Abstract
The oxidation of a key cysteine residue (Cys106) in the parkinsonism-associated protein DJ-1 regulates its ability to protect against oxidative stress and mitochondrial damage. Cys106 interacts with a neighboring protonated Glu18 residue, stabilizing the Cys106-SO2− (sulfinic acid) form of DJ-1. To study this important post-translational modification, we previously designed several Glu18 mutations (E18N, E18D, E18Q) that alter the oxidative propensity of Cys106. However, recent results suggest these Glu18 mutations cause loss of DJ-1 dimerization, which would severely compromise the protein’s function. The purpose of this study was to conclusively determine the oligomerization state of these mutants using X-ray crystallography, NMR spectroscopy, thermal stability analysis, CD spectroscopy, sedimentation equilibrium ultracentrifugation, and crosslinking. We found that all of the Glu18 DJ-1 mutants were dimeric. Thiol crosslinking indicates that these mutant dimers are more flexible than the wild-type protein and can form multiple crosslinked dimeric species due to the transient exposure of cysteine residues that are inaccessible in the wild-type protein. The enhanced flexibility of Glu18 DJ-1 mutants provides a parsimonious explanation for their lower observed crosslinking efficiency in cells. In addition, thiol crosslinkers may have an underappreciated value as qualitative probes of protein conformational flexibility.
Keywords: DJ-1, PARK7, parkinsonism, Parkinson Disease, cysteine oxidation, structure
Introduction
Mutations in the human protein DJ-1 (PARK7) cause rare forms of recessively inherited parkinsonism (Bonifati et al. 2003). Initially discovered as a Ras-dependent oncogene (Nagakubo et al. 1997), DJ-1 was independently shown to play a role in regulating RNA-protein interactions (Hod et al. 1999) and rodent fertility (Wagenfeld et al. 1998) prior to the discovery that it was a gene for parkinsonism. Loss of DJ-1 function due to knockout or point mutation sensitizes multiple cell types to oxidative stress and mitochondrial dysfunction in culture (Yokota et al. 2003, Canet-Aviles et al. 2004, Martinat et al. 2004, Im et al. 2010, Larsen et al. 2011, Giaime et al. 2012, Stefanatos et al. 2012, Shadrach et al. 2013) and in various animal model systems (Menzies et al. 2005, Meulener et al. 2005, Aleyasin et al. 2007, Billia et al. 2013).
DJ-1 is believed to be a multifunctional protein that protects cells from mitochondrially associated oxidative stress through its participation in several pro-survival pathways (Kahle et al. 2009). Although multiple activities have been proposed for DJ-1, the details of its molecular function remain incompletely understood. At the subcellular level, DJ-1 appears to play an important role in mitochondrial maintenance and function (Canet-Aviles et al. 2004, Blackinton et al. 2009, Giaime et al. 2012, Guzman et al. 2010, Hao et al. 2010, Irrcher et al. 2010, Joselin et al. 2012), which is thought to be directly relevant to parkinsonism (Cookson & Bandmann 2010, Imai & Lu 2011, de Vries & Przedborski 2013, Hauser & Hastings 2013).
At the molecular level, DJ-1 is a homodimer that contains a conserved reactive cysteine residue (Cys106) that is critical for the protein’s ability to respond to oxidative stress (Wilson et al. 2003, Canet-Aviles et al. 2004, Blackinton et al. 2009, Joselin et al. 2012). Mutation of Cys106 to other residues abrogates DJ-1-mediated protection against oxidative stress in cell culture (Canet-Aviles et al. 2004, Shadrach et al. 2013), Drosophila melanogaster (Meulener et al. 2006, Hao et al. 2010), and rat models (Aleyasin et al. 2007). Cys106 has a low pKa value (Witt et al. 2008) and is easily oxidized to Cys106-sulfinic acid (Cys-SO2−) (Canet-Aviles et al. 2004), which correlates with its ability to maintain normal mitochondrial morphology after exposure to rotenone (Blackinton et al. 2009). Therefore, the oxidative status of Cys106 has been proposed to be a key regulator of DJ-1 function (Kinumi et al. 2004, Canet-Aviles et al. 2004, Blackinton et al. 2009, Kato et al. 2013).
Interpretation of these results is complicated by the fact that mutation of Cys106 abrogates both its oxidation and any other function(s) that may require the reduced cysteine residue. In a previous combined structural and cell biological study (Blackinton et al. 2009), we addressed this complication by making conservative mutations at a nearby glutamic acid (Glu18) that interacts with reduced and oxidized Cys106. Two mutations (E18Q and E18N) allowed Cys106 to oxidize to Cys106-SO2− under physiological conditions, while another substitution (E18D) was oxidation-impaired and did not form Cys106-SO2− as readily as wild-type protein or the other Glu18 mutants. We found that the oxidation-competent forms of DJ-1 (wild-type, E18N, and E18Q) protected cells against rotenone and maintained normal mitochondrial morphology, while oxidation-impaired mutants (C106A, and E18D) did not, indicating that oxidation of Cys106 was important for these aspects of DJ-1 function. Notably, high (1.6–1.15 Å) resolution crystal structures of all of these mutants indicated that they, like the wild-type protein, were dimeric (Blackinton et al. 2009).
As interpreted, these previous observations demonstrate the importance of Cys106 oxidation in DJ-1 function (Blackinton et al. 2009). However, a recent report by Maita et al. proposed that mutations at Glu18 in human DJ-1 result in a nearly complete loss of protein dimerization detected using co-immunoprecipitation of tagged proteins from cultured cells (Maita et al. 2013). Additionally, Maita et al. found evidence of a potential functional role for monomeric DJ-1 in enhancing mitochondrial localization of the protein (Maita et al. 2013). As DJ-1 is a homodimer whose oligomerization is important for its structural integrity and cellular function (Miller et al. 2003, Moore et al. 2003, Ramsey & Giasson 2010, Repici et al. 2013), loss of dimerization resulting from mutations at Glu18 would complicate the published interpretation of prior results.
In the present study, we examine the oligomerization state of Glu18 mutants in DJ-1. We used a combination of thermal stability analysis, circular dichroism (CD) spectroscopy, sedimentation equilibrium ultracentrifugation, NMR spectroscopy, X-ray crystallography as well as in vitro and cell-based crosslinking to show that these mutant proteins are predominantly dimeric in vitro. In cultured cells, Glu18 DJ-1 mutants exist as crosslinkable dimers, but they have reduced crosslinking efficiency compared to the wild-type protein. Analysis of in vitro crosslinking experiments using the thiol-reactive crosslinker bis-malaeimidoethane (BMOE) suggests that this is due to increased conformational flexibility of otherwise dimeric Glu18 mutant proteins. We conclude that Glu18 mutants of human DJ-1 are dimeric and structurally comparable to the wild-type protein. Furthermore, this work demonstrates the utility of thiol crosslinkers as a tool for exploring protein flexibility in cases where multiple free cysteine residues with differing degrees of solvent exposure are present.
Methods
Expression and Purification of Recombinant DJ-1
A construct bearing human DJ-1 cloned between the NdeI and XhoI sites of pET15b was transformed into BL21(DE3) E. coli. This construct expresses human DJ-1 with a N-terminal hexahistidine tag that was removed using thrombin, generating a final protein with the vector-derived sequence “GSH” at its N-terminus. Protein expression and purification using Ni2+ metal affinity chromatography was performed as previously described (Lakshminarasimhan et al. 2010) except that all buffers contained freshly added 2 mM DTT. Nucleic acids, which are common contaminants in recombinant DJ-1, were removed by passage over Hi-Q anion exchange column (Bio-Rad). The final protein ran as a single band in overloaded, Coomassie blue-stained SDS-PAGE and was concentrated to 20 mg ml−1 (1 mM; ε280=4000 M−1 cm−1) with a 10 kDa MWCO regenerated cellulose membrane spin concentrator at 4°C (Millipore).
