Abstract
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression. Since the discovery of lin-4, the founding member of the miRNA family, over 360 miRNAs have been identified for Caenorhabditis elegans (C. elegans). Prediction and validation of targets are essential for elucidation of regulatory functions of these miRNAs. For C. elegans, crosslinking immunoprecipitation (CLIP) has been successfully performed for the identification of target mRNA sequences bound by Argonaute protein ALG-1. In addition, reliable annotation of the 3′ untranslated regions (3′ UTRs) as well as developmental stage-specific expression profiles for both miRNAs and 3′ UTR isoforms are available. By utilizing these data, we developed statistical models and bioinformatics tools for both transcriptome-scale and developmental stage-specific predictions of miRNA binding sites in C. elegans 3′ UTRs. In performance evaluation via cross validation on the ALG-1 CLIP data, the models were found to offer major improvements over established algorithms for predicting both seed sites and seedless sites. In particular, our top-ranked predictions have a substantially higher true positive rate, suggesting a much higher likelihood of positive experimental validation. A gene ontology analysis of stage-specific predictions suggests that miRNAs are involved in dynamic regulation of biological functions during C. elegans development. In particular, miRNAs preferentially target genes related to development, cell cycle, trafficking, and cell signaling processes. A database for both transcriptome-scale and stage-specific predictions and software for implementing the prediction models are available through the Sfold web server at http://sfold.wadsworth.org.
Keywords: microRNA, target binding site, prediction, GO analysis, developmental stage
Introduction
In animals and plants, microRNAs (miRNAs) are an abundant class of small endogenous non-coding RNAs (ncRNAs) of ~22 nucleotides (nts) in length. Since the discovery of the first miRNA lin-4 in Caenorhabditis elegans by the Ambros lab,1 tens of thousands of miRNAs have been identified and annotated in miRBase.2 miRNAs are post-transcriptional regulators of diverse developmental and physiological processes. For target recognition and regulation in animals, miRNAs guide the RNA-induced silencing complex (RISC) by binding to partially complementary sequences typically in the 3′ untranslated regions (3′ UTRs) of the mRNAs, leading to translational repression and/or mRNA destabilization.3
Identification of the targets for miRNAs is essential for understanding their regulatory functions. Genetic studies have identified numerous targets for some worm miRNAs; however, the regulatory functions for the majority of worm miRNAs are not yet understood. Computational predictions can be helpful for target elucidation. Most of the prediction algorithms have incorporated the seed rule, i.e., the target site within 3′ UTR forms Watson-Crick (WC) pairs with bases at positions 2 through 7 or 8 of the 5′ end of the miRNA.4 However, exceptions to the seed rule have been reported by both C. elegans and mammalian studies.5-12 Other proposed sequence features for enhancing targeting specificity include sequence conservation, strong base-pairing to the 3′ end of the miRNA, local AU content, and location of miRNA binding sites (near either end of the 3′ UTR is favorable).13 Furthermore, the importance of target structural accessibility for miRNA target recognition has also been demonstrated by several independent studies.14-18
In recent years, experimental target identification methods based on crosslinking immunoprecipitation (CLIP) and high-throughput sequencing have been reported for mammalian systems and C. elegans.19-21 CLIP methods involve UV irradiation for covalently crosslinking miRNA targets to the Argonaute (AGO) proteins, the catalytic components of the RISC complex. The crosslinked RNAs are treated by partial RNase digestion. The shortened RNAs are amplified by RT-PCR and then sequenced for the identification of AGO crosslinked sequences that contain miRNA binding sites. For C. elegans, CLIP-derived clusters (CDCs) of ~100 nts were identified for target binding sites of ALG-1, the AGO protein mainly responsible for miRNA function in worm. Importantly, the CLIP experiment was also performed for alg-1 genetic mutants so that the background noise in CDCs for wild-type worms could be removed. The CLIP technique not only provides high-resolution data with respect to the precise locations of the binding sites, but also is powerful for revealing the presence of “seedless” sites (non-canonical sites). In addition to data from ALG-1 CLIP, improved annotation has been established for C. elegans 3′ UTR isoforms expressed during different developmental stages, i.e., embryonic, L1, L2, L3, L4, adult hermaphrodite, and male.22,23 Moreover, developmental stage-specific expression profile of worm miRNAs has become available.24
In this work, we performed a comprehensive enrichment analysis of target site features for both seed and seedless sites identified from ALG-1 CDCs. We used enriched miRNA binding site features for the development of logistic models for prediction of miRNA binding sites. We assessed accuracy of predictions by cross validation and compared the performance with established algorithms. We used the models to make transcriptome-scale and developmental stage-specific predictions of miRNA binding sites in C. elegans. We performed a gene ontology (GO) analysis for stage-specific predictions to examine biological functions that are regulated by miRNAs during C. elegans development. For dissemination of the results, we have developed both database and software tools that are freely available to the scientific community.
