Abstract
N6-methyladenosine (m6A) is a prevalent modification of eukaryotic mRNAs. It regulates yeast cell fate and is essential to the development and fertility of metazoans. Although its presence in mRNA has been known since the early 1970s, the function of m6A remained a mystery until the spate of discoveries in the past three years. Here, we focus on the discovery of m6A “readers” (proteins that specifically recognize m6A), and their functions in tuning mRNA stability, as well as the broader significance of such m6A-dependent regulation of gene expression.
Keywords: N6-methyladenosine, RNA methylation, RNA stability, RNA-binding protein, YTH domain, YTHDF2, gene expression regulation, m6A, reversible RNA modification
Eukaryotic mRNAs (mRNAs) not only encode precise protein sequence, but also convey information for their processing, transportation, translation, and decay, thereby collectively creating a complex layer of gene regulation at the post-transcriptional stage. Known mechanisms involving RNA structure, microRNA, and translation regulation all contribute to post-transcriptional regulation of gene expression. Established regulatory modes of mRNA involve short RNA sequences in untranslated regions (UTRs) as exemplified by AU-rich element (stability), iron-responsive element (translation), zipcode element (localization), micro RNA seeding sequences (translation and stability), and the mRNA cap and polyadenylated tail. In this paper, we discuss a new mechanism of regulation: reversible internal RNA methylation.
Essential Description of m6A
N6-methyladenosine (m6A) is a major internal (non-cap) modification of eukaryotic mRNA (mRNA).1 On average, every mRNA has three m6A residues within a context of G(m6A)C (70%) or A(m6A)C (30%).2 Recent advances in m6A-sequencing technology have also revealed m6A enrichment in long exons and around the stop codon.3-5 Despite the widespread distribution of m6A sites over 7000 human transcripts, the m6A content of individual mRNAs is non-uniform; each m6A site can be non-stoichiometric while some mRNAs are free of m6A.1 m6A is post-transcriptionally installed by an m6A methyltransferase complex1,6 in coordination with other mRNA processing events, namely 3′ polyadenylation, 5′ capping, and splicing. In addition to previously identified subunit METTL3, we have recently revealed two other components of mammalian m6A methltransferase complex: METTL14 forms a stable heterodimer with METTL3 as the enzymatic core; WTAP interacts with the heterodimer to affect their methyltransferase activity inside cells.7 The discovery of two functionally significant m6A demethylases (FTO8 and AlkBH59) has defined mRNA methylation as a reversible process. METTL3 is essential for proper meiotic entry of budding yeast,10,11 the viability of plants,12 fruit flies,13 and human HeLa cells,6 while defects in FTO and AlkBH5 affect body weight and fertility respectively, thus demonstrating the physiological importance of m6A.
Considering m6A as a new layer of information on top of the primary sequence, methylation, and demethylation resemble writing and erasing. Yet a mechanism to read out the methylation information must exist. m6A can either repel RNA-binding proteins that interact with A or be recognized by methyl-specific binding proteins or “readers.” Potential readers have been suggested in early pull-down experiments using methylated RNA probes.3 We have characterized the YTH domain family proteins as m6A readers and provided the first functional role of m6A through this reading process.14 The analogy between “writer/eraser/reader” and methyltransferase/demethylase/selective binding partners, though not scientifically accurate, is advantageous in order to abstract the working pattern of all chemical modifications of DNA, mRNA, and proteins. Comparable to studies of DNA methyl-CpG binding proteins (MeCPs),15 we believe that “readers” provide clues to uncover the mechanism and cellular function associated with m6A.
YTH Domain Family as m6A-Specific Binding Proteins
The YTH domain is a newly discovered domain that binds to short, degenerated, and single-stranded RNA sequences.16 The YTH domain family has 174 members in various eukaryotic species.17 In humans, the YTH domain family (YTHDF) contains three members, YTHDF1, YTHDF2, and YTHDF3. YTHDF2 and YTHDF3 were selectively identified using synthetic RNA bait containing m6A.3 We also discovered YTHDF1 with a slightly different bait sequence. All three proteins have significantly higher affinity for methylated probes as measured by gel shift assay, and thus are m6A-specific binders.
