Abstract
The flagellum protein flagellin of Listeria monocytogenes is encoded by the flaA gene. Immediately downstream of flaA, two genes, cheY and cheA, encoding products with homology to chemotaxis proteins of other bacteria, are located. In this study we constructed deletion mutants with mutations in flaA. cheY, and cheA to elucidate their role in the biology of infection with L. monocytogenes. The ΔcheY, ΔcheA, and double-mutant ΔcheYA mutants, but not ΔflaA mutant, were motile in liquid media. However, the ΔcheA mutant had impaired swarming and the ΔcheY and ΔcheYA mutants were unable to swarm on soft agar plates, suggesting that cheY and cheA genes encode proteins involved in chemotaxis. The ΔflaA, ΔcheY, ΔcheA, and ΔcheYA mutants (grown at 24°C) showed reduced association with and invasion of Caco-2 cells compared to the wild-type strain. However, spleens from intragastrically infected BALB/c and C57BL/6 mice showed larger and similar numbers of the ΔflaA and ΔcheYA mutants, respectively, compared to the wild-type controls. Such a discrepancy could be explained by the fact that tumor necrosis factor receptor p55 deficient mice showed dramatically exacerbated susceptibility to the wild-type but unchanged or only slightly increased levels of the ΔflaA or ΔcheYA mutant. In summary, we show that listerial flaA. cheY, and cheA gene products facilitate the initial contact with epithelial cells and contribute to effective invasion but that flaA could also be involved in the triggering of immune responses.
Listeria monocytogenes is a motile, facultatively intracellular bacterium that causes food-borne infections in humans and animals, with symptoms of septicemia, meningitis, and meningoencephalitis (57). This gram-positive bacterium is widely distributed in the environment and is able to grow over a wide range of temperatures (1 to 45°C), pHs, and osmotic pressures (51). L. monocytogenes is unusual among pathogens in being able to grow at refrigeration temperatures. Contamination of cold-stored foods with L. monocytogenes has been implicated in several outbreaks of epidemic and sporadic disease (20, 50). Entry into the host normally occurs in the gut; in animal models, bacteria pass the gastrointestinal barrier and possibly penetrate the intestinal epithelial cells overlaying Peyer's patches (37, 44). The organism then disseminates to the brain and to the spleen, liver, and other lymphatic systems. The virulence of L. monocytogenes is due to its capacity to invade and multiply within host cells, including macrophages, hepatocytes, and epithelial, endothelial, and neuronal cells (10, 57). Early after internalization, bacteria disrupt the phagosomal membrane and access the cytoplasm, where they polymerize actin and spread from cell to cell (11, 55). Each step of the infectious process is dependent on the production of virulence factors, including invasion proteins (InlA and InlB), listeriolysin O, phospholipases, and ActA, which are controlled by the pleiotropic transcriptional activator PrfA (34, 39). The expression of several of the virulence genes is thermoregulated, with a higher expression at 37°C than at 20°C (17, 33). The flagellar filament of Listeria is composed of one major subunit, flagellin, which is produced and assembled at the cell surface when L. monocytogenes is grown between 20 and 25°C, whereas its production is markedly reduced at 37°C (42). Flagellin is encoded by the flaA gene, which is transcribed as a monocistronic unit (15). Immediately downstream of the flaA gene are two genes, cheY and cheA, encoding polypeptides with high homology to the chemotaxis proteins CheY and CheA of Bacillus subtilis and Escherichia coli, located in a bicistronic unit (16). Analysis of transposon-generated mutants with insertions in the promoter region of this operon suggest that cheY and cheA are involved in chemotaxis (21). The transcription of flaA, cheY, and cheA is pronounced at 25°C but nondetectable at 37°C (15, 16).
B. subtilis and E. coli CheY and CheA constitute a two-component regulatory system involved in signal transduction of chemotaxis (23, 53). In B. subtilis the autophosphorylating activity of CheA increases by binding of attractants to transmembrane receptors (24). Phosphorylated CheA donates the phosphate to the response regulator CheY (4, 24), and phosphorylated CheY interacts with the flagellar motor switch complex to induce counterclockwise (CCW) rotation of the flagella, resulting in smooth swimming behavior (4). The default clockwise (CW) rotation of the flagella in the absence of interaction of CheY-P with switch proteins is associated with tumbling (5).
Flagella and motility contribute to virulence in several bacteria, such as Campylobacter jejuni (25), Legionella pneumophila (13), Clostridium difficile (54), Helicobacter pylori (18), Aeromonas caviae (45), Salmonella enterica serovar Typhi (36), Vibrio cholerae (46), and V. anguillarum (41). Further, the flagella of bacteria such as S. enterica serovar Enteritidis and Pseudomonas aeruginosa are involved in the triggering of innate immune responses in the host, and flagellin up-regulates tumor necrosis factor alpha (TNF-α) expression (9, 58). Toll-like receptors (TLRs) are a family of cell surface receptors that are identified by a conserved cytoplasmic signaling domain. TLRs recognize pathogen-associated molecular patterns and mediate the production of cytokines necessary for the development of effective immunity (31). Flagellin from L. monocytogenes binds to TLR5 (26), which is expressed by intestinal epithelial cells, monocytes, and dendritic cells (6, 40). Likewise, TLR5 stimulates the production of TNF-α (26), a cytokine of importance in host resistance to enteric listeriosis (3). To study the role of the flagella and chemotaxis of L. monocytogenes in virulence, we constructed and phenotypically characterized defined flaA. cheY, and cheA mutants. We show that flagella and chemotaxis facilitate the adhesion to and invasion of eukaryotic host cells. Furthermore, diminished virulence of ΔflaA and ΔcheYA mutants relative to the wild type (WT) was observed in TNFR-p55−/− but not in WT mice, suggesting that flagella and cheYA-encoded polypeptides participate in the triggering of TNF secretion, leading to protection against infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The L. monocytogenes WT strain 12067, described previously (40), and its isogenic mutants (described below) were routinely grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) at 37 or 24°C. E. coli DH5α was used as the host for recombinant plasmids and was grown in Luria-Bertani broth. Strains harboring pAUL-A or pAT19 derivatives were grown in the presence of erythromycin (150 μg/ml for E. coli and 5 μg/ml for L. monocytogenes).
