Summary
Immunoevasins are key proteins employed by viruses to subvert host immune responses. Determining their high-resolution structures is key to understanding virus-host interactions towards the design of vaccines and other antiviral therapies. Mouse cytomegalovirus (MCMV) encodes a unique set of immunoevasins, the m02-m06 family, that modulates major histocompatibility complex class I (MHC-I) antigen presentation to CD8+ T cells and natural killer (NK) cells. Notwithstanding the large number of genetic and functional studies, the structural biology of immunoevasins remains incompletely understood, due largely to crystallization bottlenecks. Here we implement a new technology employing sparse NMR data and integrative Rosetta modeling to determine the structure of the m04/gp34 immunoevasin extracellular domain. The structure reveals a new β fold that is representative of the m02-m06 family of viral proteins, several of which are known to bind MHC-I molecules and interfere with antigen presentation, suggesting its role as a diversified immune regulation module.
Introduction
Cytomegaloviruses (CMV) are important models of pathogen-host interactions, widely recognized for their ability to interfere with host immune responses to accomplish the multi-faceted task of inhibiting the recognition of infected cells by CD8+ T-cells while at the same time avoiding destruction by NK cells according to the “missing-self” hypothesis (i.e. the lack of antigen-presenting MHC-I molecules on the cell surface) (Lemmermann et al., 2012). To do this, the virus maintains a series of genes encoding immune evasion/regulatory proteins (Lilley and Ploegh, 2005). In particular, the m02-m16 family is a class of early-expressed genes of mouse CMV crucial for viral survival and infectivity. However, there is no identifiable homology to any determined structure in the protein data bank (PDB), nor amino acid sequence similarity to any other protein family. A member of the m02-m06 class, the m04/gp34 protein is unique in its ability to bind MHC class-I molecules in the endoplasmic reticulum (ER) and accompany them to the cell surface (Kleijnen et al., 1997), while the closely related m06 protein binds MHC-I and directs it to the endosome (Tomas et al., 2010). Therefore it has been proposed that, by countering the MHC-retaining functions of other viral proteins (m06, and the MHC-I-like m152), m04 helps MCMV to evade the NK cell response (Babic et al., 2011; Babic et al., 2010; Holtappels et al., 2006). The combined effects of m04, m06 and m152 on CD8+ T cells and NK responses reveal a complex and still poorly understood aspect of MCMV immune evasion (Pinto et al., 2006). While the structural basis for the function of m152/gp40 immunoevasin has been previously revealed by X-ray crystallography (Wang et al., 2012), and structures of other MCMV MHC-I-like proteins have been similarly characterized (Adams et al., 2007; Berry et al., 2013; Mans et al., 2007; Natarajan et al., 2006), for other molecules a detailed picture is still lacking due to difficulties in crystallization and the large size of their in vivo functional complexes that make conventional NMR studies challenging. In particular, extensive crystallization trials in our laboratory using different constructs of m04 repeatedly failed to yield well-diffracting protein crystals.
Recent advances in computational methods using the program Rosetta (Leaver-Fay et al., 2011) allow for accurate modeling of protein structures from sparse NMR datasets containing chemical shifts (CS), RDCs and a minimal subset of the proton-proton distances used by conventional methods (Shen et al., 2008). By combining the use of CS-Rosetta with extensive deuteration of sidechain protons to improve 13C relaxation and the use of transverse relaxation optimized spectroscopy (TROSY) at high magnetic fields to improve 15N and amide 1H relaxation (Pervushin et al., 1997), the structure of larger proteins and protein complexes can now be determined by solution NMR (Raman et al., 2010; Sgourakis et al., 2011). Methyl protons are re-introduced using site-specific labeling of Ile, Leu and Val (ILV) residues (Tugarinov et al., 2006), a scheme that greatly simplifies the NMR spectra while still providing a sparse set of long-range methyl-methyl NOEs as structural restraints. In addition, the use of the resolution-adapted structural recombination (RASREC) algorithm within Rosetta greatly enhances the sampling of non-local features such as long-range contacts through β-pairings (Bradley and Baker, 2006) and allows structural convergence in systems with complicated backbone topologies and high contact order (Lange and Baker, 2012). RASREC-Rosetta offers clear advantages over conventional simulated annealing protocols, in terms of both performance and convergence: In a benchmark set of 5 proteins of sizes 15-40kDa with known crystal structures, RASREC guided by ILV data was found to outperform conventional protocols in terms of both precision (convergence) and accuracy relative to the target structure (Lange et al., 2012). This is in part due to the use of an empirically optimized all-atom energy function that defines the local hydrogen-bonding and sidechain packing features once a sufficiently converged low-resolution model of the backbone (within 2-5 Å accuracy relative to the native structure) can be obtained using a sparse network of long-range experimental restraints. Thus, while experimental data are still required to define an overall backbone fold, Rosetta alleviates the need for extensive sidechain assignments and a high density of NOE restraints (in excess of 10 restraints per residue) that is typically required by conventional protocols. For larger, more challenging systems where the sparse ILV and amide-amide NOE data are insufficient to determine conclusively the target backbone structure, obtaining RDCs from spectra gathered in multiple alignment media is a powerful way to improve further structural convergence and validate the final Rosetta models, as shown in recent RASREC applications (Rao et al., 2014; Warner et al., 2011). Recent TROSY-based methods allow quantitative RDC measurements for larger proteins at high accuracy (Fitzkee and Bax, 2010).
Here we combine these advanced NMR technologies to determine the structure of the m04 extracellular domain (m04ED). The calculations converge to a well-defined structure showing an elaborate β-sheet topology that is reminiscent, but highly divergent from the canonical Ig fold (Williams and Barclay, 1988). Sequence alignments with other members of the viral m02-m06 family show that the structural features of the m04 core are broadly conserved among members of this family, suggesting that the interactions involved in MHC-I binding are likely to be similar as well.
Results
The m04 extracellular domain binds MHC class-I molecules
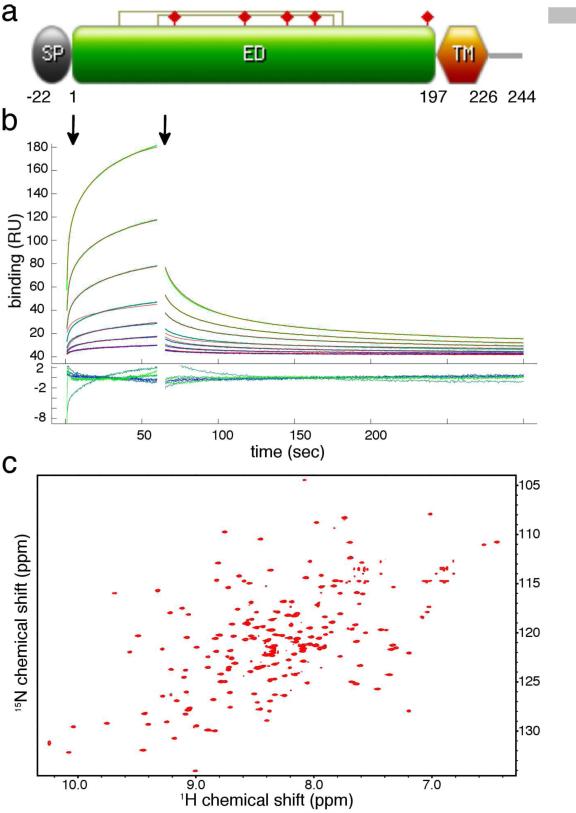
m04 was originally identified as a 34 kDa glycoprotein that co-immunoprecipitates with MHC-I molecules upon MCMV infection (Kleijnen et al., 1997). Towards determining the molecular requirements of m04 recognition by MHC, previous studies have highlighted the requirement for proper MHC folding and association with its light chain β2-microglobulin to accommodate m04 binding, suggesting that m04 interacts with properly conformed, peptide-loaded MHC-I molecules (Lu et al., 2006). This work further suggested that the transmembrane (TM) region of m04 is critical for its interaction with the MHC-I molecule H2-Kb. To test whether this is a general feature of m04/MHC interactions and to identify a minimal, functional m04 construct suitable for structural studies by NMR, we expressed the extracellular domain of the m04 protein (m04ED), excluding the signal peptide, the TM region and the intracellular C-terminal tail (Figure 1a). The protein was expressed in E.coli and therefore also lacked the post-translational glycosylations present in the wild-type protein (WT m04 has 5 consensus glycosylation sites of which 3 have been confirmed to be utilized in vivo (Kleijnen et al., 1997)). To evaluate the biological activity of recombinant m04 protein we examined its interaction with the MHC-I molecule H2-Dd by surface plasmon resonance (SPR). Preliminary SPR experiments (Figure 1b) demonstrate a direct but weak (KD ≥ 100μM) interaction between m04ED and the MHC-I molecule H2-Dd ,even in the absence of the TM region and post-translational modifications. By contrast, the MHC-I-like molecule, MULT-1, used here as a negative control, did not bind (Figure S1).