The oxidation status of all recombinant proteins was determined by electrospray mass spectrometry using an Agilent 1200 LC system coupled to the electrospray ionization (ESI) source of a Bruker Solarix – 70 hybrid FTMS mass spectrometer in the Redox Biology Center (RBC) Mass Spectrometry Core Facility at the University of Nebraska-Lincoln.. The mobile phases were water and 0.1% formic acid in 100% acetonitrile. After injection, the proteins were separated on ProSwift RP-10R column (1 × 50 mm; Thermo Scientific) with a flow rate of 20 μL min−1. The system was controlled by HyStar v.3.4.8.0 software (Bruker Daltonics). Mass spectrometry data was collected with resolving power of 200,000 in positive mode, a capillary voltage of 4,500 V, and an end plate offset of 500 V. The dry temperature was set at 180 °C. Dry gas flow was maintained 4 L min−1. Acquisition range was 600 – 3,000 m/z with 0.3 s accumulation time. Data was deconvoluted and analyzed by DataAnalysis v.4.0 software (Bruker Daltonics) and isotope distribution compared with the predicted pattern calculated using Molecular Weight Calculator v.1.0 (www.wsearch.com.au). For the proteins used in this study, wild-type, E18D, and E18Q DJ-1 were fully reduced but E18N and E18A DJ-1 contained a species that was 32 amu larger than expected, indicating probable oxidation of Cys106 to Cys106-SO2−. The fraction of oxidized protein was determined using the ratio of the peak intensity of the oxidized protein to the sum of the intensities of the oxidized and reduced species. Based on peak intensity ratios, E18N DJ-1 was 60% oxidized, while the E18A mutant was 34% oxidized. A second preparation of E18A DJ-1 was purified as above, but with 3 mM DTT in all buffers and handling quickly on ice. Fully Cys106-SO2−-oxidized E18A DJ-1 was prepared by dialyzing the protein against 25 mM HEPES pH=7.5, 100 mM KCl, oxidized on ice for 1 hour in the presence of a five-fold molar excess of H2O2, then dialysed against 25 mM HEPES pH=7.5, 100 mM KCl, 3 mM DTT. The oxidation state of the protein was confirmed by electrospray mass spectrometry as described above.
Thermal Stability Determination of DJ-1
The melting temperatures of DJ-1 variants were determined using the differential scanning fluorimetry assay (Pantoliano et al. 2001). Proteins were diluted to 1–5 mg ml−1 in storage buffer (25 mM HEPES pH 7.5, 100 mM KCl, 2 mM DTT). Sypro Orange (Life Technologies, Invitrogen) was diluted to 50, 100, 250, 500× in storage buffer and added to the samples. Samples were heated from 20–95°C at a rate of 2°C min−1 while the fluorescence emission of Sypro Orange was monitored at 575 nm in 8-strip PCR tubes with optically clear caps (Bio-Rad) in a Bio-Rad iCyclerQ real-time PCR. The data were plotted as the first derivative of fluorescence as a function of temperature, where the maximum of this function was taken as the melting temperature (Tm).
Secondary Structure Content Determination Using Far-UV Circular Dichroism
Proteins were dialyzed overnight at 4°C against 10 mM sodium phosphate pH 7.2, and then diluted in the dialysis buffer to a final concentration of 0.25 mg ml−1 (12.5 μM). CD spectra were measured using a Jasco J-815 CD spectrometer with a grating of 3400 lines cm−1 in a 1 mm pathlength quartz cuvette. Spectra were measured in continuous scanning mode at a rate of 20 nm min−1, an accumulation of four scans per spectrum, 1 nm bandwidth, data pitch of 0.1 nm, and data integration time of 1 s. The protein concentration of the sample in the CD cuvette was determined by measuring the absorbance of the sample at 205 nm (Scopes 1974):
Where c is the protein concentration in mg ml−1, A205 is the absorbance of the sample at 205 nm, and l is the path length in cm. Once the molar concentration of protein was determined using a molecular mass of, The mean molar residue ellipticity (deg cm2 dmol−1 residue−1) was calculated using the equation:
where θ(λ) is the mean residue molar ellipticity as a function of wavelength, n is the number of residues in the protein (192 in the case of tag-cleaved DJ-1), c is the concentration of the protein (M), l is the sample path length (cm), and θobs(λ) is the measured ellipticity as a function of wavelength (nm). The molecular weight of DJ-1 is 20,114 Da
Sedimentation Equilibrium Ultracentrifugation
Tag-cleaved DJ-1 proteins were dialyzed overnight at 4°C against 25 mM Tris pH 7.5, 100 mM KCl, and 2 mM DTT. The proteins were diluted to 1.0, 1.5, and 2.5 mg ml−1 in the dialysis buffer and centrifuged at 16,000×g to remove any particulate matter or aggregated protein. The samples were loaded into six-sector charcoal filled epon centerpieces fitted with sapphire windows, placed in an An-50 Ti rotor, and centrifuged at 20°C using a Beckman Coulter XL-I analytical ultracentrifuge. Three different rcfs were used: 2.3×104, 3.2×104, and 4.6×104×g. At each rotor speed, the absorption at 270 nm as a function of radius was measured at both 20 and 22 hours. The absence of any changes in these two scans indicated that equilibrium had been reached. The centrifugation data were fitted using both the monomer-dimer self-association and ideal single species models in the Origin 6.0 Beckman analysis software package using partial specific volumes of the proteins and the solvent density values calculated using SedNTerp (Laue et al. 1992).
E18A DJ-1 Crystallization, Data Collection, and Refinement
Purified tag-cleaved E18A DJ-1 was dialyzed against 25 mM HEPES, pH 7.5, 100 mM KCl, and 2 mM DTT, and centrifugally concentrated to 25 mg ml−1. Crystals were grown using sitting drop vapor equilibration by mixing 2 μl of protein and 2 μl of reservoir and equilibrating against 1.3–1.6 M sodium citrate, 50 mM HEPES, pH 7.5 at room temperature. Bipyramidal crystals in space group P3121 appeared within 3–4 days and measured ~300 μm in each dimension. The crystals were cryoprotected by serial transfer through solutions of sodium malonate pH 7.4 to a final concentration of 3.4 M (Holyoak et al. 2003), mounted onto nylon loops, and cryocooled by rapid plunging into liquid nitrogen. X-ray diffraction data were collected using a Rigaku MicroMax 007 rotating Cu anode source operating at 40 kV and 20 mA. Diffraction data were recorded with a Raxis IV++ image plate detector and the data were integrated and scaled using HKL2000 (Otwinowski & Minor 1997). Data statistics are shown in Table 1.
Table 1.
Molecular mass of DJ-1 mutants determined by sedimentation equilibrium centrifugation
| Protein | Fit molecular mass (Da) | Predicted dimeric molecular mass (Da) |
|---|---|---|
| Wild-type | 35590 | 40345 |
| E18A | 35091 | 40228 |
| E18D | 34097 | 40317 |
| E18Q | 33026 | 40343 |
| E18N | 34199 | 40315 |
| L10P | 22505 | 40312 |
The coordinates for human DJ-1 (PDB code 1P5F (Wilson et al. 2003)) were used as a starting point for stereochemically restrained coordinate and atomic displacement parameter refinement in PHENIX (Afonine et al. 2012). Rigid body refinement was followed by manual adjustments to the protein model and placement of ordered water molecules by inspection of electron density in COOT (Emsley & Cowtan 2004). Multiple cycles of stereochemically restrained refinement were performed in PHENIX using a bulk solvent correction, riding hydrogen atoms, and individual anisotropic displacement parameters. The refinement of anisotropic displacement parameters is justified by the observation-to-parameter ratio for the model and data and decreases both R and Rfree values by ~3%. Final model statistics are provided in Table 1. Structural figures were made with POVScript+ (Fenn et al. 2003).
Preparation of 15N-labeled DJ-1 Proteins
Recombinant uniformly labeled 15N DJ-1 proteins were expressed using pET15b in BL21(DE3) E. coli. Cells were grown in 100 mL LB media supplemented with 100 μg mL−1 ampicillin overnight at 37°C with shaking at 250–270 rpm. 50 mL of the overnight growth in LB was washed twice in 10 mL of 15N M9 defined media (6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1 g 15NH4Cl (Cambridge Isotope Labs, 99.9% purity), 2 mM MgSO4, 0.2% glucose, 0.1 mM CaCl2, 0.001% thiamine, and 100 μg mL−1 ampicillin) and used to inoculate 1 L of 15N labeled M9 media. The culture was incubated at 37°C with shaking at 250 rpm. Protein expression was induced at OD600=0.5 by the addition of 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) followed by 4–5 hrs of incubation at 37° C with shaking. Cells were harvested by centrifugation and cell pellets (~2 g) were flash frozen in liquid nitrogen and stored at −80°C until needed.
Uniformly 15N labeled proteins were purified using the same protocol as unlabeled recombinant proteins above and dialyzed against 25 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.5, 25 mM NaCl, 2.5 mM DTT overnight at 4°C and centrifugally concentrated to 14–20 mg ml−1. The final 15N-labeled DJ-1 proteins ran as single bands of ~20 kDa on overloaded Coomassie Blue-stained SDS-PAGE. All proteins were flash frozen in liquid nitrogen and stored at −80°C until needed.
The 15N labeling efficiency as well as oxidation status of recombinant 15N proteins were determined using electrospray mass spectrometry as described in “Expression and Purification of Recombinant DJ-1”. All proteins were >99 % uniformly 15N labeled, although only 15N-WT and 15N-E18D DJ-1 were fully reduced. Based on peak intensity ratios, E18Q was 43% oxidized, E18N 24% oxidized, E18A 64% oxidized, and E18D was completely reduced, consistent with a previous report (Blackinton et al. 2009).
