Abstract
In trypanosomes, mRNAs are processed by trans-splicing; in this process, a common exon, the spliced leader, is added to all mRNAs from a small RNA donor, the spliced leader RNA (SL RNA). However, little is known regarding how this process is regulated. In this study, we investigated the function of two serine-arginine-rich proteins, TSR1 and TSR1IP, implicated in trans-splicing in Trypanosoma brucei. Depletion of these factors by RNAi suggested their role in both cis- and trans-splicing. Microarray was used to examine the transcriptome of the silenced cells. The level of hundreds of mRNAs was changed, suggesting that these proteins have a role in regulating only a subset of T. brucei mRNAs. Mass-spectrometry analyses of complexes associated with these proteins suggest that these factors function in mRNA stability, translation, and rRNA processing. We further demonstrate changes in the stability of mRNA as a result of depletion of the two TSR proteins. In addition, rRNA defects were observed under the depletion of U2AF35, TSR1, and TSR1IP, but not SF1, suggesting involvement of SR proteins in rRNA processing.
Keywords: trypanosomes, trans-splicing, SR proteins, rRNA processing, mRNA stability, proteomics of splicing complexes
Introduction
Trypanosomes are parasitic protozoa causing infamous diseases such as African sleeping sickness (Trypanosoma brucei), Leishmaniasis, and Chagas’ disease or American trypanosomiasis (Trypanosoma cruzi). In addition, this family of organisms serves as an important model system to study the role of post-transcriptional regulation. These parasites lack conventional RNA polymerase II promoters for protein coding genes. However, histone modification was shown to play a role in the regulation of gene expression of Trypanosoma brucei.1 In these parasites, genes are transcribed as polycistronic mRNAs that are processed by concerted action of trans-splicing and polyadenylation. These processes are coupled, and perturbation of splicing signals affects the polyadenylation of the upstream gene.2-5
In trans-splicing, a common spliced leader (SL) is added to all mRNAs from a small RNA donor, the SL RNA.6-8 Several recent studies shed light on the contribution of trans-splicing and polyadenylation to the control of global gene expression, and identified alternative processing of transcripts at either their 5′ end, or in a larger number of cases, at their 3′ end.9,10 Despite these additional pathways, the most robust mechanism regulating the trypanosome transcriptome is mRNA stability.11-13
Currently, little is known regarding factors that regulate trans-splicing.13 Several RNA binding proteins that were shown to participate in splicing regulation in metazoa also exist in trypanosomes. Among these, are heterogeneous nuclear ribonucleoprotein (hnRNP) and SR proteins carrying a serine arginine motif, termed the RS domain.6-8 Polypyrimidine tract binding proteins (PTB) or hnRNP I homologs were shown to be required for trans-splicing of mRNAs carrying a C-rich polypyrimidine tract.14 The PTB proteins were also shown to regulate mRNA stability.14,15
SR proteins function in metazoa in the constitutive splicing process, but also modulate alternative splicing.16 A genome-wide survey in metazoa identified a large number of proteins carrying serine-arginine (RS) domains present in SR proteins. The SR proteins have a modular structure containing one or two copies of an RNA recognition motif (RRM) at the N terminus that provides RNA binding specificity, and a C-terminal RS domain that acts to promote protein–protein interactions that facilitate recruitment to the spliceosome.17,18 SR proteins were identified among the hundreds of proteins present in the RNA polymerase II transcription complex, and are often loaded co-transcriptionally and accompany the fully spliced mRNA to the cytoplasm.19
Since splice site consensus sequences are not sufficient to direct assembly of the spliceosome, sequences present in exons or introns such as exonic and intronic splicing enhancers (ESE and ISE), or exonic and intronic splicing silencers (ESS or ISS), are used to bind factors that regulate spliceosome assembly. SR proteins stabilize interactions between the U1 snRNP at the 5′ splice site and U2AF65 at the 3′ splice site. SR proteins are also known to bind to ESE and antagonize the activity of hnRNP proteins recognizing ESS.20 In metazoa such as C. elegans, several SR proteins are essential, but others are not.21 In mouse, many SR proteins are essential for life.22
SR proteins have other functions in addition to their role in splicing, such as nuclear export, non-sense-mediated decay, and translation. SR proteins affect translation directly and indirectly. SF2/ASF was shown to associate with polyribosomes and to enhance translation, probably via release of 4E-BP, a competitive inhibitor of cap-dependent translation.17
Recent studies support the role of SR proteins not only as splicing regulators, but also implicate these proteins in genome stability, chromatin binding, transcription elongation, mRNA stability, mRNA export, and translation (see review23).
The function of SR proteins is regulated by phosphorylation and de-phosphorylation. The RS domain is extensively phosphorylated on serine residues and this modification controls the localization of the protein. Mammalian SR proteins become dephosphorylated during the course of pre-mRNA processing, and promote mRNP transit through the nuclear pore complex.24 SR proteins associate with the exon-junction complex (EJC) and this interaction leads to RNP compaction.25 SR proteins were also recently shown to control, together with non-coding RNAs, the formation of nuclear “speckles;” these domains contain pre-mRNA processing factors and non-coding RNAs.26
The U2 auxiliary factors U2AF35 and U2AF65 also belong to the family of proteins carrying RS domains. Most recently, we demonstrated that the trypanosome homologs of these proteins not only function in splicing, but also in mRNA stabilization.27 U2AF35 was also shown to bind to ribosomes and to associate with factors involved in rRNA processing and ribosome assembly.27
Three SR proteins were identified in T. brucei, TSR1, TSR1IP, and RRM1.28-30 TSR1 contains an RS domain at its C terminus and two RRM domains at its N terminus. Using a three-hybrid system, it was demonstrated that TSR1 can bind to SL RNA in yeast.
TSR1IP was shown to interact with TSR1 by two-hybrid screen. The protein shares homology with the U1 70 kDa protein, but it is not the U1 70 kDa homolog, since a closer bona fide homolog was subsequently identified.31 TSR1IP, however, contains a domain present in U1 70 kDa of many eukaryotes that interacts with poly (A) polymerase and inhibits its activity. None of the functional experiments described above were performed in trypanosomes, and thus, the role of these proteins remains elusive.
In this study, we silenced the expression of TSR1 and TSR1IP by RNAi, and observed that these proteins affect the transcriptome not only by affecting splicing but also by controlling mRNA stability. The proteins are found mainly in nuclear speckles, but biochemical fractionation also detected the two proteins on polyribosomes. Purification of the proteins associated with these factors in Leishmania tarentolae demonstrated the co-purification not only of splicing factors and proteins involved in mRNA stability, but also of ribosomal proteins and proteins involved in rRNA processing. Indeed, silencing of TSR1 and TSR1IP as well as U2AF35 but not SF1 resulted in rRNA processing defects. No effect on rRNA transcription was observed in the TSR1 and TSR1IP silenced cells, though TSR1IP is also essential for transcription of long polymerase II transcripts. Thus, we provide evidence for the role of TSR1 and TSR1IP not only in splicing regulation but also in mRNA stability and rRNA processing. Although evidence is provided for the effect of the two TSR proteins on these cellular functions, we cannot rule out the possibility that the phenotype observed under depletion may result, in part, from secondary effects. This is especially relevant for the rRNA processing defects observed under silencing of these factors, since the defects observed may also result from effects these factors exert on the production and level of mRNA encoding for factors involved in rRNA processing. Nevertheless, the results presented in this study support our recent experiments demonstrating a robust role of splicing factors in mRNA stability, which is so far the dominant mechanism controlling mRNA abundance in these organisms.
Results
TSR1 and TSR1IP are essential splicing factors whose depletion affects both cis- and trans-splicing
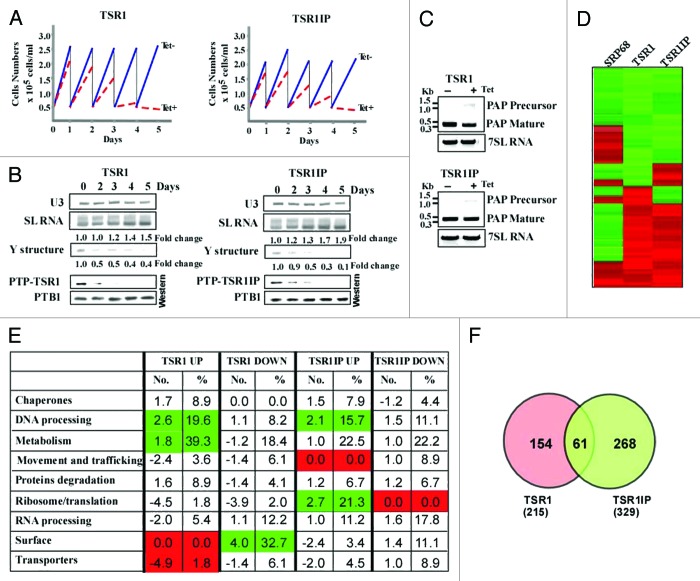
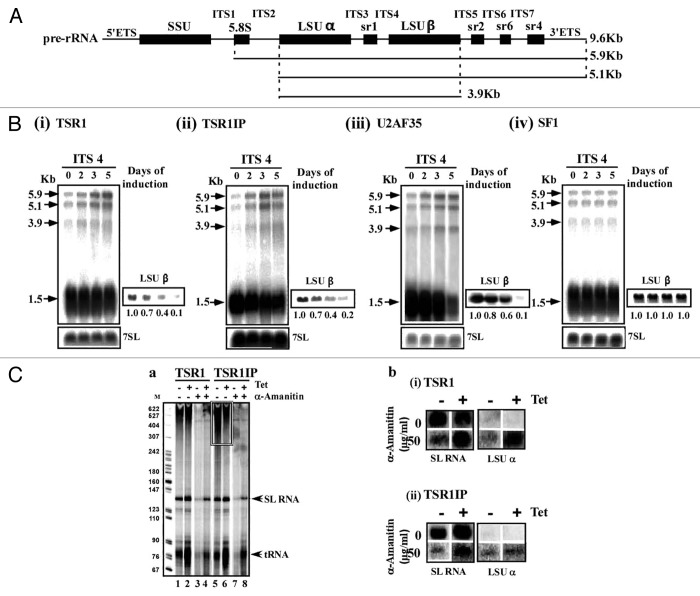
The T. brucei TSR1 and TSR1IP factors29,30 were described more than 10 y ago. However, these studies did not address the role of these proteins in vivo. To examine the role of TSR1 and TSR1IP in splicing, the genes were conditionally silenced using stem-loop constructs. The two factors are essential for life, since their silencing results in growth arrest (Fig. 1A). To evaluate the efficiency of silencing, the cells carrying the silencing constructs were tagged with the tandem affinity purification (TAP)-C epitope/TEV protease cleavage site/protein A tag known as PTP, at the authentic sites.32 We observed efficient silencing as early as the second day of induction, and complete elimination of the protein on the third day (Fig. 1B).