Genetic manipulations.
Chromosomal DNA and plasmid extractions, restriction enzyme digestions, and DNA ligations were performed by standard methods (48), using enzymes supplied by New England Biolabs. PCRs were performed with custom primers (DNA Technology, Aarhus C, Denmark) and AmpliTaq DNA polymerase from Perkin-Elmer (Branchburg, N.J.). Nucleotide sequencing was carried out using the Thermo Sequenase fluorescence labeled primer cycle-sequencing kit as specified by the manufacturer (Amersham Pharmacia Biotech, Little Chalfont, England) on an A.L.F. sequencer (Amersham Pharmacia Biotech), using fluorescein-labeled primers.
Construction of the chromosomal deletion mutants.
Deletion mutants with mutations in the flaA. cheY, and cheA genes were generated by PCR with specific primers with incorporated restriction sites (underlined) to introduce in-frame deletions. (i) To create the ΔflaA mutant, the oligonucleotide pair A (GCGGAATTCGCGCAGGTGTTGGAGCCGATG) and B (GCGGGATCCCAAACGTTCTTGCGCTTGAG) with a BamHI restriction site were used to amplify a 428-bp DNA fragment (AB) of the 5′ region of flaA (15), encoding the first 32 N-terminal amino acid residues. The oligonucleotide pair C (GCGGGATCCCAAACACCGCAAATGTTAACTC), with a BamHI restriction site, and D (TCGTCGGGTCTTTCTCAAGC) served to amplify a 798-bp DNA fragment (CD) at the 3′ region encoding the last 14 C-terminal amino acids of FlaA. The two PCR products were digested with two restriction enzymes: AB with EcoRI and BamHI and CD with HindIII and BamHI. The resulting PCR products were cloned into the temperature-sensitive vector pAUL-A (7). (ii) To create the ΔcheY mutant, the oligonucleotide pair E (GCGAAGCTTTCTCAAGAAGCAACGGAAGC) and F (GCGGGATCCATGAACATTGCATCGTCGAC), with a BamHI restriction site, were used to amplify a 609-bp DNA fragment (EF) of the 5′ region of cheY (16) encoding the first 14 N-terminal amino acid residues. The oligonucleotide pair G (GCGGGATCCGGTTTTAGAGGCGTTAGAAAAAGC), with a BamHI restriction site, and H (GCGGAATTCAATCGCTTCTCTGTCTGGCG) served to amplify a 602-bp DNA fragment (GH) at the 3′ region encoding the last 12 C-terminal amino acids of CheY. The two PCR products were digested: EF with HindIII and BamHI and GH with EcoRI and BamHI. The resulting PCR products were cloned into the vector pAUL-A. (iii) To create the ΔcheA mutant, the oligonucleotide pair I (GCGGAATTCCAAACACCGCAAATGTTAACTC) and J (GCGGGATCCTTCTCAAGCTGTAACAGATTATC), with a BamHI restriction site, were used to amplify a 796-bp DNA fragment (IJ) of the 5′ region of cheA (16) encoding the first 36 N-terminal amino acid residues. The oligonucleotide pair K (GCGGGATCCTCAGATTGCCTTTTCTGGAGC), with a BamHI restriction site, and L (GCGAAGCTTTTAGCATGCGTTTTCCTCCC) served to amplify a 857-bp DNA fragment (KL) at the 3′ region encoding the last 26 C-terminal amino acids of CheA. The two PCR products were digested: IJ with EcoRI and BamHI and KL with HindIII and BamHI. The resulting PCR products were cloned into the vector pAUL-A. (iv) To create the ΔcheYA mutant, the oligonucleotide pair M (GCGGAATTCTCTCAAGAAGCAACGGAAGC) and F (GCGGGATCCATGAACATTGCATCGTCGAC), with a BamHI restriction site, were used to amplify a 609-bp DNA fragment (MF) of the 5′ region of cheY encoding the first 14 N-terminal amino acid residues. Fragment MF and fragment KL described above were digested: MF with EcoRI and BamHI and fragment KL with HindIII and BamHI. The resulting PCR products were cloned into the vector pAUL-A. Plasmids pAUL-ΔflaA, pAUL-ΔcheY, pAUL-ΔcheA, and pAUL-ΔcheYA were transformed into L. monocytogenes 12067 by electroporation. To obtain the chromosomal in-frame deletions, recombinant clones were processed as previously described (35). The appropriate gene deletions were confirmed by PCR sequencing of chromosomal DNA from mutants (data not shown).
Complementation.
For complementation of the cheY mutant, a 796-bp DNA fragment containing the cheY gene and its promoter was amplified by oligonucleotides I and J (described above). The PCR product was digested with EcoRI and BamHI and cloned into the shuttle vector pAT19 (56), creating pcheY. Plasmid pcheY was transformed into the L. monocytogenes ΔcheY mutant by electroporation. For complementation of the cheA and cheYA mutants, a 2,685-bp DNA fragment containing the cheY and cheA genes and their promoter was amplified with oligonucleotides G (GGCGAATTCGGACGAGGGGCTTTTCTTTT) and H (GCGGGATCCCCAGGTTTCTTTTCCACTTCG). The PCR product was digested with EcoRI and BamHI and cloned into the vector pAT19, creating pcheYA. Plasmid pcheYA was transformed into L. monocytogenes ΔcheA and ΔcheYA mutants.
Motility assays.