NMR Sample Preparation, Data Collection, and Processing
Samples (500 μL) were prepared for NMR by adding D2O to a final concentration of 10% and then transferred to a 5mm, high-throughput 7″ standard series NMR tube (Norell). All 1H-15N HSQC spectra were collected from samples maintained at 303 K (30°C) using a Bruker AVANCE DRX 500 MHz spectrometer equipped with a 5 mm triple resonance, z-axis gradient cryoprobe with a cold proton channel at the University of Nebraska-Lincoln Research instrumentation facility. Spectra were acquired at spectral sweep widths of 14 and 40 ppm for the proton and nitrogen dimensions, respectively and were collected at 2000 × 256 resolution. Data processing was performed using NMRPipe (Delaglio et al. 1995) and analysis was performed using NMRView (Johnson 2004). Chemical shift assignments for wild-type DJ-1 were retrieved from the Biological Magnetic Resonance Data Bank with accession number BMRB 17507 (Malgieri & Eliezer 2008). Weighted total chemical shift changes were calculated using the equation
Where ΔδH+N is the weighted total chemical shift change, Δδ1H is the change in proton chemical shift, and Δδ15N is the change in nitrogen chemical shift. A few resonances in each mutant protein could not be matched to the corresponding residue in the wild-type protein due to spectral crowding or absence, and thus no ΔδH+N could be calculated for those residues.
In Vitro Crosslinking of Recombinant DJ-1
Recombinant proteins were exchanged into phosphate buffered saline (PBS) using desalting columns (Pierce, Zeba spin column with 7 kDa cutoff). Disuccinimidyl suberate (DSS, Pierce) and BMOE (Pierce) were resuspended in dimethylsulfoxide (DMSO) and diluted to the appropriate concentrations using PBS. Protein (75 μM) was incubated with 0, 75, 150, or 375 μM crosslinker for 30 minutes at room temperature. Reactions were quenched by the addition of NUPAGE LDS Sample buffer (Invitrogen) containing β-mercaptoethanol, then 2 μg of each protein was separated using 4–20% Criterion Precast TGX gels (Bio-Rad). Gels were stained with GelCode Blue Stain reagent (Pierce) and imaged using an Odyssey CLx infrared imaging system (LI-COR Biosciences). Additionally, BMOE crosslinked proteins were analyzed by Western blot using antibodies \against the N and C terminals of DJ-1 (Abcam ab76008 and Santa Crux sc-27006). Quantification was performed using Image Studio software (LI-COR Biosciences).
HEK293FT Transient Transfection and Plasmids
HEK293FT cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) + 10% v/v fetal bovine serum (FBS) and 0.8 × 106 cells were transfected with 2 μg of plasmid DNA using lipofectamine (Invitrogen). pDEST40 vectors containing wild type human DJ-1, E18N/D/Q, and C106A mutations have been previously described (Blackinton et al. 2009) and are available through Addgene. E18A and C106S mutants were made using site directed mutagenesis (Stratagene Quick Change II). All plasmids were sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Life Technologies).
Crosslinking DJ-1 in Living Cells
Cells were collected 2–3 days after transfection by scraping and washed 3× with ice-cold PBS. Cell pellets were resuspended in PBS containing 1 mM DSS, 1 mM BMOE, or the equivalent concentrations of DMSO for 30 min at RT. Reactions were quenched by the addition of either Tris-HCl (4 mM) or DTT (10 mM) for DSS and BMOE, respectively. Cell pellets were lysed using radioiummunoprecipitation assay buffer (25mM Tris-HCl pH 7.5, 150mM NaCl, 1% NP-40, 1% Sodium Deoxycholate, 0.1% SDS) for 30 min on ice, then cleared by centrifugation (16,000×g, 10 min, 4°C). Protein concentrations were determined using the 660 nm protein assay (Pierce). Proteins (7.5 μg) were separated on 4–20% Criterion Precast TGX gels then transferred to nitrocellulose membranes using a Bio-Rad Trans-Blot Turbo system. Blots were blocked using Odyssey blocking buffer for 1 hr at RT then probed overnight with primary antibody (Invitrogen ms-anti-V5, 1:3000). LiCor secondary antibodies were used at 1:15,000 and incubated with blots for 1 hr at RT. Blots were imaged using an Odyssey CLx infrared imaging system and quantified using Image Studio software.
Purification of 6xHis-tagged DJ-1 From HEK293FT Lysates
HEK293FT cells that had been transiently transfected with the indicated pDEST40 plasmids were collected 3 days after transfection. Cell pellets were sonicated in His Buffer (GE Lifesciences) supplemented with 20mM imidazole. Lysates were cleared by centrifugation (16,000×g, 10 min, 4°C) then incubated with histidine purification resin (Pierce) for 30 min at 4°C. The resin was washed and then bound proteins were eluted using 500 mM imidazole. Equal volumes of protein were separated using 4–20% Criterion Precast TGX gels and Western blots were performed using rb-anti-DJ-1 (Abcam ab76241, 1:2000) and ms-anti-V5 (Invitrogen, 1:2000). LiCor secondary antibodies were used at 1:15,000 and blots were imaged and quantified as described above.
Data Accessibility
The experimental structure factors and refined crystallographic model for E18A DJ-1 have been deposited with the Protein Data Bank (PDB) with accession code 4OQ4.
Results
Wild-type DJ-1 and Various Glu18 Mutants Possess Similar Thermal Stabilities and Secondary Structure
The dimerization of DJ-1 buries a total of ~2,700 Å2 of surface area with substantial hydrophobic character (Tao & Tong 2003, Honbou et al. 2003, Wilson et al. 2003, Huai et al. 2003, Lee et al. 2003). Because of this large and partially hydrophobic dimer interface, DJ-1 is overwhelmingly dimeric in solution (Wilson et al. 2003, Repici et al. 2013). The L166P and L10P parkinsonism-associated DJ-1 mutants both disrupt homodimerization, resulting in proteins that do not fold properly and are consequently unstable and aggregation-prone in solution (Macedo et al. 2003, Miller et al. 2003, Moore et al. 2003, Ramsey & Giasson 2010, Repici et al. 2013). Therefore, we expected that other mutants which disrupt DJ-1 dimerization would also result in large changes in thermal stability and secondary structural content.
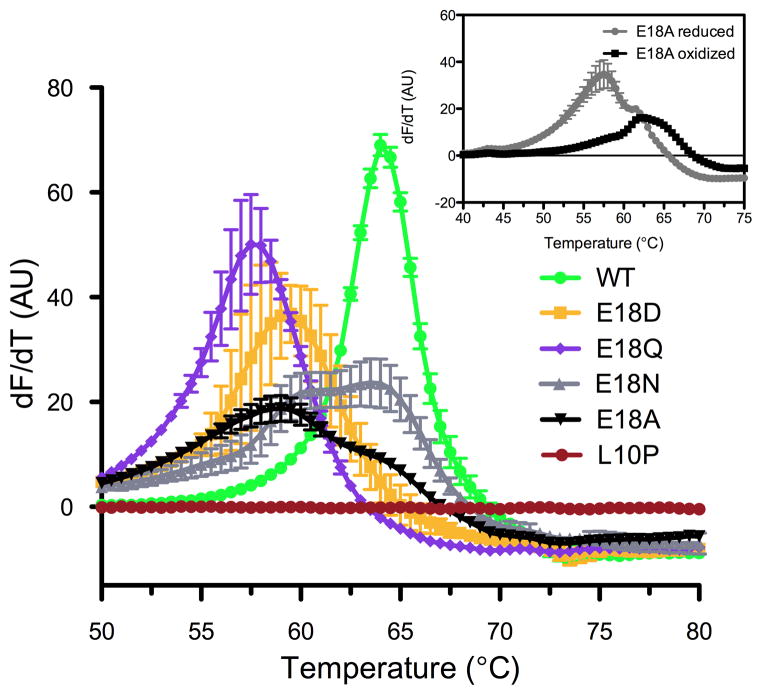
DJ-1 thermal stability was measured using differential scanning fluorimetry (Pantoliano et al. 2001) with proteins that were fully reduced at Cys106 wherever possible. Wild-type DJ-1 had a measured melting temperature (Tm) of 65°C (Fig. 1), consistent with previous reports (Hulleman et al. 2007, Lakshminarasimhan et al. 2008, Malgieri & Eliezer 2008, Lin et al. 2012). E18Q DJ-1, the most structurally conservative mutation, had a Tm of 57°C, while the E18D mutant had a Tm of 59°C. The melting transitions for E18N and E18A DJ-1 showed two peaks, corresponding to approximate Tm values of 60°C and 64°C for E18N DJ-1 and 58°C and 64°C for E18A DJ-1 (Fig. 1). The higher Tm transition in both is due to the presence of some Cys106-SO2− oxidized protein in these samples (see below). Therefore, all of the Glu18 mutants have modestly reduced thermal stabilities (~7°C) compared to the wild-type protein. In contrast, monomeric L10P DJ-1 showed no detectable unfolding transition, consistent with prior reports that it cannot homodimerize and is poorly folded. The presence of clear unfolding transitions for wild-type DJ-1 and the Glu18 mutants indicates that they are well-folded and dimeric.