Figure 1. TSR1 and TSR1IP are required for trans- and cis-splicing. (A) Growth curves of T. brucei cells silenced for TSR1 and TSR1IP. Growth of uninduced cells was compared with growth after tetracycline addition. Both uninduced and induced cultures were diluted daily to 5 × 104 cells per ml. (B) TSR1 and TSR1IP silencing affects trans- splicing. Cells expressing either the PTP-TSR1 and TSR1 silencing constructs, or PTP-TSR1IP and TSR1IP silencing constructs were silenced for the number of days indicated. Upper panel: Total RNA (10 µg) from the same cells was subjected to primer extension with an oligonucleotide complementary to the intron region of the SL RNA (S-1). Primer extension of U3 was used to determine the amount of RNA used. The products were separated on a 6% acrylamide denaturing gel. The results were quantified using ImageJ. The levels of SL RNA and Y structure intermediate are given as fold change with respect to the amount present at day 0, and were normalized to the level of U3 snoRNA. Lower panel: Proteins (from 107 cells) were extracted from the silenced cells at the time points indicated, separated on a 10% SDS polyacrylamide gel, and subjected to western analysis. Reactivity with PTB1 antibodies was used as a control for equal loading. (C) cis-splicing of PAP in TSR1 and TSR1IP silenced cells. RNA was prepared from uninduced cells, or after 2 d of induction. cDNA was prepared, and subjected to PCR amplification with oligonucleotides specific to the mature or pre-PAP transcript. The level of 7SL RNA was used to control for equal loading of cDNA. (D) Heat map of the transcriptome of TSR1, TSR1IP, and SRP68 silenced cells. Transcripts that differed from the control by > 1.5-fold change (P value < 0.05) were chosen for the analysis. Each column represents the average of seven biological replicates. The diagram represents the differential expression or fold change according to the following color scale: red, upregulated genes; green, downregulated genes (see Materials and Methods). (E) Functional enrichment analysis of gene sets of down and upregulated genes during TSR1 and TSR1IP silencing. The genes presented in Table S2 were categorized based on the gene ontology presented in the figure, as previously described.27 The fold change of each category is given, comparing the percentage of a category in the regulated genes to the percentage of the same category in the data set (Table S2). The percentage of the genes in each category among the regulated genes is also given. Green or red represent categories that were significantly enriched (Fisher exact test, one side P value < 0.05) in comparison to the whole genome annotation (in total, we identified 1493 genes in the “genome” with annotations). (F) Venn diagrams illustrating overlaps between TSR1 and TSR1IP regulated genes.
Next, the effect on trans-splicing was examined by determining the level of SL RNA and of the Y structure intermediate by primer extension using a primer that extends both the Y structure intermediate and the SL RNA. Recently, we demonstrated that silencing basal splicing factors such as U2AF35 and SF1, increases the level of SL RNA; this is accompanied by a decrease in the level of Y structure intermediate, suggesting that these factors are essential for the first step of splicing.27 An effect on the first step of splicing is evident by the decrease in the Y structure intermediate, whereas increase in the Y structure intermediate suggests effects on the second step of splicing.33 Our results (Fig. 1B) demonstrate accumulation of SL RNA and reduction in the Y structure intermediate as early as day 2 of silencing, suggesting that these factors globally affect trans-splicing at the first splicing step. The early effect on the level of SL RNA suggests a direct effect on trans-splicing. TSR1 and TSR1IP silencing affected the Y structure intermediate; however, the extent of SL RNA increase was lower compared with the effect seen in cells depleted of the basal splicing factors SF1 and U2AF35 (4.2- and 6.2-fold increase, respectively),27 suggesting that these factors affect the splicing of only a sub-set of genes, unlike the basal splicing factors mentioned above, which are believed to affect the splicing of the majority of mRNAs (details are given below). The lower increase in SL RNA could not be attributed to incomplete silencing, since the silencing observed was very efficient, based on the reduction of the tagged proteins (Fig. 1B).
Since the SR proteins seem to affect only a sub-set of genes, it was of interest to determine whether these factors are also involved in cis-splicing. To address this question, we examined the effect of silencing on production of the poly(A) polymerase transcript containing a cis-spliced intron. Silencing resulted in defects in cis-splicing (Fig. 1C), suggesting that both proteins are essential for cis-splicing, as well. The defects observed in cis-splicing are reminiscent of the perturbation observed following silencing of the U1-specific factor U1 70 kDa, which participates exclusively in cis-splicing,34 suggesting the direct role of SR proteins in splicing of the PAP1 mRNA.
Silencing of TSR1 and TSR1IP affects only a subset of genes
Based on the small effect of TSR1 or TSR1IP silencing on the level of SL RNA (Fig. 1B), we expected to observe a limited effect on the transcriptome, compared with the robust effect observed under basal splicing factor depletion.27 Cells carrying the TSR1 and TSR1IP were silenced for 3 d, and RNA was subjected to microarray analysis. In previous studies, the third day of silencing was used as a preferred time point to analyze splicing defects.27,33,35-39 Each experiment was repeated seven times (biological replicates), and two technical replicates were performed for each experiment. Normalized data were used to identify genes whose expression was significantly up-or downregulated (P value, 0.05; ANOVA) with a magnitude exceeding a cutoff of 1.5-fold. The cut-off was heuristically selected to be large enough to enable experimental validation and eliminate false positives, and yet small enough to capture the maximal number of regulated genes. The number of downregulated genes upon TSR1 and TSR1IP silencing was 113 and 179, and 102 and 150 genes were upregulated, respectively, suggesting that SR proteins regulate only a sub-set of mRNAs. Lists of the affected genes are provided in S-2. When the transcriptomes of the basal splicing factors U2AF35, U2AF65, and SF1 silenced cells were analyzed, and differences exceeding a cutoff of 1.5-fold (P value 0.05) were considered, approximately 800 (both up- and downregulated) transcripts were revealed, highlighting the stringency of the analysis, since the genome contains around 9000 trans-spliced genes.27 Given the stringent analysis performed on the basal splicing factors,27 the low number of transcripts (200–300) shown here to be affected by the SR protein depletion indicates that only a subset of transcripts are regulated by the SR proteins, confirming that SR proteins cannot be considered basal splicing factors.
Since SR proteins are known to interact with each other, sometimes forming a bridge between the 5′ and 3′ splice sites, or enhancing the binding of U2AF35/65 to the 3′ splice domain, we examined the degree of overlap between the genes affected by the two SR proteins. The heat map and Venn diagram demonstrate significant overlap between the SR regulated genes (Fig. 1D and F) (61 co-regulated genes, P value for the overlap is 7 × 10−16).
The transcriptome of cells silenced in essential genes, such as the TSR1 and TSR1IP silenced cells, might be influenced by the cell death process. To examine whether the changes observed are in part due to the perturbation induced by cell death, the transcriptome of the SR silenced cells was compared with that of SRP68 silenced cells. Silencing of the SRP68 leads to perturbations in protein translocation across the ER membrane, and eventually to cell death.40 The heat map showed no significant overlap between these three transcriptomes (Fig. 1D). As opposed to the significant correlation (0.51 [P = 0]) found between the co-regulated genes in the two TSR proteins, the correlation between co-regulated genes in SRP68 and TSR1 was 0.0073 (P = 0.865), and that between TSR1IP and SRP68 was 0.0876 (P = 0.053), demonstrating that despite the fact that silencing of each of the three genes leads to cell death, the changes in the transcriptome do not reflect any shared mechanism, but rather indicate changes that are either a consequence of the regulation exerted by these factors on gene expression (SR proteins), and/or changes that are elicited to cope with the perturbation.
The sets of genes regulated by the SR proteins are enriched with distinct biological functions. Since conventional GO analysis (http://www.geneontology.org/) provides only a very vague annotation of gene families, we manually curated a database of all genes that are present in the microarray and were affected by silencing of the basal splicing factors.27 These 1493 genes were divided into distinct functional groups that are significant for parasite homeostasis such as transporters, chaperones, protein degradation, or genes involved in metabolism, RNA processing, ribosome biogenesis, and others. To examine the validity of the enrichment of the gene families among the affected transcripts, the percentage of the genes belonging to each category among the population of the affected genes was compared with the percentage of genes in the same category in the data set described above. The fold change in gene abundance in the two data sets is presented (Fig. 1E), with enrichment as a positive value, or underrepresentation depicted as a negative value.
A striking and significant finding was that many of the TSR1 upregulated genes encode proteins that function in DNA processing and signaling. However, proteins that are involved in ribosome biogenesis were underrepresented. On the other hand, among the TSR1IP upregulated genes, are genes involved in ribosome function, translation-related functions, and DNA processing, whereas the TSR1IP downregulated genes are enriched with those lacking chaperones. In comparison, TSR1 downregulated genes are significantly enriched in signaling and surface proteins. Thus, TSR1 and TSR1IP each seem to regulate a distinct family of genes in addition to the genes that are co-regulated by both proteins.
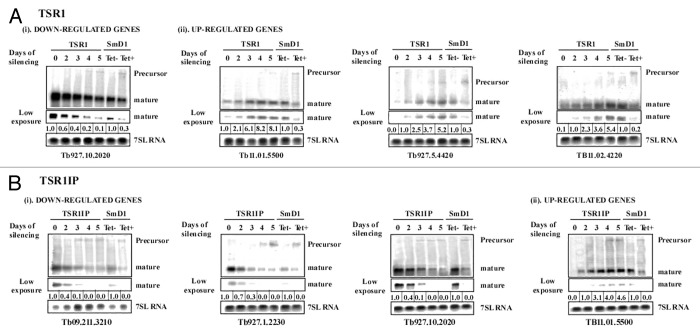
The finding that the number of upregulated genes was similar to the number of downregulated genes, suggests that the effect on the transcriptome may reflect other processes beyond splicing. Indeed, we recently demonstrated that basal splicing factors bind mature mRNAs and regulate mRNA stability.27 In the case of basal splicing factor silencing, the number of upregulated mRNAs was also similar to the number of downregulated transcripts. This result was surprising given the fact that depletion of splicing factors was expected to decrease the production of mRNA, thereby leading to downregulation of gene expression. To verify the effect of the depletion on the level of the mRNAs and its corresponding precursors, four different transcripts were analyzed over the course of silencing. The kinetics of change of mature mRNA and the corresponding precursor were subsequently analyzed by northern blotting (Fig. 2). Each northern contained, as a control, the transcript and its precursor in SmD1 silenced cells, which are severely inhibited for both cis- and trans-splicing41 (Fig. 2). The effect on gene expression was best observed after the third day of silencing (as used for the microarray experiments). Under depletion, the reduction or increase of mRNAs appeared before the accumulation of the precursor, suggesting that the effect on mature RNA is not necessarily due to the effect on splicing (Fig. 2). The level of precursor on the third day of silencing was negligible compared with that of the mature mRNAs, suggesting that the microarray results probably reflect only the level of the mRNAs and not those of their corresponding precursors.