Results
Identification of enriched target site features
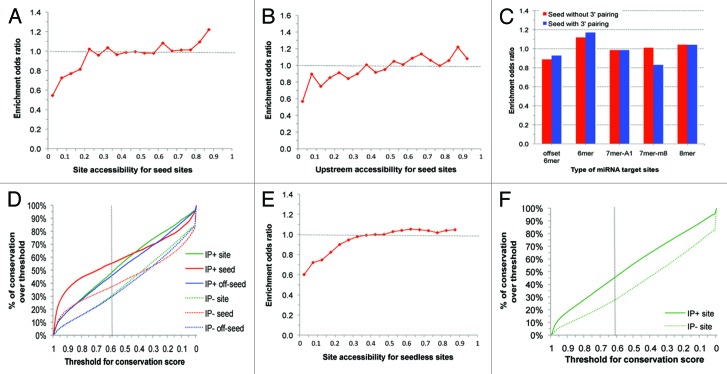
For each of the sequence, thermodynamic, and target structure features (Table 1), we performed enrichment analysis to identify features enriched in ALG-1 CDCs. Among the seed sites, 14 355 (11%) are within CDCs and referred to as the IP+ seed sites, the other 112 589 (89%) are referred to as the IP- seed sites, indicating that seed alone is a poor predictor with high false-positive rate. Features enriched for IP+ seed sites include site accessibility (Fig. 1A), upstream accessibility (window size of 10 nt, Fig. 1B), 6mer and 8mer seed (Fig. 1C), site conservation and seed conservation (Fig. 1D), seed accessibility, ΔGnucl, and ΔGhybrid. Among the seedless sites, 461 798 are in the IP+ set (within CDCs) and 3 820 416 are in the IP- set (outside CDCs). The enriched features for IP+ seedless sites include site accessibility (Fig. 1E), site conservation (Fig. 1F), upstream accessibility (10 nt), downstream accessibility (10 nt), 3′ base-pairing, ΔGnucl, and ΔGhybrid. These enriched features were used for the development of our logistic prediction models.
Table 1. Features computed for each potential miRNA binding site (seed or seedless)*.
| Type and name of feature | Description |
|---|---|
|
Sequence (1) Seed; (2) AU content; (3) Site location; (4) miRNA 3′ base pairing |
(1) Offset 6mer, 6mer, 7mer-A1, 7mer-m8 and 8mer seed sites;13 (2) percentage of AU for the block (of 5 nt, 10 nt,…, 30 nt) upstream or downstream of the binding site; (3) proximity to the ends of 3′ UTR; (4) presence of contiguous WC base pairing for miRNA nt positions 12–1713,27 |
|
Thermodynamic and target structure (1) ΔGhybrid; (2) ΔGnucl; (3) ΔGtotal; (4) Seed accessibility; (5) Site accessibility; (6) Upstream accessibility; (7) Downstream accessibility |
(1) ΔGhybrid is the measure of stability for miRNA:target hybrid as computed by RNAhybrid;26 (2) ΔGnucl measures the potential of nucleation for miRNA-target hybridization;18,36 (3) ΔGtotal measures the total energy change of the hybridization;17 (4–7) structural accessibility is evaluated by a probabilistic measure of single-strandedness for a block of nucleotides and is computed by using structures predicted by Sfold39,40 for the binding site, complementary seed region within the binding site, and for the block upstream or downstream of the binding site |
|
Conservation (1) Site conservation score; (2) Seed/off-seed conservation score |
The conservation score by the PhastCons program25 through multiple-sequence alignments of five other nematode genomes to the C. elegans genome (ce6) was used to measure conservation for a binding site, and the complementary seed and off-seed region within the binding site |
*The details of feature computation can be found in reference 34.