Figure 1.
DJ-1 with mutations at Glu18 is stably folded. Thermal stability of DJ-1 was determined using the differential scanning fluorimetry assay. In panel A, all proteins show a cooperative unfolding transition except L10P DJ-1 (red), which is a pathogenic mutation that disrupts the DJ-1 dimer. Melting temperatures (Tm) are the maximum value of the first derivative of the fluorescence signal as a function of temperature (dF/dT). The magnitude of these transitions differs due to variability in the rate of fluorescence changes during denaturation. E18A (black triangles) and E18N (grey triangles) DJ-1 have bimodal unfolding transitions, indicative of a mixture of reduced and Cys106-SO2−-containing proteins. The inset shows the thermal denaturation of reduced (grey circles) and intentionally Cys106-SO2−-oxidized E18A DJ-1 (black squares), confirming that the oxidized form is the more stable species. All data were measured in triplicate with averaged values and standard deviations indicated.
Our previous work has shown that oxidation of Cys106 to the sulfinic acid (Cys106-SO2−) results in a ~12°C stabilization of DJ-1 (Lin et al. 2012). The presence of bimodal thermal denaturation profiles for the E18N and E18A DJ-1 samples (Fig. 1) led us to suspect that the higher Tm transition in each was due to the presence of spontaneously oxidized proteins containing Cys106-SO2−. We verified this using electrospray mass spectrometry (see Methods), which showed that approximately 60% of E18N and 34% of E18A DJ-1 were present as species whose mass was 32 amu greater than expected, consistent with oxidation to the Cys106-SO2− form. The difficulty of purifying fully reduced E18N DJ-1 has been noted (Witt et al. 2008), owing to its enhanced propensity for oxidation at Cys106. In contrast, detailed biochemical characterization of E18A DJ-1 has not been previously reported. Predominantly reduced E18A DJ-1 was purified using elevated DTT (3 mM) and handling the protein on ice at all times. A portion of this protein was then oxidized to the sulfinic acid form by incubation with a five-fold molar excess of H2O2 (Blackinton et al. 2009) and the thermal denaturation of each sample was measured (Fig. 1 inset). The 6 °C increase in Tm for Cys106-SO2− E18A DJ-1 is less than the 12 °C increase observed for wild-type DJ-1 (Lin et al. 2012) (Fig. S1), indicating that the single hydrogen bond between Glu18 and Cys106-SO2 eliminated by the E18A mutation accounts for ~6 °C of thermal stabilization upon Cys106 oxidation.
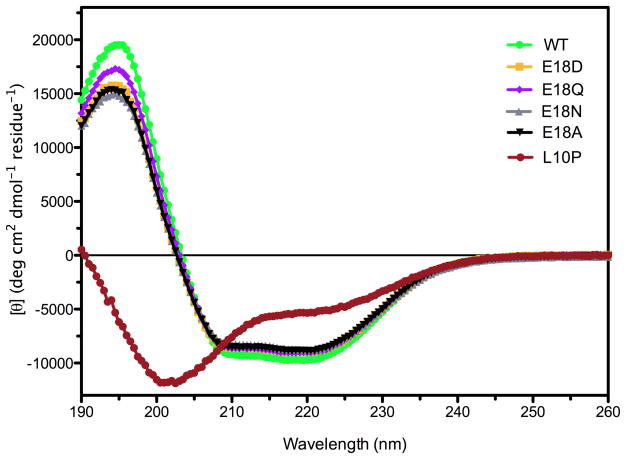
The secondary structural content of the Glu18 mutants and L10P DJ-1 was determined using far UV circular dichroism (CD) spectroscopy and compared to wild-type DJ-1. All of these proteins except dimerization-impaired L10P DJ-1 showed similar secondary structural content (Fig. 2). In contrast, the CD spectrum of L10P DJ-1 indicated a profound loss of secondary structure that accompanies loss of dimerization. Therefore, the CD and thermal stability results consistently indicate that mutations at Glu18 do not result in major loss of structural integrity in DJ-1.
Figure 2.
The secondary structural content of DJ-1 is unaffected by mutations at Glu18. Far UV circular dichroism (CD) spectroscopy was used to determine the secondary structural content of DJ-1 mutants. Wild-type (WT) DJ-1 and all of the Glu18 mutants have similar CD spectra, indicating no loss of secondary structure that would be expected to accompany changes in oligomerization. In contrast, the dimer-disrupting L10P mutation (red circles) results in profound changes in its CD spectrum.
Sedimentation Equilibrium Ultracentrifugation Indicates That Glu18 Mutants are Dimeric in Solution
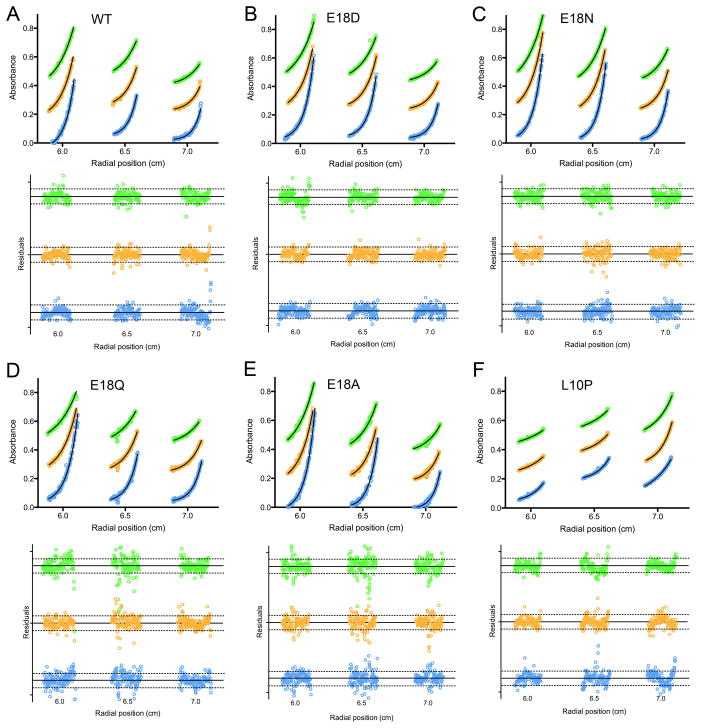
Prior atomic resolution X-ray crystallographic studies show that E18D, E18N, and E18Q DJ-1 formed dimers that were identical to wild-type DJ-1 (Blackinton et al. 2009, Witt et al. 2008). However, despite the extensive DJ-1 dimer interface, it is possible that the relatively high concentration of the protein needed for crystallization or other features of the crystallization experiment may favor artifactual dimerization of the Glu18 mutants. To address this, sedimentation equilibrium ultracentrifugation was used to determine the oligomerization states of DJ-1 mutants in solution at 50–125 μM. As seen in Fig. 3 and accompanying Table 1, all mutants except L10P sedimented similarly to dimeric wild-type DJ-1. L10P DJ-1 sedimented as a monomeric species that showed a pronounced tendency to aggregate, resulting in a poorer quality of fit as judged by the residuals (Fig. 3). For all proteins except the dimerization-impaired L10P mutant, including the wild-type protein, the molecular masses determined by centrifugation are ~4–5 kDa smaller than the predicted mass of 40 kDa for dimeric DJ-1 (Table 1). Therefore, the Glu18 mutants have comparable molecular masses to wild-type DJ-1. Attempts to fit the data using a more complex monomer-dimer self association model did not result in any improvement in the quality of fit, indicating that wild-type and Glu18 DJ-1 mutants are dimeric under these solution conditions. In contrast, L10P DJ-1 is monomeric (Table 1), as is evident from its distinct sedimentation (Fig. 3).
Figure 3.
Glu18 mutants of DJ-1 are stable dimers in solution. The molecular mass of wild-type and mutants of DJ-1 was determined by sedimentation equilibrium centrifugation. In each panel, three concentrations of protein (1.0, 1.5, and 2.5 mg ml−1 from left to right) were run at three different rcfs: 2.3×104 (green), 3.2×104 (orange), and 4.6×104 (blue) xg. Nine total datasets for each sample were globally fitted to determine the molecular mass. The weighted fit residuals are shown below each panel, with dotted lines indicating the χ2=1 region. Wild-type DJ-1 and all mutants sediment as dimers, while L10P (panel F) sediments as an aggregation-prone monomer. All fit residuals except L10P (panel F) lack systematic trends, indicating a good fit to the model. The poorer fit of L10P DJ-1 is due to the propensity of this protein to aggregate. The traces at different rotor speeds are vertically offset to improve clarity. Experimental measurements are shown as symbols and the best-fit curve in solid black lines.