Figure 2. TSR1 and TSR1IP silencing results in reduction or elevation of distinct mRNAs. (A and B) Cells expressing the RNAi silencing constructs were silenced for the indicated duration. Total RNA (20 µg) was subjected to Northern analysis with T7 transcribed antisense RNA probe specific to the gene affected. RNA from SmD1 silenced cells was used to control for the precursors that accumulate under splicing perturbations. To control for equal loading, the blot was hybridized with a random-labeled 7SL RNA probe. The results were quantified using ImageJ. The levels of mature transcript are given as fold change with respect to their level at day 0, and were normalized to the level of 7SL RNA. Short exposure of the same gel is shown below each Northern. (A) TSR1 silenced cells: (i) TSR1- downregulated genes; (ii) TSR1- upregulated genes; (B) TSR1IP silenced cells: (i) downregulated; and (ii) upregulated genes.
Splicing and polyadenylation signal properties of TSR regulated transcripts
Because splicing is globally perturbed under the depletion of the SR proteins, we sought to determine whether transcripts regulated under SR silencing exhibit any special properties in their splicing and/or polyadenylation signals. Using recent mapping of SL and poly (A) sites in T. brucei genes,9 we examined the following properties of the splicing and polyadenylation signals: (1) splicing/polyadenylation diversity, a measure of the heterogeneity in site selection; (2) polypyrimidine tract characteristics such as its length and distance to the AG dinucleotide; (3) the pyrimidine fraction upstream of the splice site and the purine fraction downstream of it; (4) the distance between a poly (A) site and a downstream splice site; and (5) the length of the 5′- and 3′-UTRs. A full description of these statistics and additional parameters are presented in S-4. Previously, we found that for genes up- and downregulated under U2AF65 and SF1 depletion, polyadenylation is more heterogeneous than in other genes; U2AF65 downregulated genes have significantly fewer pyrimidines and shorter PPTs, and U2AF35 regulated genes have less diversity in their 3′ splice sites. For the SR proteins, we also observed a number of trends. First, transcripts downregulated under TSR1IP depletion (and to some extent also under TSR1 depletion) have weaker splicing signals manifested by shorter and lower pyrimidine content of PPT, and changes in 5′-UTR length. No changes in PPT-AG distance or SL site selection diversity were observed among the regulated transcripts. For both factors, upregulated genes had longer purine-rich sequences downstream to the 3′ splice site than downregulated genes, which is likely related to the affinity of the TSR proteins for purine-rich sequences. Finally, regulated genes (mostly upregulated) had higher than expected heterogeneity in poly (A) site selection, suggesting that these factors may be involved in regulation of polyadenylation. Indeed, SR proteins were shown to regulate histone 3′ end formation in mammalian cells.42
TSR1 and TSR1IP affect the stability of mRNA
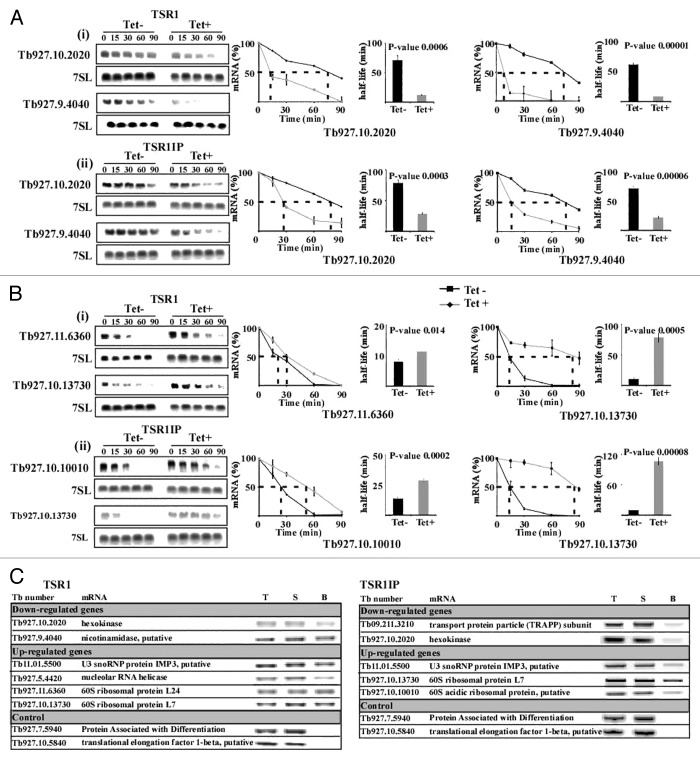
The changes in the level of mRNA upon silencing may stem from the effect of these factors on mRNA stability. To test this possibility, the half-life of mRNA was determined following treatment of cells with Actinomycin D and sinefungin to inhibit transcription and splicing, respectively before and after silencing. mRNAs upregulated or downregulated following silencing of the two factors were selected. Northern analysis was performed, and the mRNA level was measured by densitometry with the values normalized using the level of 7SL RNA. Decay of the mRNAs in un-induced cells was compared with decay in the silenced cells. For most mRNAs, half-lives were significantly altered between un-induced and silenced cells (P value < 0.005 [t test]; one-tailed, unpaired, equal variance), suggesting that changes in the level of mRNAs are correlated with changes in mRNA stability (Fig. 3).
Figure 3. Changes in stability of mRNAs upon silencing of TSR1 and TSR1IP. Uninduced and TSR1 or TSR1IP silenced cells (3 d after induction) were treated with sinefungin (2 µg/ml) and, after 10 min, with Actinomycin D (30 µg/ml). RNA was prepared at the time points indicated above the lanes, separated on a 1.2% agarose-formaldehyde gel, and subjected to northern analysis with the indicated gene-specific probes. 7SL RNA was used to control for equal loading. (A) The half-life of downregulated transcripts. (i), TSR1; (ii), TSR1IP. (B) Half-life of upregulated genes. (i) TSR1; (ii) TSR1IP. The hybridization signals were measured by densitometry. The decay curves are shown with the blots, and the half-life is illustrated by the broken lines. The decay in the absence of induction (-Tet) is indicated by a black line, and following induction (+Tet) by a gray dashed line. The experiments were repeated three times; each data point corresponds to the average, with the standard deviation indicated. The half-life was then calculated by fitting the normalized RNA levels to an exponential decay. The half-lives (averaged over the three experiments) are shown as bars with standard deviations, along with P values (t test) for the difference between the half-lives in uninduced compared with silenced cells. (C) Affinity selection of TSR1 and TSR1IP substrates using tagged proteins. Whole cell extracts were prepared from 5 × 109 cells with PTP tagged TSR1 and TSR1IP after 5 min of UV cross-linking, and the extract was subjected to affinity purification on IgG beads, as described previously.27 RNA was eluted from the beads, and cDNA was prepared from the bead eluate, 5% of the total RNA, and 5% of the RNA from the supernatant. cDNA was subjected to PCR with SL forward primer and reverse primer from the ORF of each gene (specified in Supplemental S-1). As a control, two transcripts whose levels were unchanged in the silenced cells were used. T, total RNA; S, supernatant; B, beads.
To assess the direct binding of the SR proteins to the mRNAs, the T. brucei PTP-tagged SR proteins were used to affinity select mRNAs of the regulated transcripts. To stabilize the mRNA–protein interaction by cross-linking, the cells expressing the tagged proteins were exposed to UV irradiation. Extracts were prepared from the cells, and the RNA was extracted from the affinity selected RNPs. The extracted RNA was subjected to RT-PCR using gene-specific antisense primers and an SL sense primer, which recognizes all trypanosome mRNAs. The results (Fig. 3C) demonstrate binding of TSR1 and TSR1IP to transcripts whose levels were either up- or downregulated during silencing, mostly as a result of the regulation exerted on mRNA stability. No binding was observed for substrates whose level was unaffected in the silenced cells. These results demonstrate the specific association of the SR proteins with the mature mRNAs of the transcripts whose level they regulate.
While we assume that the effect of SR depletion on stability mostly reflects direct binding of the factors to the 3′ UTR, we cannot exclude the possibility that SR depletion affects the splicing of a master regulator that influences the stability of these mRNAs.
TSR1 and TSR1IP are localized in the nuclear speckles but also co-purify with ribosomal proteins
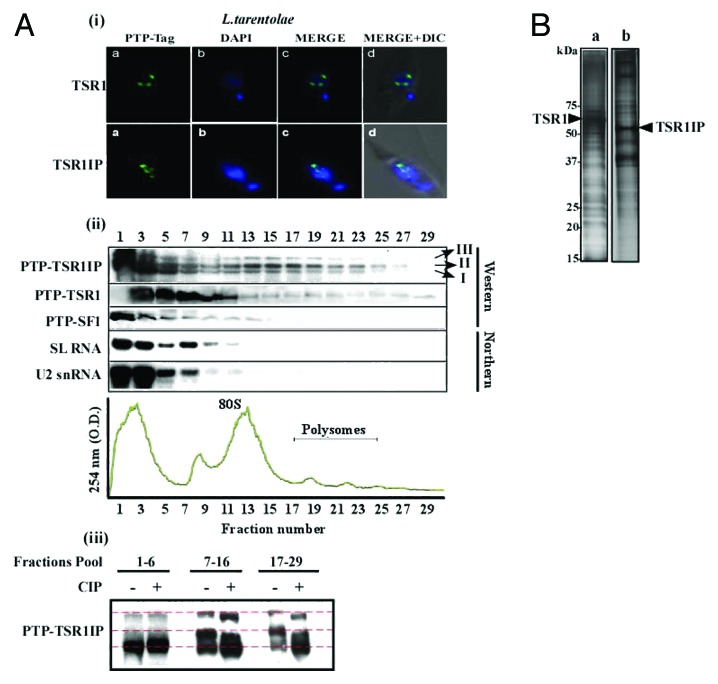
To gain further insight into the role of TSR1 and TSR1IP in the cell, the proteins associated with these factors were examined using the L. tarentolae system. Using Sm, Lsm, U1A, U2AF35, and SF1 as bait, we demonstrated the successful purification of a large amount of complexes, in quantities sufficient for mass spectrometry analysis.27,34 Since expression of the tagged protein is episomal, it was necessary to demonstrate that the subcellular localization of the tagged factor is identical to its homologous protein in T. brucei, which was tagged by PTP and incorporated at the authentic gene site. The localization of the protein was determined by immunofluorescence. The results, shown in Figure 4A-i, suggest that both the L. tarentolae and T. brucei proteins are found within nuclear speckles. Next, the distribution of L. tarentolae TSR1 and TSR1IP and SF1-tagged protein on RNP complexes was examined following fractionation on 15–50% sucrose gradients that separate single ribosomes (monosomes) from polyribosomes (Fig. 4A-ii). The samples were subjected to northern analysis, with SL and U2 snRNA probes used to locate the spliceosomal complexes among the RNPs. The results indicate that whereas SF1 was found co-migrating with spliceosomal complexes where most of the SL and U2 snRNA species fractionate, TSR1 and TSR1IP were also found on ribosomes and polysomes (Fig. 4A-ii). The fractionation of the two SR proteins resembled that of U2AF35,27 which was found not only co-migrating with spliceosomal complexes but also in complexes enriched with ribosomes, and protein factors involved in mRNA stability and regulation of translation.27
Figure 4. Localization of TSR1 and TSR1IP, their association with RNP complexes and with proteins. (A) (i) Immunofluorescence of PTP-tagged proteins in L. tarentolae. (a) Immunofluorescence of the PTP-tagged factor; (b) DAPI-stained nuclei; (c) merge of (a and b); (d) DIC merged with (c). (ii) Fractionation of tagged proteins on RNP complexes in L. tarentolae. Extract from 5 × 108 cells was layered on a continuous 15–50% (w/v) sucrose gradient in polysome buffer. Gradients were centrifuged at 4 °C for 2 h at 35 000 rpm in a Beckman SW41 rotor. Then, 1 ml fractions were collected using the ISCO gradient fractionation system. The absorbance profile at 245 nm is given and the position of 80S monosome and the polysomes are indicated (the identity was verified by the distribution of 18S and 28S rRNAs). RNA and protein were purified from the samples. The proteins were subjected to western analysis with an antibody that recognizes the PTP-tag. RNA was subjected to northern analysis with SL RNA and U2 snRNA probes. The three isoforms of the TSR1IP are indicated. (iii) De-phosphorylation of TSR1IP. Fractions obtained from the experiment described in section (ii) were pooled as indicated and subjected to de-phosphorylation with 10 units of CIP. The fractions were then incubated for 30 min at 37 °C. The proteins were separated on a 12% gel and subjected to western analysis with antibodies recognizing the PTP-tag. (B) The proteins associated with TSR1 and TSR1IP in L. tarentolae. Purification was performed using ~2 × 1011 cells, as described in Materials and Methods. The purified proteins were separated on a 12% SDS-polyacrylamide gel and stained with silver. The molecular mass markers are indicated.
Interestingly, several forms of the SR proteins were observed, especially for TSR1IP. The fast migrating form was found mostly in the top of the gradient (fractions 1–6, form I), while the slow migrating forms (form II and III) were found co-migrating with spliceosomal complexes (fractions 7–10), ribosomes (fractions 11–17), and polysomes (fractions 18–29). Since SR proteins are known to undergo phosphorylation,43 it was necessary to examine whether forms II and III are phosphorylated versions of the protein. To this end, fractions from the top of the gradient, containing ribosomes or polysomes (Fig. 4A-iii), were subjected to de-phosphorylation by treatment with calf-intestine alkaline phosphatase. The results indicate that both form II and III are phosphorylated, since their migration became faster as result of the enzyme treatment. However, the de-phosphorylated forms migrated more slowly than the un-modified form (I), suggesting that the protein may undergo additional modification(s).
Next, the complexes carrying these proteins were purified, separated on a 12% polyacrylamide gel, stained with silver (Fig. 4B), and analyzed by mass-spectrometry (Tables 1 and 2; Table S5). The list in Tables 1 and 2 includes only the non-ribosomal proteins found to interact with TSR1 and TSR1IP, whereas the ribosomal proteins are listed in S-5. TSR1 purification selected splicing factors such as U5-specific proteins, Sm proteins, SYF1 from the PRP 19 complex known as the NTC complex,34,38 and TRRM1,28 the SR protein that was previously shown to co-purify with U2AF35.27 Several helicases also co-purified with these proteins and were observed by us in the purification of Sm, Lsm, and U1A L. tarentolae complexes,34 as well as in the purification of T. brucei spliceosomal complexes.38 The purification also revealed factors involved in mRNA stability, such as UBP2.44,45 Other RNA binding proteins such as DRBD2, RBP23, poly (A) binding proteins, and ALBA protein also co-purified with TSR1. There are no functional data on DRBD2 and RBP23.46 However, ALBA proteins associate with T. cruzi and T. brucei ribosomes.47,48 Interestingly, MRP1 and MRP2 were also shown to be associated with TSR1. These proteins regulate mRNA stability of mitochondrial transcripts, and so far, no evidence exists for their role in nuclear mRNA metabolism.49,50 Although we could not rule out the possibility that these proteins are contaminants, their selective capture in this purification and not in previous purifications performed by us,34 argue that these proteins may also specifically associate with the complexes containing TSR1. A growing number of mitochondrial proteins were shown recently to have dual functions in the cell.51
Table 1. Mass spectrometric identification of TSR1 purification products (L.t).
| Protein category and accession number | Annotation | Trypanosoma brucei homolog | Number of peptides | e-value | Purifications | Reference(s) or source | |
|---|---|---|---|---|---|---|---|
| Splicing factors | |||||||
| 1 | LmjF.07.0870 | TSR1/RBSR1 | Tb927.8.900 | 42,22 | 1E-30 | NHP2-Lt, TSR1IP-Lt, U2AF35-Lt | 29 |
| 2 | LmjF.32.2200 | U5–116K/Snu114 | Tb11.01.7080 | 2 | 0.00000462 | PRP19-Tb, Lsm3-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, | 34 , 38 , 65 |
| 3 | LmjF.33.3190 | SmD2 | Tb927.2.5850 | 1 | 0.000559 | CPSF73, U1A-Lt, Lsm3-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, | 65 |
| 4 | LmjF.35.4460 | SmF | Tb09.211.1695 | 1 | 0.006573 | Lsm3-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt | 65 |
| 5 | LmjF.23.1550 | SYF1 | Tb927.5.1340 | 1 | 0.000471 | PRP19-Tb, SmD1-Tb,38 SmD3-Lt, | 34 , 38 |
| 6 | LmjF.27.2100 | TRRM1 | Tb927.2.4710 | 2 | 0.000071 | TSR1IP-Lt, U2AF65-Tb, U2AF35-Lt | 28 |
| 7 | LmjF.32.0400 | ATP-dependent RNA helicase, putative | Tb927.10.14550 | 8 | 0.0000683 | CPSF73, SMN-Lt, NHP2-Lt, PRP19-Tb, U1A-Lt, Lsm3-Lt, SmD3-Lt, TSR1IP-Lt, U2AF65-Tb, U2AF35-Lt, SF1-Lt | 34 |
| 8 | LmjF.07.0340 | ATP-dependent DEAD/H RNA helicase, putative | Tb927.8.1510 | 2 | 0.000302 | CPSF73, SMN-Lt, U1A-Lt, Lsm3-Lt, SmD3-Lt, TSR1IP-Lt, U2AF35-Lt, SF1-Lt | 34 |
| 9 | LmjF.35.3100 | ATP-dependent RNA helicase, putative | Tb09.211.3510 | 1 | 0.000318 | U1A-Lt, Lsm3-Lt, U2AF35-Lt, SF1-Lt | 34 |
| mRNA metabolism | |||||||
| 10 | LmjF.35.5040 | PABP1 | Tb09.211.0930 | 12,18 | 3.41E-09 | NHP2-Lt, U1A-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, TSR1IP-Lt, U2AF35-Lt | 82 , 83 |
| 11 | LmjF.35.4130 | PABP2 | Tb09.211.2150 | 19,25 | 1.07E-11 | NHP2-Lt, PRP19-Tb, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, TSR1IP-Lt, U2AF35-Lt | 84 |
| 12 | LmjF.25.0080 | PABP | ** | 6,9 | 7.22E-12 | NHP2-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, TSR1IP-Lt, U2AF35-Lt | GeneDB |
| 13 | LmjF.35.2200 | DRBD2 | Tb09.211.4540 | 5 | 0.0000665 | NHP2-Lt, TSR1IP-Lt, U2AF35-Lt | 85 |
| 14 | LmjF.25.0500 | UBP2 | Tb11.03.0580 | 1 | 0.000466 | 86 - 88 | |
| 15 | LmjF.17.0550 | RBP23 | Tb927.10.11270 | 1 | 0.001262 | 46 , 85 | |
| 16 | LmjF.27.1110 | MRP1 | Tb11.55.0009 | 4 | 0.000000627 | NHP2-Lt, U2AF35-Lt | 49 , 50 |
| 17 | LmjF.09.1120 | MRP2 | Tb11.01.4860 | 3 | 0.000123 | TSR1IP-Lt, U2AF35-Lt | 49 , 89 |
| 18 | LmjF.13.0450 | Alba2 | Tb11.02.2030 | 1 | 0.00752 | 47 | |
| 19 | LmjF.25.0540 | RAP55 | Tb11.03.0530 | 1 | 0.009507 | Lsm3-Lt, SmD3-Lt, SF1-Lt | 34 |
| 20 | LmjF.35.0370 | DHH1 | Tb927.10.3990 | 1 | 0.00125 | CPSF73, SMN-Lt, U1A-Lt, Lsm3-Lt, SmD3-Lt, TSR1IP-Lt, U2AF35-Lt, SF1-Lt | 90 |
| Ribosomal biogenesis and assembly proteins | |||||||
| 21 | LmjF.15.1380 | NOP58 | Tb09.160.3820 | 8 | 1.28E-08 | SMN-Lt, SmD3-Lt, U2AF35-Lt | 55 |
| 22 | LmjF.21.1760 | CBF5 | Tb927.10.170 | 5,4 | 0.00000576 | NHP2-Lt, U2AF35-Lt | 54 |
| 23 | LmjF.15.1470 | SNU13 | Tb09.160.3670 | 2 | 0.0000017 | SMN-Lt, NHP2-Lt, Lsm3-Lt, SmD3-Lt, U2AF35-Lt | 55 |
| 24 | LmjF.10.0210 | SIK1/ NOP56 | Tb927.8.3750 | 3 | 0.0000179 | NHP2-Lt, U2AF35-Lt | 55 |
| 25 | LmjF.32.0750 | NRBD2/P37 | Tb11.01.5590 | 3 | 0.001042 | TSR1IP-Lt, U2AF35-Lt | 91 |
| 26 | LmjF.33.1870 | NOG1 | Tb11.02.0620 | 3 | 0.00000422 | U2AF35-Lt | 92 |
| Helicases | |||||||
| 27 | LmjF.22.1500 | ATP-dependent DEAD/H RNA helicase, putative | Tb927.6.740 | 3 | 0.000409 | U2AF35-Lt | GeneDB |
| 28 | LmjF.27.1300 | hypothetical protein, conserved (p58) | Tb11.46.0009 | 13 | 0.0000128 | NHP2-Lt, TSR1IP-Lt, U2AF35-Lt | GeneDB |
| Hypothetical proteins | |||||||
| 29 | LmjF.36.1580 | hypothetical protein, conserved | Tb927.10.6000 | 2 | 0.010697 | SF1-Lt | GeneDB |
| 30 | LmjF.28.2330 | hypothetical protein, conserved | Tb11.01.3480 | 1 | 0.000000199 | PRP19-Tb, SmD3-Lt, | 34 |
| 31 | LmjF.18.0300 | hypothetical protein, conserved | Tb927.10.13800 | 1 | 0.007004 | SmD3-Lt, | 34 |
| 32 | LmjF.35.5390 | hypothetical protein, conserved | Tb927.4.310 | 1 | 0.005636 | TSR1IP-Lt, U2AF35-Lt, SF1-Lt | GeneDB |
L. tarentolae proteins were identified by mass spectroscopy as described in Materials and Methods. Each protein is described by its accession number (http://www.genedb.org), annotation, T. brucei homolog, numbers of peptides, other purifications revealing the same protein, and reference. Proteins were grouped into splicing factors, mRNA metabolism, ribosomal biogenesis and assembly proteins, helicases, and hypothetical proteins. The e-value reflects the probability of identification of the protein by MS data.
Table 2. Mass spectrometric identification of TSR1IP purification products (L.t.).
| Protein category and accession number | Annotation | Trypanosoma brucei homolog | Number of peptides | e-value | Purifications | Reference(s) or source | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Splicing factors | ||||||||||||
| 1 | LmjF.34.0495 | TSR1IP | Tb927.10.2910 | 12 | 0.00000464 | SF1-Lt | 30 | |||||
| 2 | LmjF.07.0870 | TSR1/RBSR1 | Tb927.8.900 | 6 | 5.87E-07 | NHP2-Lt, TSR1-Lt, U2AF35-Lt | 29 | |||||
| 3 | LmjF.27.2100 | TRRM1 | Tb927.2.4710 | 2 | 0.005601 | TSR1-Lt, U2AF65-Tb, U2AF35-Lt | 28 | |||||
| 4 | LmjF.32.0400 | ATP-dependent RNA helicase, putative | Tb927.10.14550 | 6 | 1.42E-08 | CPSF73, SMN-Lt, NHP2-Lt, PRP19-Tb, U1A-Lt, Lsm3-Lt, SmD3-Lt, TSR1-Lt, U2AF65-Tb, U2AF35-Lt, SF1-Lt | 34 | |||||
| 5 | LmjF.07.0340 | ATP-dependent DEAD/H RNA helicase, putative | Tb927.8.1510 | 2 | 0.00000798 | CPSF73, SMN-Lt, U1A-Lt, Lsm3-Lt, SmD3-Lt, TSR1-Lt, U2AF35-Lt, SF1-Lt | 34 | |||||
| mRNA metabolism | ||||||||||||
| 6 | LmjF.35.5040 | PABP1 | Tb09.211.0930 | 1 | 0.000261 | NHP2-Lt, U1A-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, TSR1-Lt, U2AF35-Lt | 82 , 83 | |||||
| 7 | LmjF.35.4130 | PABP2 | Tb09.211.2150 | 6 | 0.000178 | NHP2-Lt, PRP19-Tb, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, TSR1-Lt, U2AF35-Lt | 84 | |||||
| 8 | LmjF.25.0080 | PABP | ** | 3 | 0.000073 | NHP2-Lt, SmB-Tb,65 SmD1-Tb,38 SmD3-Lt, TSR1-Lt, U2AF35-Lt | GeneDB | |||||
| 9 | LmjF.35.2200 | DRBD2 | Tb09.211.4540 | 2 | 0.008095 | NHP2-Lt, TSR1-Lt, U2AF35-Lt | 85 | |||||
| 10 | LmjF.09.1120 | MRP2 | Tb11.01.4860 | 1 | 0.006042 | TSR1-Lt, U2AF35-Lt | 49 , 89 | |||||
| 11 | LmjF.19.1270 | RNA binding protein, putative | Tb927.10.15870 | 2 | 0.00000954 | GeneDB | ||||||
| 12 | LmjF.35.0370 | DHH1 | Tb927.10.3990 | 1 | 0.002744 | CPSF73, SMN-Lt, U1A-Lt, Lsm3-Lt, SmD3-Lt, TSR1-Lt, U2AF35-Lt, SF1-Lt | 90 | |||||
| Ribosomal biogenesis and assembly proteins | ||||||||||||
| 13 | LmjF.32.0750 | NRBD2/P37 | Tb11.01.5590 | 1 | 0.0000288 | TSR1-Lt, U2AF35-Lt | 91 | |||||
| Helicases | ||||||||||||
| 14 | LmjF.27.1300 (p58) | hypothetical protein, conserved | Tb11.46.0009 | 2 | 0.000324 | NHP2-Lt, TSR1-Lt, U2AF35-Lt | GeneDB | |||||
| Hypothetical proteins | ||||||||||||
| 15 | LmjF.35.5390 | hypothetical protein, conserved | Tb927.4.310 | 1 | 0.005682 | TSR1-Lt, U2AF35-Lt, SF1-Lt | GeneDB | |||||
| 16 | LmjF.35.4870 | hypothetical protein, conserved | Tb09.211.1230 | 1 | 0.0000148 | SF1-Lt | GeneDB | |||||
| 17 | LmjF.08.0340 | hypothetical protein, conserved | Tb927.5.3330 | 1 | 0.007234 | SF1-Lt | GeneDB | |||||
L. tarentolae proteins were identified by mass spectroscopy as described in Materials and Methods. Each protein is identified by its accession number (http://www.genedb.org), annotation, T. brucei homolog, numbers of peptides, other purifications revealing the same protein, and reference. Proteins were grouped into splicing factors, mRNA metabolism, ribosomal biogenesis and assembly proteins, helicases and hypothetical proteins. The e-value reflects probability of identification of the protein by the MS data.
Interestingly, the non-spliceosomal proteins associated with TSR proteins differed from those associated with SF1 and U2AF35. For instance, RBP29 and DRBD10, as well as PUF proteins, were shown to associate with SF1 and U2AF35, and UBP1 and RBP29 were shown to associate with U2AF35,27 whereas TSR proteins were shown to associate mostly with RBP26 and UBP2. The DRBD2 protein was present in both the U2AF35 purification and the TSR purification. Both TSR1 and TSR1IP selected tens of ribosomal proteins, translation factors, and metabolic enzymes, suggesting that these proteins are associated with ribosomes. Many of the metabolic proteins detected here were previously shown to be associated with translating ribosomes in T. cruzi,47 as well as with U2AF35.27
Interestingly, TSR1IP was shown to interact with SF1, and TSR1 with U2AF35. Both proteins were shown to interact with the third SR protein, TRRM1, as was U2AF35.27 In other eukaryotes, SR proteins were shown mainly to interact with U2AF35.52
The ribosomal proteins associated with the SR proteins are listed in S-5, and this list resembles the list observed under U2AF35 purification, but not that observed with SF1 purification,27 nor with the purification of SmD1, Lsm, or U1A.34 All these purifications were performed using the same method. Thus, mass-spectrometry data and the fractionation (Fig. 4A-ii) of the SR proteins indicate the specific association of these factors with ribosomes and polysomes.
Depletion of TSR1 and TSR1IP but not of SF1 affects rRNA processing
Co-purification of U2AF35 revealed the association of the factor with proteins involved in rRNA processing.27 The co-purification of TSR1 and TSR1IP also revealed the selection of nucleolar proteins, especially snoRNP proteins, and of factors such as NRBD2/p37 and NOG1 that are involved in rRNA processing.53 This association may suggest the direct involvement of these factors in ribosome biogenesis.
To examine the possible role of these factors and of U2AF35 in rRNA processing, rRNA processing defects were monitored during the silencing of four splicing factors (U2AF35, SF1, and the two SR proteins). rRNA processing in trypanosomes requires several cleavages; trypanosome-specific cleavages generate the two LSU subunits, LSUα, and LSUβ, releasing the srRNA fragments sr1, 2, 4, and 6. Cleavage at the internal spacer 1 (ITS1) releases a 5.9 Kb precursor that is further cleaved at ITS2 and ITS5 to form the 3.9 Kb precursor (depicted in Fig. 5A). To examine the possible effect on TSR protein depletion on rRNA, a northern analysis was performed with an ITS4 probe that detects processing of the large rRNA subunit. Indeed, accumulation of precursors (3.9, 5.1, and 5.9 Kb) and reduction of mature rRNA were detected as early as the second day of silencing in U2AF35, TSR1, and TSR1IP, but not in SF1 silenced cells (Fig. 5B), suggesting a primary defect in rRNA processing. Although the data presented indicate a primary effect of U2AF35 and TSR proteins on rRNA processing, the possibility that this profound phenotype may also result from a secondary effect cannot be excluded, due to the fact that factors involved in rRNA processing are among the SR regulated transcripts. Note, however, that factors involved in rRNA processing were upregulated under TSR depletion. The rRNA processing defects are severe, to the same extent observed when the snoRNA core protein CBF5 or NOP1 are silenced.54,55
Figure 5. The role of splicing factors in rRNA processing. (A) Schematic representation of the rRNA precursor. The positions of intronic sequences; external transcribed spacers (ETS), and internal transcribed spacers (ITS) are indicated flanking the mature rRNA, containing the small subunit (SSU) as well as the coding sequences (black bars). The structure of the precursors (9.6, 5.9, 5.1, and 3.9 Kb) is depicted. (B) Effect of silencing of splicing factors on rRNA processing. Total RNA was extracted from cells carrying the construct during silencing. RNA (10 μg) was separated on a 1.2% agarose gel containing 2.2 M formaldehyde and probed with an ITS4 probe. The 7SL RNA probe was used to control for equal loading. Marker size is indicated in Kb. Short exposure of the blot hybridized with the LSUβ probe is presented on the right side of the blot. The positions of the precursors are indicated by arrows. (i) TSR1, (ii) TSR1IP, (iii) U2AF35, and (iv) SF1. (C) TSR1 and TSR1IP silencing has no effect on rRNA transcription. (a). Nascent RNA synthesis in permeable cells before and after silencing of TSR1 and TSR1IP. Permeable cells were prepared from the same number of cells carrying the silencing construct, without induction (-Tet) or after tetracycline induction for 3 d (+Tet), in the presence or absence of 50 μg/ml α-amanitin, as described in Materials and Methods. The RNA was fractionated on a 6% (w/v) denaturing gel. The identity of the RNAs is indicated. The location of heterogeneous mRNAs is indicated in boxes. Lanes 1 and 5, RNA from uninduced cells; lanes 3 and 7, RNA from uninduced cells treated with 50 μg/ml α-amanitin; lanes 2 and 6, RNA from silenced cells; lanes 4 and 8, RNA from silenced cells treated with 50 μg /ml α-amanitin. (b) Slot-blot analysis of transcripts synthesized in permeable cells. RNA was prepared from permeable cells, as described in (a), and was used for hybridization with a blot containing DNA encoding for the genes, as indicated.
The robust phenotype observed on rRNA processing might be due to an effect of these factors on transcription. SR proteins interact directly with polymerase II as well as with certain transcription factors.56 To examine the effect of SR proteins on transcription, a permeable cell system was used.57 This system is able to detect the transcription of SL RNA, mRNA, rRNA, and tRNA. Cells silenced for TSR1 and TSR1IP (un-induced or after 3 d of silencing) were made permeable in the absence or presence of 50 µg/ml α-amanitin. At this α-amanitin concentration, polymerase II but not polymerase I transcription is inhibited.57 The RNA extracted from the different permeabilized cells was analyzed on a denaturing gel, or used for hybridization with gene probes for rRNA and SL RNA (as a control).
The results (Fig. 5C-a) demonstrate efficient synthesis of the SL RNA, tRNAs, and long heterogeneous polymerase II transcripts (at the top of the gel) that disappear in cells treated with α-amanitin. Interestingly, the silencing of TSR1IP affected the accumulation of polymerase II transcripts (at the top of the gel), suggesting that this factor might participate in regulating polymerase II transcription in trypanosomes, as in higher eukaryotes (Fig. 5C-a, lane 6). The inhibition of polymerase II transcription by α-amanitin enables the detection of transcripts that may be generated from the rRNA locus (top of the gel). To verify that rRNA is synthesized in the permeable cells, and to examine whether SR protein silencing affects polymerase I transcription, the RNA from the permeable cells was used in slot blot hybridization (Fig. 5C-b). Nascent-transcribed rRNA was detected only in the presence of α-amanitin. However, no effect on rRNA transcription could be observed as a result of TSR1 or TSR1IP silencing, suggesting that rRNA transcription is not affected in the SR protein silenced cells. The reduction in the production of mature rRNA observed in SR protein silenced cells must therefore result only from inhibition of processing, and not of transcription.
The finding that proteins involved in rRNA processing associate with the TSR proteins (Tables 1 and 2) further supports the direct role of these proteins in rRNA processing.
Discussion
In this study, we investigated the role of the two T. brucei SR proteins in regulating splicing, mRNA stability, and their possible involvement in rRNA processing. Transcriptome analysis of silenced cells suggests the existence of only a sub-set of targets regulated by these SR proteins, indicating that these factors are not general splicing factors, but rather, are specific to defined substrates. Analysis of the proteins associated with these SR proteins supports their role not only in splicing, but also in mRNA stability and in rRNA processing. In addition, these factors may also regulate translation, since their hyper-phosphorylated form seems to fractionate with ribosomes and polyribosomes, and to co-purify with ribosome-associated proteins.
Factors involved in splicing regulation in metazoa and the role of SR proteins in trypanosomes
Genome-wide studies mapping SL addition sites suggested extensive alternative splicing changes throughout the lifecycle of the parasite.10,58 Alternative splicing was also recently shown to control protein localization, enabling the generation of two isoforms of tRNA-synthetase, a mitochondrial, and a cytoplasmic enzyme.59 trans-splicing must therefore be a tightly regulated process to generate this rich repertoire of alternatively spliced forms. However, very little is known about factors that participate in such regulation. Early studies from our group suggested that PTB proteins are involved in trans-splicing of a distinct subset of transcripts bearing a C-rich polypyrimidine tract.14 Recently, we found that hnRNP F/H might be a strong candidate for mediating stage-specific splicing regulation, since the protein is differentially regulated and highly expressed in the bloodstream form. In addition, the protein recognition site, AAGAA, is located around the 3′ splice site and the poly (A) site.60 In this study, we found that substrates downregulated by TSR1 and TSR1IP silencing contain weaker splicing signals such as shorter and pyrimidine-poor PPTs, and that the regulated genes have more heterogeneous poly (A) site selection. RNA-seq of these transcriptomes may reveal additional substrates, especially non-abundant transcripts that are not detected by the microarray analysis but also have weak splicing signals, and hence, rely on SR proteins for their splicing.
Recent studies from mammalian cells using CLIP suggest that binding sites of SRSF-3 and -4 were more abundant in exons. However, binding to introns was also observed in domains close to the splice site, suggesting their role in splicing.42 CLIP of TSR1 and TSR1P will be necessary to determine the binding of these factors to intron and exon domains.
Of special interest is the finding that mammalian SRSF-3 and -4 regulate mRNAs involved in RNA metabolism, including spliceosomal functions,42 and that human SFRS1 also shows high enrichment of binding to mRNAs involved in RNA metabolism, such as SR proteins, and proteins involved in mRNA and rRNA processing.61 Similarly, in trypanosomes, TSR1IP upregulated transcripts were enriched with genes involved in ribosome function and translation.
The role of TSR1 and TSR1IP in mRNA stability
The involvement of splicing factors in functions other than splicing was reported recently for the basal splicing factors, U2AF35/65 and SF1. It was demonstrated that these factors affect mRNA stability most probably due to binding of these factors to the 3′ UTR.27 It was argued that in trypanosomes, such interactions might be prevalent due to two major specific characteristics of gene expression. First, trans-splicing and polyadenylation are linked.2-5 In trypanosomes, trans-splicing determines which poly(A) site will be selected, and hence, the size of the 3′ UTR. Second, extensive alternative polyadenylation was observed, with over 400 cases already described in the procyclic stage.9 Thus, alternative polyadenylation may generate transcripts that contain splicing signals at their 3′ UTR that bind these basal splicing factors.
Although we argued that the effect of these basal splicing factors on mRNA stability emerges from the direct binding of the splicing factor to mature mRNA, and demonstrated this mechanism for U2AF65, we must also consider the possibility that the observed changes in mRNA stability may stem not only from the direct binding of the splicing factors to the 3′ UTR, but rather, from secondary perturbations. CLIP-Seq analyses of these TSR proteins and basal splicing factors are in progress and should shed light on the binding site of these proteins either near the 3′ splice site (affecting splicing) or at the 3′UTR, affecting stability and possibly translation. Interestingly, despite the fact that the CLIP studies of SR proteins observed binding to the 3′ UTR,62,63 only a single study in mammalian cells demonstrated the role of these factors in mRNA stability.64
Purification of complexes containing the splicing factors supports their role in mRNA decay and translation
Our mass spectrometry data identified several RNA binding proteins that may also function in mRNA stability and even translational control. Trypanosome spliceosomal complexes were comprehensively identified by purifying Sm proteins in both T. brucei38,65 and Leishmania.34 Previously, we observed that affinity selection of proteins associated with the basal splicing factors failed to detect core components of the spliceosome, such as Sm proteins, and U snRNP-specific proteins. However, such proteins were selected with TSR1. These results may indicate that the spliceosomal complexes carrying the basal factors as well as SR proteins are fragile and dissociate during purification. However, it is also possible that the level of spliceosomal complexes containing these factors is low compared with storage sub-spliceosomal complexes that include these factors. Indeed, studies in mammals identified a large pool of extra-spliceosomal complexes in nuclear speckles.66
The large number of ribosomal proteins selected with TSR1 and TSR1IP supports the association of these factors with ribosomes. As described in the Introduction, SR proteins were shown to both directly and indirectly affect translation.17
The phosphorylation state of the SR proteins has been under extensive investigation in many systems. The results presented here suggest the presence of three distinct TSR1IP forms. The two forms (II and III) are present on spliceosomal complexes, but also on ribosomes and polysomes. It was previously demonstrated that phosphorylation of SR proteins is required for the interaction between SR proteins; for instance, the phosphorylation of ASF/SF2 is essential for the interaction with the U1 snRNP 70 kDa protein.67 However, it was also reported that after spliceosome assembly, de-phosphorylation is required for the transition to the catalytic step of splicing; thus, both phosphorylated and dephosphorylated forms of proteins are expected in spliceosomes.67 De-phosphorylation is also required for the association of the SR proteins with the export machinery.68
It was of great interest to find that TSR1IP (forms II and III) is associated with both ribosomes and polyribosomes. These two forms are phosphorylated, as their migration changed as a result of de-phosphorylation. Despite the demonstration that SR proteins enhance translation, the status of the phosphorylation of the SR protein associated with translation was not previously addressed.69
However, TSR1P must undergo additional post-translational modification(s), since de-phosphorylation did not convert form III to I. Indeed, SR proteins undergo acetylation, methylation, and also ubiquitinylation (Ub), as well as modification by the small Ubiquitin-like modifier (SUMO).23 Ub conjugation to SR proteins was suggested to serve as regulatory signal, rather than to direct the protein for degradation.23
Involvement of splicing factors in rRNA processing
The association described above of TSR1 and TSR1IP with nucleolar proteins, especially snoRNP proteins, and also with factors such as NRBD2/p37 and NOG1, may suggest the direct involvement of these factors in ribosome biogenesis. It was previously reported that the yeast PRP43 helicase involved in splicing is also directly involved in rRNA processing.70 The association of TSR1, TSR1IP, and U2AF35 with proteins implicated in rRNA processing and the direct and robust effect of the depletion on rRNA production indicate the direct role of these SR proteins and U2AF35 in rRNA processing. However, in addition, to their primary effect, depletion of rRNA processing factors which are TSR targets can also indirectly affect rRNA processing. Interestingly, depletion of the TSR proteins led to upregulation of mRNAs coding for factors involved in rRNA processing, suggesting a possible feedback loop to compensate for the loss of the SR proteins. Indeed, in mammalian cells, SR proteins also regulate both the level of proteins involved in rRNA processing, as well as the level of snoRNA.42 However, this is the first study to demonstrate the direct association of SR proteins with proteins involved in rRNA processing and to demonstrate rRNA processing defects in SR silenced cells. Of note, is the recent finding that trans-splicing occurs in the nucleolus and may participate in rRNA processing.71 Most recently, the two T. brucei RNA binding proteins, PUF7 and PUF10, were reported to localize to the nucleolus, and were shown to be essential for rRNA processing together with BOP1 and NRG.72 These four proteins exhibit several interactions. However, the most interesting finding regarding these proteins is that their depletion results in an increased level of mRNA that encodes GPEET procyclin surface protein, under conditions in which GPEET is normally repressed. NRG1 was also shown to directly bind to mRNAs, suggesting that in trypanosomes the two processes of mRNA metabolism and ribosomal processing are linked.72 The linkage between nucleolar function and splicing is intriguing, since both these processes determine the growth rate and proliferation of these parasites.
In sum, this study describes the role of TSR proteins in RNA processing. These proteins are among the first SR proteins that emerged in evolution, since trypanosomes diverged early in the eukaryotic lineage, long before baker’s yeast (Saccharomyces cerevisiae), which lack SR proteins.16 Despite the fact that trypanosomes are ancient eukaryotes, the function of their two SR protein share remarkable resemblance to their metazoal counterparts; in addition, their regulated substrates, as in mammals, are ribosomal proteins and factors involved in splicing and RNA processing. We are only at the earliest stages of a full understanding of the interplay between the processes regulated by SR proteins.
Materials and Methods
Cell growth and transfection
Procyclic T. brucei strain 29-13, which carries integrated genes for the T7 polymerase and the tetracycline repressor, was grown in SDM-79 medium supplemented with 10% fetal calf serum in the presence of 50 μg/ml hygromycin and 15 μg/ml G418. Cells were transfected as previously described.41
Construction of RNAi constructs
Stem-loop constructs were generated to silence TSR1 and TSR1IP using primers listed in S-1, as described.73 The constructs expressing dsRNA were linearized with EcoRV. The expression of dsRNA was induced using 8 µg/ml tetracycline.
Tagging of the splicing factors in L. tarentolae and T. brucei
For generation of the tagged constructs, genes were amplified using the primers listed in S-1. The fragments were cloned into the pSNSAP1 vector (kindly provided to us by Dr Larry Simpson, UCLA). The cloned vector (20 μg) was transfected into L. tarentolae and selected using neomycin resistance.74 To generate PTP-tagged constructs in T. brucei that encode a triple tag composed of the ProtC binding site, TEV protease recognition site, and protein A, the gene of interest was amplified with primers listed in S-1 and cloned into the PTP vector.31
Purification of the complexes associated with the SR proteins
Tandem affinity purification was performed from whole L. tarentolae cell extracts, as described.34 The proteins were analyzed by mass-spectrometry as follows: The samples were digested by trypsin, analyzed by LC-MS/MS on LTQ-Orbitrap (Thermo), and identified by Sequest 3.31 software against the GeneDB L. major-specific database (http://www.genedb.org).
Northern blots and primer extension analysis
Primer extension was performed as previously described.75,76 The extension products were analyzed on 6% acrylamide denaturing gels. For northern analysis, total RNA was extracted, separated on agarose-formaldehyde gel, and analyzed using RNA probes. Primers are listed in List S1.
Western blot analysis
Whole cell lysates (107 cells) were fractionated by SDS-PAGE, transferred to PROTRAN membranes (Whatman, BA-83 10401387), and reacted with anti rabbit IgG or anti PTB1 antibodies. The bound antibodies were detected with goat anti-rabbit immunoglobulin G (IgG) coupled to horseradish peroxidase, and were visualized by ECL (Amersham Biosciences, RPN2106).
Immunofluorescence assay
PTP-tagged TSR1 and TSR1IP cells were washed with PBS, mounted on poly-l-lysine-coated slides, fixed in 8% formaldehyde, and immunofluorescence was performed as described77 using FITC-conjugated anti-rabbit IgG. The cells were visualized with a Zeiss LSM 510 META inverted microscope.
mRNA stability analysis
Uninduced cells, and cells 3 d after induction (1.5 × 109 cells) were concentrated and resuspended into 25 ml of SDM-79 medium. Cells were aliquotted into five batches and incubated at 27 °C for 30 min. Cells were pre-treated with 2 µg/ml sinefungin (Sigma) for 10 min, and then with 30 µg/ml Actinomycin D (Sigma). RNA was isolated and subjected to northern analysis, as previously described.14 Each experiment was repeated three times. The RNA level was normalized to 1 at t = 0, and the decay was fitted to Exp[-R*t]. R was calculated by regressing -ln([normalized RNA level]) on t (in minutes), using ordinary least squares without the intercept term. The half-life was then calculated as ln(2)/R.
Microarray analysis
Total RNA was isolated from uninduced cells, and from silenced cells after 3 d of induction. Total RNA was labeled using the Ambion Amino Allyl MessageAmp II aRNA kit (Ambion). DNA microarrays were obtained through NIAID's Pathogen Functional Genomics Resource Center (managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by the J. Craig Venter Institute), hybridized using the Gene Expression hybridization kit (Agilent Technologies), and processed as previously described in detail.14 The data from all arrays were first subjected to Normexp-Background correction78 and Loess within array normalization79 using the Bioconductor Limma package.80 The subsequent analysis was performed using Partek® Genomics SuiteTM software, version 6.6 (© 2012 Partek Inc.). Normalized data from seven biological replicates were analyzed to identify genes whose expression was up- or downregulated by an arbitrary cutoff of at least 1.5-fold, and had P value < 0.05 in all replicates when testing for differential expression (ANOVA-test). Heat maps were generated using Euclidean distance as a similarity measure. An annotation table was prepared after annotating regulated genes manually, assisted by GeneDB (http://tritrypdb.org/tritrypdb/).
Fractionation of tagged L. tarentolae proteins on RNP complexes
Extracts were prepared from L. tarentolae cells expressing the PTP-tagged proteins, as previously described34 but using the polysome buffer (150 mM KCl, 4 mM MgCl2, 20 mM Tris pH [7.5], 1 mM DTT, and 10 μg/ml leupeptin). Cycloheximide (50 μg/ml) was added to the cells prior to collecting the cells by centrifugation. Extract was prepared as previously described.34 The lysate was cleared at 50 000 × g for 30 min, and centrifuged through a 15–50% (w/v) sucrose gradient at 4 °C for 2 h at 35 000 rpm in a Beckman SW41 rotor.
De-phosphorylation of TSR proteins
Fractions from the gradient were pooled in groups and treated with 1 µl of CIP (Calf Intestinal Alkaline Phosphatase [10 units/µl, NEB]) in 100 µl reaction volume (final concentration 0.1 units/µl) followed by incubation at 37 °C for 30 min, according to the manufacturer’s recommendations. Pooled fractions (CIP treated/untreated) were then analyzed on a 12% acrylamide gel.
Cell permeabilization
The permeabilization procedure was similar to that described by Tschudi and Ullu.81 The only deviation from the published protocol is that transcription buffer TB × 1 (150 mM sucrose, 20 mM potassium glutamate, 10 mM HEPES-KOH [pH 7.9], 2.5 mM MgCl2, 1 mM dithiotheritol, 10 μg/ml leupeptin) was used. Un-induced or cells after 2 d of silencing were permeabilized. Transcription was performed either in the presence or absence of 50 µg/ml-amanitin. RNA was extracted from the cells and fractionated on a 6% denaturing gel or subjected to slot blot hybridization. For slot-blot analysis of the RNA synthesized in permeable cells, plasmid DNA was used. Hybridization was performed at 55–60 °C in 60% (v/v) formamide, 2 × SSC (0.3 M sodium chloride, 0.03 M sodium citrate), 100 μg/ml salmon sperm DNA, and 0.1% (w/v) Sarcosyl, with the entire RNA fraction extracted from permeable cells. After hybridization, filters were washed twice in 2 × SSC and 0.1% (w/v) SDS at 65 °C for 30 min.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work was supported by the Deutsche Forcshungsgemeinschaft (DFG) via DIP and the Israel Science Foundation. Michaeli S holds the David and Inez Myers Chair in RNA silencing of diseases. Carmi S thanks the Human Frontier Science Program for financial support. We thank Dr Mark Katzenellenbogen for his initial analysis of the microarray data.
References
- 1.Figueiredo LM, Cross GA, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol. 2009;7:504–13. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 2.Hug M, Hotz HR, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol Cell Biol. 1994;14:7428–35. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 4.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 5.Vassella E, Braun R, Roditi I. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 1994;22:1359–64. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Günzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell. 2010;9:1159–70. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–40. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–74. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 9.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–57. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Clayton C. The regulation of trypanosome gene expression by RNA-binding proteins. PLoS Pathog. 2013;9:e1003680. doi: 10.1371/journal.ppat.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 2011;27:23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern MZ, Gupta SK, Salmon-Divon M, Haham T, Barda O, Levi S, Wachtel C, Nilsen TW, Michaeli S. Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA. 2009;15:648–65. doi: 10.1261/rna.1230209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estévez AM. The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 2008;36:4573–86. doi: 10.1093/nar/gkn406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Twyffels L, Gueydan C, Kruys V. Shuttling SR proteins: more than splicing factors. FEBS J. 2011;278:3246–55. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 18.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 19.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–81. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 20.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–211. doi: 10.1017/S1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longman D, Johnstone IL, Cáceres JF. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–37. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möröy T, Heyd F. The impact of alternative splicing in vivo: mouse models show the way. RNA. 2007;13:1155–71. doi: 10.1261/rna.554607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risso G, Pelisch F, Quaglino A, Pozzi B, Srebrow A. Regulating the regulators: serine/arginine-rich proteins under scrutiny. IUBMB Life. 2012;64:809–16. doi: 10.1002/iub.1075. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–5. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 25.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–64. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu XD, Dundr M, Prasanth KV. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell. 2012;23:3694–706. doi: 10.1091/mbc.E12-03-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta SK, Carmi S, Waldman Ben-Asher H, Tkacz ID, Naboishchikov I, Michaeli S. Basal splicing factors regulate the stability of mature mRNAs in trypanosomes. J Biol Chem. 2013;288:4991–5006. doi: 10.1074/jbc.M112.416578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manger ID, Boothroyd JC. Identification of a nuclear protein in Trypanosoma brucei with homology to RNA-binding proteins from cis-splicing systems. Mol Biochem Parasitol. 1998;97:1–11. doi: 10.1016/S0166-6851(98)00118-2. [DOI] [PubMed] [Google Scholar]
- 29.Ismaïli N, Pérez-Morga D, Walsh P, Mayeda A, Pays A, Tebabi P, Krainer AR, Pays E. Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol Biochem Parasitol. 1999;102:103–15. doi: 10.1016/S0166-6851(99)00091-2. [DOI] [PubMed] [Google Scholar]
- 30.Ismaïli N, Pérez-Morga D, Walsh P, Cadogan M, Pays A, Tebabi P, Pays E. Characterization of a Trypanosoma brucei SR domain-containing protein bearing homology to cis-spliceosomal U1 70 kDa proteins. Mol Biochem Parasitol. 2000;106:109–20. doi: 10.1016/S0166-6851(99)00205-4. [DOI] [PubMed] [Google Scholar]
- 31.Palfi Z, Schimanski B, Günzl A, Lücke S, Bindereif A. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 2005;33:2493–503. doi: 10.1093/nar/gki548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schimanski B, Nguyen TN, Günzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005;4:1942–50. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang XH, Liu Q, Liu L, Tschudi C, Michaeli S. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol Biochem Parasitol. 2006;145:29–39. doi: 10.1016/j.molbiopara.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Tkacz ID, Gupta SK, Volkov V, Romano M, Haham T, Tulinski P, Lebenthal I, Michaeli S. Analysis of spliceosomal proteins in Trypanosomatids reveals novel functions in mRNA processing. J Biol Chem. 2010;285:27982–99. doi: 10.1074/jbc.M109.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tkacz ID, Lustig Y, Stern MZ, Biton M, Salmon-Divon M, Das A, Bellofatto V, Michaeli S. Identification of novel snRNA-specific Sm proteins that bind selectively to U2 and U4 snRNAs in Trypanosoma brucei. RNA. 2007;13:30–43. doi: 10.1261/rna.174307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tkacz ID, Cohen S, Salmon-Divon M, Michaeli S. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei. Mol Biochem Parasitol. 2008;160:22–31. doi: 10.1016/j.molbiopara.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Ruan JP, Shen S, Ullu E, Tschudi C. Evidence for a capping enzyme with specificity for the trypanosome spliced leader RNA. Mol Biochem Parasitol. 2007;156:246–54. doi: 10.1016/j.molbiopara.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luz Ambrósio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RM, Günzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot Cell. 2009;8:990–1000. doi: 10.1128/EC.00075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaé N, Wang P, Gu T, Hühn M, Palfi Z, Urlaub H, Bindereif A. Essential role of a trypanosome U4-specific Sm core protein in small nuclear ribonucleoprotein assembly and splicing. Eukaryot Cell. 2010;9:379–86. doi: 10.1128/EC.00353-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lustig Y, Goldshmidt H, Uliel S, Michaeli S. The Trypanosoma brucei signal recognition particle lacks the Alu-domain-binding proteins: purification and functional analysis of its binding proteins by RNAi. J Cell Sci. 2005;118:4551–62. doi: 10.1242/jcs.02578. [DOI] [PubMed] [Google Scholar]
- 41.Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J Biol Chem. 2003;278:51469–78. doi: 10.1074/jbc.M308997200. [DOI] [PubMed] [Google Scholar]
- 42.Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapra AK, Ankö ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–90. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 44.Hartmann C, Benz C, Brems S, Ellis L, Luu VD, Stewart M, D’Orso I, Busold C, Fellenberg K, Frasch AC, et al. Small trypanosome RNA-binding proteins TbUBP1 and TbUBP2 influence expression of F-box protein mRNAs in bloodstream trypanosomes. Eukaryot Cell. 2007;6:1964–78. doi: 10.1128/EC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartmann C, Clayton C. Regulation of a transmembrane protein gene family by the small RNA-binding proteins TbUBP1 and TbUBP2. Mol Biochem Parasitol. 2008;157:112–5. doi: 10.1016/j.molbiopara.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Wurst M, Robles A, Po J, Luu VD, Brems S, Marentije M, Stoitsova S, Quijada L, Hoheisel J, Stewart M, et al. An RNAi screen of the RRM-domain proteins of Trypanosoma brucei. Mol Biochem Parasitol. 2009;163:61–5. doi: 10.1016/j.molbiopara.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Alves LR, Avila AR, Correa A, Holetz FB, Mansur FC, Manque PA, de Menezes JP, Buck GA, Krieger MA, Goldenberg S. Proteomic analysis reveals the dynamic association of proteins with translated mRNAs in Trypanosoma cruzi. Gene. 2010;452:72–8. doi: 10.1016/j.gene.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Mani J, Güttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Späth G, Roditi I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vondrusková E, van den Burg J, Zíková A, Ernst NL, Stuart K, Benne R, Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem. 2005;280:2429–38. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- 50.Read LK, Göringer HU, Stuart K. Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins and gRNA domains but does not require preedited mRNA. Mol Cell Biol. 1994;14:2629–39. doi: 10.1128/MCB.14.4.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta. 2011;1808:1012–20. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–70. doi: 10.1016/0092-8674(93)90316-I. [DOI] [PubMed] [Google Scholar]
- 53.Michaeli S. rRNA Biogenesis in Trypanosomes. In: Bindereif A, ed. RNA Metabolism in Trypanosomes. Belin, Heidelberg: Springer-Verlag, 2012:123-48. [Google Scholar]
- 54.Barth S, Hury A, Liang XH, Michaeli S. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J Biol Chem. 2005;280:34558–68. doi: 10.1074/jbc.M503465200. [DOI] [PubMed] [Google Scholar]
- 55.Barth S, Shalem B, Hury A, Tkacz ID, Liang XH, Uliel S, Myslyuk I, Doniger T, Salmon-Divon M, Unger R, et al. Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryot Cell. 2008;7:86–101. doi: 10.1128/EC.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee KM, Tarn WY. Coupling pre-mRNA processing to transcription on the RNA factory assembly line. RNA Biol. 2013;10:380–90. doi: 10.4161/rna.23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ullu E, Tschudi C. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 1990;18:3319–26. doi: 10.1093/nar/18.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rettig J, Wang Y, Schneider A, Ochsenreiter T. Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res. 2012;40:1299–306. doi: 10.1093/nar/gkr794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta SK, Kosti I, Plaut G, Pivko A, Tkacz ID, Cohen-Chalamish S, Biswas DK, Wachtel C, Waldman Ben-Asher H, Carmi S, et al. The hnRNP F/H homologue of Trypanosoma brucei is differentially expressed in the two life cycle stages of the parasite and regulates splicing and mRNA stability. Nucleic Acids Res. 2013;41:6577–94. doi: 10.1093/nar/gkt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corioni M, Antih N, Tanackovic G, Zavolan M, Krämer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2011;39:1868–79. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palfi Z, Jaé N, Preusser C, Kaminska KH, Bujnicki JM, Lee JH, Günzl A, Kambach C, Urlaub H, Bindereif A. SMN-assisted assembly of snRNP-specific Sm cores in trypanosomes. Genes Dev. 2009;23:1650–64. doi: 10.1101/gad.526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rino J, Desterro JM, Pacheco TR, Gadella TW, Jr., Carmo-Fonseca M. Splicing factors SF1 and U2AF associate in extraspliceosomal complexes. Mol Cell Biol. 2008;28:3045–57. doi: 10.1128/MCB.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–67. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci U S A. 2004;101:9666–70. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–68. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Combs DJ, Nagel RJ, Ares M, Jr., Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–34. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayer MG, Santos MG, Silva MF, Floeter-Winter LM. Footprints of a trypanosomatid RNA world: pre-small subunit rRNA processing by spliced leader addition trans-splicing. Mem Inst Oswaldo Cruz. 2012;107:522–31. doi: 10.1590/S0074-02762012000400013. [DOI] [PubMed] [Google Scholar]
- 72.Schumann Burkard G, Käser S, de Araújo PR, Schimanski B, Naguleswaran A, Knüsel S, Heller M, Roditi I. Nucleolar proteins regulate stage-specific gene expression and ribosomal RNA maturation in Trypanosoma brucei. Mol Microbiol. 2013;88:827–40. doi: 10.1111/mmi.12227. [DOI] [PubMed] [Google Scholar]
- 73.Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–9. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 74.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–24. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hury A, Goldshmidt H, Tkacz ID, Michaeli S. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot Cell. 2009;8:56–68. doi: 10.1128/EC.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang XH, Liu L, Michaeli S. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J Biol Chem. 2001;276:40313–8. doi: 10.1074/jbc.M104488200. [DOI] [PubMed] [Google Scholar]
- 77.Lustig Y, Sheiner L, Vagima Y, Goldshmidt H, Das A, Bellofatto V, Michaeli S. Spliced-leader RNA silencing: a novel stress-induced mechanism in Trypanosoma brucei. EMBO Rep. 2007;8:408–13. doi: 10.1038/sj.embor.7400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 79.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–73. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 80.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 81.Tschudi C, Ullu E. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell. 1990;61:459–66. doi: 10.1016/0092-8674(90)90527-L. [DOI] [PubMed] [Google Scholar]
- 82.Bates EJ, Knuepfer E, Smith DF. Poly(A)-binding protein I of Leishmania: functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res. 2000;28:1211–20. doi: 10.1093/nar/28.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Batista JA, Teixeira SM, Donelson JE, Kirchhoff LV, de Sá CM. Characterization of a Trypanosoma cruzi poly(A)-binding protein and its genes. Mol Biochem Parasitol. 1994;67:301–12. doi: 10.1016/0166-6851(94)00133-2. [DOI] [PubMed] [Google Scholar]
- 84.Hotchkiss TL, Nerantzakis GE, Dills SC, Shang L, Read LK. Trypanosoma brucei poly(A) binding protein I cDNA cloning, expression, and binding to 5 untranslated region sequence elements. Mol Biochem Parasitol. 1999;98:117–29. doi: 10.1016/S0166-6851(98)00156-X. [DOI] [PubMed] [Google Scholar]
- 85.De Gaudenzi J, Frasch AC, Clayton C. RNA-binding domain proteins in Kinetoplastids: a comparative analysis. Eukaryot Cell. 2005;4:2106–14. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D’Orso I, Frasch AC. TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J Biol Chem. 2001;276:34801–9. doi: 10.1074/jbc.M102120200. [DOI] [PubMed] [Google Scholar]
- 87.D’Orso I, Frasch AC. TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and Poly(A)-binding proteins forming a ribonucleoprotein complex. J Biol Chem. 2002;277:50520–8. doi: 10.1074/jbc.M209092200. [DOI] [PubMed] [Google Scholar]
- 88.Volpon L, D’Orso I, Young CR, Frasch AC, Gehring K. NMR structural study of TcUBP1, a single RRM domain protein from Trypanosoma cruzi: contribution of a beta hairpin to RNA binding. Biochemistry. 2005;44:3708–17. doi: 10.1021/bi047450e. [DOI] [PubMed] [Google Scholar]
- 89.Blom D, Burg Jv, Breek CK, Speijer D, Muijsers AO, Benne R. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 2001;29:2950–62. doi: 10.1093/nar/29.14.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holetz FB, Correa A, Avila AR, Nakamura CV, Krieger MA, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem Biophys Res Commun. 2007;356:1062–7. doi: 10.1016/j.bbrc.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 91.Prohaska K, Williams N. Assembly of the Trypanosoma brucei 60S ribosomal subunit nuclear export complex requires trypanosome-specific proteins P34 and P37. Eukaryot Cell. 2009;8:77–87. doi: 10.1128/EC.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jensen BC, Wang Q, Kifer CT, Parsons M. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J Biol Chem. 2003;278:32204–11. doi: 10.1074/jbc.M304198200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.