argues in favor of dynamically pleomorphic endosomal compartments, as proteins and lipids are recycled through the cell. Endosomal compartments change in protein composition, lipid framework, as well as internal cargo and pH during trafficking.36,42 Here we observed both location and time-dependent associations of Rab11-FIPs with PS-containing membranes that are likely mediated by both direct and indirect interactions. The selective nature of Rab11-FIP associations with PS-containing compartments supports the concept that dynamic associations between Rab11-FIPs and phospholipid-containing domains regulates cargo sorting in the Rab11-dependent recycling system.
The current study has demonstrated selective associations among Rab11-FIPs with LactC2-labeled membranes that have important implications in Rab11-dependent vesicle recycling. The data presented here confirm previous findings that PS is likely an integral component of endosomal compartments occupied by Rab11 and Rab11-FIPs during vesicle recycling.23 The development of molecular probes that detect specific lipids22 including PS,23 PI(3)P,43 PI(3,5)P,44 and cholesterol45 have made it possible to visualize these dynamic trafficking processes in the context of a lipid framework. The presence of selective associations and discrete domains indicates potential sorting points within the Rab11-containing recycling system, as well as intersections among multiple pathways. Future investigations regarding the specific mechanisms that permit Rab11-FIP interactions with various phospholipids and the modes by which they regulate the cargoes that pass through related compartments will help determine the progression of these processes. All of these findings implicate a dynamic set of interactions among protein and phospholipid regulators to define the complexities of trafficking through the recycling system.
Materials and Methods
Plasmids and expression vectors
The preparation of the following plasmids has previously been described: mCherry-Rab5a,46 mCherry-Rab8a,47 EGFP-Rab11a,26 EGFP-Rab11-FIPs (FIP1A, FIP1B, FIP1C),48 EGFP-Rab11-FIP2 and EGFP-Rab11-FIP3,13 EGFP-Rab11-FIP2ΔC2,12 EGFP-MyosinVb-tail.49 EGFP-Rab11-FIP5 was a gift from R. Prekeris at the University of Colorado. Preparation of mCherry Rab7a was prepared by cloning Rab7a from EGFP-Rab7a, a gift from A. Waddinger-Ness at the University of New Mexico, using EcoR1 and Sal1 restriction sites. Preparation of mCherry-Rab10a was completed courtesy of Dr. Joseph Roland using EcoR1 and Sal1 restriction sites. Preparation of mCherry-Rab11a was performed by cloning Rab11a from EGFP-Rab11a using EcoRI and Sal1 restriction sites. Preparation of EGFP-Rab11-FIP1CΔC2 was completed by Dr. Janice Williams in the Goldenring laboratory by amplification of FIP1C residues 126–549 from EGFP-Rab11-FIP1C and ligated back into EGFP vector using EcoR1 and BamH1 restriction sites. Preparation of Venus-Rab11-FIP1A were performed by Dr. Jenny Schafer by cloning FIP1A from EGFP-Rab11-FIP1A using Sal1 and BamH1 restriction sites. Preparation of the Cerulean-Rab11-FIP3 was performed by cloning Cerulean50 into EGFP vector following removal of EGFP using Nhe1 and BsgR1 restrictions sites. EGFP-LactC2 vector was purchased from Addgene (plasmid 2285) and was originally developed by S Grinstein at the University of Toronto.23 The LactC2 gene was subsequently cloned into mCherry-plasmid using BglII and EcoRI restrictions sites courtesy of Dr. Jessica Maszerik in the laboratory of M Tyska at Vanderbilt University.
Single and Dual Expression Live Cell Imaging
HeLa cells were maintained in RPMI media (Cellgro 15–040-CV) with 10% fetal bovine serum (PAA Laboratories Inc. A15–704) in 35-mm glass bottom dishes (Matek P35G-0–14-C) for at least one day. Cells were transfected using Effectene (Qiagen 301425) and 200ng of DNA for each construct used. HeLa cells were transfected with EGFP-LactC2 and mCherry-Rab constructs, EGFP-Rab11-FIP and mCherry-LactC2 constructs for at least 8 h. Cells were then imaged in a 37 °C, 5% CO2 chamber using a 100x oil immersion objective (1.4 numerical aperture) on a Deltavision deconvolution microscope (Applied Precision) and a Photometrics CoolSNAP HQ2 camera. Images sizes were collected at 512x512 pixels at a minimum of every 2 s for at least 1 min at exposures sufficient to achieve a minimum 5 to 1 signal to noise ratio. All files were deconvolved using the Applied Precision Softwork package.