1H-15N HSQC NMR Spectra of Glu18 Mutants are Similar to Dimeric Wild-type DJ-1
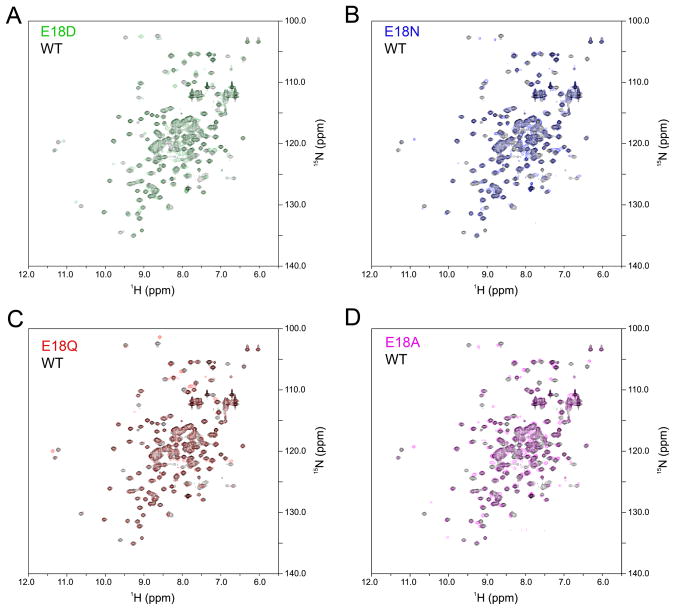
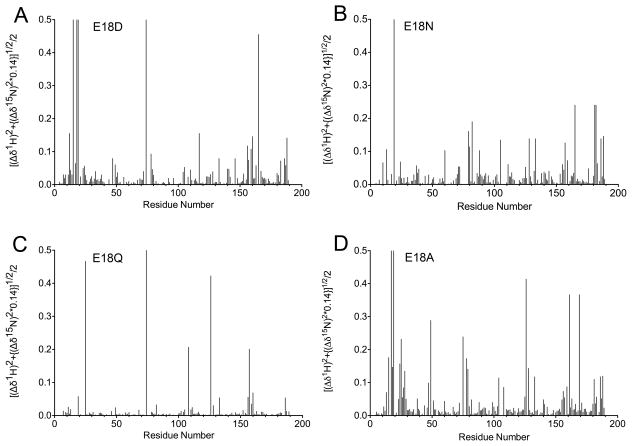
Two-dimensional NMR 1H-15N heteronuclear single quantum coherence (HSQC) spectra were collected for wild-type DJ-1 and the four Glu18 mutants (E18A, E18D, E18N, E18Q) to determine their oligomerization states in solution. Because NMR chemical shifts are extremely sensitive to the local environments of resonating nuclei, any change in DJ-1 oligomerization state would result in significant changes in the 1H-15N HSQC spectrum of the protein. Figure 4 shows the wild-type DJ-1 HSQC spectrum in black with the Glu18 mutants overlaid with the indicated colors. The 1H-15N HSQC spectrum of wild-type DJ-1 agrees well with a previously published 1H-15N HSQC spectrum for DJ-1 (Malgieri & Eliezer 2008). All four Glu18 mutant spectra are very similar to that of wild-type DJ-1, confirming that these mutations result in only small perturbations to the structure of dimeric DJ-1 (Fig. 4). As expected, the substitutions at Glu18 result in changes near the site of mutation compared to wild-type DJ-1 (Fig. 5). The regions most impacted by substitution at residue 18 are residues 10–30, 70–80, 120–130, and 155–170. Apart from residues 120–130, all of these regions are near Glu18 in the crystal structure of DJ-1 and thus chemical shift perturbations in these areas are expected. The stretch of residues from 120–130 is near Cys106, which hydrogen bonds to Glu18. Based on the number and magnitude of the chemical shift changes, E18Q is the least disruptive mutation, E18D and E18N DJ-1 are intermediate, and E18A is the most disruptive (Fig. 5). However, E18A still exhibits high spectral similarity to wild-type DJ-1. Because NMR reports on the most abundant species in solution, the low signal-to-noise features of spectra were inspected to determine if a minor species with a different spectrum indicative of major oligomeric changes could be detected. No evidence for a population of monomeric DJ-1 could be found in any spectrum.
Figure 4.
NMR demonstrates that wild-type and the Glu18 mutants are dimeric in solution. 1H-15N heteronuclear single quantum coherence (HSQC) spectra were collected with 15N labeled DJ-1. In all panels, the HSQC spectrum of wild-type DJ-1 is shown in black. All Glu18 mutant spectra superimpose well with the wild-type spectrum, indicating that all proteins are in the same oligomerization state.
Figure 5.
Glu18 mutations result in localized NMR chemical shift perturbations. Chemical shift changes were calculated by comparing the wild-type and Glu18 mutant 1H-15N HSQC spectra. These chemical shift changes were calculated as shown on the Y-axis. The regions most affected by substitution at residue 18 are residues 10–30, 70–80, 120–130, and 155–170. All of these regions except 120–130 are in close proximity to residue 18 in the DJ-1 crystal structure. Weighted chemical shift changes were plotted using PRISM (GraphPad).
The Crystal Structure of E18A DJ-1 Confirms a Dimeric Protein With an Oxidized Cys106 Residue
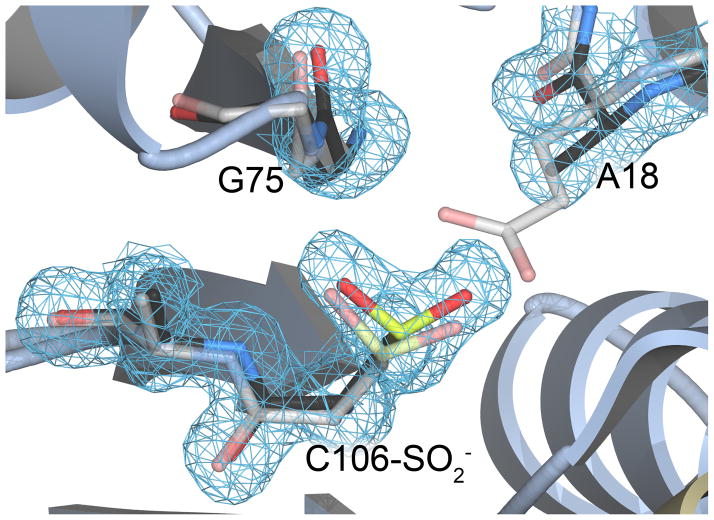
Maita et al. (Maita et al. 2013) reported the first study of the E18A DJ-1 mutant, which they made in addition to the E18D, E18Q, and E18N mutations initially designed and studied by us (Witt et al. 2008, Blackinton et al. 2009). Unlike the E18D, E18Q, and E18N mutants, no crystal structure has been reported for E18A DJ-1. We determined the crystal structure of E18A DJ-1 at 1.49 Å resolution. Like previous structures of DJ-1 in space group P3121, E18A DJ-1 crystallizes as a dimer with one monomer in the asymmetric unit. The dimer is generated by crystallographic symmetry and is the form of the protein that exists in solution (see above), burying ~2800 Å2 of total surface area at the dimer interface (Krissinel & Henrick 2007). The structure of E18A DJ-1 is essentially identical (Cα RMSD=0.2 Å) to that of oxidized wild-type DJ-1 (PDB code 1SOA). Consistent with the thermal denaturation evidence for spontaneously oxidized E18A DJ-1 (Fig. 1), we find that Cys106 in the crystal structure is oxidized to Cys106-SO2− (Fig. 6). The spontaneous oxidation of Cys106 is commonly observed in crystal structures of DJ-1, reflecting the enhanced reactivity of this residue (Canet-Aviles et al. 2004, Blackinton et al. 2009). The Cys106-SO2− in E18A DJ-1 indicates that the stabilizing hydrogen bond donated by the protonated sidechain of Glu18 in wild-type DJ-1 is not absolutely required for Cys106 oxidation.
Figure 6.
Cys106 is oxidized to the cysteine-sulfinic acid in crystalline E18A DJ-1. The environment around Cys106 is shown in the 1.49 Å resolution crystal structure of E18A DJ-1 (black carbon atoms). The structure of wild-type DJ-1 (grey carbon atoms, semi-transparent) is superimposed, showing the location of Glu18 and its contacts with surrounding residues. The oxidation of Cys106 to the sulfinic acid is well-supported by 2mFo-DFc electron density contoured at 1.0 σ (blue).