Figure 1. Enrichment of representative site features: (A) site accessibility for seed sites; (B) upstream accessibility (window size of 10 nt) for seed sites; (C) type of miRNA target seed sites; (D) percentage (Y-axis) of sites/seed/off-seed regions with conservation scores greater than or equal to a pre-specified threshold (X-axis), in the IP+ seed set or the IP- seed set (dashed line corresponding to a threshold of 0.57 previously used for defining conservation25,41); (E) site accessibility for seedless sites; (F) percentage (Y-axis) of seedless sites with conservation scores greater than or equal to a pre-specified threshold (X-axis), in the IP+ set or the IP- set.
miRNA binding site prediction and performance evaluation
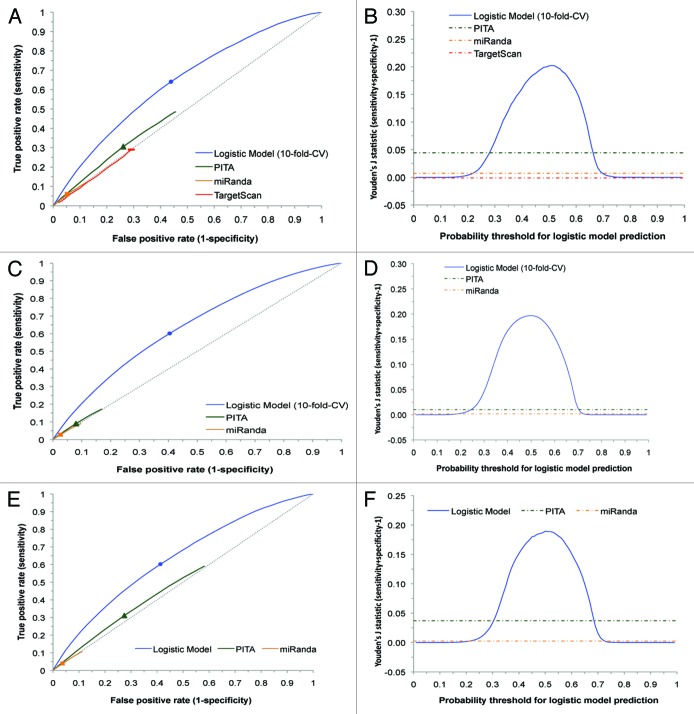
For performance evaluation, we constructed a receiver operator characteristic (ROC) curve for plotting the true positive rate (TPR = sensitivity) against the false positive rate (FPR = 1-specificity) by varying the threshold of a prediction score, e.g., logistic probability of our model, context score of TargetScan,27 energy score of miRanda,28 or PITA.16 The Youden’s J statistic29 computed by (TPR−FPR) was used as the overall measure of performance.
For seed sites in the 3′ UTRs, we compared our predictions with TargetScan, miRanda, and PITA. At a comparable FPR level, our logistic model has a substantially higher TPR than TargetScan, miRanda, and PITA (Fig. 2A). For a logistic probability threshold of 0.5, the improvement by the logistic model on Youden’s J statistic is about 0.15 over PITA, 0.19 over miRanda, and 0.20 over TargetScan (Fig. 2B). Among the three established algorithms, TargetScan is the worst performer. This is because TargetScan is primarily based on the predictions of 7mer and 8mer seed sites, and only 8mer is marginally enriched in the IP+ set (Fig. 1C).
Figure 2. Performance comparison of logistic models with three established algorithms for site predictions in 3′ UTRs (dashed diagonal line for random predictions). ROC curve and Youden’s J statistic are shown for the predictions of seed sites (A and B), seedless sites (C and D), and seedless site with one G•U pair or one mismatch within seed complementary region (E and F). The color-matched dots on ROC curves correspond to a logistic probability threshold of 0.5. The rectangle, triangle and square correspond to the best-performing score threshold (according to Youden’s J statistic) for TargetScan, PITA and miRanda, respectively.
For seedless site prediction, we only compared our model with PITA and miRanda, as TargetScan does not predict seedless sites. The predictions are similar to seed site predictions in the trends of receiver operating characteristic (ROC) curves and the Youden’s J statistic, with a higher degree of improvement (Fig. 2C and D). In particular, for the class of seedless sites with one G•U pair or one mismatch in the seed complementary region, our logistic model was also found to have major improvement over PITA and miRanda (Fig. 2E and F).