Structured Illumination Microscopy (SIM)
HeLa cells expressing full-length or truncated Rab11-FIPs with mCherry-Lact C2 were prepared as described above. Cells were cultured on 35mm glass bottom plates (Matek P356–0-14-C) and transfected with 200ng of the respective DNA in RPMI (Cellgro 15–040-CV) media. Images were collected using 30–100 ms exposure times with an Applied Precision/GE OMX structured illumination microscope51 with an APO 60x objective (1.49 NA), sCMOS camera, and processed using the Softworx package. Colocalization analysis of SIM data was performed using Image J Colocalization tool52 and statistical analysis was conducted using a two tailed, unpaired Student’s t test with a threshold of 0.05.
Isolation and Western Blotting of EGFP-Rab11-FIPs
Preparation of HEK cell lysates for western blot analysis was conducted as previously described.25 Briefly, HEK-293 cells co-transfected with EGFP empty vector or EGFP-Rab11-FIP chimeras (200 ng) and mCherry-LactC2 (200 ng) were scraped on ice into 1 mL of 30 mM Tris, 150 mM sodium acetate, and 20 mM magnesium acetate (Buffer A). Cells were sheared open by passing cells 50 times through a 27 gauge needle and lysates for detergent preparation had 1% Triton X-100 added at this time for 30 min at 4 °C. Resulting supernatant was added to GFP binding protein (GBP)53 on agarose beads equilibrated in Buffer A. Equal amounts of protein from each condition were added to beads and rotated at 4 °C overnight then beads were resuspended in 30 µL Laemmli buffer.54 All samples were boiled and resolved on 10% SDS-PAGE gel, transferred to nitrocellulose membrane (Whatman 10 401 196) and probed for mCherry using a rabbit anti-DsRed (Clontech 632–496) antibody. Signals were detected using Pierce ECL Western Blotting Substrate (Product 32106) and exposure to and development of Kodak film (864–6770). Membranes were probed a second time using a primary rabbit anti-EGFP (ab290 Abcam) antibody at 1:2500 in 5% milk in 0.1% TBS-Tween-20 and a Trueblot anti-rabbit IgG HRP (eBioscience 18–8816–33) rabbit secondary at 1:5000 in 5% milk in 0.1%TBS-Tween-20. Membranes were washed and developed as above.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant RO1 DK48370 to JRG and the NWB was supported by T32 CA106183. Live cell imaging studies were performed in the Vanderbilt Digital Histology Shared Resource and the Epithelial Biology Center. Confocal fluorescence imaging was performed through the use of the VUMC Cell Imaging Shared Resource supported by the Vanderbilt Digestive Disease Research Center and the Vanderbilt-Ingram Comprehensive Cancer Center and National Institute of Health (NIH) Grants CA68485, DK20593, DK58404 and HD15052. The authors would like to thank Dr. Rytis Prekeris at the University of Colorado for providing the EGFP-Rab11-FIP5/Rip11 construct and Dr. Waddinger-Ness from the University of New Mexico for the gift of the EGFP-Rab7a construct.
References
- 1.Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–29. doi: 10.1016/0092-8674(90)90369-P. [DOI] [PubMed] [Google Scholar]
- 2.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–59. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–14. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 5.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 6.Lee MT, Mishra A, Lambright DG. Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic. 2009;10:1377–89. doi: 10.1111/j.1600-0854.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ullrich O, Reinsch S, Urbé S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–24. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prekeris R, Klumperman J, Scheller RHA. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell. 2000;6:1437–48. doi: 10.1016/S1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- 9.Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem Biophys Res Commun. 2002;299:770–9. doi: 10.1016/S0006-291X(02)02720-1. [DOI] [PubMed] [Google Scholar]
- 10.Schonteich E, Wilson GM, Burden J, Hopkins CR, Anderson K, Goldenring JR, Prekeris R. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J Cell Sci. 2008;121:3824–33. doi: 10.1242/jcs.032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsay AJ, McCaffrey MW. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J Biol Chem. 2002;277:27193–9. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- 12.Hales CM, Vaerman JP, Goldenring JR. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J Biol Chem. 2002;277:50415–21. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- 13.Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–75. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- 14.Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;334:661–5. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- 15.Cho W. Membrane targeting by C1 and C2 domains. J Biol Chem. 2001;276:32407–10. doi: 10.1074/jbc.R100007200. [DOI] [PubMed] [Google Scholar]
- 16.Rizo J, Südhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–82. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 17.Lindsay AJ, McCaffrey MW. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J Cell Sci. 2004;117:4365–75. doi: 10.1242/jcs.01280. [DOI] [PubMed] [Google Scholar]