Figure 1.
Biophysical characterization of m04ED for structural studies. (a) Domain organization of full-length m04/gp34, indicating the positions of signal peptide (SP), extracellular domain (ED) and transmembrane domain (TM). Disulfide bonds are indicated with connected lines, and glycosylation sites with filled red prisms. (b) SPR binding sensograms collected using immobilized m04ED (WT) with increasing concentrations of H2-Dd flow-through, as outlined in Experimental Procedures. The start of the injection (association) and wash out (dissociation) phases are indicated with vertical arrows. The data were fit using EVILFIT (Svitel et al., 2003) (thin red lines – Kd ~395μM). Residual errors in the fit are shown in the inset. As a negative control the MHC-I-like molecule MULT-1 was injected over the same SPR surface (Figure S1). (c) TROSY-HSQC 1H-15N correlation spectra of m04ED −C7S recorded at 900 MHz, 12 °C, pH 6.5.
This is the first direct measurement of the MHC/m04 interaction using a biophysical technique. Previous studies demonstrated the requirement of the m04 transmembrane region for efficient m04/MHC interaction in detergent cell lysates (Lu et al., 2006). Taken together, these results support a model of weak interaction between the lumenal domains when strongly coupled via their transmembrane domains.
NMR backbone relaxation rates identify a well-ordered structural core
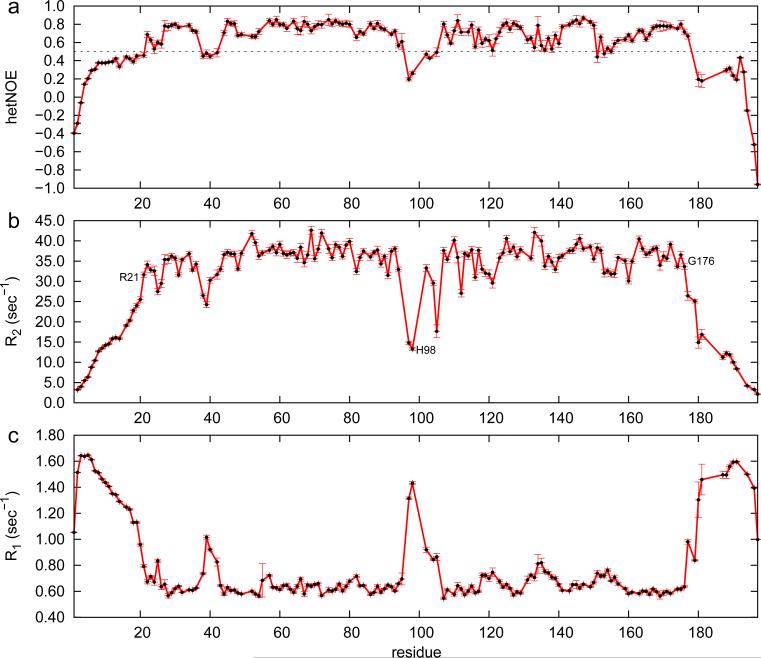
The optimized m04ED construct shows well-dispersed 2D 1H-15N HSQC spectra indicative of a folded, stable protein core (Figure 1c). We used backbone relaxation experiments to probe the extent of backbone conformational dynamics on the fast (ps-ns) timescale (Kay et al., 1989) . The resulting 15N R1 and R2 (Figure 2b, c) relaxation rates combined with the heteronuclear 15N-{1H} NOE (Figure 2a) indicate the presence of a well-ordered structural core spanning residues 21-176, that includes a flexible loop at residues 95-102. The N-terminal region (residues 1-20) is highly mobile on the nanosecond timescale, as evidenced by decreased 15N-{1H} NOE, increased R1 and decreased R2 relaxation rates. This highly charged segment (11/20 charged residues) includes 3.5 turns of a regular α-helix for residues 6-19, as indicated by the analysis of backbone chemical shifts using the program TALOS-N (Shen and Bax, 2013), and the presence of short-range dαN(i,i+3) and dNN(i,i+1) NOE patterns (Wuthrich, 1986), confirming the α-helical structure. The elevated dynamic characteristics and the absence of long-range HN-HN or HN-CH3 NOEs to core residues suggest that the N-terminal helix interacts only transiently with the core. Similarly, the C-terminal residues 177-197 are highly mobile on the ps-ns timescale, as indicated by the 15N relaxation data. This region is found primarily in a disordered loop/coil conformation that, like the N-terminus, does not participate in strong packing interactions with the core structure. Taken together, these results reveal the presence of a well-ordered protein fold for residues 21-176, flanked by two terminal capping sequences. We therefore focus on determining the structure of this m04ED core region, to elucidate its fold and identify structural features that could serve as possible binding sites for MHC-I molecules.
Figure 2.
15N amide relaxation rates and 15N-{1H} NOE ratios for the full-length m04ED sequence, recorded at 600 MHz 1H frequency. R2 values were obtained from measured R1ρ rates after correction for off-resonance effects (Massi et al., 2004). Key residues that demarcate the structural core (21-176) and flexible loop region (95-102) are highlighted on the plots. A dotted line in (a) indicates the 0.5 15N-{1H} NOE cutoff value used to define rigid and dynamic structural elements within the m04ED molecule.
Rosetta structure determination using RDCs and sparse ILV NOE data
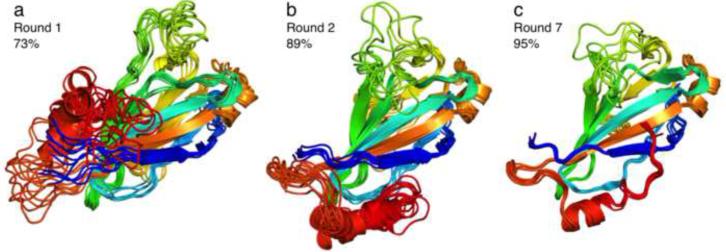
Towards obtaining a converged structure of the m04ED core, we performed a number of iterative CS-Rosetta calculations by progressively increasing the number of experimental restraints supplied (Table 1, Figure 3). First, using only the backbone chemical shifts (CS) and 23 long-range HN-HN NOEs (Table 1, #1, Figure 3a) we obtained a preliminary structure of the m04ED core, in which the first 130 residues (residues 21-150) converged to a well-defined fold. This fold consists of a 5-stranded antiparallel β-sheet platform (with the β-pairing topology β1 – β9 – β4 – β5 – β6), a shorter antiparallel 4-stranded sheet (β2 – β3 – β8 – β7) packing underneath the upper side of the platform, and connecting loops of different lengths, with a 6-residue helical segment located in the β8 – β9 loop that connects the bottom and top β-sheets (as outlined in Figure 6d). Two disulfide bonds at residues 26 to 147 (β1-β9) and 47 to 142 (β3-β9) stabilize the tertiary fold. Analysis of the 13Cα and 13Cβ chemical shifts of the four Cys residues confirms their oxidized state. While the chemical shifts help define the local backbone conformation, the long-range HN-HN NOEs constrain the β-pairing topology. Several salient features of the structure, including a β-bulge centered at Asp72 of strand β4 and a bend at Leu88 of strand β5, are supported by the NOE and CS data and confirm the power of the Rosetta structure determination protocol for this highly sparse NOE dataset (1.5 long-range restraints per 10 residues). The remaining 26 C-terminal residues of the core sequence (151-176) were poorly converged in the first round of models, as no long-range HN-HN NOEs could be identified for this part of the sequence. Analysis of the backbone chemical shifts using TALOS-N (Shen and Bax, 2013) indicates that this segment contains three turns of an α-helix (residues 160-171).