Next-generation sequencing and advances in RNA–protein complex isolation technique have greatly empowered transcriptome-wide studies of RNA-binding proteins. By an integration of photoactivatable–ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP),18 ribosome profiling,19 and mRNA lifetime profiling results, current conclusions about YTHDF2 are: (1) YTHDF2 indeed binds m6A inside the cell since YTHDF2-binding sites agree well with m6A sites; (2) there is no direct interaction between YTHDF2 and polysomes; (3) the major function of YTHDF2 is to accelerate the decay of its targets (by roughly 30% as determined by YTHDF2 knockdown); (4) the YTH domain at the C terminus of YTHDF2 is sufficient to recognize m6A while the N-terminal domain involves localizing the RNA to processing bodies where RNA decay can occur.20 Results using a reporter system show that binding of YTHDF2 to its RNA target takes place in parallel with or at a late stage of deadenylation, which is a prerequisite step of eukaryotic mRNA decay.21 Taken together, these results have led to a mechanistic model in which YTHDF2 transduces m6A code into an RNA turnover signal by its modular structure and then conveys its bound RNA to decay machinery.
Recent studies of YTHDF2 homolog in yeasts may support a conserved role of the YTH domain protein in controlling mRNA turnover.5,22,23 Mmi1, the YTHDF2 homolog in fission yeast (Schizosaccharomyces pombe), selectively degrades meiotic mRNA transcripts by RNA surveillance machinery during vegetative growth.22 In budding yeast (Saccharomyces cerevisiae), m6A conditionally accumulates during sporulation induced by nutrition starvation.24 Ydr374c (Pho92 or MRB1), the corresponding YTH domain protein in budding yeast, has also been shown as a m6A reader protein.5 It regulates the stability of Pho4 mRNA, an important transcription factor in the phosphate signal transduction (PHO) pathway, by binding to its 3′UTR in a phosphate-dependent manner.23 Mechanistically, Pho92 interacts with the Pop2-Ccr4-Not deadenylation complex, which coincides with our observation that YTHDF2 co-localizes with Pop2 (CNOT7) in human HeLa cells. Hence, these studies in yeast shed light on a possible role of m6A in nutrition metabolism via regulation of mRNA stability.
m6A-Dependent Control of mRNA Stability
To better understand the effect of m6A, it is worth discussing how m6A-dependent control of mRNA stability compares to other means of gene expression regulation. Based on transcriptome-wide measurements of RNA levels from cell populations, temporal mRNA level changes in response to external stimuli was suggested to be primarily determined by the change of transcription rate.25 However, the change of mRNA degradation rate is important in order to define sharp responses. We have also observed that transcripts bearing m6A have inherently shorter lifetimes than non-targets.26 Genes can be roughly classified into housekeeping genes whose protein production is always in demand, or regulatory genes whose protein production is conditional or strictly controlled at low abundance. Given that some mRNAs encoding housekeeping genes are free of m6A (globin, histone)1 and that YTHDF2 RNA targets are enriched with regulatory genes such as transcription factors,14 it is possible that m6A represents one mechanism that imposes precise control over the expression of those regulatory genes.