- 18.Rainero E, Caswell PT, Muller PA, Grindlay J, McCaffrey MW, Zhang Q, Wakelam MJ, Vousden KH, Graziani A, Norman JC. Diacylglycerol kinase α controls RCP-dependent integrin trafficking to promote invasive migration. J Cell Biol. 2012;196:277–95. doi: 10.1083/jcb.201109112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vance JE. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J Lipid Res. 2008;49:1377–87. doi: 10.1194/jlr.R700020-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Vance JE, Steenbergen R. Metabolism and functions of phosphatidylserine. Prog Lipid Res. 2005;44:207–34. doi: 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Bohdanowicz M, Grinstein S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol Rev. 2013;93:69–106. doi: 10.1152/physrev.00002.2012. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Zheng H. Illuminating the lipidome to advance biomedical research: peptide-based probes of membrane lipids. Future Med Chem. 2013;5:947–59. doi: 10.4155/fmc.13.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–3. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 24.Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol. 2012;13:463–70. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- 25.Baetz NW, Goldenring JR. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol Biol Cell. 2013;24:643–58. doi: 10.1091/mbc.E12-09-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Provance DW, Jr., Mercer JA, Bähler M, Goldenring JR. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12:1843–57. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–75. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem Biophys Res Commun. 2002;292:909–15. doi: 10.1006/bbrc.2002.6736. [DOI] [PubMed] [Google Scholar]
- 29.Meyers JM, Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J Biol Chem. 2002;277:49003–10. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- 30.Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem. 2002;277:12190–9. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- 31.Kelly EE, Horgan CP, Adams C, Patzer TM, Ní Shúilleabháin DM, Norman JC, McCaffrey MW. Class I Rab11-family interacting proteins are binding targets for the Rab14 GTPase. Biol Cell. 2010;102:51–62. doi: 10.1042/BC20090068. [DOI] [PubMed] [Google Scholar]
- 32.Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, et al. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog. 2013;9:e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donaldson JG. Arfs, phosphoinositides and membrane traffic. Biochem Soc Trans. 2005;33:1276–8. doi: 10.1042/BST20051276. [DOI] [PubMed] [Google Scholar]
- 34.Shin OH, Ross AH, Mihai I, Exton JH. Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J Biol Chem. 1999;274:36609–15. doi: 10.1074/jbc.274.51.36609. [DOI] [PubMed] [Google Scholar]
- 35.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9:833–45. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;2:721–30. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- 37.Barr FA. Review series: Rab GTPases and membrane identity: causal or inconsequential? J Cell Biol. 2013;202:191–9. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. Phosphatidylserine dynamics in cellular membranes. Mol Biol Cell. 2012;23:2198–212. doi: 10.1091/mbc.E11-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naslavsky N, Weigert R, Donaldson JG. Characterization of a nonclathrin endocytic pathway: membrane cargo and lipid requirements. Mol Biol Cell. 2004;15:3542–52. doi: 10.1091/mbc.E04-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 41.Araç D, Chen X, Khant HA, Ubach J, Ludtke SJ, Kikkawa M, Johnson AE, Chiu W, Südhof TC, Rizo J. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–17. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 42.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–88. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci U S A. 2013;110:21165–70. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maxfield FR, Wüstner D. Analysis of cholesterol trafficking with fluorescent probes. Methods Cell Biol. 2012;108:367–93. doi: 10.1016/B978-0-12-386487-1.00017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ducharme NA, Ham AJ, Lapierre LA, Goldenring JR. Rab11-FIP2 influences multiple components of the endosomal system in polarized MDCK cells. Cell Logist. 2011;1:57–68. doi: 10.4161/cl.1.2.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–37. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin M, Goldenring JR. The Rab11-FIP1/RCP gene codes for multiple protein transcripts related to the plasma membrane recycling system. Biochim Biophys Acta. 2006;1759:281–95. doi: 10.1016/j.bbaexp.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Roland JT, Lapierre LA, Goldenring JR. Alternative splicing in class V myosins determines association with Rab10. J Biol Chem. 2009;284:1213–23. doi: 10.1074/jbc.M805957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–9. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 51.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–7. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 52.Manders EEM, Verbeek FJ, Aten JA. Measurement of co-localisation of objects in dual-colour confocal images. J Microsc. 1993;169:375–82. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 53.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–9. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.