Table 1.
Summary of iterative CS-Rosetta structure calculations
| # | Experimental restraints used | Converged residues* | Fconverged | <ERosetta>** | <Qwork> | <Qfree>*** | Ct-helix+ |
|---|---|---|---|---|---|---|---|
| 1 | NOEamide | 24-94, 102-117, 123-149 | 73% | −212±4 | 1.43 | 1.23 | Top++ |
| 2 | NOEamide, RDC1 | 23-94, 105-170 | 89% | −224±2 | 0.40 | 0.68 | Under |
| 3 | NOEamide, RDC1, RDC2 | 21-95, 103-170 | 91% | −231±4 | 0.37/0.41 | N/A | Under |
| 4 | NOEamide, NOEmet-amide, RDC1 | 21-94, 103-174 | 94% | −225±3 | 0.38 | 0.58 | Under |
| 5 | NOEamide, NOEmet-amide,NOEmet-met | 21-94, 102-117, 123-172 | 90% | −230±3 | 0.54 | 0.80 | Under |
| 6 | NOEamide, NOEmet-amide,NOEmet-met, RDC1 | 21-94, 103-174 | 94% | −229±1 | 0.44 | 0.60 | Under |
| 7 | NOEamide, NOEmet-amide,NOEmet-met, RDC1, RDC2 | 21-94, 103-175 | 95% | −236±4 | 0.36/0.40 | N/A | Under |
Below 3 Å backbone RMSD in the 10 lowest-energy models
Average and S.D. computed over the 10 lowest-energy models, in Rosetta Energy Units
Computed using an independent RDC dataset not used in the structure calculations. N/A indicates that all available RDC data were used as restraints.
Placement of the C-terminal α-helical segment (residues 160-171) relative to the extended β-sheet platform
This segment is not converged within 3 Å backbone RMSD
Figure 3.
Convergence of the m04ED core structure at different phases of the structure determination process. Ribbon diagrams of the 10 lowest-energy models at selected stages of CS-Rosetta/RASREC structure calculations outlined in Table 1. Each ensemble is superimposed on the coordinates of the lowest-energy model and colored blue to red from N- to C-terminus. The fraction of residues converged within 3 Å backbone RMSD is indicated in each diagram. The final ensemble of (c) is converged to an average backbone RMSD of 0.85 Å, excluding the flexible loop region spanning residues 95-102.
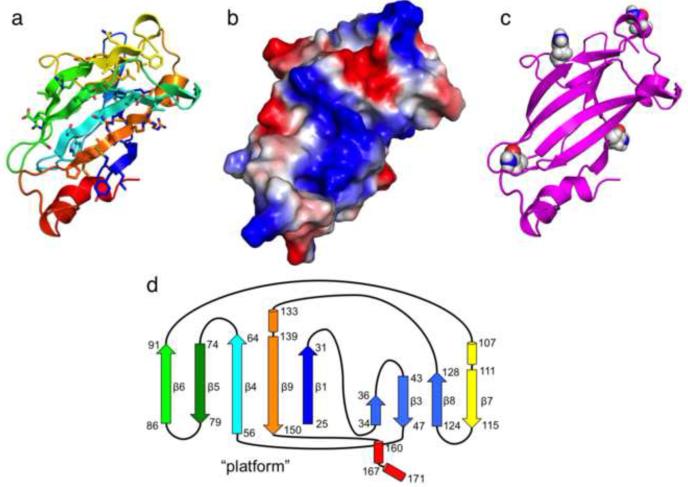
Figure 6.
m04ED core structure summary
(a) m04ED core NMR structure indicating the position of side chains on the conserved β-sheet platform. The extended loop residues (95-102) have been omitted for improved viewing. (b) Vacuum electrostatics surface computed using APBS (Baker et al., 2001), shown in the same molecular orientation of the platform as in (a). (c) High-confidence predicted glycosylation sites using NetNGlyc (Blom et al., 2004) (d) β-sheet connectivity diagram for the m04ED core structure. β–strands are drawn as arrows, and short α-helices as cylinders. The position of the “platform” β–sheet from panel (a) is indicated on the diagram.
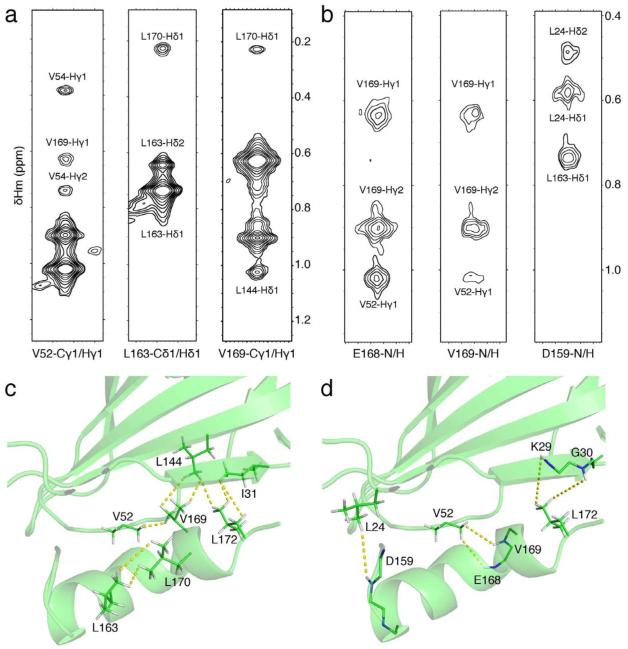
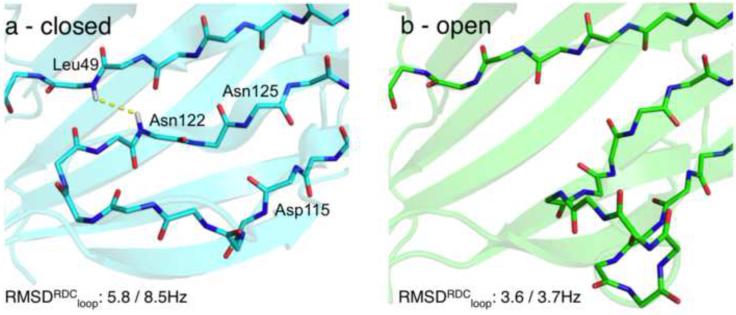
To better define the conformation of the C-terminal helix, we measured RDCs in two alignment media (Pf1 phage and positively charged gel) that report on the orientation of N-H vectors with respect to an overall alignment frame (Bax and Grishaev, 2005). The two datasets are complementary in the sense that the two alignment tensors show a normalized scalar product (Sass et al., 2000) of only 0.51 (corresponding to a 60° angle in 5-dimensional alignment tensor space), meaning that the two measurements are quite independent from one another. Additionally, the linear correlation between the raw RDC datasets is 0.36, further suggesting that the two datasets are for all practical purposes sufficiently independent (Figure S2c) (Tolman and Ruan, 2006). Inspection of the RDCs for core residues 160-171 (Figure S2a, b) reveals a kink in the C-terminal helix, centered at Ser167, in which the helical axis changes orientation by an angle of ~60°. This trend is consistent between the two RDC datasets and is further supported by the backbone chemical shift data. We then performed two sets of calculations (Table 1 #2,3) using the RDCs in addition to the HN-HN NOEs and backbone chemical shifts. In the first run we used only RDCs from the Pf1-aligned sample, while reserving the second RDC dataset, measured in positively charged gel, for validation. The calculations resulted in a better converged C-terminus (backbone RMSD within 3Å), in which the kinked α-helix packs underneath strands β1 and β9 of the lower end of the extended β-sheet platform, with hydrophobic contacts mediated by the side chains of Met162, Leu163, Met166, Val169, Leu170 and Leu172 (Figure 3b). This placement of the C-terminal kinked helix cross-validates well with respect to the second, unused RDC dataset (Qfree (Cornilescu et al., 1998) of 0.68 versus 1.23 for the first-round models (constructed with no RDC restraints), corresponding to a Pearson’s linear correlation coefficient Rp of 0.72 (Cornilescu and Bax, 2000)). However, even with the inclusion of the second RDC dataset as weak restraints in the calculation, the convergence of the C-terminal helix was insufficient to define an atomic model (i.e. with a backbone RMSD of <2Å), due to the lack of long-range NOEs for the helical residues 160-171 (Table 1, #3). We then recorded additional 3D NOESY spectra using a sparsely labeled ILV sample with the aim to improve structural convergence for the C-terminal segment and to allow further validation of the structure. Careful analysis of the spectra revealed a total of 19 long-range HN-CH3 NOEs that were included in the subsequent round of calculations (Table 1, #4). This set includes five restraints from Asp159, Val168, Leu169 and Leu172 that link the C-terminal helix to the main core of the structure (Figure 4b, d), while the remaining 14 restraints validate the β-sheet topology obtained in the previous models. The resulting models show improved convergence of the C-terminus (backbone RMSD within 1.5Å), and RDC cross-validation statistics (Qfree = 0.58 / Rp = 0.82). Finally, we identified 25 long-range CH3-CH3 NOEs in the corresponding methyl NOESY spectra, 6 of which map to the C-terminal α-helix (Figure 4a, c), validating the previous round of