Swarming on soft agar was analyzed as described by Kathariou et al. (30), with some modifications. Individual colonies were transferred to a petri plate containing tryptic soy broth (Difco) with 0.25% agar, and the motility plates were incubated for 24 h at 24 or 37°C. The low density of the agar allowed the bacteria to move within the agar, forming a halo of growth around the point of inoculation. Motility was also examined by phase-contrast microscopy at a magnification of ×1,000, using an Axiolab microscope (Carl Zeiss, Oberkochen, Germany) and a hanging-drop preparation from a liquid culture of L. monocytogenes grown at 24°C (optical density at 600 nm [OD600], 0.8). The behavior of the bacteria was assessed qualitatively. Linear advances were regarded as swimming, and stationary rotations were regarded as tumbling.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
To analyze total cell lysates of L. monocytogenes strains, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were performed as previously described (15). Bacteria were grown to late logarithmic growth phase (OD600, 0.8), and cell lysates of L. monocytogenes were made by sonication of cells. The samples were run on a 12.5% polyacrylamide gel and incubated with a monoclonal antibody against L. monocytogenes 4b flagellin, kindly provided by W. Donachie, Moredun Research Institute, Edinburgh, Scotland.
Electron microscopy.
Bacteria from liquid cultures incubated at 24°C (OD600, 0.8) were washed twice in physiological saline buffer. A drop of bacterial suspension was placed on a carbon-coated grid. After 90 s, the excess was carefully removed and the preparations were negatively stained in 2% uranyl acetate for 45 s. Air-dried grids were examined in a Philips CM100 transmission electron microscope at 80 kV.
Infection of cell cultures.
Determination of cell association and invasion of L. monocytogenes was performed as described by Larsen et al. (32), with some modifications. Enterocyte-like Caco-2 cells obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) were cultured in Eagle's minimal essential medium (MEM) enriched with Glutamax and HEPES (Invitrogen, Tåstrup, Denmark) and supplemented with 20% heat-inactivated (30 min at 56°C) fetal calf serum (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), and 0.5 ml of gentamicin (50 mg/ml) (Invitrogen) at 37°C under 5% CO2. The cells were used at passages 10 to 15. They were trypsinized, and the cell concentration was adjusted to 5 × 105 cells per ml. They were grown without gentamicin to a monolayer (68 to 72 h at 37°C) in Eagle's MEM supplemented with 10% heat-inactivated fetal calf serum and 0.1 mM nonessential amino acids. For infection experiments, overnight bacterial cultures were diluted 25-fold and grown at 24 or 37°C with agitation until the OD600 reached 0.8. Bacteria were harvested, adjusted to approximately 3 × 106 bacteria per ml, and added to wells of 12-well plates, resulting in a multiplicity of infection of about 30 bacteria per cell. Following 1 h of incubation at 37°C, the cells were washed three times with phosphate-buffered saline (PBS). To kill extracellular bacteria, 2 ml of MEM with gentamicin (10 μg/ml) was added to the wells, and the mixture was then incubated for 1 h at 37°C under 5% CO2. The cells were washed three times and lysed by adding 1 ml of 0.1% Triton X-100. The number of viable bacteria released from the cells was assessed on agar plates. In some experiments, bacteria were centrifuged on the cells at 1,000 × g for 1.5 min. The CFU after the first 1 h of incubation reflects the bacteria associated with the monolayer (attached on the surface or at some stage of internalization). The CFU 1 h after the addition of gentamicin in the extracellular environment represents the viable bacteria that are internalized by the mammalian cells. All parameters were assayed in duplicate. Each experiment was repeated twice. The data were analyzed statistically by Student's t test.
Mouse infections.
Overnight cultures of bacteria were diluted 25-fold and incubated at 24 or 37°C with agitation to an OD600 of 0.8. Bacteria were diluted in PBS prior to infection. Eight-week-old female BALB/C mice (Taconic M & B, Ry, Denmark) were infected intragastrically with 2 × 109 bacteria (five mice per group). Seven-week-old female C57BL/6 mice (WT) and TNFR-p55−/− mice (43), bred in our animal facilities, were infected intragastrically with 4 × 108 bacteria (six per group). The mice were sacrified at different times after infection, and the spleens and livers were removed and homogenized in PBS containing 0.1% Triton X-100. Tenfold serial dilutions of the lysates in PBS were plated on BHI agar plates. Colonies were counted after overnight incubation at 37°C. The data were analyzed statistically by Student's t test.
RESULTS
Construction and characterization of defined L. monocytogenes flaA. cheY. cheA, and cheYA mutants.
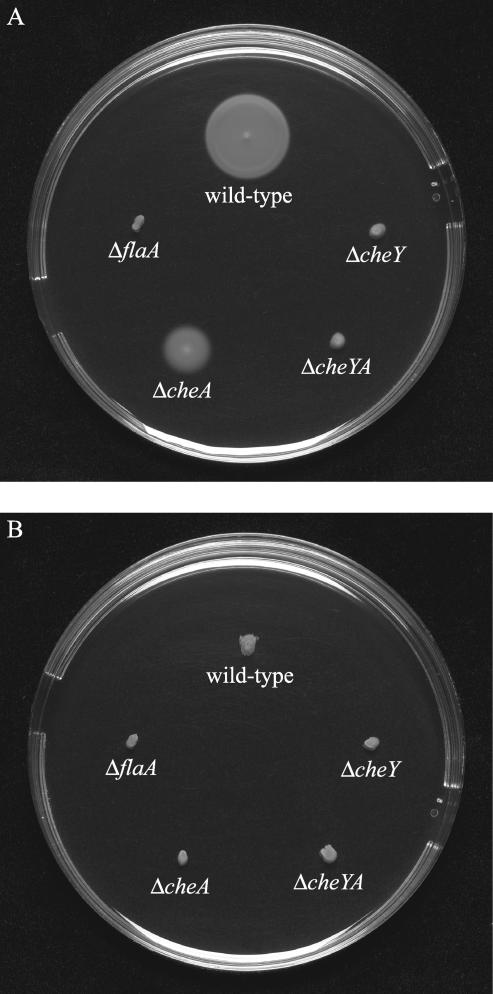
To analyze the involvement of the flaA. cheY, and cheA gene products in the virulence of L. monocytogenes, we constructed mutant strains carrying in-frame deletions in each of the flaA. cheY, and cheA genes (ΔflaA, ΔcheY, and ΔcheA) as well as an in-frame deletion in both the cheY and cheA genes (ΔcheYA). These chromosomal mutants were obtained by allelic exchange in L. monocytogenes strain 12067. This WT strain was chosen for construction of mutants since the only flaA. cheY, and cheA sequences available at the time this project was initiated were those from L. monocytogenes strain 12067 (15, 16). The phenotype of the mutants were analyzed by semisolid swarm plate assays (Fig. 1A). After incubation at 24°C, the WT strain showed good swarming, with concentric rings that increased with the period of incubation. The ΔflaA mutant did not show swarming and produced small, compact colonies with sharp boundaries. The ΔcheY and the ΔcheYA mutants formed a small fuzzy-looking swarm ring, while the colonies of the ΔcheA mutant showed swarming, but the swarm ring was much smaller than that of the WT, indicating that cheY is required for swarming and that cheA is contributing to a lesser degree. At 37°C, none of the strains showed swarming (Fig. 1B). None of the four mutants were affected for growth rate in BHI medium (data not shown).