At the transcriptional level, transcriptional factors (TFs) recognize the genomic sequence. At the same time, DNA methylations or other forms of chemical modifications exist in high eukaryotes that specific reader proteins can recognize in order to exert additional regulation of gene expression (Fig. 1). Nature seems to use the same “trick” to gain additional control of gene expression at the mRNA level. Various RNA-binding proteins, parallel with TFs on DNA, exist to affect transport, storage, splicing, and stability of mRNAs. Now, we show that reader proteins also exist to recognize internal mRNA methylation, which provide additional complexity as well as the means to affect eventual protein production (Fig. 1). Chemical methylations can be added or removed on mRNA.6-9 Therefore, these m6A codes are dynamic, reversible, and dependent on cell type and state. Such a feature of m6A might be extremely important during dynamic cell differentiation process, such as embryonic13 and neuronal27 development. It is highly possible that m6A codes can function in concert with all other RNA sequence elements as well as RNA-binding proteins, again analogous to the interplays between TFs and 5mC reader proteins on DNA.28
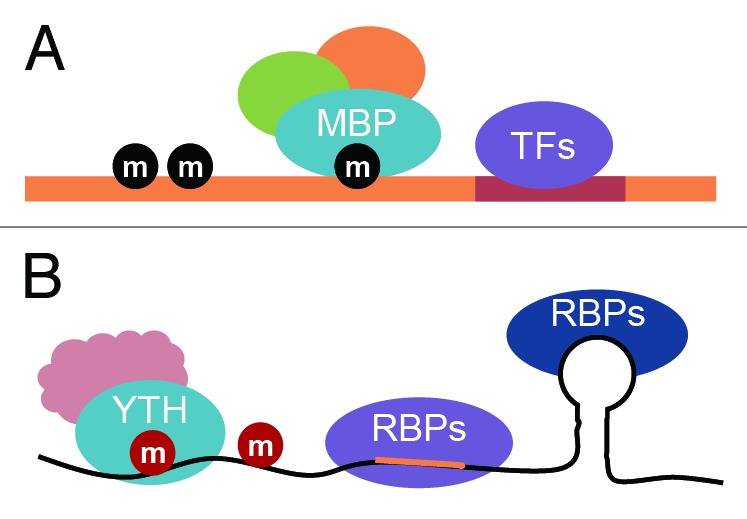
Figure 1. Specific binding proteins recognize DNA and RNA methylations. (A) The methyl-binding proteins (MBP) recognize mammalian DNA methylation (black ball). The figure also illustrates the methylation state with binding of MBP and proteins (green and orange) that interact with MBP as well as the various transcriptional factors (purple) that control transcription. (B) The YTH domain family proteins (blue) selectively recognize internal RNA methylation (red ball). The fate of mRNA is controlled by the interplay of methyl-specific binding proteins, protein factors (pink) that interact with YTH domain family proteins, and other RNA-binding proteins (RBPs, dark blue and purple) that recognize RNA sequence and/or structure.
Future Directions
YTHDF1 and YTHDF3 have already been identified as m6A-specific binding proteins;14 therefore, functional characterizations of these two YTHDF proteins are ongoing. Given the high-sequence similarities between YTHDFs, it is possible that these proteins are degenerate to some extent. We have shown that YTHDF2 mediates the translocation of m6A-containing RNA from the translatable pool to non-ribosomal mRNA–protein particles; however, we do not know if m6A-containing RNAs are targeted by YTHDF2 before any translation or after they exit translation. In addition, various RNA-binding proteins coat mRNAs; it is therefore tempting to envision that m6A readers work in concert with other sequence-specific RNA-binding proteins to collectively decide the fate of mRNA. It will be valuable to characterize the protein interactome of YTHDF2 as well as the other two YTHDF proteins.
Finally, knockdown of YTHDF2 causes reduced viability of HeLa cells. However, without knockout model organisms such as knockout mouse the exact physiological function of YTHDF2 remains unknown. Future investigations in this area are necessary. The discovery and characterization of m6A reader proteins represent critical steps in order to understand reversible m6A-dependent regulation at the RNA level,26 but there are still vast knowledge gaps between molecular details and the biological necessity of m6A that require further research and connection.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by National Institutes of Health GM071440 (He C). We thank Ian A Roundtree and Sarah F Reichard for editing the manuscript.
References
- 1.Tuck MT. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int J Biochem. 1992;24:379–86. doi: 10.1016/0020-711X(92)90028-Y. [DOI] [PubMed] [Google Scholar]
- 2.Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–6. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 3.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 4.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–21. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–47. [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–86. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–88. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci U S A. 2011;108:14855–60. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–34. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH, et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285:14701–10. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci. 2002;27:495–7. doi: 10.1016/S0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 18.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–8. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiriart E, Vavasseur A, Touat-Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HJ, Jeong SJ, Kim KN, Baek IJ, Chang M, Kang CM, Park YS, Yun CW. A novel protein, Pho92, has a conserved YTH domain and regulates phosphate metabolism by decreasing the mRNA stability of PHO4 in Saccharomyces cerevisiae. Biochem J. 2014;457:391–400. doi: 10.1042/BJ20130862. [DOI] [PubMed] [Google Scholar]
- 24.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–18. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–42. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014 doi: 10.1038/nrg3724. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 27.Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16:1042–8. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 28.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]


