Abstract
MicroRNAs (miRNAs) are ubiquitous gene regulators that modulate essential cellular processes at the post-transcriptional level. In metazoans and their viruses, most miRNAs are produced from hairpin-containing primary transcripts that are sequentially cleaved by nuclear Drosha and cytoplasmic Dicer. In the last decade, alternative mechanisms that bypass either the Drosha or Dicer cleavage step have emerged, increasing the complexity of the miRNA regulatory network. Here, we highlight non-canonical pathways that generate miRNAs using a variety of molecular machineries that play fundamental roles in the biogenesis and processing of other classes of cellular RNAs.
Keywords: microRNA biogenesis, Microprocessor, Dicer, exportin-5, mirtron, m7G-capped pre-miRNA, AGO2 slicer activity, viral microRNA, tRNaseZ, Integrator
Introduction
The discovery of microRNAs (miRNAs) has profoundly changed our understanding of how gene expression can be regulated at the post-transcriptional level. These ~22-nucleotide (nt) RNA molecules associate with Argonaute (AGO) proteins to form the functional cores of RNA-induced silencing complexes (RISCs) and interact with target mRNAs (mRNAs) through imperfect Watson-Crick base-pairing. Specific targeting is most frequently conferred by the seed region at the 5′ end of the miRNA (nt 2–8) pairing with specific sequences in the 3′ untranslated region (UTR) of an mRNA.1 RISC binding leads to translation repression and/or destabilization and degradation of the targeted mRNA.2-4 More than half of human genes are estimated to be regulated by miRNAs and there are more than a thousand annotated miRNAs genes in the human genome.5,6 Therefore, miRNAs contribute significantly to cellular pathways including differentiation, proliferation, and apoptosis, which are critical to human development and disease. It is not surprising that miRNA expression profiles are altered in a variety of disease states, including cancer.7,8
Given that miRNAs play essential roles in fine-tuning the gene-regulatory network, it should be possible to manipulate miRNA production to combat various diseases. To this end, substantial effort has been devoted to elucidating the molecular mechanisms by which these small molecules are generated. It is now clear that most animal miRNAs are produced from RNA hairpin structures embedded in long primary transcripts through sequential cleavage in the nucleus and cytoplasm by two different RNase III-class enzymes (detailed below). Yet, not long after the canonical miRNA biogenesis pathway was established, reports of a variety of alternative pathways that bypass one of the RNase III cleavage steps began to emerge. This versatility in miRNA biogenesis may permit expression of particular miRNAs in different developmental stages or altered cell states to achieve differential gene regulation. Recently, we discovered yet another novel miRNA biogenesis pathway that not only bypasses nuclear cleavage by Drosha, but also utilizes a different mode of nuclear-cytoplasmic transport; it produces mature miRNAs exclusively from one arm of the precursor (pre-)miRNA hairpin.9
Here, we present additional data supporting our recent finding of a variant miRNA biogenesis pathway. We also review presently known miRNA biogenesis pathways in animals and their viruses. In plants, miRNA biogenesis also relies on RNase III-family enzymes but is substantially different from animal miRNA biogenesis. Readers are referred to recent reviews for details.10,11