FIG. 1.
Swarming of L. monocytogenes mutants in semisolid agar. The L. monocytogenes WT strain 12067 and the ΔflaA, ΔcheY, ΔcheA, and ΔcheYA mutants were stabbed into semisolid agar plates (tryptic soy broth plus 0.25% agar). The plates were incubated at 24°C (A) or 37°C (B) for 24 h.
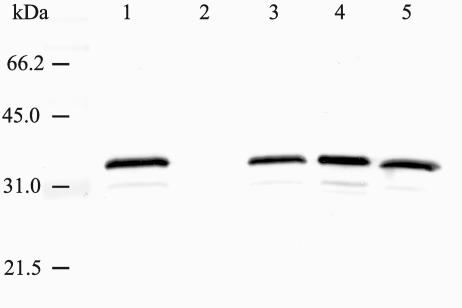
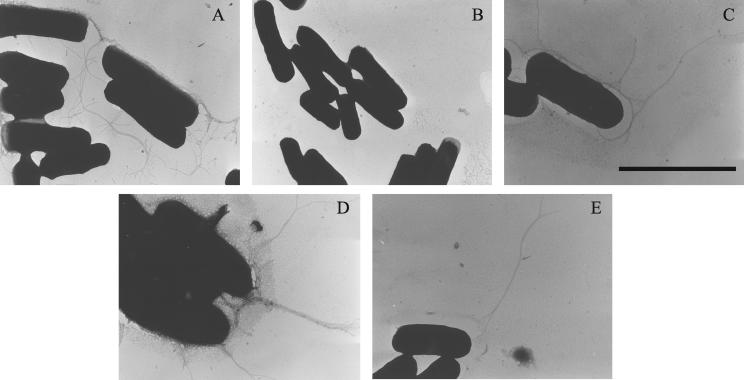
To determine whether the deletions in cheY and cheA had effects on flagellin production, we performed Western blot analysis with a monoclonal antibody directed against the flagellin protein of L. monocytogenes. The ΔcheY, ΔcheA, and ΔcheYA mutants and WT bacteria showed similar flagellin levels (Fig. 2). Microscopic examination of cells grown in liquid culture to late-logarithmic phase showed that the ΔcheY, ΔcheA, and ΔcheYA mutants all had flagella (Fig. 3). As expected, no flagella and flagellin were detected in the ΔflaA mutant as analyzed by electron microscopy or Western blotting, respectively (Fig. 2 and 3). Phase-contrast microscopy revealed that the ΔcheY, ΔcheA, and ΔcheYA mutants were all motile and showed only tumbling behavior whereas the WT showed tumbling and smooth swimming.
FIG. 2.
Western blot analysis of whole-cell lysates of the ΔflaA, ΔcheY, ΔcheA, and ΔcheYA mutants grown at 24°C (5 × 107 CFU). Samples were run on a 12.5% polyacrylamide gel and incubated with a monoclonal antibody specific for the L. monocytogenes 4b flagellin. Lanes: 1, L. monocytogenes 12067 (WT); 2, ΔflaA mutant; 3, ΔcheY mutant; 4, ΔcheA mutant; 5, ΔcheYA mutant. Molecular mass standards are shown on the left.
FIG. 3.
Electron micrographs of the flagellated L. monocytogenes WT strain (A), the nonflagellated ΔflaA mutant (B), the ΔcheY mutant (C), the ΔcheA mutant (D), and the ΔcheYA mutant (E). Bacteria were grown at 24°C to late logarithmic growth phase, applied to carbon-coated grids, shadowed with 2% uranyl acetate, and examined under a transmission electron microscope. The scale bar, shown in panel C and valid for all panels, represents 1.5 μm (B), 1 μm (A, C, and E), and 0.5 μm (D).
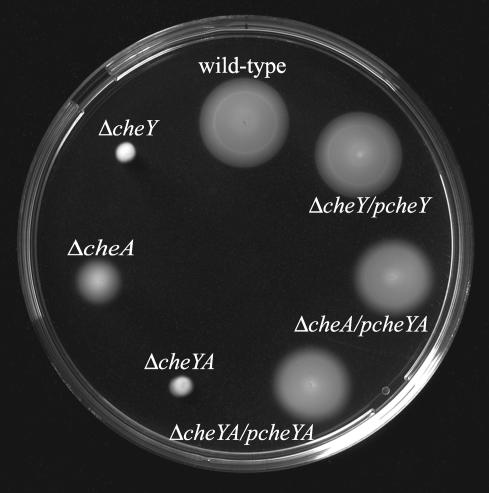
For complementation of the ΔcheY mutant, the vector pAT19 containing the cheY gene and its promoter region (pcheY) was introduced into the ΔcheY mutant. For complementation of the ΔcheA and the ΔcheYA mutants, pAT19 containing the cheY and cheA genes and their promoter (pcheYA) was introduced into the ΔcheA and ΔcheYA mutants. As controls, the WT and the ΔcheY, ΔcheA, and ΔcheYA mutants carrying the vector without inserts were used. On semisolid agar, swarming of the complemented mutants was restored (Fig. 4). Phase-contrast microscopy demonstrated that the three complemented strains showed both tumbling and smooth swimming like the WT. None of the phenotypes were restored by the plasmid vector alone.