In Vitro Crosslinking Indicates Increased Flexibility of Glu18 Mutant Dimers
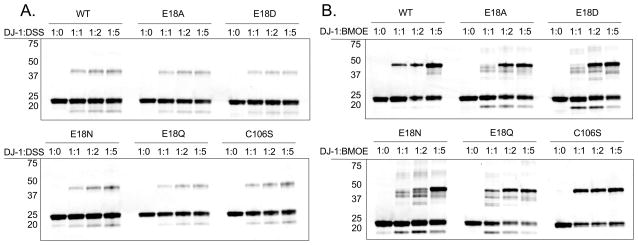
The ability of DJ-1 to be chemically crosslinked as a dimer was used as an additional probe of its solution oligomeric state. Two homobifunctional crosslinkers were used: the primary amine-reactive DSS and thiol-reactive BMOE. DSS is expected to form several potential crosslinks involving multiple possible pairs of lysine residues in the DJ-1 dimer. As shown in Fig. 7, all recombinant DJ-1 proteins form DSS-crosslinked dimers that are resolved by SDS-PAGE, as well as uncrosslinked monomers and a faster-migrating species that is likely the intramolecular crosslinked DJ-1 monomer (Hulleman et al. 2007). Importantly, the presence of monomer bands indicates that crosslinking did not go to completion, which is typical but is not proportional to the amount of monomer in solution because samples were resolved on denaturing gels.
Figure 7.
In vitro crosslinking of recombinant proteins. Coomassie stained SDS-PAGE gels containing tag-cleaved recombinant wild type DJ-1 and mutants that were exposed to increasing amounts of the amine-crosslinker DSS (Panel A), or the sulfhydryl crosslinker BMOE (Panel B). Molar ratios of DJ-1 monomer to crosslinker are shown at the top of each lane. Approximate molecular masses for the marker are shown on the left of each gel. Monomeric DJ-1 migrates with an apparent molecular mass of ~22 kDa, while the dimeric protein migrates with an approximate mass of 45 kDa. The multiple dimeric crosslinked species of the Glu18 mutants are due to crosslinks involving distinct pairs of cysteine residues and indicate enhanced flexibility of these dimeric proteins that is not observed for wild-type or C106S DJ-1. Similar results were obtained in three independent experiments for each crosslinker.
In contrast to DSS, BMOE can form a more limited number of crosslinks involving only the three cysteine residues per monomer of DJ-1, Cys46, Cys53, and Cys106. Based on the crystal structure of DJ-1, the Cys53-Cys53 dyad formed by dimerization of DJ-1 has a Sγ-Sγ separation of 3.5 Å, closer than the reported crosslinking range of 6–11 Å for BMOE (Green et al. 2001). The proximity of these two thiols in the DJ-1 dimer and their high degree of solvent accessibility makes the Cys53-Cys53 pair the most likely target of BMOE crosslinking. Therefore, BMOE is potentially a sensitive probe of the conformation and solution oligomerization state of DJ-1. As expected, BMOE crosslinked the DJ-1 dimer (Fig. 7B). In the Glu18 mutants, however, there are several faster-migrating species below the dominant dimer band but above the monomer band (Fig. 7B). These faster-migrating dimer species likely correspond to crosslinks within the DJ-1 dimer that involve multiple cysteine residues. With three cysteine residues per DJ-1 monomer, there are a total of 13 distinct crosslinked dimeric species that can theoretically form, not all of which may be distinguishable by gel electrophoresis. Western blotting confirms that each band is DJ-1 (Fig. S2) and they cannot be proteolytic fragments as they are not present at higher molar ratios of BMOE to DJ-1 (Fig. 7B) and they are detected by antibodies directed at both N- and C-terminal eptiopes (Fig. S2). The faster-migrating dimeric crosslinked species are totally absent in the C106S mutant (Figure 7B), indicating that Cys106 may be involved in forming additional crosslinks in the mutants. The 10.3 Å distance between Cys106 and Cys46 in the crystal structure of the DJ-1 dimer is at the far end of the reported BMOE crosslinking range (Green et al. 2001), although Cys46 is completely buried in the crystal structure and thus should not be readily crosslinked. Therefore, the DJ-1 dimer must transiently sample conformations that differ significantly from the crystal structure to allow additional crosslinks involving Cys106 and Cys46 to form. The multiple dimeric BMOE-crosslinked species formed by Glu18 mutants demonstrate that these mutations increase the flexibility of the DJ-1 dimer without resulting in complete dissociation of the oligomer.
Cellular Crosslinking Efficiency is Reduced for Glu18 Mutant DJ-1
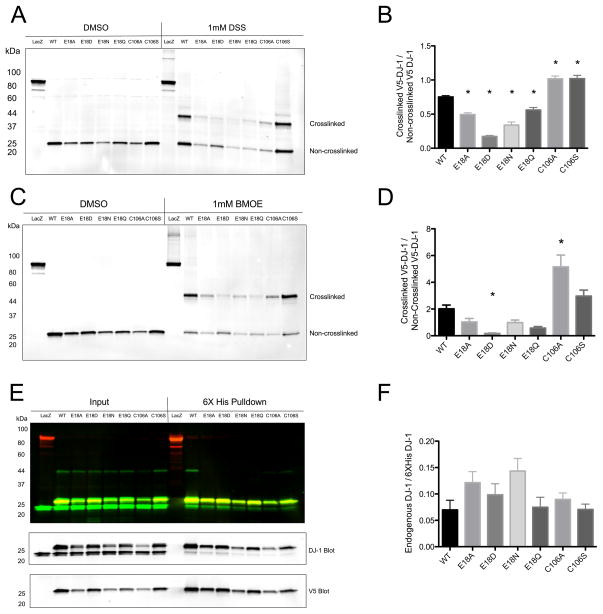
We next tested the ability of DJ-1 to be crosslinked in a cellular context using C-terminally 6xHis-V5-tagged DJ-1 protein that was transiently expressed in HEK293FT cells. Both DSS and BMOE resulted in formation of dimeric species for all proteins, confirming that DJ-1 and the Glu18 mutants exist at least in part as dimers in cells (Fig. 8A,B). However, compared to the wild-type and C106S mutant proteins, the cellular crosslinking efficiency was diminished for the Glu18 mutants. This trend was observed for both DSS and BMOE and does not correlate strongly with loaded protein amount (linear regression R2 = 0.1544, p=0.0781, n=7 DJ-1 variants measured in triplicate).
Figure 8.
DJ-1 crosslinking and heterodimer formation in HEK293FT cells. In Panel A, HEK293FT cells transiently transfected with 6xHis-V5 tagged wild type DJ-1 and mutants were exposed to the amine crosslinker DSS and V5-tagged protein was analyzed by Western blot. Crosslinked dimers of ~45 kDa are apparent for all proteins, but are less prominent for the Glu18 mutants. In Panel B, DSS crosslinking efficiency was quantified using densitometry (n=6 independent samples, * denotes p < 0.05 versus WT, one-way ANOVA followed by Bonferroni’s multiple comparison test). Panel C shows V5 Western blot analysis of proteins from HEK293FT cells transiently transfected with 6xHis-V5 tagged proteins and exposed to the sulfhydryl crosslinker BMOE. Crosslinked dimeric species for all DJ-1 proteins are evident at ~45 kDa. In Panel D, the crosslinking efficiency of BMOE was quantified using densitometry (n=9, * denotes p < 0.05 versus WT, one-way ANOVA followed by Bonferroni’s multiple comparison test). In Panel E, V5-6xHis tagged DJ-1 proteins were purified from HEK293FT lysates using Ni2+ affinity resin and then Western blots were probed with anti-V5 (red) and anti-DJ-1 (green) antibodies to examine heterodimer formation. In Panel F, the amounts of endogenous wildtype DJ-1 that co-purified with tagged exogenous proteins are quantified (n=6, one-way ANOVA p = 0.0546). All V5-6xHis-DJ-1 proteins are able to form mixed heterodimers with endogenous DJ-1 with comparable efficiency.
We next examined the ability of exogenous DJ-1 mutants to form heterodimers with endogenous DJ-1 in cells. 6xHis-V5 tagged DJ-1 was transiently expressed in cells and the His-tagged proteins were purified using Ni2+-affinity resin (Fig. 8D,E). Endogenous DJ-1 was co-purified with all forms of 6xHis tagged Glu18 mutants but not with the negative control protein LacZ. Quantification showed that all the Glu18 mutants were able to form heterodimers with endogenous DJ-1 with efficiencies that are similar to the wild-type protein. Taken together, the cellular crosslinking and 6xHis tag pulldown data demonstrate that Glu18 mutants of DJ-1 are able to form both homo- and heterodimers when expressed in human cells.
Discussion
DJ-1 is a dimeric cytoprotective protein whose oligomerization state is important for its stability and function. Here, we used a variety of in vitro biophysical techniques and crosslinking in living cells examine a series of designed mutations at Glu18, the residue that stabilizes Cys106 oxidation. Given the conservative character of most of these mutations and their previously reported dimeric crystal structures, it would be surprising if they resulted in a total loss of protein dimerization as recently proposed (Maita et al. 2013). Except for the dimer-disrupting mutant L10P, all of the DJ-1 proteins examined here consistently showed evidence for dimer formation. We, and others, have previously reported that E18N and E18Q DJ-1 were protective against oxidative stress and mitochondrial damage, while the oxidation-impaired E18D mutant was not (Blackinton et al. 2009, Cao et al. 2014). Furthermore, consistent with our prior work (Blackinton et al. 2009) but differing from a recent report (Maita et al. 2013), we observed consistently enhanced susceptibility of Cys106 to oxidation in E18N and E18A DJ-1 but a reduced propensity for cysteine oxidation in E18D DJ-1 during routine mass spectrometric characterization of the recombinant proteins used in this study. Our current results therefore suggest that the effects of E18 mutants are not due to loss of dimerization, but instead to changes in the oxidative propensity of Cys106.
Despite biophysical evidence that the Glu18 mutants of DJ-1 are predominantly dimeric, including at moderate concentrations of protein in solution, we replicated a central experimental finding of Maita et al. that cellular crosslinking efficiency of these mutants is reduced compared to wild-type protein (Maita et al. 2013). This effect is observed for both DSS and BMOE, excluding the possibility of an artifact arising from the particular chemistry used to generate the crosslink. We propose that the appearance of multiple crosslinked dimeric species for the recombinant Glu18 mutants at lower ratios of BMOE to protein, which are not detected for the wild-type or C106S proteins, demonstrates that the Glu18 mutant dimers have greater flexibility than wild-type DJ-1. This enhanced flexibility allows for crosslinks involving transiently exposed Cys46 and Cys106 residues to form in addition to the dominant Cys53-Cys53 crosslink. Crosslinks involving either Cys106 or Cys46 would require DJ-1 to transiently adopt conformations that differ significantly from that observed in the crystal structure. The involvement of Cys106 in these additional crosslinks is suggested by the absence of these additional crosslinked species for that mutant. We and others have observed that mutation of Cys106 results in stabilization of DJ-1 (Hulleman et al. 2007), which may also contribute to the absence of additional crosslinked dimeric species for C106S DJ-1 (Fig. S1). Parsimony suggests that Cys106 mutants are unlikely to be more dimeric than wild-type DJ-1, which is already highly dimeric. Taken together with our biophysical characterization of DJ-1 variants, we propose that the simplest interpretation of the in vitro crosslinking data is that differences between mutants are influenced largely by flexibility rather than changes in the monomer-dimer equilibrium.
Although the results of this study are apparently at variance with the prior report indicating that Glu18 mutants of DJ-1 are predominantly monomeric (Maita et al. 2013), all oligomeric proteins exist in equilibrium with some concentration of their monomeric forms. The concentration of monomeric DJ-1 might be significant in some subcellular environments or for certain modified forms of DJ-1. However, monomeric DJ-1 is poorly folded and thus not cytoprotective based on prior characterization of monomeric parkinsonism-associated mutants L10P and L166P (Miller et al. 2003, Moore et al. 2003, Ramsey & Giasson 2010). Furthermore, close plant homologues of human DJ-1 feature two tandemly repeated DJ-1 domains that compose a pseudo-dimeric fusion protein (Oh et al. 2004). Because such dual DJ-1 domain fusion proteins cannot dissociate into monomers, they provide strong phylogenetic evidence against the relevance of monomeric DJ-1 at least in plants. The creation and characterization of an artificial dual domain human DJ-1 fusion protein could open an informative avenue for further investigation.
The absence of multiple crosslinked species in cells suggests either that DJ-1 is less flexible in the cellular environment or that the other species are degraded in vivo. Our data do not distinguish between these possibilities, although we note that crosslinking is a non-equilibrium technique whose outcome depends on the kinetics of the crosslinking event and the timescale of the conformational sampling. Kinetic effects may explain why dimeric species are observed only for lower ratios of BMOE to DJ-1 in vitro, and the longer timescale of crosslinking in cells may explain why a predominant dimer is seen in cells.
Although disulfide-based methods have been used to probe the conformations of some proteins, the utility of BMOE or other thiol crosslinkers as probes of conformational flexibility has not been previously reported to the best of our knowledge. Specialized crosslinkers have been used to probe protein conformational heterogeneity in some systems (Li et al. 2012), although thiol crosslinkers such as BMOE offer the possibility to apply this method more broadly to the large class of proteins containing free (rather than disulfide-linked) cysteines. Cysteine residues constitute only ~2.3% of amino acids and thus generate few distinct crosslinked species, which can then be resolved in SDS-PAGE gels. Because the crosslinker must have access to a pair of free cysteine residues, the number and identity of these crosslinks are sensitive to changes in cysteine solvent accessibility that occur during protein motion. The power of crosslinking to probe protein flexibility is greatly enhanced by the ability to specifically identify which residues are crosslinked using mass spectrometry and could be applied to DJ-1 (or other systems) in the future.
Supplementary Material
Table 2.
Crystallographic data and model statistics
|
Data collection
| |
| X-ray source | Rigaku MicroMax 007 |
| X-ray wavelength (Å) | 1.54 |
| Space group | P3121 |
| Cell dimensions | |
| a=b, c (Å) | 74.68, 74.85 |
| α=β, γ | 90, 120 |
| Molecules in ASU | 1 |
| Wilson B factor (Å2) | 17.3 |
| Resolution (Å) a | 32–1.49 |
| No. reflections | 39842 (3916) |
| Completeness (%) | 99.8 (100) |
| Redundancy | 6.7 (6.2) |
| Rmerge b | 0.085 (0.634) |
| 〈 I 〉/〈 σ(I) 〉 | 21.5 (2.7) |
| CC1/2 | (0.795) |
|
| |
| Refinement | |
|
| |
| Rworkc; Rfreed; Ralle (%) | 11.8, 13.9, 11.9 |
| No. of protein atoms | 1430 |
| No. of solvent atoms | 231 |
| No. of heteratoms | 1 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (deg.) | 1.316 |
| Ramachandran plot favored; allowed; forbidden (%) | 99. 0.5, 0.5 |
Values in parentheses are for highest-resolution shell (1.54–1.49 Å).
, where i is the ith observation of a reflection with indices h,k,l and angle brackets indicate the average over all i observations.
, where Fchkl is the calculated structure factor amplitude with index h,k,l and Fohkl is the observed structure factor amplitude with index h,k,l.
Rfree is calculated as Rwork, where the Fohkl are taken from a test set comprising 5% of the data (1894 reflections) that were excluded from the refinement.
Rall is calculated as Rwork, where the Fohkl include all measured data (including the Rfree test set).
Acknowledgments
We thank Dr. Jiri Adamec (University of Nebraska-Lincoln) of the Redox Biology Center Mass Spectrometry Core Facility, Dr. Robert Powers (University of Nebraska-Lincoln) and Sara Basiaga with assistance in collection of the NMR data, and Dr. Jiusheng Lin (University of Nebraska-Lincoln) for helpful discussions. This work was supported, in part, by National Institutes of Health Grant R01 GM092999 (to M. A. W.). This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (D.N.H. and M.R.C.).
