Abstract
Objective
To determine the extent of mitochondrial DNA (mtDNA) damage in systemic lupus erythematosus (SLE) patients compared to healthy subjects and to determine the factors associated with mtDNA damage among SLE patients.
Methods
A cross-sectional study was performed in 86 SLE patients (per American College of Rheumatology classification criteria) and 86 healthy individuals matched for age and gender. Peripheral blood mononuclear cells (PBMCs) were collected from subjects to assess the relative amounts of mtDNA damage. Quantitative polymerase chain reaction assay was used to measure the frequency of mtDNA lesions and mtDNA abundance. Socioeconomic-demographic features, clinical manifestations, pharmacologic treatment, disease activity, and damage accrual were determined. Statistical analyses were performed using t test, pairwise correlation, and Pearson’s chi-square test (or Fisher’s exact test) as appropriate.
Results
Among SLE patients, 93.0% were women. The mean (SD) age was 38.0 (10.4) years and the mean (SD) disease duration was 8.7 (7.5) years. SLE patients exhibited increased levels of mtDNA damage as shown by higher levels of mtDNA lesions and decreased mtDNA abundance as compared to healthy individuals. There was a negative correlation between disease damage and mtDNA abundance and a positive correlation between mtDNA lesions and disease duration. No association was found between disease activity and mtDNA damage.
Conclusion
PBMCs from SLE patients exhibited more mtDNA damage compared to healthy subjects. Higher levels of mtDNA damage were observed among SLE patients with major organ involvement and damage accrual. These results suggest that mtDNA damage have a potential role in the pathogenesis of SLE.
Keywords: Systemic lupus erythematosus, disease activity, disease damage, mitochondrial DNA, mitochondrial dysfunction, oxidative stress
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease of unknown etiology characterized by dysfunction of T and B cells, the formation of antinuclear antibodies and immune complexes, inflammation, and organ dysfunction1. Although both genetic and environmental factors are involved in the pathogenesis of SLE, the molecular mechanisms contributing to disease pathology and progression remain unclear. A leading hypothesis is that oxidative stress and mitochondrial dysfunction play a significant role in SLE pathogenesis.
Mitochondrial dysfunction has been associated with aging and diverse human diseases such as diabetes and neurodegenerative disorders2–4, and recently to autoimmunity5. The link of mitochondria with autoimmune diseases may be related to their role in triggering apoptosis and the activation and proliferation of peripheral lymphocytes6, processes that require the generation of ATP by functional mitochondria. Moreover, mitochondria are the principal sources of endogenous radical oxygen species (ROS) generated as by-products of oxidative phosphorylation7, which in turn modulate T cell activation, cytokine production and cell survival1,16,8,9. However, ROS can inflict damage to DNA, lipids and proteins10 and lead to decreased mitochondrial function and tissue dysfunction in SLE11. Mitochondrial dysfunction in SLE is associated with increased ROS generation, increased mitochondrial transmembrane potential, reduced generation of ATP and increased mitochondrial mass6,8,12.
The mammalian mitochondrial DNA (mtDNA) is localized in the mitochondrial matrix. MtDNA is a 16.5 kb double-stranded, closed-circular molecule that encodes 13 proteins of the electron transport chain (ETC) that are essential to the process oxidative phosphorylation and thus, for ATP production13,14. MtDNA is highly vulnerable to ROS-induced oxidative damage15, probably owing to the limited actions of mtDNA repair mechanisms and its close exposure to a major intracellular ROS generation site. As a result, mtDNA is at risk for oxidative damage and consequently it may contribute to altered energy metabolism and tissue dysfunction. Furthermore, mtDNA variants in SLE have been associated with mitochondrial dysfunction, increased levels of oxidative stress and disease susceptibility16,17.
One mechanism that may be contributing to mitochondrial dysfunction and thus, to the pathogenesis of SLE, is oxidative stress. Levels of lipid peroxidation and protein oxidation products are significantly increased in serum from SLE patients as compared to healthy subjects, and correlate with disease activity18–22. In addition, urine levels of lipid peroxidation products are elevated in SLE patients23 and are associated with higher disease activity and fatigue24. Levels of protein carbonyls are higher in lupus patients and correlate with the severity of the disease25,26. Furthermore, ROS and lipid peroxidation products are significantly increased in SLE lymphocytes and levels of glutathione are reduced in SLE patients versus healthy controls27. Consistent with increased oxidative stress levels in SLE, the activity of antioxidant enzymes is significantly reduced in lupus patients compared with healthy controls18,20,22. However, the role of oxidative damage to the mtDNA in the pathogenesis and progression of SLE remains unexplored. We hypothesized that mtDNA damage plays a critical role in the pathogenesis of SLE. To address this hypothesis, first, we compared the extent of mtDNA damage in peripheral blood mononuclear cells (PBMCs) between SLE patients and healthy individuals, and second, we determined the association of mtDNA damage with major organ involvement, disease activity, and damage accrual among SLE patients.
Methods
Patient Population
A cross-sectional study was performed in 86 adult Puerto Rican patients with SLE and 86 healthy individuals matched for age and sex. Patients were enrolled from November 2009 to May 2010. SLE patients were recruited at the lupus clinics of the University of Puerto Rico Medical Sciences Campus in San Juan, Puerto Rico. All SLE patients were ≥21 years of age and fulfilled at least four of the eleven American College of Rheumatology (ACR) classification criteria for SLE28,29. The study was approved by the Institutional Review Board of the University of Puerto Rico Medical Sciences Campus Human Research Protection Office and all subjects gave written informed consent.
Prior to study visit, patients had routine visits at 3-month intervals. Additional visits were scheduled as needed per disease activity or complications. At each routine visit, a structured questionnaire was completed for each patient to gather information about demographic parameters, health-related behaviors, clinical manifestations, laboratory tests (complete blood cell count, comprehensive metabolic panel, urine analysis, erythrocyte sedimentation rate (ESR), lipid panel, anti-dsDNA antibodies, and C3 and C4 complement levels) pharmacologic treatment, disease activity and disease damage. For all patients, a lupus autoantibody panel was performed at the time of SLE diagnosis.