FIG. 4.
Swarming of complemented ΔcheY, ΔcheA, and ΔcheYA mutants in semisolid agar. The ΔcheY, ΔcheA, and ΔcheYA mutants with plasmids carrying the cheY (pcheY) or cheY/cheA (pcheYA) genes were stabbed into a semisolid agar plate. The WT strain and the ΔcheY, ΔcheA and ΔcheYA mutants carrying the parent vector pAT19 served as controls. The plate was incubated at 24°C for 30 h.
Effects of mutations in flagellar and chemotaxis genes on bacterial association with cells.
The ability of the ΔflaA, ΔcheY, ΔcheA, and ΔcheYA mutants to associate with and invade the human enterocyte-like cell line Caco-2 was analyzed. Caco-2 cell association was significantly reduced for the mutants grown at 24°C compared to the WT strain (10.6 to 12.7% of control values; P < 0.001) (Table 1). Likewise, invasion by the mutant bacteria was significantly reduced in comparison to that of the WT strain (P < 0.001), with levels from 1.4 to 7.0% of the wild-type values. To further determine if the reduced invasion capacity of the ΔflaA and ΔcheYA mutants was due to impaired locomotion, the bacteria were centrifuged onto the monolayers prior to the invasion assay. Centrifugation increased cell association to 78.8 and 75.8% of the WT values, respectively, indicating that flagella and chemotaxis are important to ensure that bacteria contact host cells. However, cell association was still lower than that of the WT strain (P < 0.005) (Table 1).
TABLE 1.
Association and invasion of Caco-2 cells by wild-type L. monocytogenes 12067 and the isogenic flagellum and chemotaxis mutants
| Growth temp (°C) and strain | Without centrifugation
|
With centrifugation
|
||
|---|---|---|---|---|
| No.a (%)b of cell-associated bacteria (CFU/ml) | No.a (%) of invading bacteria (CFU/ml) | No. (%) of cell-associated bacteria (CFU/ml) | No. (%) of invading bacteria (CFU/ml) | |
| 24 | ||||
| WT | (3.3 ± 0.1) × 106 (100) | (2.7 ± 0.3) × 105 (100) | (6.6 ± 0.5) × 106 (100) | (1.4 ± 0.4) × 106 (100) |
| ΔflaA mutant | (3.5 ± 0.6) × 105c (10.6) | (3.8 ± 0.5) × 103c (1.4) | (5.2 ± 0.4) × 106d (78.8) | (3.6 ± 0.9) × 105d (25.7) |
| ΔcheY mutant | (4.2 ± 0.2) × 105c (12.7) | (1.2 ± 0.2) × 104c (4.4) | ND | ND |
| ΔcheA mutant | (4.0 ± 0.3) × 105c (12.1) | (1.9 ± 0.1) × 104c (7.0) | ND | ND |
| ΔcheYA mutant | (4.1 ± 0.1) × 105c (12.4) | (1.6 ± 0.2) × 104c (5.9) | (5.0 ± 0.4) × 106d (75.8) | (8.4 ± 0.6) × 105d (60.0) |
| 37 | ||||
| WT | (2.6 ± 0.2) × 105 (100) | (1.2 ± 0.3) × 105 (100) | ND | ND |
| ΔflaA mutant | (2.4 ± 0.2) × 105 (NS)e (92.3) | (1.1 ± 0.1) × 105 (NS) (91.7) | ND | ND |
| ΔcheYA mutant | (2.2 ± 0.1) × 105d (84.6) | (9.3 ± 0.7) × 104 (NS) (77.5) | ND | ND |
Association and invasion assays were performed without or with centrifugation of the bacteria onto the monolayer. Bacteria were grown in BHI broth at either 24 or 37°C, and invasion assays carried out as described in Materials and Methods. Each experiment was repeated twice, and the mean number of associated (both attached and intracellular) and invading bacteria were recorded. Results are given as mean ± standard deviation.
The values for association and invasion were normalized to the values for the WT strain (under identical culture and centrifugation conditions), which were arbitrarily set at 100.
Significantly different from the WT strain under identical culture and centrifugation conditions (P < 0.001).
Significantly different from the WT strain under identical culture and centrifugation conditions (P < 0.005).
NS, not significantly different from the WT strain under identical culture and centrifugation conditions.
Cell association of the ΔcheYA mutant grown at 37°C was only slightly reduced as compared to that of the WT strain. No differences in cell association between the WT strain and the ΔflaA mutant grown at 37°C were observed. The mutant and WT strains grown at 37°C showed similar invasion ability (Table 1).
Virulence of flaA and cheYA mutants in WT and TNFR-p55−/− mice.
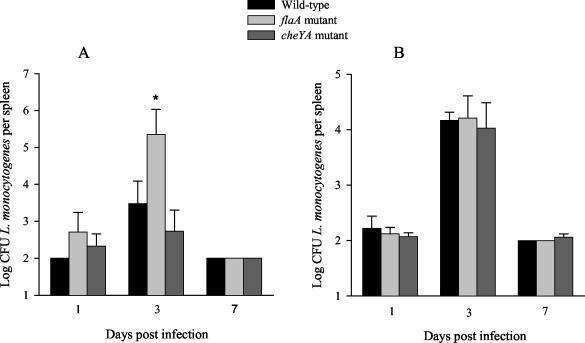
To analyze the effect of deleting the flaA and cheYA genes on the virulence of L. monocytogenes, bacterial levels in mice intragastrically (i.g.) infected with ΔflaA and ΔcheYA mutants were compared to those infected with the WT strain. In a first series of experiments, we observed that spleens of BALB/c mice 3 days after i.g. infection with the ΔflaA mutant showed 100-fold larger bacterial numbers than did the spleens of mice infected with the WT strain (Fig. 5A). Spleens of mice infected i.g. with the ΔcheYA or WT L. monocytogenes strain showed similar bacterial levels. No differences in bacterial numbers in spleens were observed when the ΔflaA, and ΔcheYA mutants and WT L. monocytogenes were grown at 37°C (Fig. 5B). These results indicate that abolished expression of the flaA gene increases the virulence of L. monocytogenes in BALB/c mice compared to the flagellated WT strain and the ΔcheYA mutant.