models (Table 1, #5, 6, 7). All NOE cross-peaks with a peak signal to noise greater than 10, that also showed a mirror peak in the case of HN(N)H, (H)NNH, H(C)CH and (H)CCH 3D NOESY spectra, were manually assigned and classified to either intra-residue, sequential or medium-range (that further validated the backbone and methyl resonance assignments), and long-range corresponding to a sequence separation greater than 5 residues. While the short to medium-range NOEs were consistent with the first-round Rosetta models, inclusion of such restraints in the calculations does not lead to any gains in convergence of the local backbone structure that is already heavily constrained by the chemical shift data. Including all the available long-range NOEs, backbone chemical shifts and RDC data (Table 1, #7) led to a structural ensemble (Figure 3c) that is converged to within a backbone RMSD of 0.85 Å, excluding the loop region spanning residues 95-102 (Figure S3b). This loop is highly mobile on the ps-ns timescale, as evidenced by the reduced 15N-{1H} NOE values (Figure S3a), and further supported by the near-zero RDC values (Figure S2a, b). We find that the use of RDC restraints strongly improves convergence for the backbone core spanning residues 21-176 (95%, with the remaining 5% of the sequence corresponding to the flexible loop at residues 95-102), including a short, ordered loop at residues 118-122 (Table 1 - compare #5 with #6,7). The open, type-I’ conformation of the loop (Figure 5b) is supported by analysis of the backbone chemical shifts (Shen and Bax, 2012), and further validated by the absence of a long range HN-HN NOE between Leu49 and Asn122 that would be expected for an alternative closed conformation sampled in the RDC-free calculations (Figure 5a). The final full-atom models (Table 1, #7, Table 2 and Figure 3c) have good packing and structural statistics (assessed separately using MOLPROBITY (Chen et al., 2010)) while satisfying all experimental NOE restraints and showing good fits to both RDC datasets (Figure S4a,b).
Figure 4.
ILV NOE structural restraints used to determine the placement of the C-terminal helix.
Representative 1H-1H strips from the CH3-CH3 (a) or HN-CH3 (b) NOESY 3D 1H-separated spectra, recorded using an ILV-labeled sample. Structure diagrams indicating the position of long-range CH3-CH3 (c) and HN-CH3 NOEs (d) involving at least one residue from the C-terminal helix, residues 159-172. The same 1H ppm y-scale is used for all NOE strips within (a) and (b), as indicated in the axis of the last strip from each group. The strips were drawn at the ppm values corresponding to the methyl (a) or amide (b) atom pair assignments indicated below each panel, with each x-axis centered on the Hm (a) or HN (b) assigned chemical shift values. In panel (a), the first and second most intense peaks correspond to the diagonal (self-peak) and second methyl group within the same spin system, respectively. The assignments of each additional NOE cross peak are further indicated. The spectra were contoured using 10 levels spaced by factors of 1.4, starting at a height equal to 2 times the background noise. The lowest-energy model in the final NMR ensemble was used in panels (c), (d).
Figure 5.
Possible loop conformations for residues D115-1N125. (a) Closed and (b) Open loop conformations sampled in Rosetta structure calculations using only the sparse NOE distance restraints (a) or NOEs supplemented by amide RDCs from two alignment media (b). The RMS of the residual χ2 in the RDC structure fits (Pf1 / Gel) is indicated for each conformation. A strong HN-HN NOE cross-peak between the well-resolved resonances of Leu49 and Asn122 (indicated with a yellow line in (a)) expected in the "closed" conformation was not observed in the H(N)NH and (H)NNH NOESY spectra, further supporting the "open" loop conformation. While the NOE data alone are insufficient to define the loop, convergence to the “open” conformation is obtained when RDC restraints are included in addition to NOEs.
Table 2.
NMR restraints and structural statistics
| NMR distance, orientation & dihedral angle restraints |
|
|---|---|
| Distance restraints | |
| Total NOE (long-range) | 67 |
| HN-HN | 23 |
| HN-CH3* | 19 |
| CH3-CH3* | 25 |
| Total dihedral angle restraints++ | 292 |
| Φ | 146 |
| Ψ | 146 |
| Total RDC restraints | 231 |
| dataset 1 - Pf1 | 119 |
| dataset 2 – gel | 112 |
| Structural statistics**+ | |
| RDC Q-factors | |
| dataset 1 - Pf1 | 0.36 |
| dataset 2 – gel | 0.40 |
| Average RMSD (Å)** | |
| Backbone | 0.85 |
| All heavy atom | 1.25 |
used to bias the selection of 3mer and 9mer backbone fragments, as outlined in Experimental Procedures
not stereo-specifically assigned
computed over the 10 lowest-energy structures, excluding the loop region at residues 95-105
no NOE violations (distance > 4.0 Å between interacting sites) present in the final ensemble
The m04 core structure exemplifies a highly diverse Ig-based immunoevasin fold
m04 does not show any homology to non-viral proteins by amino acid sequence comparisons or to any sequence of known structure in the PDB. At first sight, the m04 core resembles the fold of the immunoglobulin superfamily (Ig), that can be further divided into 4-9 main structural classes (Bateman et al., 1996; Halaby et al., 1999) depending on overall size, strand connectivity and loop size. A Dali (Holm and Rosenstrom, 2010) search using the NMR structure as a target suggests a statistically significant structural similarity (Z-score ≤ 5.0) to a variety of Ig-like molecules (Igs) such as various antibody chains (eg. PDB ID: 1AXS-chain B) and HLA class-II Ig-like domains (eg. 4H1L-chain H). Moreover, the disulfide bond at C142 – C47 with Trp62 packing against it and the β strand arrangement are clearly indicative of an Ig fold. However, a closer look at the structural alignments reveals major differences from the canonical Ig superfamily.