Variables
Variables from sociodemographic, health-related behaviors, clinical, immunologic and pharmacologic domains were studied in SLE patients. Sociodemographic factors health-related behaviors and body mass index (BMI) were determined in healthy subjects. Socioeconomic-demographic parameters included age, gender, years of education, family income, and type of health insurance (private vs. government). Health-related behavior features included cigarette smoking. For lupus patients age at diagnosis was defined as the time at which a patient met ACR criteria for SLE and disease duration was defined as time interval between SLE diagnosis and study visit.
The clinical domain included the assessment of SLE manifestations, immunologic abnormalities, comorbidities, disease activity, disease damage, and pharmacologic therapy. Cumulative SLE clinical manifestations were determined as defined in the ACR classification criteria for SLE28,29. The following serologic tests were determined: anti-nuclear (ANA), anti-double stranded DNA (dsDNA), anti-Smith (Sm), anti- small nuclear ribonucleoparticle (snRNP), anti-Ro (SSA), anti- La (SSB), and antiphospholipid antibodies, and serum complements (C3 and C4). Selected comorbidities were recorded including diabetes mellitus, hypertension, coronary artery disease, cerebrovascular events, peripheral artery disease, venous thromboembolism, and hypothyroidism. Disease activity was determined with the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) (30) and the Systemic Lupus Disease Activity Measure (SLAM)31,32. Disease damage was assessed with the Systemic Lupus International Collaborating Clinics/American College of Rheumatology (SLICC) Damage Index (SDI)33. Cumulative and current exposure to glucocorticoids, hydroxychloroquine, cyclophosphamide, methotrexate, mycophenolate mofetil, azathioprine, angiotensin converting enzyme (ACE) inhibitors/angiotensin receptor blockers, statins, and aspirin was also examined.
Major organ involvement attributed to SLE was defined as the presence of renal disease (proteinuria > 0.5gm/24 hours, cellular casts, lupus nephritis by biopsy, glomerular filtration rate <50%, or end-stage renal disease), central nervous system involvement (seizures, psychosis, cognitive impairment/organic brain syndrome or cerebrovascular accidents) cardiac involvement (lupus-associated cardiomyopathy or valvular disease, cardiac tamponade, or large pericardial effusions) or pulmonary disease (pulmonary fibrosis or pulmonary hypertension).
Assessment of mitochondrial DNA damage and mtDNA relative abundance/copy number by Quantitative PCR
DNA isolation, quantitation, mtDNA damage and abundance analyses were performed as previously described by our laboratory34.35. For assessing DNA damage we used samples showing high molecular weight genomic DNA with no evidence of degradation products. To measure mtDNA damage we employed a quantitative PCR assay that relies on the principle that oxidative lesions present in the DNA will interrupt the movement of the thermostable polymerase resulting in decreased replication of the DNA amplicon. Consequently, the presence of DNA lesions results in a decrease in the amplification of the fragment of interest. An 8.9 kb human mitochondrial fragment was amplified using the MasterAmp™ Extra-long PCR reagents (Epicentre) with an initial denaturation for 45 seconds at 94 °C followed by 23 cycles of 94 °C and 15 seconds denaturation and anneling/extension steps at 68 °C for 12 minutes. A final extension reaction was performed at 72 °C for 10 minutes. The following primer nucleotide sequences were used for the amplification of the 8.9 kb human mitochondrial fragment: 5’-CT AAG CCT CCT TAT TCG AGC CGA-3’ (5999-sense) and 5’-TTT CAT CAT GCG GAG ATG TTG GAT GG-3’ (14841-antisense). The amplification of a 117 bp mtDNA fragment was performed to measure the levels of mtDNA molecules/abundance. The likelihood of an oxidative lesion being introduced into such a small fragment is low and therefore, the amplification is independent of the presence of lesions, thus providing a measure of mtDNA abundance. The 117 bp mtDNA fragment was amplified using an initial denaturation at 94 °C for 1 minute followed by 18 cycles of denaturation for 1 minute at 94 °C, annealing/extension at 66 °C for 45 seconds and extension at 72 °C for 45 seconds. The nucleotide sequences used for the amplification of the 117 bp mitochondrial fragment were the following: 5’-CAT GCA AGC ATC CCC GTT CC-3’ (694-sense) and 5’-CTG TTT CCC GTG GGG GT GTG -3’ (817-antisense). Relative copy numbers were calculated comparing PBMCs from SLE patients to those of the healthy individuals.
Calculations of mtDNA lesion frequency
The DNA lesion frequency per strand was calculated using the Poisson equation as we previously described36. Because the amplification of the DNA fragment is directly proportional to the fraction of undamaged DNA templates, the average lesion frequency per DNA per strand can be calculated as λ = −ln AL/AH, where AL and AH represent the amount of amplification product of the DNA from the lupus patients and the age-matched healthy subjects, respectively. We corrected for possible differences in the amounts of mtDNA molecules by normalizing the amplification of the 8.9 kb mtDNA fragment compared with those of the 117 bp mtDNA fragment. Levels of mtDNA lesions were calculated by comparing PBMCs from SLE patients to the age-matched healthy controls.
Statistical analysis
Descriptive statistics were used to portray the study using the mean (standard deviation, SD) or median (25th– and 75th percentiles) for continuous data; frequencies and proportions were used for categorical data. Comparisons between cases and controls were done using the paired t-test (or Wilcoxon signed-rank test as appropriate) and by McNemar’s X2 test result for matched data. Association between mtDNA damage and SLE activity and damage scores were evaluated as continuous variables using pairwise correlation. Association between mtDNA damage and other parameters such as clinical manifestations, comorbid conditions and treatment modalities were done using unpaired t test statistics (or Mann-Whitney U test in the case of non-Gaussian distribution) and Pearson’s chi-square test (or Fisher’s exact test) as appropriate. Statistical significance was set at p<0.05. The statistical analysis was performed using the statistical software STATA version 12 (STATA Corp, College Station, TX).
Results
The sociodemographic features, health-related behaviors and BMI of SLE patients and healthy individuals are shown in Table 1. SLE patients had a lower level of education (13.7 versus 16.4 years of education, p<0.001) and were less likely to have a private health insurance (20.9% versus 81.6%, p<0.001) when compared to healthy individuals. BMI was higher among SLE patients than controls (28.3 vs. 25.9, p=0.019). No significant differences were found for age, gender, and cigarette smoking.
Table 1.
Socio-demographic features in SLE patients and healthy subjects
| Variable | Healthy subjects n=86 |
SLE patients n=86 |
P |
|---|---|---|---|
| Gender, % female | 80 (93.0) | 80 (93.0) | >0.999 |
| Age, mean (SD) years | 38.0 (10.4) | 37.9 (11.4) | 0.954 |
| Education, mean (SD) years | 16.4 (2.8) | 13.7 (2.8) | <0.001 |
| Insurance, n (%) | |||
| Private | 58 (81.6) | 18 (20.9) | <0.001 |
| Government | 13 (18.3) | 67 (78.8) | |
| Smoking (ever), n (%) | 8 (9.3) | 14 (16.3) | 0.228 |
| Body mass index, mean (SD) kg/m2 | 25.9 (6.1) | 28.3 (6.9) | 0.019 |
SD: Standard Deviation
Of 86 patients with SLE, 80 were females (93%). The mean (SD) age was 37.9 (11.4) years (Table 1). The disease duration, cumulative clinical manifestations, serological features, pharmacologic treatment, disease activivity and damage accrual of SLE patients are depicted in Table 2. The mean (SD) disease duration was 8.7 (7.5) years. Cutaneous manifestations, arthritis and lymphopenia were the most common clinical manifestations. Thirty-eight (41.2%) patients had major organ involvement. Hypertension was the most frequent comorbidity. Corticosteroid and hydroxychloroquine were the most common medications (current and cumulative) used by these patients. The mean (SD) SLEDAI score was 2.3 (3.6) (range 0–18). Forty-nine (56.9%) patients had SLEDAI score of 0; 24 (27.9%) patients exhibited a SLEDAI score of 1–4; 34 (39.5 %) patients had a score of 5–11 and only 3 (3.5%) patients had a SLEDAI ≥12. The mean (SD) SLAM score was 4.9 (3.4) (range of 0–15). Eight (9.3%) patients had SLAM score of 0; 43 (50%) patients had a SLAM score of 1–5 and 35 (40.7 %) patients showed a score of greater than 6. The mean (SD) SDI score was 1.2 (1.5) (range 0–7). Forty-two (48.8 %) patients had a SDI score of 0; 35 (40.7%) patients had an SDI score of 1–3, and nine (10.5%) patients had a SDI score greater than 3.
Table 2.
Clinical manifestations, serologic features, pharmacologic treatment, disease activity and damage accrual of SLE patients (n=86).
| Features | |
|---|---|
| Disease duration, mean (SD) years | 8.7(7.5) |
| Clinical manifestations*, n (%) | |
| Malar rash | 57 (67.1) |
| Photosensitivity | 67 (78.8) |
| Oral ulcers | 30 (35.3) |
| Arthritis | 66 (78.6) |
| Pericarditis | 9 (10.6) |
| Pleuritis | 12 (14.1) |
| Proteinuria (≥0.5g/d) | 40 (46.5) |
| Cellular casts | 9 (10.6) |
| Psychosis | 6 (7.0) |
| Seizures | 11 (12.8) |
| Hemolytic anemia | 7 (8.1) |
| Leukopenia (<4k) | 39 (45.3) |
| Lymphopenia (<1.5) | 55 (64.0) |
| Thrombocytopenia | 17 (19.8) |
| Major organ involvement, n (%) | 38 (41.2) |
| Serologic features | |
| ANA | 79 (97.5) |
| Anti-dsDNA antibodies | 68 (81.0) |
| Anti-Smith antibodies | 17(26.6) |
| Lupus anticoagulant | 8 (15.4) |
| Anticardiolipin antibodies (IgG) | 19 (30.2) |
| Anticardiolipins antibodies (IgM) | 10 (16.4) |
| Comorbidities | |
| Diabetes mellitus | 8 (9.3) |
| Hypertension | 44 (51 .2) |
| Stroke | 6 (7.0) |
| Angina | 2 (2.3) |
| Myocardial infarction | 1 (1.2) |
| Venous thrombosis (peripheral) | 6 (7.0) |
| Venous thrombosis (visceral) | 3 (3.5) |
| Arterial/Vascular (peripheral) | 1 (1.2) |
| Hypothyroidism | 12 (14.0) |
| Pharmacologic treatment | |
| Hydroxychloroquine | 62 (72) |
| Glucocorticoids oral | 67 (78) |
| Azathioprine | 26 (30) |
| Cyclophosphamide | 7 (8) |
| Mycophenolate mofetil | 8 (9) |
| Methotrexate | 2 (2) |
| ACE inhibitors | 40 (46) |
| Statins | 15 (17) |
| Aspirin | 17 (19) |
| SLEDAI at study visit, mean score (SD) | 2.3 (3.6) |
| SLAM, at study visit, mean score (SD) | 4.9 (3.4) |
| SDI, mean score (SD) | 1.2 (1.5) |
As defined by the American College of Rheumatology classification criteria for systemic lupus erythematosus.
SD: Standard deviation; ANA: Antinuclear antibodies; ACE: Angiotensing converting enzyme; SLEDAI: Systemic Lupus Erythematosus Disease Activity; SLAM: Systemic Lupus Erythematosus Measure; SDI: Systemic Lupus International Collaborating Clinics/American College of Rheumatology (SLICC) Damage Index
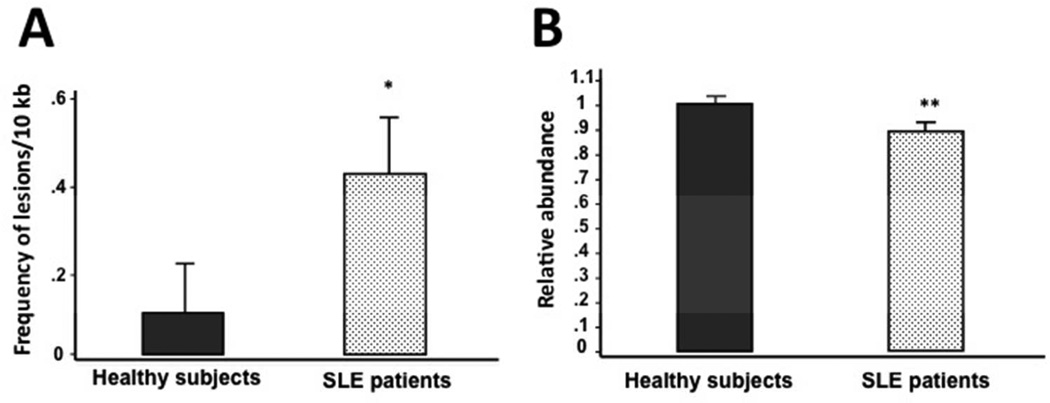
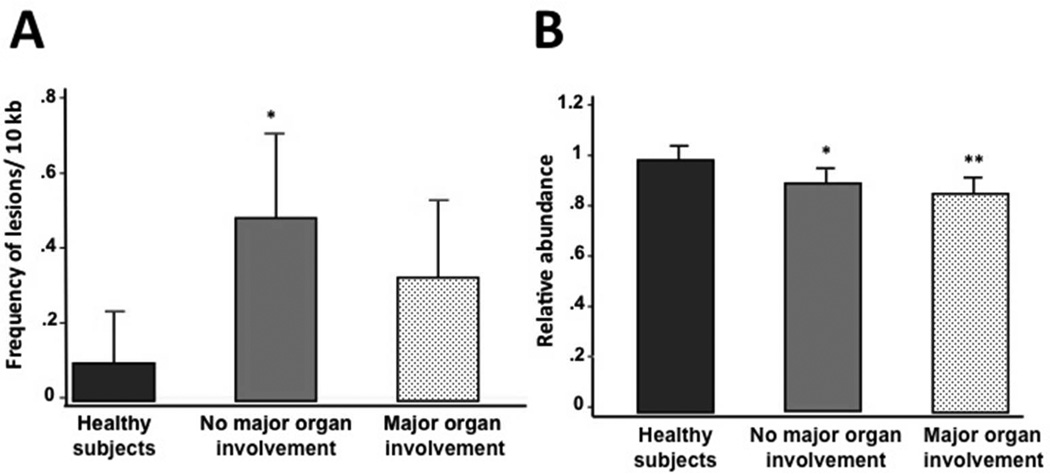
To determine if mtDNA damage is associated with SLE, we utilized QPCR to measure the extent of mtDNA oxidative damage and mtDNA abundance in PBMCs from SLE patients and healthy individuals. The QPCR assay measures oxidative damage as it detects most of the oxidative lesions produced by ROS (such as abasic sites and single-strand breaks), that represent blocks to the thermostable polymerase. SLE patients exhibited increased levels of mtDNA damage as shown by significantly higher levels (p=0.002) of mtDNA lesions compared to healthy subjects (0.41 lesions/10 kb/strand versus 0.10 lesions/10 kb/strand, respectively) (Figure 1A). Next we measured the abundance of mtDNA molecules and found a significant 11% decrease in mtDNA abundance as compared to healthy individuals (0.89 ± 0.14 versus 1.00 ± 0.17, (p<0.001) (Figure 1B). In addition, patients with no major organ involvement showed a significant (p=0.003) 20 % increase in the levels of mtDNA lesions compared to healthy individuals (0.489 lesions versus 0.101, respectively) (Figure 2A). Interestingly, patients with no major and major organ involvement showed a significant 10 % (0.906 ± 0.151) and 13 % reduction (0.87 ± 0.131), respectively, in the mtDNA abundance relative to healthy controls (Figure 2B). Among SLE patients, levels of mtDNA lesions as well as the mtDNA abundance were not significantly different in those with major organ involvement compared to patients with no major organ involvement (Figure 2A and B). However, lupus patients with major organ involvement showed a tendency to have further reductions of mtDNA abundance compared to patients with no major organ involvement (Figure 2B).
Figure 1. Mitochondrial DNA damage increases in peripheral blood mononuclear cells of SLE patients.
(A) Frequency of mtDNA lesions per 10 kb per strand. (B) Relative abundance of mtDNA molecules. Statistical differences were observed for mtDNA frequency of lesions (*p=0.002) and mtDNA abundance (**p<0.001) between healthy individuals (n=86) and SLE patients (n=86).
Figure 2. Mitochondrial DNA damage in SLE patients with (n=38) and without (n=48) major organ involvement compared with healthy subjects (n=86).
(A) Frequency of mtDNA lesions per 10 kb per strand is higher in patients with no major organ involvement. Statistical differences were observed in the number of mtDNA lesions between healthy subjects and SLE patients without major organ involvement (pairwise comparison test: *p=0.003), whereas no differences were found between healthy subjects and SLE patients with major organ involvement (pairwise comparison test: p=0.205). (B) Relative abundance of mtDNA molecules is lower in SLE patients with major organ involvement. Levels of mtDNA abundance were also significantly different between the groups (healthy subjects vs. SLE patients without major organ involvement (pairwise comparison test: *p=0.002) and healthy subjects vs. SLE patients with major organ involvement (pairwise comparison test: **p<0.001).
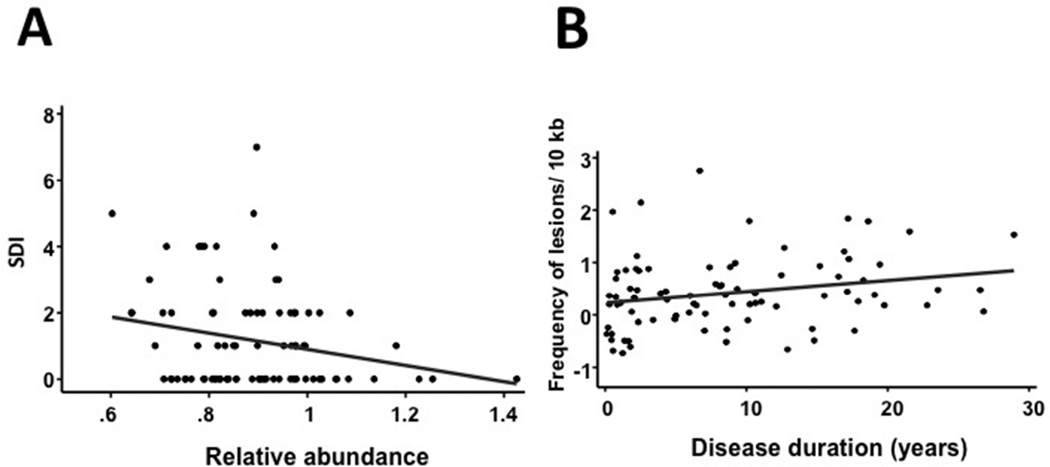
There was a significant negative correlation between disease damage, as determined by SDI, and mtDNA abundance (Figure 3A; r= −0.229, p=0.034), but no association was found between SDI and the levels of mtDNA lesions (data not shown). In addition, there was a positive correlation between disease duration and the number of mtDNA lesions (Figure 3B; r=0.231; p=0.034), but no association was found between disease duration and mtDNA abundance (data not shown). The number of mtDNA lesions and abundance were not associated with sociodemographic features, clinical manifestations, serologic markers, comorbidities, pharmacologic agents, and disease activity (by SLEDAI and SLAM) (data not shown).
Figure 3. Relationship between mtDNA abundance and Lupus Severity Index (SDI) and between the frequency of mtDNA lesions and disease duration.
(A) We found a negative correlation between the relative abundance of mtDNA molecules and SDI. This difference was statistically significant (Kruskal-Wallis test: r=−0.229; p=0.034). (B) We found a positive correlation between the number of mtDNA lesions and disease duration. The correlation observed was statistically significant (pairwise comparison test: r=0.2306; p=0.0338).
Discussion
Several discoveries in recent years have established that the pathogenesis of SLE comprises multiple mechanisms leading to immunological abnormalities. Although mitochondrial dysfunction has emerged as an important event associated with SLE and other chronic degenerative conditions6,8, the causative mechanisms leading to mitochondrial dysfunction in autoimmune diseases have not been completely elucidated. In this study, we show that SLE patients exhibit significant levels of mitochondrial damage in the form of increased levels of mtDNA lesions and mtDNA depletion compared to healthy individuals. Among lupus patients, those with damage accrual and major organ involvement were more likely to have lower abundance of mtDNA molecules. In addition, we found a correlation between levels of mtDNA lesions and disease duration.
Previous reports show increased levels of oxidative stress or ROS-mediated damage to lipids and proteins in SLE18–22, 25–27, 37, however, these studies did not examine the effects of ROS on the mitochondrial genome. QPCR analysis showed a substantial increase in mtDNA damage in PBMCs from SLE patients, shown as a significant increase in the frequency of mtDNA lesions. Our results are in accordance with previous observations of increased levels of the oxidative lesion 8-oxo-deoxyguanosine (8-oxodG) in nuclear DNA from lymphocytes and polymorphonuclear leukocytes38 and in urine39 in SLE patients compared to healthy subjects. However, in these studies the authors did not determined levels of mtDNA damage. The increased levels of mtDNA lesions we observed are consistent with lupus patients exhibiting increased levels of oxidative stress and with mitochondria as important targets of ROS damage in SLE, as levels of glutathione and the activity of antioxidant enzymes are significantly reduced in SLE patients versus healthy controls18,20,22,27 and ROS levels are increased21,27.
The observation that levels of mtDNA lesions were significantly higher in lupus patients without major organ involvement relative to healthy controls supports the idea that lupus patients are undergoing a chronic state of oxidative stress and inflammatory processes leading to increased ROS generation. However, patients with major organ involvement showed decreased levels of lesions in the mtDNA compared to those with no major organ distress. It is possible that the significant increase in mtDNA damage we observe may trigger apoptosis of lupus PBMCs in patients with major organ involvement, thus resulting in decreased detection of mtDNA lesions. This idea is in accordance with the observation that SLE PBMCs exhibit increased apoptosis that correlate with disease activity40. Moreover, single-strand breaks in the mtDNA, a common oxidative lesion41,42 may induce apoptosis of mammalian cells. Tann and collaborators showed that the accumulation and persistence of single-strand breaks in the mtDNA induces mitochondrial dysfunction and apoptosis in human cell lines43.
Interestingly, all lupus patients (independently of major organ involvement) showed a sustained loss of mtDNA molecules compared to healthy controls. The significant reduction in mtDNA abundance we observed in lupus patients compared to healthy controls further supports our hypothesis that oxidative damage to the mtDNA may be contributing to the progression of the disease, as loss of mtDNA molecules may result from oxidative damage. Indeed, treatment with hydrogen peroxide results in a significant loss of mtDNA molecules and reduced mitochondrial function in an embryonic cell line44. In another study, although no significant differences were found in the numbers of mtDNA copies in lupus patients compared to healthy individuals, mtDNA copy numbers decreased among lupus patients with low SLEDAI scores relative to patients exhibiting medium and high scores45. Thus, our findings suggest that mtDNA damage in the form of oxidative lesions or mtDNA depletion may be contributing to SLE pathogenesis.
Multiple studies have shown that mitochondrial hyperpolarization and ATP depletion occur in SLE6,8. The increased damage to the mtDNA and the loss of mtDNA molecules we observe in our lupus cohort may contribute to lupus-associated mitochondrial dysfunction. The increased levels of mtDNA lesions and mtDNA depletion may contribute to impaired mtDNA replication causing defective synthesis of the mitochondrial ETC protein subunits, both resulting in ATP depletion/mitochondrial dysfunction. A recent study found that the expression of mtDNA-encoded genes (involved in ATP synthesis) are downregulated in SLE patients46 suggesting that reduced expression of mtDNA-encoded genes and ATP depletion may contribute to the pathology of SLE. Along this line is the observation that fibromyalgia patients exhibit a close association between mitochondrial dysfunction and reduced levels of coenzyme Q10, a mitochondrial electron carrier and antioxidant47,48.
Our results showing a negative association between damage accrual (SDI) and mtDNA depletion suggest that the loss of mtDNA molecules may be contributing to mitochondrial dysfunction and tissue damage in SLE. In addition, the frequency of mtDNA lesions is increased with longer disease duration suggesting that lupus patients are chronically exposed to oxidative processes that in turn lead to persistent mtDNA lesions. Alternatively, oxidative lesions to the mitochondrial genome may not be appropriately repaired in lupus patients. It has been shown that PBMCs from SLE patients exhibit an impaired ability to repair hydrogen peroxide-induced oxidative damage to the nuclear DNA39,49. Defining whether mtDNA repair contributes to the increase in mtDNA lesion frequency we observe in SLE patients is an important future direction.
Our data did not reveal any association between disease activity (determined by the SLEDAI and SLAM) and mtDNA abundance or mtDNA lesion number. It is possible that our findings were influenced by the fact that around 50% of the SLE patients enrolled in this study had a SLEDAI of 0 and up to an 85% SLE patients had a SLEDAI <5, indicative of mild disease activity. Similar results were found for disease activity measured by the SLAM. Thus, the majority of our patients was in clinical remission or had mild disease activity at enrollment. These findings are in agreement with studies that have measured oxidative stress in SLE patients, which did not find an increase in oxidative markers in patients with stable or mild disease (24). Conversely, other studies found a correlation between numbers of mtDNA copies (46) and oxidative stress markers only in patients with higher levels of disease activity18,19,22,25,37,50. In addition, increased ROS generation51 and glutathione depletion27 were positively associated with disease activity.
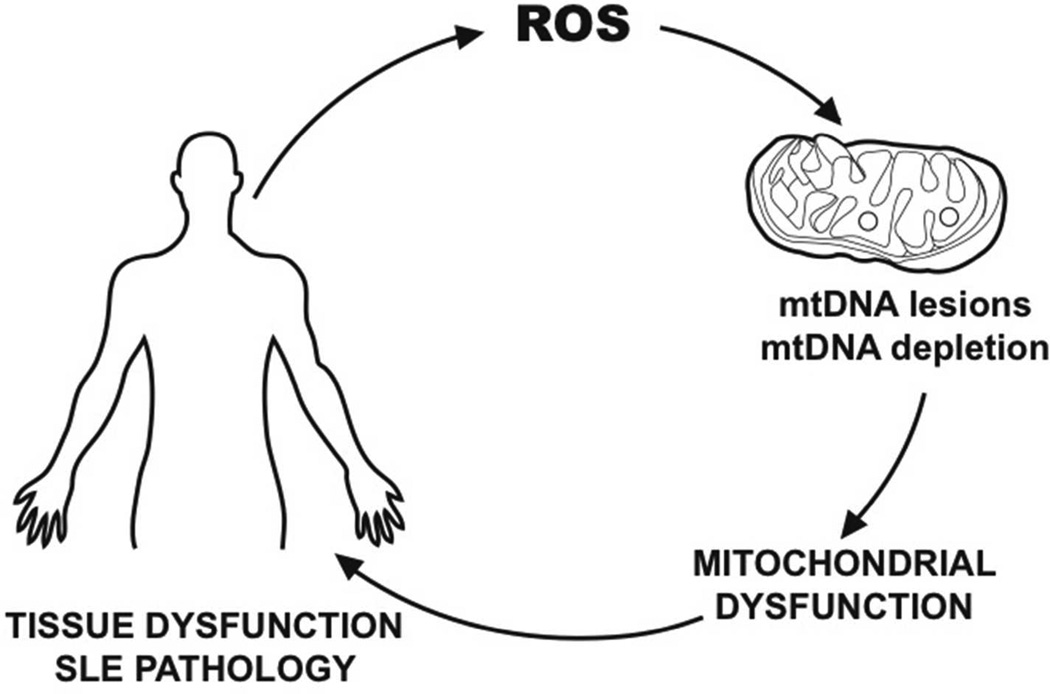
Taken together our results suggest that damage to the mitochondrial genome in the form of mtDNA lesions and/or mtDNA depletion are potential biomarkers to evaluate severity of disease/accrual damage. Moreover, our results further suggest that early interventions to prevent mtDNA depletion and mtDNA lesions may be relevant to treat SLE. We propose a model in which increased levels of mtDNA damage may play a role in SLE pathogenesis (Figure 4). Oxidative stress may lead to increased levels of lesions in the mtDNA and to reduced abundance of mtDNA molecules. Thus, mtDNA damage in the form of lesions and mtDNA depletion may lead to mitochondrial transmembrane hyperpolarization, ATP reduction, further ROS generation, that in turn lead to tissue dysfunction/damage in SLE, thus, contributing to the progression and severity of the disease.
Figure 4. Proposed model of mitochondrial DNA damage and SLE pathogenesis.
Our results are consistent with a model of mtDNA damage involving increased oxidative stress in SLE, increased frequency of mtDNA lesions, depletion of mtDNA molecules, altered mitochondrial bioenergetics, tissue dysfunction and disease progression.
Acknowledgements
The authors would like to acknowledge Amarylis Irizarry Hernández, Graphic Designer at the University of Puerto Rico Medical Sciences Campus School of Medicine, for the artwork. This work was supported by grants from the National Center for Research Resources (U54 RR026139-01A1, 2G12RR003051); the National Institute on Minority Health and Health Disparities (8U54 MD007587-03, 2G12MD007600); and the National Center for Advancing Translational Sciences (TL1TR000145) from the National Institutes of Health.
References
- 1.Perl A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity. 2010 Feb;43(1):1–6. doi: 10.3109/08916930903374741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma ZA, Zhao Z, Turk J. Mitochondrial Dysfunction and β-Cell Failure in Type 2 Diabetes Mellitus. Exp Diabetes Res. 2012 doi: 10.1155/2012/703538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowell BB, Shulman GI. Mitochondrial Dysfunction and Type 2 Diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 4.Shukla V, Mishra SK, Pant HC. Oxidative stress in neurodegeneration. Adv Pharmacol Sci. 2011 doi: 10.1155/2011/572634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat Rev Rheumatol. 2013 Oct 8; doi: 10.1038/nrrheum.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gergely P, Jr, Grossman C, Niland B, Puskas F, Neupane H, Allam F, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46(1):175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J. 1973;134(3):707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perl A, Gergely P, Jr, Nagy G, Koncz A, Banki K. Mitochondrial hyperpolarization: a checkpoint of T-cell life, death and autoimmunity. Trends Immunol. 2004;25(7):360–367. doi: 10.1016/j.it.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kesarwani P, Murali AK, Al-Khami AA, Mehrotra S. Redox regulation of T-cell function: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal. 2013;18(12):1497–1534. doi: 10.1089/ars.2011.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29(3–4):222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 11.Perl A, Gergely P, Jr, Banki K. Mitochondrial dysfunction in T cells of patients with systemic lupus erythematosus. Int Rev Immunol. 2004;23(3–4):293–313. doi: 10.1080/08830180490452576. [DOI] [PubMed] [Google Scholar]
- 12.Nagy G, Barcza M, Gonchoroff N, Phillips PE, Perl A. Nitric oxide-dependent mitochondrial biogenesis generates Ca2 signaling profile of lupus T cells. J Immunol. 2004;173(6):3676–3683. doi: 10.4049/jimmunol.173.6.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- 14.Attardi G. The elucidation of the human mitochondrial genome. Bioessays. 1986;5(1):34–39. doi: 10.1002/bies.950050111. [DOI] [PubMed] [Google Scholar]
- 15.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vyshkina T, Sylvester A, Sadiq S, Bonilla E, Canter JA, Perl A, et al. Association of common mitochondrial DNA variants with multiple sclerosis and systemic lupus erythematosus. Clin Immunol. 2008;129(1):31–35. doi: 10.1016/j.clim.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jönsen A, Yu X, Truedsson L, Nived O, Sturfelt G, Ibrahim S, et al. Mitochondrial DNA polymorphisms are associated with susceptibility and phenotype of systemic lupus erythematosus. Lupus. 2009;18(4):309–312. doi: 10.1177/0961203308097477. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X, Chen F. The effect of lipid peroxides and superoxide dismutase on systemic lupus erythematosus: a preliminary study. Clin Immunol Immunopathol. 1992;63(1):39–44. doi: 10.1016/0090-1229(92)90091-2. [DOI] [PubMed] [Google Scholar]
- 19.Grune T, Michel P, Sitte N, Eggert W, Albrecht-Nebe H, Esterbauer H, et al. Increased levels of 4-hydroxynonenal modified proteins in plasma of children with autoimmune diseases. Free Radic Biol Med. 1997;23(3):357–360. doi: 10.1016/s0891-5849(96)00586-2. [DOI] [PubMed] [Google Scholar]
- 20.Kurien BT, Scofield RH. Free radical mediated peroxidative damage in systemic lupus erythematosus. Life Sci. 2003;73(13):1655–1666. doi: 10.1016/s0024-3205(03)00475-2. [DOI] [PubMed] [Google Scholar]
- 21.Mansour RB, Lassoued S, Gargouri B, El Gaïd A, Attia H, Fakhfakh F. Increased levels of autoantibodies against catalase and superoxide dismutase associated with oxidative stress in patients with rheumatoid arthritis and systemic lupus erythematosus. Scand J Rheumatol. 2008;37(2):103–108. doi: 10.1080/03009740701772465. [DOI] [PubMed] [Google Scholar]
- 22.Shah D, Kiran R, Wanchu A, Bhatnagar A. Oxidative stress in systemic lupus erythematosus: Relationship to Th1 cytokine and disease activity. Immunol Lett. 2010;129(1):7–12. doi: 10.1016/j.imlet.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Iuliano L, Praticò D, Ferro D, Pittoni V, Valesini G, et al. Enhanced lipid peroxidation in patients positive for antiphospholipid antibodies. Blood. 1997;90(10):3931–3935. [PubMed] [Google Scholar]
- 24.Avalos I, Chung CP, Oeser A, Milne GL, Morrow JD, Gebretsadik T, et al. Oxidative stress in systemic lupus erythematosus: relationship to disease activity and symptoms. Lupus. 2007;16(3):195–200. doi: 10.1177/0961203306075802. [DOI] [PubMed] [Google Scholar]
- 25.Morgan PE, Sturgess AD, Davies MJ. Increased levels of serum protein oxidation and correlation with disease activity in systemic lupus erythematosus. Arthritis Rheum. 2005;52(7):2069–2079. doi: 10.1002/art.21130. [DOI] [PubMed] [Google Scholar]
- 26.Morgan PE, Sturgess AD, Davies MJ. Evidence for chronically elevated serum protein oxidation in systemic lupus erythematosus patients. Free Radic Res. 2009;43(2):117–127. doi: 10.1080/10715760802623896. [DOI] [PubMed] [Google Scholar]
- 27.Shah D, Sah S, Wanchu A, Wu MX, Bhatnagar A. Altered redox state and apoptosis in the pathogenesis of systemic lupus erythematosus. Immunobiology. 2013;218(4):620–627. doi: 10.1016/j.imbio.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 29.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 30.Uribe AG, Vilá LM, McGwin G, Jr, Sanchez ML, Reveille JD, Alarcón GS. The Systemic Lupus Activity Measure-revised, the Mexican Systemic Lupus Erythematosus Disease Activity Index (SLEDAI), and a modified SLEDAI-2K are adequate instruments to measure disease activity in systemic lupus erythematosus. J Rheumatol. 2004;31(10):1934–1940. [PubMed] [Google Scholar]
- 31.Liang MH, Socher SA, Larson MG, Schur PH. Reliability and validity of six systems for the clinical assessment of disease activity in systemic lupus erythematosus. Arthritis Rheum. 1989;32(9):1107–1118. doi: 10.1002/anr.1780320909. [DOI] [PubMed] [Google Scholar]
- 32.Liang MH, Fortin PR, Isenberg DA, Snaith L. Quantitative clinical assessment of disease activity in systemic lupus erythematosus: progress report and research agenda. Rheumatol Int. 1991;11(3):133–136. doi: 10.1007/BF00304502. [DOI] [PubMed] [Google Scholar]
- 33.Gladman DD, Goldsmith CH, Urowitz MB, Bacon P, Fortin P, Ginzler E, et al. The Systemic Lupus International Collaborating Clinics/American College of Rheumatology (SLICC/ACR) Damage Index for Systemic Lupus Erythematosus International Comparison. J Rheumatol. 2000;27(2):373–376. [PubMed] [Google Scholar]
- 34.Castro M del R, Suarez E, Kraiselburd E, Isidro A, Paz J, Ferder L, et al. Aging increases mitochondrial DNA damage and oxidative stress in liver of rhesus monkeys. Exp Gerontol. 2012;47(1):29–37. doi: 10.1016/j.exger.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siddiqui A, Rivera-Sánchez S, Castro M del R, Acevedo-Torres K, Rane A, Torres-Ramos CA, et al. Mitochondrial DNA damage is associated with reduced mitochondrial bioenergetics in Huntington's disease. Free Radic Biol Med. 2012;53(7):1478–1488. doi: 10.1016/j.freeradbiomed.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22(2):135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, Pierangeli SS, Papalardo E, Ansari GA, Khan MF. Markers of oxidative and nitrosative stress in systemic lupus erythematosus: correlation with disease activity. Arthritis Rheum. 2010;62(7):2064–2072. doi: 10.1002/art.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bashir S, Harris G, Denman MA, Blake DR, Winyard PG. Oxidative DNA damage and cellular sensitivity to oxidative stress in human autoimmune diseases. Annals of the Rheumatic Diseases. 1993;52(9):659–666. doi: 10.1136/ard.52.9.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lunec J, Herbert K, Blount S, Griffiths HR, Emery P. 8-Hydroxydeoxyguanosine. A marker of oxidative DNA damage in systemic lupus erythematosus. FEBS Lett. 1994;348(2):131–138. doi: 10.1016/0014-5793(94)00583-4. [DOI] [PubMed] [Google Scholar]
- 40.Emlen W, Niebur J, Kadera R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J Immunol. 1994;152(7):3685–3692. [PubMed] [Google Scholar]
- 41.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33(1):1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 42.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 43.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, et al. Apoptosis induced by persistent single-strand breaks in mitochondrial genome. J Biol Chem. 2011;286(37):31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furda AM, Marrangoni AM, Lokshin A, Van Houten B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair (Amst) 2012;11(8):684–692. doi: 10.1016/j.dnarep.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H-T, Lin CS, Chen WS, Liao HT, Tsai CY, Wei YH. Leukocyte mitochondrial DNA alteration in systemic lupus erythematosus and its relevance to the susceptibility to lupus nephritis. Int J Mol Sci. 2012;13(7):8853–8868. doi: 10.3390/ijms13078853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HM, Sugino H, Aoki C, Nishimoto N. Underexpression of mitochondrial-DNA encoded ATP synthesis-related genes and DNA repair genes in systemic lupus erythematosus. Arthritis Res Ther. 2011;13(2):R63. doi: 10.1186/ar3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordero MD, De Miguel M, Moreno Fernández AM, Carmona López IM, Garrido Maraver J, Cotán D, et al. Mitochondrial dysfunction and mitophagy activation in blood mononuclear cells of fibromyalgia patients: implications in the pathogenesis of the disease. Arthritis Res Ther. 2010;12(1):R17. doi: 10.1186/ar2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cordero MD, De Miguel M, Carmona-López I, Bonal P, Campa F, Moreno-Fernández AM. Oxidative stress and mitochondrial dysfunction in fibromyalgia. Neuro Endocrinol Lett. 2010;31(2):169–173. [PubMed] [Google Scholar]
- 49.Kurien BT, Scofield RH. Lipid peroxidation in systemic lupus erythematosus. Indian J Exp Biol. 2006;44(5):349–356. [PubMed] [Google Scholar]
- 50.Hassan SZ, Gheita TA, Kenawy SA, Fahim AT, El-Sorougy IM, Abdou MS. Oxidative stress in systemic lupus erythematosus and rheumatoid arthritis patients: relationship to disease manifestations and activity. Int J Rheum Dis. 2011;14(4):325–331. doi: 10.1111/j.1756-185X.2011.01630.x. [DOI] [PubMed] [Google Scholar]
- 51.Shah D, Wanchu A, Bhatnagar A. Interaction between oxidative stress and chemokines: possible pathogenic role in systemic lupus erythematosus and rheumatoid arthritis. Immunobiology. 2011;216(9):1010–1017. doi: 10.1016/j.imbio.2011.04.001. [DOI] [PubMed] [Google Scholar]