FIG. 5.
CFU in the spleens of i.g. infected BALB/c mice. L. monocytogenes WT and the ΔflaA and ΔcheYA mutants were grown at either 24°C (A) or 37°C (B) before being used to infect BALB/c mice (five mice per group). The mice were sacrified at the indicated time points after infection, and their spleens were homogenized and plated. The means and standard errors of the mean are shown. An asterisk indicates a significant difference in the log CFU of the mutant compared to the WT (P < 0.05 as determined by Student's t test). A representative of two independent experiments is shown.
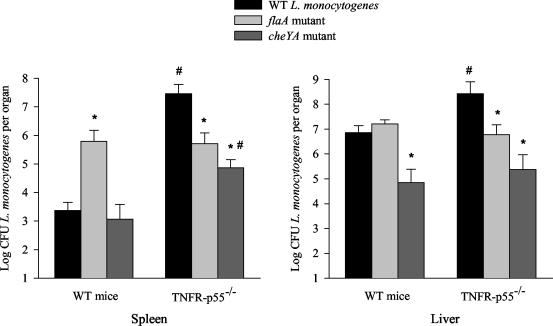
To address whether TNF-mediated protective immune responses are involved in the discrepancy between invasion and virulence of ΔflaA and ΔcheYA mutants, the bacterial load in mice deficient in TNFR-p55, the main TNF receptor, was measured. TNFR-p55−/− mice have been previously shown to possess a dramatically increased susceptibility to L. monocytogenes (43, 47). The susceptibility of TNFR-p55−/− mice to WT L. monocytogenes and to the ΔflaA and ΔcheYA mutants grown at 24°C was compared. The spleens and livers from TNFR-p55−/− mice showed a dramatically increased number of WT L. monocytogenes bacteria compared to the same organs of C57BL/6 control mice (Fig. 6). Differences in bacterial numbers in organs from control and TNFR-p55−/− mice were significantly smaller when mice were infected with ΔflaA or ΔcheYA mutants. Moreover, bacterial numbers of the ΔflaA mutant recovered from the spleens of control and TNFR-p55−/− mice were similar (Fig. 6). Also, similar levels of ΔflaA and ΔcheYA in the livers of TNFR-p55−/− mice and WT mice were found (Fig. 6). This suggests that flagellin and cheYA-encoded polypeptides participate in bacterial invasion but the presence of flagella is deleterious to systemic infection, which seems to be dependent on the TNF-p55 receptor.
FIG. 6.
CFU after i.g. infection of TNFR-p55−/− and WT mice. L. monocytogenes WT and mutant strains were inoculated i.g. into WT or TNFR-p55−/− mice (six mice per group). Three days after infection, the mice were sacrified and bacterial loads (CFU) in their spleens and livers were measured. The means and standard errors of the mean are shown. The asterisk indicates a significant difference in the log CFU of a bacterial mutant relative to the WT (P < 0.05 as determined by Student's t test). The pound sign indicates a significant difference between two groups of mice infected with the same bacterial strain (P < 0.05 as determined by Student's t test).
DISCUSSION
In this work we characterized the flagellin gene, flaA, and the two chemotaxis genes, cheY and cheA, of L. monocytogenes through the construction of deletion mutants and compared their virulence with that of WT L. monocytogenes in vitro and in vivo.
Phenotypic characterization of the mutants showed that the deletion mutant with a mutation in flaA was completely defective in the production of flagella and motility. Analysis of the mutants with deletions in cheY and cheA showed impaired swarming and tumbling behavior during motility. Trans-complementation of the ΔcheY, ΔcheA, and ΔcheYA mutants restored swarming and smooth swimming behavior, indicating that the mutations had no polar effects. In B. subtilis. cheY and cheA null mutants are tumbly, in contrast to E. coli, where cheY and cheA null mutants exhibit a smooth-swimming phenotype (5, 22, 28). These data suggest that the cheY and cheA gene products of L. monocytogenes contribute to chemotactic signal transduction, as suggested from the amino acid sequence similarity (16), and that the mechanism by which they confer their control may be similar to the mechanism in B. subtilis.
The flagellum (ΔflaA) and chemotaxis (ΔcheY, ΔcheA, ΔcheYA) mutants had a reduced capacity to associate with cells when the bacteria were grown at 24°C, and centrifugation of bacteria onto cells could only partly complement this. In accordance with observations that the production of flagella is markedly reduced at 37°C (42), the ability of the aflagellate ΔflaA and chemotaxis ΔcheYA mutants to associate with cells was only slightly decreased when they were grown at 37°C prior to infection. These findings suggest that flagella and chemotaxis are important to ensure that L. monocytogenes contacts host cells, as also suggested by Flanary et al. (21). The fact that centrifugation did not completely revert the phenotype suggests a role of flagella in adhesion or invasion other than locomotion. On centrifugation, the ΔcheYA mutant was more invasive than the ΔflaA mutant, probably due to the presence of flaA. It has been suggested that components of bacterial flagella can act as adhesins and mediate the binding to host cells and mucosal surfaces (2, 59). Similar effects of mutations in motility-related genes on cell association have been shown for other bacteria. For example, mutations in the Yersinia enterocolitica flhDC or fliA genes, which are transcriptional regulators of the flagellar regulon and are required for the expression of motility (29, 61), affect the effective migration of bacteria to the host cells (60), and in S. enterica serovar Enteritidis the flagella assist in colonization of epithelial cells by enabling motility rather than providing an adhesin (12). The importance of chemotaxis of L. monocytogenes in the interactions with cells has been previously studied by using a chemotaxis mutant of L. monocytogenes strain NCTC 10527 with a transposon insertion in the promoter region of the cheYA operon. This strain was impaired in cell association but not in invasion of fibroblasts (21).