While the Ig fold is typically characterized by seven strands on two sheets (SCOP (Andreeva et al., 2008)), the m04 core structure has a total of nine strands, four of which (β2 – β31 – β8 – β7) are short (4-5 residues each) and form one layer of the β-sandwich, while five strands of 6-12 residues in length (β1 – β9 – β4 – β5 – β6) form an extended β-sheet platform on the other side of the sandwich (Figure 6d). The five-stranded platform provides a nucleus beneath which the smaller four-stranded sheet packs at its upper end. The kinked C-terminal helix supports the lower end of the larger five-stranded platform. In addition, the strand connectivity is clearly different from the closest Ig fold, found in the antibody variable (V) domains (typically an A(β1), B(β2), E(β5), D(β4) sheet packed against C(β3), F(β6), G(β7). Furthermore the spacing between the Cys residues forming the disulfide bond that links the two β-layers (typically between the B and F strands) (Cys47, Cys142) is significantly larger than found in Igs (95 vs 55-75 residues), and connects β3 to β9. Finally, m04 has one additional disulfide bond connecting Cys26 (β1) and Cys147 (β9). These features, specifically the secondary structural elements forming the hydrophobic core of the fold and the second disulfide, are highly conserved in the m02-m06 family and among different MCMV strains (Corbett et al., 2007) (Figure 7). In particular, this family preserves the NAXWXXE/HW motif (in strand β4) throughout a large number of laboratory and wild-derived isolates of MCMV. This segment is a central element of the m04 fold, likely to play a key role in stabilizing the core structure, that provides a scaffold for the grafting of various loop sequences among different family members. Taken together, these results suggest that the m04 structure is likely a product of convergent evolution towards an Ig-like structure rather than a true structural homolog of Igs.
Figure 7.
Multiple sequence alignment of m04ED with the predicted extracellular domains of other m02-m06 family members. The approximate positions of conserved secondary structure elements are highlighted on the top of each alignment block (β1-β9: β-strands, red ovals: α-helices). The position of the two disulfide bonds on the m04ED structure is indicated with capital C letters and connecting dashed lines, while high-confidence predicted glycosylation sites on the m04ED sequence are indicated with stars. Protein sequences for m02 (UniProt ID: YP_214010) m03 (YP_214011) m05 (YP_214013), and m06 (YP_214014) were all taken from the Smith MCMV genome: NC_004065. The sequence of m03.5 (ABM74010), not present in the Smith strain was taken from strain G1F.
To identify the ER-lumenal domains of these proteins from the full-length protein sequence, we first identified the end of the signal peptide using the SignalP (www.cbs.dtu.dk/services/SignalP/) and Phobius (http://phobius.sbc.su.se/) servers, then identified the start of transmembrane domains using TMpred (www.ch.embnet.org/software/TMPRED_form.html). Sequence alignments were constructed using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2/) and colored by sequence similarity using BoxShade (www.ch.embnet.org/software/BOX_form.html).
Discussion
This description of the solution structure of the m04/gp34 glycoprotein extracellular domain reveals the power of a hybrid methodology that combines multidimensional NMR with sparse labeling and data collection techniques and integrative computational modeling (Ward et al., 2013). The value of progressive addition of experimental datasets in achieving a converged set of minimum energy structures is documented here, and highlights the importance of including ILV NOEs and RDCs in the proper placement of structural elements. In particular, these distance and orientational restraints permitted the placement of the C-terminal α-helical segment to a defined position underlying a β-sheet platform, and precise modeling of a second loop conformation. The dynamic aspects of the m04ED structure revealed by NMR relaxation experiments, notably the highly mobile amino and carboxy-terminal segments as well as the central loop region, may relate to the function of the molecule in its ability to interact with a number of diverse MHC-I molecules as well as the difficulty in identifying suitable crystallization conditions.
The overall structure of m04ED reveals a novel β topology, punctuated with connecting regions containing both coil and helix, distantly related to the Ig fold. This core β structure permits variation in the connecting loops as evidenced by the amino acid sequence diversity observed in other members of the m02-m06 viral protein family, and should prove useful in designing crystallizable variants towards determining the structures of other family members. The closely related m06 also carries out an immunomodulatory function and binds MHC-I molecules (Reusch et al., 1999). Amino acid sequence analysis of the extended family that includes m02-m16 has suggested that all of these genes encode structurally related proteins, and the structure of m04ED provides a toehold in deriving their structures. Whether the structural similarities carry over to related functions or the m04 structure serves as a robust scaffold for loop variability offering novel function remains to be determined.
m04/gp34 is post-translationally processed in the ER and contains at least three N-asparaginyl-linked carbohydrate moieties (Kleijnen et al., 1997). We have determined the NMR structure of m04 using a bacterially expressed construct, so it is important to consider the predicted location of the N-asparaginyl sites in the context of this structure. An analysis of the amino acid sequence of m04 using the NetNGlyc 1.0 Server (Blom et al., 2004) identifies seven potential N-X-S/T sites in m04ED, of which five, at positions 5, 32, 55, 116 and 131, are likely to be used. The core of m04ED contains four of these sites, and the locations of the Asn residues on the amino acid sequence alignment and on the ribbon diagram of the structure are shown in Figure 6c and Figure 7. All of the glycosylation sites are located in accessible loop regions and are not expected to interfere with the proper folding of the core structure.
Immunoprecipitation experiments of MCMV infected cells demonstrate that m04 is the major component associated with MHC-I molecules (Kleijnen et al., 1997). Biochemical experiments indicate the importance of a functional peptide loading apparatus (including transporter associated with antigen processing (TAP) and tapasin) as well as the transmembrane region of m04 for the association (Lu et al., 2006). Our experiments, employing a truncated m04 construct lacking the transmembrane and cytoplasmic regions of the protein, indicate that the m04ED is sufficient for a transient interaction with MHC-I, and that the m04ED may contribute to interaction with the lumenal domain of assembled MHC-I heterotrimers (MHC-I heavy chain, β2m, and peptide). The apparent low affinity of the m04ED interaction with soluble MHC-I contrasts with the results of immunoprecipitation experiments of virus-infected or transfectant cell lysates, where m04 is a predominant MHC-I-associated molecule (Kleijnen et al., 1997; Lu et al., 2006). These differences may also result from the clear differences in measurement of association of molecules constrained by two-dimensional membrane surfaces as compared to three-dimensional solution measurements. Alternatively, in addition to the demonstrated role of the transmembrane regions, we cannot rule out the participation of additional cellular or viral components in this interaction. Inspection of the structure and consideration of other molecules that interact with MHC-I and MHC-I-like molecules (T cell receptors, CD8 co-receptor, NK receptors, other viral immunoevasins including m152, US2, CV203, E3-19K glycoprotein) prompts us to speculate on possible m04ED sites that might participate in MHC-I interactions. Examination of the surface electrostatic potential of m04ED (Figure 6b) suggests that the large basic groove along the platform of the five-stranded β-sheet formed by Arg143 and Arg145 on β9 and Arg89 on β6 may accommodate an extended acidic region of the MHC-I, like the conserved region observed on many MHC-I α3 domains, and particularly notable on H2-Dd (Achour et al., 1998; Li et al., 1998; Wang et al., 2002). The MHC footprint on m04 and vice versa can be mapped using high-resolution NMR as shown previously for interactions involving the T cell receptor (Duchardt et al., 2007; Varani et al., 2007), an approach that is currently pursued in our laboratory using labeled H2-Dd.
The novelty of the m04 fold, the lack of homologous amino acid sequences in any genomic database (including genomes of the closely related rat CMV (Vink et al., 2000)), and the preservation of a range of related family members in a number of different mouse CMV isolates raise the question as to whether the m02-m06 family has arisen uniquely in the mouse CMVs and rapidly expanded because of its utility, or that CMVs of other species lost members of the family because of lack of selective advantage. For example, the closely related K181, G4 and Smith MCMV strains all have functional m02-m06 genes while strains G1F and MI6A have substituted m03.5 for m03 and strains W8211 and W8 have both m03 and m03.5 in addition to m02, m04-m06 (Corbett et al., 2007). The availability of this first structure of an m02 family paradigm may not only contribute to a detailed understanding of the basis of its MHC regulatory function, but may provide insight into the expansion and contraction of immunomodulatory viral gene families (Smith et al., 2013).
Experimental Procedures
Sample preparation for SPR and NMR
DNA encoding m04 was PCR-amplified from virus containing cell culture supernatants and has the sequence of the K181 laboratory strain of MCMV (Uniprot ID A2Q6L0), with the additional mutations: I31V, I44V. m04ED was expressed in E. coli as insoluble inclusion bodies, denatured in 6M guanidine-HCl, refolded in vitro, and purified by size exclusion and ion exchange chromatographies. The final sample conditions for NMR were 0.5 mM m04ED, 50 mM NaCl in 20 mM NaH2PO4 buffer, pH 6.5. The quality of the 2D 1H-15N HSQC correlation spectra was further improved by the single C7S mutation, leading to well-resolved spectra free of conformational exchange line broadening. However, protein degradation and aggregation limited sample life to 3-5 weeks, even while keeping the sample at a relatively low temperature of 12 °C during NMR data collection. The extracellular domain of H2-Dd and mouse β2-microglobulin for SPR studies were expressed, refolded with the synthetic decamer peptide p18-I10 (RGPGRAFVTI) and purified as described previously (Wang et al., 2009).
SPR measurements
Surface plasmon resonance (SPR) binding studies were carried out on a Biacore T100 System (GE Healthcare Life Sciences) in which a CM5 Sensor chip was derivatized with 6300 resonance units (RU) of recombinant m04ED. The purified m04ED protein was coupled to the dextran surface of a CM5 sensor chip at a density of 1000 RU, following standard N-hydroxysuccinimide (NHS) / N-ethyl-N'-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) activation and covalent coupling at pH 5.0 (Corr et al., 1993). Graded concentrations of H2-Dd/p18-I10 (Figure 1b) or MULT-1, used here as a negative control, (Figure S1), prepared and purified as outlined above, were then injected over the m04 surface in HBST buffer (HBS-EP, 10 mM HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% surfactant P20). For MULT1, concentrations were 38.76 μM, 19.4 μM, 9.7 μM, 4.85 μM, 2.4 μM and 1.2 μM; for H2-Dd/p18I10/β2m, 21.5 μM, 10.75 μM, 5.4 μM, 2.7 uM, 1.35 μM, 0.67 μM, and .335 μM. Analyte proteins were offered to the coupled surfaces at a flow rate of 30 μl/min, for 60 sec, at which time washout was initiated and carried out for 600 sec. Regeneration of the m04ED surface was carried out with a 30 sec pulse of 0.5M NaCl followed by re-equilibration for 100 sec in running buffer. Data were corrected for background binding to a mock coupled surface, and analyzed with Evilfit (Svitel et al., 2003).
NMR backbone, sidechain assignments, backbone relaxation rates and ILV NOE measurements
All experiments were recorded at a temperature of 12 °C using 600 MHz, 800 MHz and 900 MHz cryoprobe-equipped Bruker spectrometers. We used an array of standard triple-resonance assignment experiments (HNCO, HN(CA)CO, HNCA and HNCACB) supplemented with H(N)NH and (H)NNH 3D NOESY datasets, recorded at 600 MHz using a mixing time of 250 ms. All resulting spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with Sparky (Goddard and Kneller). Orthogonal projections from each 3D dataset were extracted based on the centered HNCO peak positions and visualized using the strip-plotting interface in nmrDraw (scroll.tcl), obtaining a highly consistent network of final assignments, with a completeness >95% (excluding Pro residues). The strip-plot visualization allowed identification of 23 strong long-range HN-HN NOEs, sufficient to define a protein fold for the m04ED core, as outlined in detail in the structure calculation section.
To assign selectively labeled ILV methyls, we used the SIM-HMCM(CGCBCA)CO (Tugarinov et al., 2014) and HMCM(CG)CBCA (Tugarinov and Kay, 2003) triple-resonance spectra recorded at 600 MHz that link the methyl resonances to the previously established backbone C' and Cβ/Cα resonance assignments, respectively. These experiments were performed using a deuterated sample that was specifically 13C-labelled at the sidechains of ILV residues such as to yield a linear spin system (Tugarinov et al., 2006). In this manner we obtained complete assignments for all 66 labeled Cδ1, Cδ1/Cδ2 and Cγ1/Cγ2 methyls in the m04 sequence (containing 2 Ile, 16 Leu and 16 Val residues respectively), that were identified in a high-resolution methyl HMQC spectrum obtained at 900 MHz using a separate ILV sample that was 13C-labeled only at the methyl carbon atoms (Figure S5). Finally, using the same sample, we recorded two complementary 3D methyl-to-amide NOESY spectra at 900 MHz and two methyl-to-methyl NOESY datasets at 600 MHz using 250 ms and 200 ms mixing times, respectively. The processed methyl NOE data further validated the methyl assignments and provided at total of 44 long-range contacts (19 HN-CHM3 and 25 CH3-CH3) that were manually picked and readily assigned using Sparky (Goddard and Kneller).
Backbone amide 15N R1 relaxation rates and heteronuclear NOE ratios were measured from a perdeuterated, amide 1H sample using TROSY-readout methods (Lakomek et al., 2012). R2 rates were obtained from rotating-frame R1ρ rates (Massi et al., 2004) measured under a spin-lock field strength of 2 kHz, after correction for the 15N off-resonance, tilted field. Uncertainties in the R1 and R1ρ measurements were estimated from the spectral noise levels using 21 Monte Carlo simulations, while uncertainties in the heteronuclear NOE ratios were propagated directly from the noise levels in the reference and attenuated spectra. All backbone relaxation spectra were recorded at 600 MHz.
RDC measurements
We measured RDCs under two different alignment conditions. Initially, we explored a 15 mg/ml Pf1 phage sample, but this resulted in strong over alignment of m04 due to electrostatic interactions of the positively charged protein (+8) with the highly negatively charged phage particles. To circumvent this problem we decreased the Pf1 concentration to 7.5 mg/ml, while also increasing the salt concentration to 200 mM. Under these conditions, Pf1 is in the paranematic phase region, where the alignment strength (measured by the residual 2H quadrupole splitting of the lock solvent) scales with the strength of the magnetic field (Zweckstetter and Bax, 2001), which was further used as a handle to adjust the alignment strength to a desirable range while retaining good spectral resolution. ARTSY (Fitzkee and Bax, 2010) spectra were recorded at 600 MHz, leading to a 28 Hz range of final RDC values. We obtained a complementary RDC dataset in positively charged 5.5% w/v acrylamide/bis-acrylamide stretched gel (Chou et al., 2001) (containing 6%wt/v DADMAC-14) by recording ARTSY spectra at 800 MHz, leading to a 26Hz range of RDC values (Figure S2). As a figure of merit for the goodness-of-fit of the models to the experimental RDC data, we used the Q factor (Clore and Garrett, 1999; Cornilescu et al., 1998), that reports the deviation of the back-calculated RDCs from the raw data, relative to a range of RDCs estimated from a randomly distributed set of vectors assuming an alignment tensor of known Da and R parameters:
where Da and R refer to the magnitude and rhombicity of the alignment tensor , and Dcalc and Dobs are the calculated and observed dipolar couplings, respectively. Qfree is the Q-factor computed for a linearly independent RDC dataset, not used in the structure calculations. The Pearson’s linear correlation coefficient Rp is defined as: where cov(Dcalc, Dobs) is the covariance between the individual computed and calculated RDCs, and σDcalc, σDobs are the standard deviations of the experimental and computed RDCs, respectively.