The Canonical miRNA Biogenesis Pathway
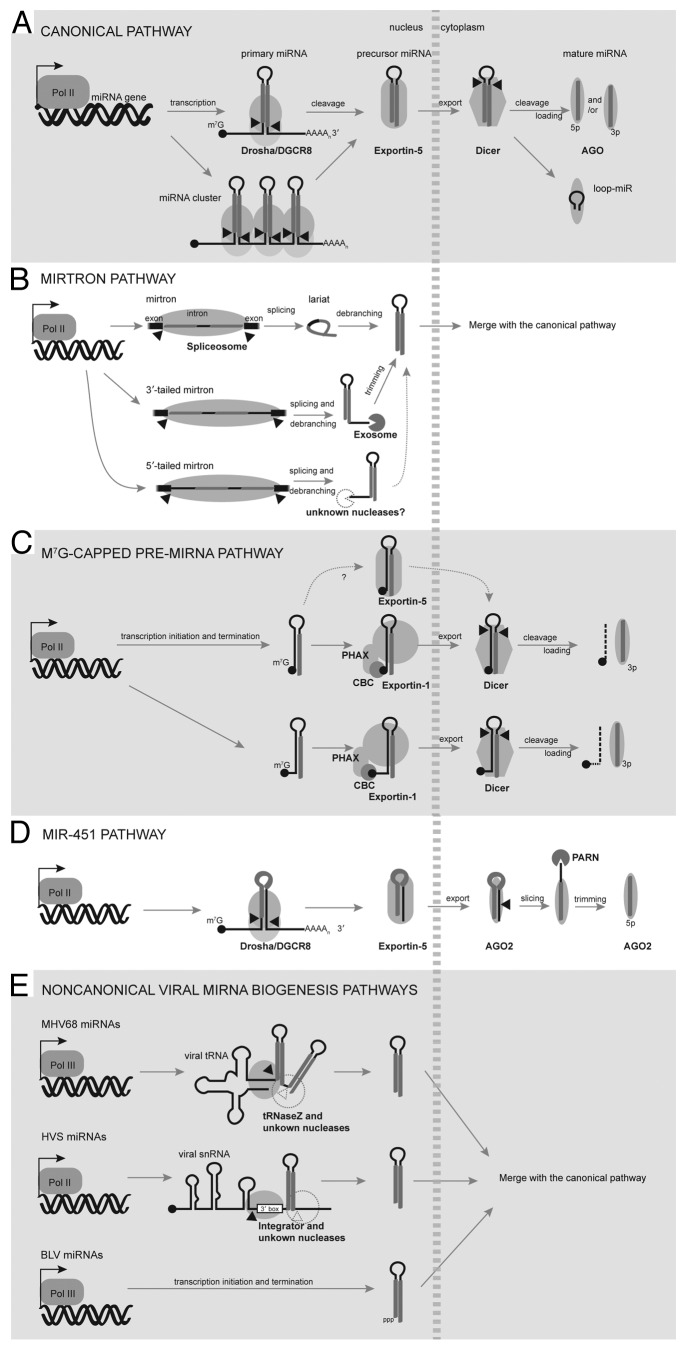
In metazoans, canonical miRNA biogenesis starts with the transcription of long primary (pri-)miRNA transcripts that contain one or a cluster of hairpin structures harboring mature miRNA sequences (Fig. 1A). Pri-miRNAs are typically transcribed by RNA polymerase (pol) II,12,13 with a few exceptions that may be transcribed by RNA pol III.14-16 Pol II transcription generally allows more elaborate regulation and tissue- or cell type-specific expression. Following Pol II transcription, pri-miRNAs are 5′ 7-methylguanosine (m7G)-capped and polyadenylated at their 3′ ends, similar to mRNAs. In fact, many miRNA hairpins are embedded in the introns or exons of mRNAs or other long non-coding (lnc) RNAs, conferring dual functions on these primary transcripts.17,18 Other miRNA hairpins are located in intergenic regions, driven by independent RNA pol II promoters.
Figure 1. MiRNA biogenesis pathways in animals and their viruses. (A) The canonical pathway. Pri-miRNAs containing a single or a cluster of hairpins are typically transcribed by Pol II, 5′-capped, and 3′-polyadenylated. The Microprocessor complex (Drosha and DGCR8) cleaves pri-miRNAs to release the pre-miRNA hairpins, which are exported by XPO5 and processed by Dicer into mature miRNA duplexes in the cytoplasm. One miRNA strand is preferably selected by AGO to form RISC. In certain cases, the loop of the pre-miRNA hairpin may be incorporated into RISC. (B) The mirtron pathway. Microprocessor-independent mirtrons are directly generated by splicing of short introns and the resulting lariats are debranched to form pre-miRNA hairpins. Mirtrons could have tails on either end of the pre-miRNA hairpins. 3′-tailed mirtrons are further trimmed by the exosome, while the nuclease that processes 5′-tailed mirtrons remains elusive. (C) The m7G-capped pre-miRNA pathway. M7G-capped pre-miRNAs are directly transcribed by Pol II, bypassing Microprocessor cleavage. The presence of the 5′ cap directs pre-miRNAs to the PHAX-XPO1 export pathway, known to function in the export of snRNAs. However, except for the 5′-tailed variants, the m7G-capped pre-miRNAs retain the ability to be exported by XPO5. After Dicer cleavage, the 5′-capped 5p-miRNA is unable to associate effectively with AGO, resulting in the production of only 3p-miRNPs. (D) The miR-451 pathway. Drosha cleavage of pri-miR-451 releases an unusual pre-miRNA that is too short to be processed by Dicer. Instead, AGO2 cleaves the 3p-arm of pre-miR-451 and PARN trims to generate a 5p-miRNA. (E) Drosha-independent miRNA biogenesis in animal viruses. MHV68 pri-miRNAs are tRNA-pre-miRNA chimeras that are processed by tRNaseZ at the 5′ end of the first pre-miRNA hairpin. The enzyme that separates the two pre-miRNAs is unknown. HVS pri-miRNAs are snRNA-pre-miRNA chimeras that are processed by Integrator to release the pre-miRNA. The 3′-end formation mechanism for HVS pre-miRNAs remains elusive. BLV miRNAs are derived from pre-miRNAs that are directly transcribed by Pol III as endogenous shRNAs. All viral pre-miRNAs described here are exported by XPO5 and processed by Dicer.
In the nucleus and often co-transcriptionally, pri-miRNA hairpins are recognized and cleaved by the Microprocessor complex, minimally composed of Drosha, an RNase III enzyme, and DGCR8 (DiGeorge critical region 8), an RNA binding protein that is also known as Pasha (Partner of Drosha) in invertebrates.19-23 The Microprocessor counts 10–11 base pairs from the base of the pri-miRNA stem-loop and cleaves to release the 55–70 nt pre-miRNA hairpin. Both the lower stem and the flanking single-stranded region are required for Drosha cleavage.24,25 The terminal loop of the pri-miRNA also influences Drosha cleavage in some cases.26,27 This cleavage yields a pre-miRNA with a 5′ monophosphate group and a 2-nt 3′-end overhang, characteristic of RNase III enzyme processing products.
Once produced, the pre-miRNAs are bound by Exportin-5 (XPO5) in the presence of its Ran-GTP co-factor and exported into the cytoplasm through the nuclear pore complex.28-30 The association between pre-miRNA and XPO5 requires recognition of the mini-helix structure and the 2-nt 3′-end overhang.29,31-33 Conversely, 5′ overhangs on pre-miRNAs are inhibitory for XPO5 binding, and hence, prevent pre-miRNA nuclear-cytoplasmic export.29,32 Interestingly, XPO5 also protects pre-miRNAs from nuclease attack, as knocking down XPO5 does not result in pre-miRNA accumulation in the nucleus.28,32 Structural work has confirmed an extensive interaction between pre-miRNA and XPO5 that would be expected to counteract nuclease digestion.33
Once in the cytoplasm, GTP hydrolysis leads to dissociation of the pre-miRNA from XPO5. Subsequently, another RNase III enzyme, Dicer, cleaves off the loop of the pre-miRNA hairpin to produce a ~22 bp mature miRNA duplex, with 2-nt 3′-end overhangs on both strands.34-37 Dicer protein recognizes structural features to ensure accurate and efficient cleavage: counting ~22 nt from both the 5′ monophosphate and the 3′-end overhang of the pre-miRNA hairpin.38-40 Recently, it was reported that the distance from the Dicer cleavage site to the pre-miRNA loop is also critical for accurate processing.41 Different from Drosha cleavage, human Dicer alone is sufficient to carry out accurate and efficient cleavage reactions in vitro. Although not essential for activity, RNA binding proteins including TRBP (TAR RNA binding protein) and PACT (protein activator of PKR) associate with Dicer in vivo, facilitating assembly of the miRNA into RISC (see below).42-44 In contrast, Drosophila Dicer-1 requires the RNA binding protein Loquacious (Loqs) for pre-miRNA processing.45-47
The miRNA duplex resulting from Dicer cleavage is incorporated into AGO proteins dependent on the RISC loading complex, which contains Dicer, AGO, and RNA binding proteins such as TRBP/PACT in humans and Loqs in Drosophila.42,43,48,49 In humans, there are four AGO proteins (AGO1–4), all of which can bind miRNAs and regulate gene expression. However, only AGO2 possesses catalytic slicer activity. Flies have two AGO proteins, with AGO1 mediating miRNA function.50 After the miRNA duplex is loaded, one strand is preferentially selected to form the functional miRNA–RISC complex (miRISC), while the other strand is displaced and degraded.51-53 The duplex strand with relatively weak base-pairing at its 5′ end is preferentially selected and designated as the guide strand or miRNA;54,55 the other strand is termed the passenger strand or miRNA*. However, because strand selection is not a stringent process, recent deep sequencing studies have identified substantial numbers of functional miRNA* strands in miRISCs.56-58 Moreover, since equal numbers of miRNAs can theoretically be derived from either arm of the miRNA duplex, it is difficult to distinguish miRNA from miRNA* and miRNAs are also named depending on their stem-loop arm of origin (miR-5p or -3p). Recently, it was demonstrated that the loop region of specific pre-miRNAs can also be incorporated into AGOs and direct target repression, suggesting that endogenous loading of AGO proteins does not occur exclusively via miRNA duplexes.59,60 Mutational analysis revealed that effective loop-miR loading is determined by a combination of features including the pre-miRNA stem and loop sequences, as well as length (~21 nt).59
The Mirtron Pathway
The first example to challenge the paradigm that all miRNAs are synthesized in the same way came from a class of Drosha-independent pre-miRNAs called “mirtrons” (pre-miRNAs/introns) in Drosophila and C. elegans (Fig. 1B).61,62 As their name implies, these pre-miRNAs are directly generated by the splicing machinery. Mutating the 5′ and 3′ splice sites abolishes both pre-miRNA and miRNA production from a mirtron,61,62 demonstrating a requirement for splicing. Importantly, mirtron hairpins are ~10 bp shorter than canonical pri-miRNA hairpins and can therefore bypass Drosha cleavage.62 Once an intron is spliced out by the spliceosome, it requires the debranching enzyme (Ldbr) to resolve the lariat and fold into the hairpin secondary structure needed for downstream steps in miRNA biogenesis. The pre-miRNA then undergoes XPO5-dependent export and merges with the canonical pathway.
It was hypothesized that Drosophila and C. elegans contain relatively short introns that are suitable for evolving of pre-miRNA-like molecules. Later, hundreds of mirtrons were found in the short introns of vertebrates and even rice genomes.63-69 Moreover, variant mirtrons that contain either a 5′- or 3′-end extension were discovered.61,64,65 In other words, pre-miRNA hairpins can reside at either end of an intron, with the 5′ or 3′ end the pre-miRNA hairpin directly generated by splicing. In the case of a 3′-tailed mirtron in Drosophila, the tail of miR-1017 is ~100 nt long and removed by the exosome, a major 3′-5′ exonuclease acting in RNA turnover in eukaryotic cells.70 In contrast, 5′-end extended mirtrons are predominantly found in vertebrates, with the identity of the nucleases that process this class of mirtrons into the pre-miRNA molecules remaining elusive. It is not yet known whether XRN-1 or XRN-2, the major 5′-3′ exonucleases, is responsible for this trimming. Curiously, one report indicates that two predicted human mirtrons (miR-1225 and 1228) are splicing-independent, despite the fact that both ends of the predicted pre-miRNAs match splice sites.71 These so called “simtrons” (splicing-independent mirtrons) are also independent of DGCR8, XPO5, Dicer, and AGO2 for their biogenesis, but the production of mature miRNAs is reduced upon overexpression of a dominant negative Drosha. Further investigation is required to define this pathway.
The m7G-capped Pre-miRNA Pathway
Another group of Drosha-independent pre-miRNAs is directly transcribed by RNA pol II and contains a m7G cap at its 5′ ends (Fig. 1C).9 Initial indications that these miRNAs might be derived directly from stand-alone pre-miRNAs came from their insensitivity to knockdown of core components of the Microprocessor complex (Drosha or DGCR8) in mouse cells.65,72 In addition, 5p-miRNAs derived from these pre-miRNAs are significantly underrepresented in miRNA sequencing (miRNA-seq) analyses, which require a 5′ monophosphate on the RNA molecule for inclusion in the miRNA-seq library. These results implied that the 5′ ends of these pre-miRNAs were generated by transcription initiation and lack 5′ monophosphate. Therefore, together with other pre-miRNAs whose ends are defined by transcription initiation/termination, these miRNA precursors were named endogenous short hairpin RNAs (endo-shRNAs), analogous to exogenously expressed shRNAs driven by the U6 (RNA pol III) promoter.65
We recently characterized a subset of endo-shRNAs, designated m7G-capped pre-miRNAs, which are directly transcribed by RNA pol II (Fig. 1C).9 Their cap structures are added and co-transcriptionally bound by the cap binding complex (CBC) and PHAX (phosphorylated adaptor for RNA export), which was previously assigned to the export of small nuclear RNAs (snRNAs).73 Therefore, like snRNAs, m7G-capped pre-miRNAs are exported by the Exportin-1 (XPO1) pathway. Interestingly, Xenopus oocyte microinjection assays showed that saturating amounts of m7G-capped pre-miRNAs not only inhibit XPO1-dependent U1 snRNA export, but also canonical pre-miRNA export, indicating that capped pre-miRNAs can alternatively interact with XPO5. Thus, if a particular m7G-capped pre-miRNA is not efficiently bound by CBC/PHAX, it could perhaps be exported by XPO5 through the canonical pathway. Another unique feature of m7G-capped pre-miRNAs is that they produce only 3p-miRNAs. This is due to the inefficient incorporation of an m7G-capped 5p-miRNA into AGO. Hence, devising Pol II-driven shRNA constructs to deliver single (3p-) small interfering RNAs (siRNAs) for targeted gene silencing should minimize the off-target effects of undesired 5p-siRNAs.
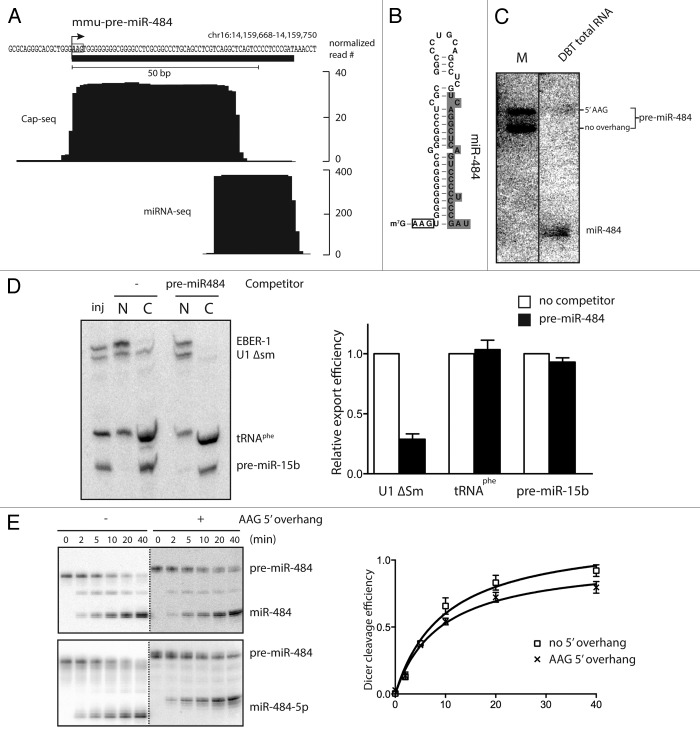
Analysis of small hairpin RNAs selected by the m7G cap-binding protein eIF4E (small RNA cap-seq) revealed that the 5′ ends of most m7G-capped pre-miRNAs map to the base of the hairpin. An exception is miR-484, whose pre-miRNA contains a 4-nt extension (counting the m7G cap, Fig. 2A and B). To confirm these extensions, we performed northern blot analysis of total RNAs extracted from the mouse astrocyte DBT cell line and detected endogenous pre-miR-484 migrating at the position of a 5′-extended hairpin marker (Fig. 2C). Several lines of evidence suggest that pre-miR-484 is exported and processed as a 5′-extended m7G-capped pre-miRNA. First, Xenopus oocyte injection confirmed that pre-miR-484 export is XPO1-dependent, as saturating amounts of pre-miR-484 inhibit XPO1-dependent U1 snRNA export (Fig. 2D). However, unlike m7G-capped pre-miRNAs that do not contain a 5′-tail, pre-miR-484 does not compete with canonical pre-miRNA export (Fig. 2D). This is consistent with the fact that a 5′-end extension on a pre-miRNA inhibits its binding to XPO5.29,32 Second, miR-484-5p is underrepresented in miRNA seq data, suggesting that it retains the m7G cap.65 Finally, the 5′-tailed pre-miR-484 is a Dicer substrate, albeit the dicing efficiency is slightly lower than that of a pre-miRNA hairpin without a 5′-end extension (Fig. 2E). Dicer cleavage generates a ~25-nt m7G-capped 5p miRNA, which is expected to be inefficiently bound by AGO proteins.
Figure 2. Mouse pre-miR-484 is a 5′-tailed m7G-capped pre-miRNA. (A) The histogram shows cap-seq reads mapping to the mmu-miR-484 locus. TSS and transcription directionality are indicated by a black arrow. The height of each bar is proportional to the normalized read number (raw read number per million mapped reads). Cap-seq and miRNA-seq data were taken from refs. 9 and 66, respectively. (B) Mouse pre-miR-484 is illustrated with miR-484 highlighted in gray. (C) Northern blot probed for miR-484 of 10 µg total RNA isolated from mouse astrocyte DBT cells. M, in vitro-transcribed m7G-capped pre-miR-484 markers with or without a 3-nt AAG 5′-end extension. (D) Xenopus oocyte microinjection assay. A mixture of 1–10 fmoles 32P-labeled EBER-1, U1∆Sm, tRNAᵖʰᵉ, and pre-miR-15b with or without 1 pmole of unlabeled pre-miR-484 (m7G-capped and containing 5′-AAG) was injected into the nuclei of X. laevis oocytes. After 2-h incubation, six oocytes were manually dissected, and RNAs from the nucleus (N) and cytoplasm (C) equivalent to one oocyte were extracted and analyzed on an 8M urea-15% polyacrylamide gel. inj, injected material. The bar graph shows relative RNA export efficiency (cytoplasmic/total) of the indicated RNAs with pre-miR-484 competition (black bars) compared with no competition (white bars). Error bars represent standard deviation from two experiments. (E) In vitro-transcribed pre-miR-484 with or without a 3-nt AAG 5′-end extension was incubated with recombinant human Dicer (a generous gift from Dr Jennifer Doudna) for various times and analyzed by northern blot to detect miR-484 (top panel) or miR-484-5p (lower panel), using 32P-labeled DNA oligos complementary to miR-484 or miR-484-5p, respectively. The blot shows cleavage of pre-miR-484 without the 5′-end extension (☐) or pre-miR-484 (x) by purified human Dicer at each time point. Error bars represent standard deviations in two experiments. The Materials and Methods used here were essentially the same as described in reference 9.
Interestingly, a new group of miRNAs derived from the transcription start sites (TSS-miRNAs) of protein coding genes has been identified recently.74 Similar to miRNAs derived from m7G-capped pre-miRNAs, the synthesis of TSS-miRNAs is Drosha/DGCR8-independent but Dicer-dependent, and 3′-end formation of the pre-miRNAs is linked to RNA pol II promoter proximal pausing. Although TSS-pre-miRNAs appear comparable to 5′-tailed m7G-capped pre-miRNAs, the capped 5′-end extension may be removed by exonuclease activities, allowing these intermediates to merge with the canonical biogenesis pathway. This model is supported by the fact that many TSS-5p-miRNAs contain 5′-monophosphates. What is not clear is whether all TSS-miRNA precursors share exactly the same start site with annotated protein-coding mRNAs. Because polynucleotide phosphorylase (PNPase) treatment in our small RNA cap-seq procedure preferentially selects shRNAs over mRNAs, it would be interesting to examine TSS-miRNA loci by small RNA cap-seq to see if a subset of TSS-miRNAs is derived from m7G-capped pre-miRNAs.
The miR-451 Pathway
To date, miR-451, an erythropoietic miRNA conserved in vertebrates, is the only Dicer-independent miRNA identified (Fig. 1D).75-77 Pri-miR-451 is cleaved by the Microprocessor as in the canonical pathway. However, the resulting pre-miRNA is only 17 base-pairs long and is therefore too short to be recognized by Dicer. Moreover, the mature miR-451 sequence includes the loop region of the pre-miRNA, arguing against a Dicer cleavage model. Indeed, Dicer knockout in either mouse ES cells or zebrafish diminished overall miRNA production, while levels of miR-451 remained unaffected.75,76 Intriguingly, AGO2 slicer activity is required for miR-451 biogenesis, as introduction of catalytically inactive AGO2 could rescue neither miR-451 expression in AGO2-knockout cells nor related developmental defects in AGO2-null animals.75,76 In lieu of Dicer cleavage, pre-miR-451 (after Drosha cleavage and XPO-5-mediated export) is directly loaded into AGO2, which cleaves between base pairs 10 and 11 on the 3p arm. The resulting ~30-nt AGO2-cleaved pre-miR-451 can then undergo poly(A)-specific ribonuclease (PARN)-mediated trimming of the remaining 3p arm, yielding a ~23 nt 5p-miRNA.78 However, trimming seems to be dispensable for target silencing in vivo, suggesting that a miRNA longer than 22 nt is functional in RISC. Similar to m7G-capped shRNAs, shRNA vectors designed to have features of the pre-miR-451 pathway could be used to deliver a single 5p-siRNA for gene silencing.
Drosha-Independent Pathways in Viruses
Given that miRNAs play fundamental roles in gene regulation, it is not surprising that many viruses express their own miRNAs to regulate host and viral genes during the viral life cycle. Like the majority of host miRNAs, most viral miRNAs utilize the canonical host pathway for biogenesis. However, some viruses are very creative in inventing their own ways of making miRNAs.
In two Herpesviruses, viral pre-miRNA hairpins reside directly downstream of another class of non-coding RNAs (ncRNAs) to form ncRNA-pre-miRNA chimeras (Fig. 1E). These chimeric pri-miRNAs are cleaved first by the 3′-end processing machinery of the ncRNA to separate the pre-miRNA from the ncRNA, replacing Drosha cleavage.79,80 In murine γ-herpesvirus 68 (MHV68), host tRNaseZ cleaves transfer RNA(tRNA)-pre-miRNA chimeras;79 in Herpesvirus saimiri (HVS), the host Integrator complex cleaves snRNA-pre-miRNA chimeras.80 In the MHV68 tRNA-pre-miRNA chimeras, there are usually two pre-miRNA hairpins, with the yield of miRNAs from the first hairpin being greater.79,81,82 The presence of a poly-U stretch at the end of the second pre-miRNA suggests that the 3′ end is defined by Pol III termination. However, the enzymatic activity that separates the two pre-miRNAs remains elusive. Similarly, the detailed 5′- and 3′-end formation mechanisms for HVS pre-miRNAs are uncharacterized: the Integrator complex potentially cleaves upstream of the 3′ box, a conserved sequence essential for snRNA 3′-end processing,83 but the resulting 5′-extended pre-miRNA may be trimmed by either the Integrator itself or other exonucleases. In both cases, viral pre-miRNAs are exported by XPO5 and undergo canonical miRNA biogenesis thereafter.79,80 Intriguingly, similar tRNA-pre-miRNA structures were identified in the mouse genome by computational analysis and confirmed by northern blotting, suggesting that the virus has acquired this unusual miRNA biogenesis pathway from the host.82 However, no snRNA-pre-miRNA chimeric RNAs have been identified in primates.
In a retrovirus, bovine leukemia virus (BLV), a subgenomic region produces several RNA pol III-dependent miRNAs (Fig. 1E).84 Such an arrangement avoids detrimental Drosha-mediated cleavage of the BLV genomic RNA. BLV pre-miRNAs appear to be directly transcribed as endo-shRNAs, mimicking U6 promoter-driven exogenous shRNAs. Such pre-miRNAs are structurally similar to canonical pre-miRNAs generated by Drosha cleavage and should be exported by XPO5 and cleaved by Dicer.
Conclusion and Perspectives
Here we have summarized the current understanding of both canonical and alternative miRNA biogenesis pathways that produce miRNAs from dedicated miRNA genes (Fig. 1). Although also dependent on Dicer action, the biogenesis of endogenous small interfering RNAs (endo-siRNAs), which are derived from long endogenous double-stranded RNAs or from long hairpin RNAs in Drosophila or mouse oocytes, are beyond the scope of this review.65,85-89 In addition, Dicer-dependent or -independent miRNA-like small RNAs are reported as being derived from other ncRNAs, such as snoRNAs, tRNAs, and vault RNAs, thereby bypassing Drosha cleavage.65,90-94
Although noncanonical miRNAs make up only a small fraction of the total miRNAs, their biogenesis pathways are conserved in different organisms. For example, the mirtron mechanism is found in almost all animals examined and one candidate was even found in rice, although not a single mirtron-derived mature miRNA is conserved in sequence across different metazoan lineages.63 The Dicer-independent pathway for generating the blood-specific miR-451 is conserved from zebrafish to human.75,76 Finally, m7G-capped pre-miRNAs can be found in all mammals.9 The conservation of mechanisms for the noncanonical miRNA pathways points to important biological significance. One possible advantage is to allow spatial and temporal expression of specific miRNAs in the absence of canonical miRNA biogenesis factors under certain conditions. It will be interesting to ask if there are consequences of forcing the production of noncanonical miRNAs through the canonical pathway.
Remarkably, Pol III- (U6) or Pol II- (U1) promoter driven exogenous shRNAs and tRNaseZ-dependent shRNAs were artificially constructed preceding the discovery of their natural counterparts.95-97 Recently, several other artificial miRNA biogenesis pathways have been devised, providing proof-of-principle evidence that yet additional pathways may exist in nature. For example, RNase III-independent miRNA biogenesis was achieved in mammalian cells by combining the tRNaseZ- or Integrator-dependent pathway with the AGO2-dependent pathway.98 Another example is the discovery that cytoplasmic RNA viruses, such as Sindbis virus and tick-born encephalitis virus, can be engineered to produce functional miRNAs.99,100 Artificially inserted pri-miRNAs are successfully processed despite the fact that these viruses replicate exclusively in the cytoplasm, perhaps suggesting a noncanonical miRNA processing pathway. Therefore, many other unexpected miRNA biogenesis mechanisms may be discovered in the years to come.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Xinguo Chen and Sandra Wolin for mouse astrocyte DBT cell line, Mingfeng Li for bioinformatics analysis, and Angela Miccinello for editorial assistance. We apologize to authors whose relevant primary publications were not cited due to space limitations. This work was supported by grants CA16038 and GM026154 from the National Institutes of Health. Xie M is a Fellow of the Leukemia and Lymphoma Society (Grant no. 5416-13). Steitz JA is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 4.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–93. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 9.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Šestan N, Steitz JA. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–80. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–99. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 15.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–21. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 20.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–48. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–97. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 30.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gwizdek C, Ossareh-Nazari B, Brownawell AM, Doglio A, Bertrand E, Macara IG, Dargemont C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J Biol Chem. 2003;278:5505–8. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- 32.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–85. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–9. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 34.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 35.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 36.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–71. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 40.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–5. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–11. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–9. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 49.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–90. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cenik ES, Zamore PD. Argonaute proteins. Curr Biol. 2011;21:R446–9. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 51.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 52.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 53.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–20. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 55.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 56.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–26. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–56. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev. 2013;27:778–92. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winter J, Link S, Witzigmann D, Hildenbrand C, Previti C, Diederichs S. Loop-miRs: active microRNAs generated from single-stranded loop regions. Nucleic Acids Res. 2013;41:5503–12. doi: 10.1093/nar/gkt251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–64. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 2012;40:438–48. doi: 10.1093/nar/gkr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012;22:1634–45. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–65. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–7. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–40. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–60. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–98. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 74.Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–34. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–8. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–26. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–42. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cazalla D, Xie M, Steitz JA. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell. 2011;43:982–92. doi: 10.1016/j.molcel.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diebel KW, Smith AL, van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA. 2010;16:170–85. doi: 10.1261/rna.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol. 2010;84:10344–53. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985;4:1827–37. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci U S A. 2012;109:3077–82. doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–7. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 86.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–6. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 89.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 91.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–60. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–71. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 95.Medina MF, Joshi S. RNA-polymerase III-driven expression cassettes in human gene therapy. Curr Opin Mol Ther. 1999;1:580–94. [PubMed] [Google Scholar]
- 96.Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–8. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Denti MA, Rosa A, Sthandier O, De Angelis FG, Bozzoni I. A new vector, based on the PolII promoter of the U1 snRNA gene, for the expression of siRNAs in mammalian cells. Mol Ther. 2004;10:191–9. doi: 10.1016/j.ymthe.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 98.Maurin T, Cazalla D, Yang S, Jr., Bortolamiol-Becet D, Lai EC. RNase III-independent microRNA biogenesis in mammalian cells. RNA. 2012;18:2166–73. doi: 10.1261/rna.036194.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–74. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rouha H, Thurner C, Mandl CW. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010;38:8328–37. doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]