Abbreviations
- BMOE
bis-malaeimidoethane
- DSS
Disuccinimidyl suberate
- DTT
dithiothreitol, HSQC, heteronuclear single quantum coherence
- IPTG
isopropyl β-D-thiogalactopyranoside
- MES
2-(N-morpholino)ethanesulfonic acid
- TLS
translation-libration-screw
Footnotes
The authors declare no conflicts of interest.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr D, Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleyasin H, Rousseaux MWC, Phillips M, et al. The Parkinson’s disease gene DJ-1 is also a key regulator of stroke-induced damage. Proc Natl Acad Sci USA. 2007;104:18748–18753. doi: 10.1073/pnas.0709379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billia F, Hauck L, Grothe D, Konecny F, Rao V, Kim RH, Mak TW. Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc Natl Acad Sci USA. 2013;110:6085–6090. doi: 10.1073/pnas.1303444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, Wilson MA. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J Biol Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Canet-Aviles RM, Wilson MA, Miller DW, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Ying M, Xie N, et al. The oxidation states of DJ-1 dictate the cell fate in response to oxidative stress triggered by 4-HPR: autophagy or apoptosis. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR, Bandmann O. Parkinson’s disease: insights from pathways. Hum Mol Genet. 2010;19 (R1):R21–27. doi: 10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RL, Przedborski S. Mitophagy and Parkinson’s disease: be eaten to stay healthy. Mol Cell Neurosci. 2013;55:37–43. doi: 10.1016/j.mcn.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D, Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
- Giaime E, Yamaguchi H, Gautier CA, Kitada T, Shen J. Loss of DJ-1 does not affect mitochondrial respiration but increases ROS production and mitochondrial permeability transition pore opening. PloS One. 2012;7:e40501. doi: 10.1371/journal.pone.0040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci USA. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser DN, Hastings TG. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease and monogenic parkinsonism. Neurobiol Dis. 2013;51:35–42. doi: 10.1016/j.nbd.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hod Y, Pentyala SN, Whyard TC, El-Maghrabi MR. Identification and characterization of a novel protein that regulates RNA-protein interaction. J Cell Biochem. 1999;72:435–444. [PubMed] [Google Scholar]
- Holyoak T, Fenn TD, Wilson MA, Moulin AG, Ringe D, Petsko GA. Malonate: a versatile cryoprotectant and stabilizing solution for salt-grown macromolecular crystals. Acta Crystallogr D, Biol Crystallogr. 2003;59:2356–2358. doi: 10.1107/s0907444903021784. [DOI] [PubMed] [Google Scholar]
- Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, Ariga H, Inagaki F. The crystal structure of DJ-1, a protein related to male fertility and Parkinson’s disease. J Biol Chem. 2003;278:31380–31384. doi: 10.1074/jbc.M305878200. [DOI] [PubMed] [Google Scholar]
- Huai Q, Sun Y, Wang H, Chin LS, Li L, Robinson H, Ke H. Crystal structure of DJ-1/RS and implication on familial Parkinson’s disease. FEBS Lett. 2003;549:171–175. doi: 10.1016/s0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- Hulleman JD, Mirzaei H, Guigard E, Taylor KL, Ray SS, Kay CM, Regnier FE, Rochet JC. Destabilization of DJ-1 by familial substitution and oxidative modifications: implications for Parkinson’s disease. Biochemistry. 2007;46:5776–5789. doi: 10.1021/bi7001778. [DOI] [PubMed] [Google Scholar]
- Im JY, Lee KW, Junn E, Mouradian MM. DJ-1 protects against oxidative damage by regulating the thioredoxin/ASK1 complex. Neurosci Res. 2010;67:203–208. doi: 10.1016/j.neures.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Lu B. Mitochondrial dynamics and mitophagy in Parkinson’s disease: disordered cellular power plant becomes a big deal in a major movement disorder. Curr Opin Neurobiol. 2011;21:935–941. doi: 10.1016/j.conb.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I, Aleyasin H, Seifert EL, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Joselin AP, Hewitt SJ, Callaghan SM, Kim RH, Chung YH, Mak TW, Shen J, Slack RS, Park DS. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- Kahle PJ, Waak J, Gasser T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic Biol Med. 2009;47:1354–1361. doi: 10.1016/j.freeradbiomed.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Kato I, Maita H, Takahashi-Niki K, Saito Y, Noguchi N, Iguchi-Ariga SM, Ariga H. Oxidized DJ-1 inhibits p53 by sequestering p53 from promoters in a DNA-binding affinity-dependent manner. Mol Cell Biol. 2013;33:340–359. doi: 10.1128/MCB.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinumi T, Kimata J, Taira T, Ariga H, Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Madzelan P, Nan R, Milkovic NM, Wilson MA. Evolution of new enzymatic function by structural modulation of cysteine reactivity in Pseudomonas fluorescens isocyanide hydratase. J Biol Chem. 2010;285:29651–29661. doi: 10.1074/jbc.M110.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Maldonado MT, Zhou W, Fink AL, Wilson MA. Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry. 2008;47:1381–1392. doi: 10.1021/bi701189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NJ, Ambrosi G, Mullett SJ, Berman SB, Hinkle DA. DJ-1 knock-down impairs astrocyte mitochondrial function. Neuroscience. 2011;196:251–264. doi: 10.1016/j.neuroscience.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe AJ, Horton JC, editors. The Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- Lee SJ, Kim SJ, Kim IK, et al. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J Biol Chem. 2003;278:44552–44559. doi: 10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- Li H, Wells SA, Jimenez-Roldan JE, Romer RA, Zhao Y, Sadler PJ, O’Connor PB. Protein flexibility is key to cisplatin crosslinking in calmodulin. Protein Sci. 2012;21:1269–1279. doi: 10.1002/pro.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Prahlad J, Wilson MA. Conservation of oxidative protein stabilization in an insect homologue of parkinsonism-associated protein DJ-1. Biochemistry. 2012;51:3799–3807. doi: 10.1021/bi3003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo MG, Anar B, Bronner IF, Cannella M, Squitieri F, Bonifati V, Hoogeveen A, Heutink P, Rizzu P. The DJ-1(L166P) mutant protein associated with early onset Parkinson’s disease is unstable and forms higher-order protein complexes. Hum Mol Genet. 2003;12:2807–2816. doi: 10.1093/hmg/ddg304. [DOI] [PubMed] [Google Scholar]
- Maita C, Maita H, Iguchi-Ariga SM, Ariga H. Monomer DJ-1 and its N-terminal sequence are necessary for mitochondrial localization of DJ-1 mutants. PloS One. 2013;8:e54087. doi: 10.1371/journal.pone.0054087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgieri G, Eliezer D. Structural effects of Parkinson’s disease linked DJ-1 mutations. Protein Sci. 2008;17:855–868. doi: 10.1110/ps.073411608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinat C, Shendelman S, Jonason A, Leete T, Beal MF, Yang L, Floss T, Abeliovich A. Sensitivity to oxidative stress in DJ-1-deficient dopamine neurons: an ES-derived cell model of primary Parkinsonism. PLoS Biol. 2004;2:e327. doi: 10.1371/journal.pbio.0020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol. 2005;15:1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Meulener M, Whitworth AJ, Armstrong-Gold CE, Rizzu P, Heutink P, Wes PD, Pallanck LJ, Bonini NM. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr Biol. 2005;15:1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Meulener MC, Xu K, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc Natl Acad Sci USA. 2006;103:12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DW, Ahmad R, Hague S, et al. L166P mutant DJ-1, causative for recessive Parkinson’s disease, is degraded through the ubiquitin-proteasome system. J Biol Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson’s disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, IguchiAriga SMM, Ariga H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- Oh KJ, Park YS, Lee KA, Chung YJ, Cho TJ. Molecular characterization of a thiJ-like gene in Chinese cabbage. J Biochem Mol Biol. 2004;37:343–350. doi: 10.5483/bmbrep.2004.37.3.343. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pantoliano MW, Petrella EC, Kwasnoski JD, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- Ramsey CP, Giasson BI. L10p and P158DEL DJ-1 mutations cause protein instability, aggregation, and dimerization impairments. J Neurosci Res. 2010;88:3111–3124. doi: 10.1002/jnr.22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repici M, Straatman KR, Balduccio N, Enguita FJ, Outeiro TF, Giorgini F. Parkinson’s disease-associated mutations in DJ-1 modulate its dimerization in living cells. J Mol Med (Berl) 2013;91:599–611. doi: 10.1007/s00109-012-0976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes RK. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974;59:277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Shadrach KG, Rayborn ME, Hollyfield JG, Bonilha VL. DJ-1-dependent regulation of oxidative stress in the retinal pigment epithelium (RPE) PloS One. 2013;8:e67983. doi: 10.1371/journal.pone.0067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanatos R, Sriram A, Kiviranta E, Mohan A, Ayala V, Jacobs HT, Pamplona R, Sanz A. dj-1beta regulates oxidative stress, insulin-like signaling and development in Drosophila melanogaster. Cell Cycle. 2012;11:3876–3886. doi: 10.4161/cc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Tong L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson’s disease. J Biol Chem. 2003;278:31372–31379. doi: 10.1074/jbc.M304221200. [DOI] [PubMed] [Google Scholar]
- Wagenfeld A, Gromoll J, Cooper TG. Molecular cloning and expression of rat contraception associated protein 1 (CAP1), a protein putatively involved in fertilization. Biochem Biophys Res Commun. 1998;251:545–549. doi: 10.1006/bbrc.1998.9512. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc Natl Acad Sci USA. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt AC, Lakshminarasimhan M, Remington BC, Hasim S, Pozharski E, Wilson MA. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47:7430–7440. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental structure factors and refined crystallographic model for E18A DJ-1 have been deposited with the Protein Data Bank (PDB) with accession code 4OQ4.