Iterative CS-Rosetta structure calculations
We used the recently developed RASREC protocol (Lange and Baker, 2012), guided by the backbone chemical shifts, a total of 67 long-range NOE distance restraints (23 HN-HN, 25 CH3-CH3 and 19 HN-CH3) and 231 RDCs from two alignment media as N-H vector orientation restraints. In summary, the approach is based on several cycles of Monte Carlo-based trials, which include the sampling of backbone fragments (3-mers and 9-mers) and β-strand pairings. Structural features that consistently lead to optimization of a target function (defined as the sum of the Rosetta energy and the experimental NOE and RDC score terms) are recombined during 6 generations of iterative structure calculations. In a series of benchmark calculations with targets of known structures, the protocol showed improved sampling efficiency over both standard CS-Rosetta structure calculations (Shen et al., 2008) and conventional NMR structure determination programs (Herrmann et al., 2002) for β-rich proteins with complicated topologies (Lange et al., 2012).
While the backbone chemical shifts are used to guide the selection of backbone fragments from high-resolution structures in the PDB (Vernon et al., 2013), long-range NOEs help define the overall protein fold and β-strand connectivity. RDCs are used as bond vector orientation restraints relative to an overall alignment frame to better define the local backbone structure, particularly in loop regions that have few or no long-range NOEs. The calculations are heavily restrained by the experimental data during the low-resolution backbone search. However, the final placement of side chain rotamers and fine-tuning of the backbone torsion angles is guided primarily by the Rosetta energy function (Leaver-Fay et al., 2011) with almost no bias from the experimental score terms.
NOEs were implemented as flat-bottom restraints with an upper limit of 4.0 Å and an exponential penalty function. In the absence of stereospecific methyl proton assignments, all pairwise combinations of protons within each pair of interacting methyl sites were averaged as r−6 to compute an effective NOE distance (Nilges, 1993). In all calculations, the connectivity of disulfide bonds was also used as an input restraint using an orientation-dependent potential (Raman et al., 2009). All calculations were carried out in 200 threads of an SGI UV2000 cluster using Intel Xeon E5-4640 processors at 2.40 GHz with 1024 GB of memory, customized to accommodate the MPI interface required by RASREC. Typical calculation runtimes under these conditions were 10-12 h. The final 10 lowest-energy models were deposited in the PDB (ID 2MIZ).
Supplementary Material
Highlights.
The m04 protein interacts with MHC-I molecules to interfere with antigen processing
Difficulties in crystallization of m04 have hindered structural characterization
Using sparse NMR data and integrative modeling we have determined the m04ED structure
The structure reveals a novel fold representative of the m02-m06 viral protein family
Acknowledgements
We thank Dr. Oliver Lange for providing consult with setting up the RASREC-Rosetta protocol and Dr. Vitali Tugarinov for assistance with the SIM-HMCM(CGCBCA)CO and HMCM(CG)CBCA triple-resonance experiments. We are grateful to Drs. Nathan May for preparing multiple sequence alignments of the m02 family and to Drs Yang Shen and Lishan Yao for preliminary work on determining optimal m04 sample conditions, as well as to Dr. Manqing Hong for providing the MULT-1 protein sample. This work is supported by the NIDDK and NIAID intramural research programs and the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achour A, Persson K, Harris RA, Sundback J, Sentman CL, Lindqvist Y, Schneider G, Karre K. The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition. Immunity. 1998;9:199–208. doi: 10.1016/s1074-7613(00)80602-0. [DOI] [PubMed] [Google Scholar]
- Adams EJ, Juo ZS, Venook RT, Boulanger MJ, Arase H, Lanier LL, Garcia KC. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci U S A. 2007;104:10128–10133. doi: 10.1073/pnas.0703735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic M, Krmpotic A, Jonjic S. All is fair in virus-host interactions: NK cells and cytomegalovirus. Trends Mol Med. 2011;17:677–685. doi: 10.1016/j.molmed.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic M, Pyzik M, Zafirova B, Mitrovic M, Butorac V, Lanier LL, Krmpotic A, Vidal SM, Jonjic S. Cytomegalovirus immunoevasin reveals the physiological role of "missing self" recognition in natural killer cell dependent virus control in vivo. J Exp Med. 2010;207:2663–2673. doi: 10.1084/jem.20100921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Eddy SR, Chothia C. Members of the immunoglobulin superfamily in bacteria. Protein Sci. 1996;5:1939–1941. doi: 10.1002/pro.5560050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A, Grishaev A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol. 2005;15:563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Berry R, Ng N, Saunders PM, Vivian JP, Lin J, Deuss FA, Corbett AJ, Forbes CA, Widjaja JM, Sullivan LC, et al. Targeting of a natural killer cell receptor family by a viral immunoevasin. Nat Immunol. 2013;14:699–705. doi: 10.1038/ni.2605. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bradley P, Baker D. Improved beta-protein structure prediction by multilevel optimization of nonlocal strand pairings and local backbone conformation. Proteins. 2006;65:922–929. doi: 10.1002/prot.21133. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J Biomol NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- Clore GM, Garrett DS. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. Journal of the American Chemical Society. 1999;121:9008–9012. [Google Scholar]
- Corbett AJ, Forbes CA, Moro D, Scalzo AA. Extensive sequence variation exists among isolates of murine cytomegalovirus within members of the m02 family of genes. J Gen Virol. 2007;88:758–769. doi: 10.1099/vir.0.82623-0. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Bax A. Measurement of proton, nitrogen, and carbonyl chemical shielding anisotropies in a protein dissolved in a dilute liquid crystalline phase. Journal of the American Chemical Society. 2000;122:10143–10154. [Google Scholar]
- Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. Journal of the American Chemical Society. 1998;120:6836–6837. [Google Scholar]
- Corr M, Boyd LF, Padlan EA, Margulies DH. H-2Dd exploits a four residue peptide binding motif. The Journal of experimental medicine. 1993;178:1877–1892. doi: 10.1084/jem.178.6.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Duchardt E, Sigalov AB, Aivazian D, Stern LJ, Schwalbe H. Structure induction of the T-cell receptor zeta-chain upon lipid binding investigated by NMR spectroscopy. Chembiochem. 2007;8:820–827. doi: 10.1002/cbic.200600413. [DOI] [PubMed] [Google Scholar]
- Fitzkee NC, Bax A. Facile measurement of (1)H-(1)5N residual dipolar couplings in larger perdeuterated proteins. J Biomol NMR. 2010;48:65–70. doi: 10.1007/s10858-010-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY3.
- Halaby DM, Poupon A, Mornon JP. The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 1999;12:563–571. doi: 10.1093/protein/12.7.563. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtappels R, Gillert-Marien D, Thomas D, Podlech J, Deegen P, Herter S, Oehrlein-Karpi SA, Strand D, Wagner M, Reddehase MJ. Cytomegalovirus encodes a positive regulator of antigen presentation. J Virol. 2006;80:7613–7624. doi: 10.1128/JVI.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Huppa JB, Lucin P, Mukherjee S, Farrell H, Campbell AE, Koszinowski UH, Hill AB, Ploegh HL. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 1997;16:685–694. doi: 10.1093/emboj/16.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakomek NA, Ying J, Bax A. Measurement of (1)(5)N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange OF, Baker D. Resolution-adapted recombination of structural features significantly improves sampling in restraint-guided structure calculation. Proteins. 2012;80:884–895. doi: 10.1002/prot.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci U S A. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmermann NA, Fink A, Podlech J, Ebert S, Wilhelmi V, Bohm V, Holtappels R, Reddehase MJ. Murine cytomegalovirus immune evasion proteins operative in the MHC class I pathway of antigen processing and presentation: state of knowledge, revisions, and questions. Med Microbiol Immunol. 2012;201:497–512. doi: 10.1007/s00430-012-0257-y. [DOI] [PubMed] [Google Scholar]
- Li H, Natarajan K, Malchiodi EL, Margulies DH, Mariuzza RA. Three-dimensional structure of H-2Dd complexed with an immunodominant peptide from human immunodeficiency virus envelope glycoprotein 120. J Mol Biol. 1998;283:179–191. doi: 10.1006/jmbi.1998.2091. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol Rev. 2005;207:126–144. doi: 10.1111/j.0105-2896.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Lu X, Kavanagh DG, Hill AB. Cellular and molecular requirements for association of the murine cytomegalovirus protein m4/gp34 with major histocompatibility complex class I molecules. J Virol. 2006;80:6048–6055. doi: 10.1128/JVI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans J, Natarajan K, Balbo A, Schuck P, Eikel D, Hess S, Robinson H, Simic H, Jonjic S, Tiemessen CT, et al. Cellular expression and crystal structure of the murine cytomegalovirus major histocompatibility complex class I-like glycoprotein, m153. J Biol Chem. 2007;282:35247–35258. doi: 10.1074/jbc.M706782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi F, Johnson E, Wang C, Rance M, Palmer AG., 3rd NMR R1 rho rotating-frame relaxation with weak radio frequency fields. Journal of the American Chemical Society. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Hicks A, Mans J, Robinson H, Guan R, Mariuzza RA, Margulies DH. Crystal structure of the murine cytomegalovirus MHC-I homolog m144. J Mol Biol. 2006;358:157–171. doi: 10.1016/j.jmb.2006.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilges M. A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins. 1993;17:297–309. doi: 10.1002/prot.340170307. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AK, Munks MW, Koszinowski UH, Hill AB. Coordinated function of murine cytomegalovirus genes completely inhibits CTL lysis. J Immunol. 2006;177:3225–3234. doi: 10.4049/jimmunol.177.5.3225. [DOI] [PubMed] [Google Scholar]
- Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, et al. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327:1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei JM, Kim D, Kellogg E, DiMaio F, Lange O, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins-Structure Function and Bioinformatics. 2009;77:89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao T, Lubin JW, Armstrong GS, Tucey TM, Lundblad V, Wuttke DS. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proc Natl Acad Sci U S A. 2014;111:214–218. doi: 10.1073/pnas.1316453111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch U, Muranyi W, Lucin P, Burgert HG, Hengel H, Koszinowski UH. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. The EMBO journal. 1999;18:1081–1091. doi: 10.1093/emboj/18.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J Biomol NMR. 2000;18:303–309. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- Sgourakis NG, Lange OF, DiMaio F, Andre I, Fitzkee NC, Rossi P, Montelione GT, Bax A, Baker D. Determination of the structures of symmetric protein oligomers from NMR chemical shifts and residual dipolar couplings. J Am Chem Soc. 2011;133:6288–6298. doi: 10.1021/ja111318m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Identification of helix capping and b-turn motifs from NMR chemical shifts. J Biomol NMR. 2012;52:211–232. doi: 10.1007/s10858-012-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, McWhorter AR, Shellam GR, Redwood AJ. The genome of murine cytomegalovirus is shaped by purifying selection and extensive recombination. Virology. 2013;435:258–268. doi: 10.1016/j.virol.2012.08.041. [DOI] [PubMed] [Google Scholar]
- Svitel J, Balbo A, Mariuzza RA, Gonzales NR, Schuck P. Combined affinity and rate constant distributions of ligand populations from experimental surface binding kinetics and equilibria. Biophys J. 2003;84:4062–4077. doi: 10.1016/S0006-3495(03)75132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman JR, Ruan K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem Rev. 2006;106:1720–1736. doi: 10.1021/cr040429z. [DOI] [PubMed] [Google Scholar]
- Tomas MI, Kucic N, Mahmutefendic H, Blagojevic G, Lucin P. Murine cytomegalovirus perturbs endosomal trafficking of major histocompatibility complex class I molecules in the early phase of infection. J Virol. 2010;84:11101–11112. doi: 10.1128/JVI.00988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. Journal of the American Chemical Society. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Venditti V, Marius Clore G. A NMR experiment for simultaneous correlations of valine and leucine/isoleucine methyls with carbonyl chemical shifts in proteins. J Biomol NMR. 2014;58:1–8. doi: 10.1007/s10858-013-9803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Bankovich AJ, Liu CW, Colf LA, Jones LL, Kranz DM, Puglisi JD, Garcia KC. Solution mapping of T cell receptor docking footprints on peptide-MHC. Proc Natl Acad Sci U S A. 2007;104:13080–13085. doi: 10.1073/pnas.0703702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon R, Shen Y, Baker D, Lange OF. Improved chemical shift based fragment selection for CS-Rosetta using Rosetta3 fragment picker. J Biomol NMR. 2013;57:117–127. doi: 10.1007/s10858-013-9772-4. [DOI] [PubMed] [Google Scholar]
- Vink C, Beuken E, Bruggeman CA. Complete DNA sequence of the rat cytomegalovirus genome. J Virol. 2000;74:7656–7665. doi: 10.1128/jvi.74.16.7656-7665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Whitman MC, Natarajan K, Tormo J, Mariuzza RA, Margulies DH. Binding of the natural killer cell inhibitory receptor Ly49A to its major histocompatibility complex class I ligand. Crucial contacts include both H-2Dd AND beta 2-microglobulin. J Biol Chem. 2002;277:1433–1442. doi: 10.1074/jbc.M110316200. [DOI] [PubMed] [Google Scholar]
- Wang R, Natarajan K, Margulies DH. Structural basis of the CD8 alpha beta/MHC class I interaction: focused recognition orients CD8 beta to a T cell proximal position. Journal of immunology. 2009;183:2554–2564. doi: 10.4049/jimmunol.0901276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Natarajan K, Revilleza MJ, Boyd LF, Zhi L, Zhao H, Robinson H, Margulies DH. Structural basis of mouse cytomegalovirus m152/gp40 interaction with RAE1gamma reveals a paradigm for MHC/MHC interaction in immune evasion. Proc Natl Acad Sci U S A. 2012;109:E3578–3587. doi: 10.1073/pnas.1214088109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AB, Sali A, Wilson IA. Biochemistry. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner LR, Varga K, Lange OF, Baker SL, Baker D, Sousa MC, Pardi A. Structure of the BamC two-domain protein obtained by Rosetta with a limited NMR data set. J Mol Biol. 2011;411:83–95. doi: 10.1016/j.jmb.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily--domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley; 1986. [Google Scholar]
- Zweckstetter M, Bax A. Characterization of molecular alignment inaqueous suspensions of Pf1 bacteriophage. J Biomol NMR. 2001;20:365–377. doi: 10.1023/a:1011263920003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.