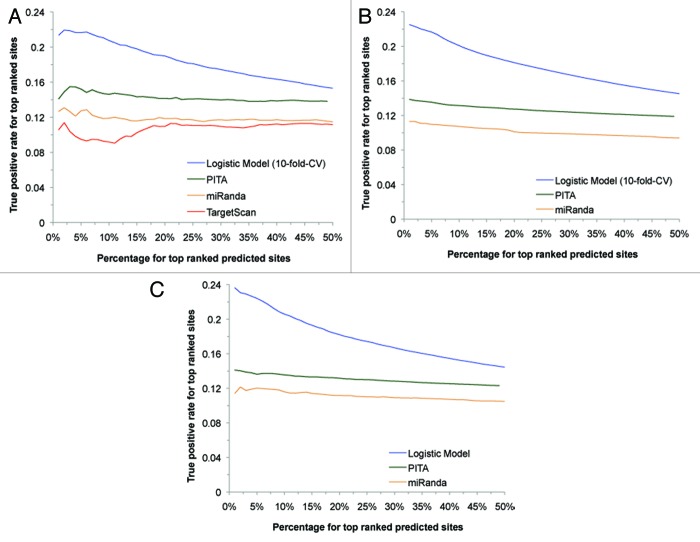
Since top-ranked predictions are of high interest for experimental validation, we compared the true positive rates of top-ranked predictions (top 1% to 50%) by our logistic models, TargetScan, PITA, and miRanda. For a given set of predicted top-ranked sites, the true positive rate is computed by the number of true positive sites (residing in ALG-1 CDCs) divided by the total number of the top-ranked sites. For seed sites in the 3′ UTRs, our logistic model was found to have a substantially higher true positive rate than PITA, miRanda, and TargetScan, especially for highly ranked predictions (Fig. 3A). E.g., for top 2% predictions, the true positive rate of the logistic model is about 7% higher than PITA, 9% higher than miRanda, and 11% higher than TargetScan. For either all seedless sites or the class of seedless sites with one G•U pair or one mismatch in the seed complementary region, we also observed substantial improvements by our logistic model over PITA and miRanda (Fig. 3B and C).
Figure 3. True positive rate comparison for top-ranked (from top 1% to top 50%) predictions of seed sites (A), seedless sites (B), seedless sites with one G•U pair or one mismatch in the seed complementary region (C) in 3′ UTRs.
Transcriptome-scale and stage-specific predictions
We applied our logistic models to transcriptome-scale predictions of miRNA binding sites in C. elegans. This included 368 miRNAs in miRBase Release 192 and 24 503 3′ UTR isoforms.22 We predicted 429 072 seed sites and 14 921 597 seedless sites. Each of these sites either resides within a CDC or has a logistic probability above 0.5. The miRNAs and the 3′ UTR isoforms are expressed in different developmental stages, i.e., embryonic, L1, L2, L3, L4, adult hermaphrodite, and male. Accordingly, we further processed transcriptome-scale predictions for stage-specific predictions by collecting predicted sites for co-expressed miRNA:3′ UTR pairs in each of the seven stages.
Stage-specific patterns of miRNA:target interactions during development
We next investigated the stage-specific patterns of miRNA:target interactions during C. elegans development. We first defined a positive miRNA:target interaction if the miRNA:target pair has at least one seed site with a logistic probability above 0.5, or one seedless site with a probability above 0.6. From all of positive miRNA:target interactions for each developmental stage, we assembled the set of the targeted genes and the set of their regulating miRNAs. For a more stringent definition of a positive interaction, we also used a probability of above 0.6 for a seed site and above 0.7 for a seedless site. For either definition, every abundant miRNA for each of the seven stages has at least one target. Among all of the 119 miRNAs expressed in at least one stage, 84 (~70.6%) are shared by all the seven stages, which is consistent with the previous observation that most miRNAs are present at steady-state levels during C. elegans development.30 For probabilities of 0.5 and 0.6, we identified 4583, 3895, 4540, 4143, 4330, 2086, and 3741 target genes for embryonic, L1, L2, L3, L4, adult hermaphrodite, and male stages, respectively. Among 7745 genes targeted by miRNAs in at least one stage, 1062 (~13.7%) are shared by all seven stages. For probabilities of 0.6 and 0.7, we identified 2736, 2379, 2869, 2577, 2625, 1205, and 2133 target genes for embryonic, L1, L2, L3, L4, adult hermaphrodite, and male stages, respectively. Among 4633 genes targeted by miRNAs in at least one stage, 644 (~13.9%) are shared by all seven stages. The high number of common targets is consistent with the previous observation that many miRNA targets seem to be stably and continuously regulated during C. elegans development.31
For each stage, using the common targets, we identified the set of remaining targets that were not shared by all seven stages. To explore functional themes among the common target genes and the remaining target gene sets, we searched for enriched GO annotations, focusing on GO terms with P value (by hyper-geometric distribution) under 0.01 and percentage (number of genes associated with the GO term divided by total number of genes of interest) above 0.01 (Tables S1 and S2).
Analysis of targets common to all seven stages
For the two definitions of positive interactions, the enriched GO terms for the common target gene set are highly overlapped so that the key findings are the same as summarized below. For biological processes, the most enriched GO terms are related to development, cell cycle, or trafficking, e.g., nematode larval development, embryonic development ending in birth or egg hatching, growth, positive regulation of growth rate, body morphogenesis, locomotion, and positive regulation of locomotion. For molecular functions, the most enriched GO terms are related to protein–nucleic acid and protein–protein interactions in cell signaling processes, e.g., structural constituent of ribosome, nucleotide binding, GTPase activity, and ATP binding. This is consistent with a previous conclusion that miRNAs preferentially target genes involved in signaling processes during C. elegans development.31 For cellular components, the most enriched GO terms are related to cytopoiesis and protein synthesis, e.g., cytoplasm, intracellular, ribosome, and ribonucleoprotein complex.
Analysis of remaining targets for each of seven stages
For the two definitions of positive interactions, the enriched GO terms for the remaining target set for each of the seven stages are largely overlapped such that we have the same conclusions below. For the remaining target gene sets, temporal pattern transition of miRNA:target interactions is evident from the enriched GO terms of each stage (Tables S1 and S2), further supporting that the miRNA-mediated regulation network is highly dynamic during C. elegans development.31 Interestingly, the L1 and male stages have substantially fewer enriched GO terms than other stages, and a large portion of these terms are not enriched in the preceding or following stage. This suggests that the embryo to L1, the L1 to L2, and the L4 to male transitions involve substantial changes in miRNA-mediated regulation of gene expression. This conclusion for the embryo to L1 and the L1 to L2 transition is consistent with a previous insight suggested by stage-specific miRNA expression profiles.31 Although a majority of enriched GO terms are preserved over the L2 to L3, the L3 to L4, and the L4 to adult hermaphrodite transition, a minority of enriched GO terms vary in these transitions, thereby indicating dynamic temporal patterns of miRNA:target interactions during development. The contrast between substantial changes in the L4 to male transition and the relatively minor changes in the L4 to adult hermaphrodite transition indicates that genes related to hermaphrodite genitalia development and sex differentiation are under strong miRNA regulation, e.g., those genes associated with the enriched GO term of negative regulation of vulval development (Table S1). Moreover, for the remaining target gene set for each of the seven stages, enriched GO terms are mainly for biological processes and rarely for cellular components (Tables S1 and S2). The GO terms for biological processes are largely related to development, cell cycle, or trafficking, and the GO terms for molecular function are largely related to protein–nucleic acid and protein–protein interactions in cell signaling processes. These observations are the same as the common target gene set.
Database and software tools
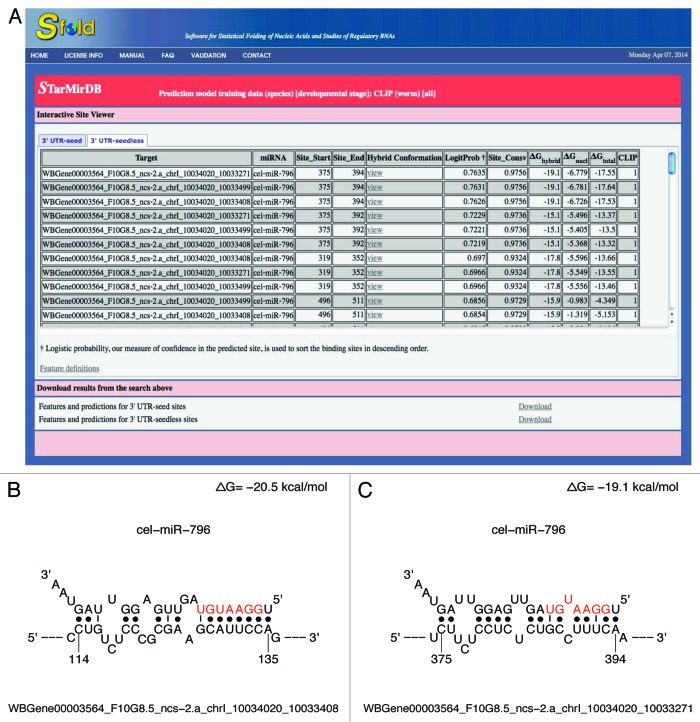
For dissemination of the results, transcriptome-scale and stage-specific predictions are freely available from STarMirDB (http://sfold.wadsworth.org/starmirDB.php), a web searchable database, with an indicator showing whether a site is supported by the ALG-1 CLIP study. We have also implemented the prediction models into the STarMir module of the Sfold web applications37 (http://sfold.wadsworth.org/cgi-bin/starmirWeb.pl), allowing users to submit any miRNA and mRNA sequences for prediction of miRNA binding sites by the models. Information for predicted sites includes site features, a logistic probability as a measure of confidence, and a high-resolution diagram of hybrid conformation. As an illustration of the output from a database search, using cel-miR-796 and ncs-2 (or WBGene00003564) as search keywords returns a list of seed sites and seedless sites for miR-796 on different ncs-2 3′ UTR isoforms for various developmental stages (Fig. 4A). For example, for isoform WBGene00003564_F10G8.5_ncs-2.a_chrI_10034020_10033271, one 7mer-A1 seed site, and one seedless site have high conservation scores of 0.9658 and 0.9756, and logistic probabilities of 0.67 and 0.76, respectively. The hybrid diagrams for two example sites are shown in Figure 4B and C, where the seed region (nt 2–8) of the miRNA is shown in red color.
Figure 4. (A) STarMirDB search result for binding sites of cel-miR-796 on ncs-2 3′ UTR isoforms, with “3′ UTR-seedless” option selected for output display; (B) hybrid diagram of a seed site; (C) hybrid diagram of a seedless site.
Discussion
Among various types of seed sites, mammalian miRNA targeting studies13,27,32,33 established that 8mer seed is the most effective, followed by 7mer-m8, 7mer-A1, 6mer, and offset 6mer. Our analysis of five mammalian CLIP data sets also supported the effectiveness of 8mer and 7mer seed sites.34 In contrast, from our enrichment analysis of the worm ALG-1 CLIP data, 6mer was found to be the only substantially enriched seed type (Fig. 1C). This is a somewhat surprising finding, however, it is consistent with two observations on miRNA:target interactions in C. elegans. For numerous genetically verified targets, e.g., lin-41, RAS, and pha-4 targeted by let-7,6,9,10 and lin-14 targeted by lin-4,35 a perfect seed match is absent at the functional sites of interaction. In a study on the interaction between lsy-6 and its target cog-1, it was shown by single nucleotide mutations that G•U base-pairing in the seed region could be well tolerated. The miRNA machinery in C. elegans may be more tolerant of G•U base-pairing and mismatches in the seed region. Furthermore, the number of enriched features and the enrichment signals are generally less than those observed in our mammalian study. These necessitate development of prediction models specific for C. elegans.
Our prediction approach computes a logistic probability as a measure of confidence for predictions. The predicted sites with higher probabilities are expected to have a greater chance for positive experimental validation. In particular, the top-ranked predictions by our approach have higher likelihood to be positive than the top-ranked predictions of the established algorithms (Fig. 3). Our approach is a successful application of our previous methodology34 to C. elegans ALG-1 CLIP data, and was shown to provide new models specifically for improved predictions of miRNA binding sites in C. elegans.
The ALG-1 CLIP data are mainly for the L4 stage. However, because the miRNA machinery is expected to be the same throughout the C. elegans development, rules on miRNA targeting learned from the L4 stage can be generalized to other stages for binding site predictions. Stage-specific CLIP data would be ideal, however, such data are not available.
Our GO analysis revealed a dynamic stage-dependent pattern of miRNA:target interactions. In addition to significant changes at the embryo to L1 and the L1 to L2 transitions that confirm a previous observation,31 we observed a significant change at the L4 to male transition and relatively minor changes at the L2 to L3, the L3 to L4, and the L4 to adult hermaphrodite transitions. These further support the previous conclusion that miRNA-mediated regulation is stable and continuous via coordinately targeting or avoiding genes involved in certain biological functions during C. elegans development.31 Furthermore, we observed that genes under strong miRNA regulation during development include those related to biological processes of development, cell cycle, trafficking, and sex differentiation, and those related to molecular functions of protein–nucleic acid and protein–protein interactions in cell signaling processes. The observation on cell signaling processes further supports a previous conclusion.31
For C. elegans, currently the only available CLIP data set is from the ALG-1 study.19 Therefore, model testing by other independent C. elegans CLIP data are not yet possible. Nevertheless, the cross validation strategy enables an assessment of the generalizability of our model to other independent data sets. The CLIP technique provides information on miRNA binding sites, but not functional outcomes (e.g., readout from a reporter) due to miRNA binding. Accordingly, our CLIP-derived models are limited to predictions of miRNA binding sites. Extension of the models for predicting functional outcomes will require analysis and modeling of high-throughput functional data.
In conclusion, we have performed a comprehensive enrichment analysis of data from the ALG-1 CLIP study. The findings allowed us to develop new models for the prediction of worm miRNA binding sites. The advantage of our approach is the utilization of recent experimental data for ALG-1 CLIP, 3′ UTR annotation, and stage-specific expression of miRNAs and mRNAs. In performance evaluation via cross validation on the ALG-1 CLIP data, the models were found to offer major improvements over established algorithms. For future studies, independent CLIP data (preferably for multiple C. elegans developmental stages) would allow inter-data set validation. Extensive experimental testing of model predictions would be needed to further assess predictive improvements. Useful information from a CLIP study is limited to abundantly expressed miRNAs and transcripts in the experimental system. Our analysis of ALG-1 CLIP data included 113 abundant miRNAs and 3093 genes in worm. The prediction models enabled transcriptome-scale predictions for 368 miRNAs and 24 503 3′ UTR isoforms, as well as stage-specific predictions for seven C. elegans developmental stages. The stage-specific GO analysis revealed that miRNAs regulate diverse biological functions during C. elegans development. The software and database tools will greatly complement the ALG-1 CLIP data for studies of miRNA regulation in C. elegans.
Materials and Methods
Description and processing of C. elegans CLIP data
The C. elegans CLIP study identified about 4800 CDCs that are unique to the wild-type (WT) worms at the fourth larval stage (L4 stage), in comparison to clusters from ALG-1 genetic mutant worms.19 These CDCs represent 3093 genes, about 20% of protein-coding genes expressed in L4 stage. Based on the annotation from a previous study38 and the NCBI map viewer for ce6 genome, we compiled 13 356 3′ UTR isoform sequences (length ≥ 30 nts) for all expressed transcripts in the CLIP experiment. The 113 most abundant miRNAs (sequencing read number ≥ 10) for the WT worms were used in our analysis.