Spleens from i.g. infected BALB/c and C57BL/6 mice contained larger numbers of the ΔflaA mutant than of the WT strain, whereas spleens from mice infected with ΔcheYA and WT L. monocytogenes strains contained similar bacterial numbers, indicating that the presence of flagella is deleterious to systemic infection. In fact, L. monocytogenes is able to repress flagellar expression at 37°C (42). This temperature regulation might serve as a mechanism to evade recognition by the innate immune system in the host. Interestingly, when comparing L. monocytogenes with the nonpathogenic but closely related species L. innocua, Kathariou et al. (30) found that the repression of flagellin in L. innocua was not as strong as the repression in L. monocytogenes. The discrepancy of the in vivo virulence test of the ΔflaA mutant with the in vitro invasion experiments with this mutant can be reconciled through the observation that the ΔflaA mutant were present at lower levels than WT bacteria in TNFR-p55−/− mice. The data suggest that the flagella of L. monocytogenes are important for triggering TNF-mediated growth control of L. monocytogenes. These data correspond to previous reports that flagella from gram-negative bacteria such as S. enterica serovar Enteritidis, P. aeruginosa, and Y. enterocolitica induce the production of cytokines, e.g., TNF-α and interleukin-1 (8, 9, 58). Interestingly, it has been shown that two regions in the conserved N- and C-terminal parts of the flagellin of E. coli are required for interleukin-8 release and TLR5 activation in a human epithelial cell line (14). Likewise, it was demonstrated that the N- and C-terminal regions of S. enterica serovar Dublin flagellin induce TNF-α production in monocyte cell lines (19). The N- and C-terminal regions are conserved across bacteria, and a considerable degree of homology was also found to these regions of the flagellin of L. monocytogenes (15), suggesting a potential role of those conserved domains of flagellin of L. monocytogenes in activation of the innate immune response. Indeed, the TLR5 recognition site of flagellin has been mapped to highly conserved N- and C-terminal amino acids which are present in flagellin of L. monocytogenes (52). Moreover, a recent study, based on conformation analysis, has predicted that a region in the N-terminal domain of flagellin of L. monocytogenes is involved in binding to TLR5. This region overlaps with the N-terminal binding site (27). However, mice deficient in the adaptor protein MyD88, which is required for TLR signaling (1, 38), contained only slightly augmented bacterial numbers of WT L. monocytogenes in their spleens (unpublished results), indicating that TNF-mediated control of L. monocytogenes is at least partially MyD88 independent.
Factors other than temperature can stimulate the expression of L. monocytogenes flagellin, which is present in trace amounts at 37°C (42). Indeed, flagellin expression is regulated by osmolarity (49). Thus, we propose that the observed effect of flagella and chemotaxis on cell invasion and the suspected role of flagella in TNF induction may be relevant to the biology of infection with L. monocytogenes.
Acknowledgments
This work was supported by a grant (53-00-0338) from the Danish Agricultural and Veterinary Research Council and a grant (711,1213/97) from SJFR, the Swedish Medical Research Council, Q-Med AB, and The Foundation for Knowledge and Competence Development, Stockholm, Sweden.
We thank T. Chakraborty (Institut für Medizinische Mikrobiologie, Justus-Liebig-Universität, Giessen, Germany) for the kind gift of the pAUL-A plasmid. We also thank Michael Engelbrecht Nielsen for assistance with various aspects of this project and Jannie Jørgensen and Jan Pedersen for excellent technical assistance.
Editor: V. J. DiRita
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beretich, G. R., Jr., P. B. Carter, and E. A. Havell. 1998. Roles for tumornecrosis factor and gamma interferon in resistance to enteric listeriosis. Infect.Immun. 66:2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, D. S., R. B. Bourret, M. L. Kirsch, and G. W. Ordal. 1993. Purification and characterization of Bacillus subtilis CheY. Biochemistry 32:9256-9261. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, D. S., and G. W. Ordal. 1991. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J. Biol. Chem. 266:12301-12305. [PubMed] [Google Scholar]
- 6.Cario, E., and D. K. Podolsky. 2000. Differential alteration in intestinal epithelial cell expression of Toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 68:7010-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty T., M. Leimeister-Wächter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciacci-Woolwine, F., I. C. Blomfield, S. H. Richardson, and S. B. Mizel. 1998. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect. Immun. 66:1127-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciacci-Woolwine, F., P. F. McDermott, and S. B. Mizel. 1999. Induction of cytokine synthesis by flagella from gram-negative bacteria may be dependent on the activation or differentiation state of human monocytes. Infect. Immun. 67:5176-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cossart, P., and M. Lecuit. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 17:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabiri, G. A., J. M. Sanger, D. A. Portnoy, and F. S. Southwick. 1990. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc. Natl. Acad. Sci. USA 87:6068-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 13.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donelly, M. A., and T. S. Steiners. 2002. Two nonadjacent regions in enteroaggregative Escherichia coli flagellin are required for activation of toll-like receptor 5. J. Biol. Chem. 277:40456-40461. [DOI] [PubMed] [Google Scholar]
- 15.Dons, L., O. F. Rasmussen, and J. E. Olsen. 1992. Cloning and characterization of a gene encoding flagellin of Listeria monocytogenes. Mol. Microbiol. 6:2919-2929. [DOI] [PubMed] [Google Scholar]
- 16.Dons, L., J. E. Olsen, and O. F. Rasmussen. 1994. Characterization of two putative Listeria monocytogenes genes encoding polypeptides homologous to the sensor protein CheA and the response regulator CheY of chemotaxis. DNA Sequence 4:301-311. [DOI] [PubMed] [Google Scholar]