Overview of framework for CLIP data analysis, prediction model training, and testing
For the development of new prediction models specific for worm using the C. elegans ALG-1 CLIP data, we adopted the core methodology that was found to be successful in our recent mammalian study.34 First, the potential miRNA binding sites were predicted by RNAhybrid program.26 Those sites residing within an ALG-1 CDC were classified as the IP+ sites, while the remaining sites were categorized as the IP- sites. Second, the site features described in Table 1 were computed for each seed or seedless site. For each feature, the degree of enrichment was measured by the ratio of the odds of occurrence in the IP+ set to that of IP- set. A ratio above one indicates enrichment of the feature, whereas a ratio below one indicates depletion. Enriched features were considered important for miRNA targeting in worm, and were used for the development of prediction models, one for seed sites and one for seedless sites. Third, nonlinear logistic regression was used for model development. The logistic model outputs a probability as a measure of confidence for a predicted site. Finally, the standard 10-fold cross validation strategy was used to test the performance of our prediction models on the ALG-1 CLIP data. According to this strategy, the data set was randomly divided into 10 subsets of equal size. During each of the 10 iterations, one subset was set aside for performance testing of the model trained on the collection of the other nine subsets. This model-training-testing process was repeated 10 times, and the mean prediction accuracy is averaged over the 10 iterations. Although currently only one CLIP data set is available for C. elegans, the cross validation strategy enables an assessment of the generalizability of our model to other independent data sets. TargetScan,27 PITA,16 and miRanda,28 which can be run locally with available software, were used for comparison. The computational procedures here are essentially the same as previously used.34
Transcriptome prediction
We used our models for transcriptome-scale predictions of worm miRNA binding sites. To this end, we included all 368 worm miRNAs in miRBase Release 19,2 and considered the recently annotated 3′ UTRs.22 We compiled 24 503 3′ UTR isoforms (sequence length ≥ 30 nts) for ~13 830 genes in WormBase WS190. These 3′ UTR isoforms are frequently and differentially expressed during different C. elegans developmental stages.
Stage-specific prediction and GO analysis
From the 24 503 3′ UTR isoforms with stage-specific expression information, we assembled 8806 isoforms for the embryonic stage, 6568 for the L1 stage, 7341 for the L2 stage, 6495 for the L3 stage, 7049 for the L4 stage, 3213 for the adult hermaphrodite stage, and 6738 for the male stage. Among the 368 worm miRNAs, we assembled 97 abundant miRNAs for the embryonic stage, 97 for the L1 stage, 101 for the L2 stage, 99 for the L3 stage, 100 for the L4 stage, 101 for the adult hermaphrodite stage, and 109 for the male stage. A miRNA is considered abundant if its read number is at least 10 from the sequencing analysis.24 For each developmental stage, we assembled stage-specific predictions for co-expressing miRNAs and mRNA isoforms, and performed GO analysis. The GO association file was downloaded from WormBase FTP with version WS190.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The Computational Molecular Biology and Statistics Core at the Wadsworth Center is acknowledged for supporting computing resources for this work. The authors thank Marco Mangone and the lab of Gene W Yeo for providing additional data. This work was supported in part by National Science Foundation (DBI-0650991 to Ding Y), National Institutes of Health grant (GM099811 to Ding Y), and Nafosted Fund of Vietnam (102.03-2010.04 to Long D).
Author Contributions
Ding Y conceived and supervised the study. Liu C and Mallick B complied and processed the data from C. elegans studies on ALG-1 CLIP, 3′ UTR annotation and stage-specific expression of miRNAs and 3′ UTR isoforms, and performed feature enrichment analysis. Liu C developed prediction models, performed model validation, performance comparison, and GO analysis. Rennie W performed both database and software implementation. Kanoria S performed testing of database interface. Long D and Wolenc A wrote the initial software for the computation of several target site features, and Carmack C provided hardware and system support for both cluster computing and the Sfold web server. Liu C and Ding Y wrote the paper with contributions from all authors. All authors read and approved the final manuscript.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–93. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 6.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 2004;18:132–7. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–51. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 8.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–30. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell. 2009;35:610–25. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods. 2013;10:253–5. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 15.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci U S A. 2005;102:4006–9. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 17.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–94. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 18.Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–9. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–9. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3’UTRs. Science. 2010;329:432–5. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato M, de Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Hammell M, Kudlow BA, Ambros V, Han M. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development. 2009;136:3043–55. doi: 10.1242/dev.039008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Mallick B, Long D, Rennie WA, Wolenc A, Carmack CS, Ding Y. CLIP-based prediction of mammalian microRNA binding sites. Nucleic Acids Res. 2013;41:e138. doi: 10.1093/nar/gkt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–50. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 36.Long D, Chan CY, Ding Y. Analysis of microRNA-target interactions by a target structure based hybridization model. Pacific Symposium on Biocomputing 2008; 13:64-74. [PubMed] [Google Scholar]
- 37.Rennie WA, Liu C, Carmack CS, Wolenc A, Kanoria S, Lu J, Long D, Ding Y. STarMir: a web server for prediction of microRNA binding sites. Nucleic Acids Res. 2014 doi: 10.1016/j.cell.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–66. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Y, Lawrence CE. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 2001;29:1034–46. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.