- 17.Dramsi, S., C. Kocks, C. Forestier, and P. Cossart. 1993. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator PrfA. Mol. Microbiol. 9:931-941. [DOI] [PubMed] [Google Scholar]
- 18.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaves-Pyles, T. D., H. R. Wong, K. Odoms, and R. B. Pyles. 2001. Salmonella flagellin-dependent proinflammatory responses are localized to the conserved amino and carboxyl regions of the protein. J. Immunol. 167:7009-7016. [DOI] [PubMed] [Google Scholar]
- 20.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanary, P. L., R. D. Allen, L. Dons, and S. Kathariou. 1999. Insertional activation of the Listeria monocytogenes cheYA operon abolishes response to oxygen gradients and reduces the number of flagella. Can. J. Microbiol. 45:646-652. [PubMed] [Google Scholar]
- 22.Fuhrer, D. K., and G. W. Ordal. 1991. Bacillus subtilis CheN, a homolog of CheA, the central regulator of chemotaxis in Escherichia coli. J. Bacteriol. 173:7443-7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrity, L. F., and G. W. Ordal. 1995. Chemotaxis in Bacillus subtilis: how bacteria monitor environmental signals. Pharmacol. Ther. 68:87-104. [DOI] [PubMed] [Google Scholar]
- 24.Garrity, L. F., and G. W. Ordal. 1997. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology 143:2945-2951. [DOI] [PubMed] [Google Scholar]
- 25.Grant, C. C. R., M. E. Konkel, W. Cieplak, Jr., and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 27.Jacchieri, S. G., R. Torquato, and R. R. Brentani. 2003. Structural study of binding of flagellin by toll-like receptor 5. J. Bacteriol. 185:4243-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapatral, V., J. W. Olson, J. C. Pepe, V. L. Miller, and S. A. Minnich. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19:1061-1071. [DOI] [PubMed] [Google Scholar]
- 30.Kathariou, S., R. Kanenaka, R. D. Allen, A. K. Fok, and C. Mizumoto. 1995. Repression of motility and flagellin production at 37°C is stronger in Listeria monocytogenes than in the nonpathogenic species Listeria innocua. Can. J. Microbiol. 41:572-577. [DOI] [PubMed] [Google Scholar]
- 31.Kirschning, C. J., and S. Bauer. 2001. Toll-like receptors: cellular signal transducers for exogenous molecular patterns causing immune responses. Int. J. Med. Microbiol. 291:251-260. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, C. N., B. Nørrung, H. M. Sommer, and M. Jakobsen. 2002. In vitro and in vivo invasiveness of different pulsed-field gel electrophoresis types of Listeria monocytogenes. Appl. Environ. Microbiol. 68:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leimeister-Wächter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, S. L., T. Ezaki, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect. Immun. 56:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marco, A. J., N. Prats, J. A. Ramos, V. Briones, M. Blanco, L. Dominguez, and M. Domingo. 1992. A microbiological, histopathological and immunohistological study of the intragastric inoculation of Listeria monocytogenes in mice. J. Comp. Pathol. 107:1-9. [DOI] [PubMed] [Google Scholar]
- 38.Medzhitov, R., and C. Janeway, Jr. 2000. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 173:89-97. [DOI] [PubMed] [Google Scholar]
- 39.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vázquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 40.Muzio, M., N. Polentarutti, D. Bosisio, P. P. Manoj Kumar, and A. Mantovani. 2000. Toll-like receptor family and signalling pathway. Biochem. Soc. Trans. 28:563-566. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, R., D. L. Milton, and H. Wolf-Watz. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 42.Peel, M., W. Donachie, and A. Shaw. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. J. Gen. Microbiol. 134:2171-2178. [DOI] [PubMed] [Google Scholar]
- 43.Pfeffer, K., T. Matsuyama, T. M. Kündig, A. Wakeham, K. Kishihara, A. Shahinian, K. Wiegmann, P. S. Ohashi, M. Krönke, and T. W. Mak. 1993. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 73:457-467. [DOI] [PubMed] [Google Scholar]
- 44.Pron, B., C. Boumaila, F. Jaubert, S. Sarnacki, J.-P. Monnet, P. Berche, and J.-L. Gaillard. 1998. Comprehensive study of the intestinal stage of listeriosis in a rat ligated ileal loop system. Infect. Immun. 66:747-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabaan, A. A., I. Gryllos, J. M. Tomás, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to Hep-2 cells. Infect. Immun. 69:4257-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson, K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothe, J., W. Lesslauer, H. Lötscher, Y. Lang, P. Koebel, F. Köntgen, A. Althage, R. Zinkernagel, M. Steinmetz, and H. Bluethmann. 1993. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature 364:798-802. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sanchez-Campillo, M., S. Dramsi, J. M. Gómez-Gómez, E. Michel, P. Dehoux, P. Cossart, F. Baquero, and J. C. Pérez-Díaz. 1995. Modulation of DNA topology by flaR, a new gene from Listeria monocytogenes. Mol. Microbiol. 18:801-811. [DOI] [PubMed] [Google Scholar]
- 50.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seeliger, H. P. R., and D. Jones. 1986. Genus Listeria pirie, 1940, p. 1235-1245. In P. H. A. Sneath, N. S. Mair, N. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 52.Smith, K. D., E. Andersen-Nissen, F. Hayashi, K. Strobe, M. A. Bergman, S. L. Barrett, B. T. Cookson, and A. Aderem. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4:1247-1253. [DOI] [PubMed] [Google Scholar]
- 53.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 54.Tasteyre, A., M.-C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 57.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyant, T. L, M. K. Tanner, and M. B. Sztein. 1999. Salmonella typhi flagella are potent inducers of proinflammatory cytokine secretion by human monocytes. Infect. Immun. 67:3619-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 60.Young, G. M., J. L. Badger, and V. L. Miller. 2000. Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect. Immun. 68:4323-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed]