Abstract
Autophagy is a homeostatic process that is important for degrading protein aggregates, nutrient deposits, dysfunctional organelles and several signaling molecules. p62/sequestosome-1 is a protein that binds to several autophagy substrates, such as ubiquitinated proteins, damaged mitochondria and signaling molecules such as an Nrf2 inhibitor Keap1, promoting their autophagic degradation. Sestrin2, a stress-inducible protein, has been recently shown to bind to p62 and promote autophagic degradation of such p62 targets. Because Sestrin2 is a metabolic regulator that suppresses diverse age- and obesity-associated pathologies, the autophagy-controlling function of Sestrin2 may be important for its other physiological functions. However, the molecular mechanism of how Sestrin2 can promote clearance of p62-associated proteins has been unclear. Here we show that Sestrin2 physically associates with ULK1 and p62 to form a complex, in which both Sestrin2 and p62 become phosphorylated by ULK1 at multiple sites. Ser403 of p62, whose phosphorylation is known to promote autophagic degradation of p62 and its targets, is among the sites phosphorylated by ULK1. ULK1-mediated p62 phosphorylation was facilitated by Sestrin2 in cells as well as in vitro kinase assays. Consistent with this finding, oligomycin-induced energy deprivation, which strongly activates ULK1, provoked a robust Ser403 phosphorylation of p62 in wild-type (WT) mouse embryonic fibroblasts (MEF). However, in ULK1/2- and Sestrin2-deficient MEF, oligomycin-induced p62 phosphorylation was dramatically attenuated, suggesting that endogenous Sestrin2-ULK1/2 mainly mediates p62 phosphorylation in response to energetic stress. Taken together, this study identifies ULK1 as a new p62 Ser403 kinase and establishes Sestrin2 as a promoter of ULK1-mediated p62 phosphorylation.
Keywords: Autophagy, ULK1, p62, Sestrin2, phosphorylation
Introduction
Autophagy is a cellular process critical for digesting nutrient deposits, eliminating toxic protein inclusion and dysfunctional organelles and regulating signal transduction machinery by selective degradation of signaling molecules [1]. Autophagy occurs constitutively, but can also be induced in response to cellular stresses in order to maintain metabolic homeostasis [2]. p62/sequestosome-1 protein is an important adaptor protein that binds to diverse autophagy substrates, such as ubiquitinated proteins, damaged mitochondria, microorganism-containing endosomes and an Nrf2 inhibitor Keap1, and promotes their autophagic degradation [3, 4]. p62-mediated degradation of Keap1 leads to activation of Nrf2, which is a transcription factor that promotes expression of various antioxidant molecules to defend cells against oxidative damages [5, 6].
Sestrin2 is a stress-inducible molecule that is important for maintenance of metabolic homeostasis [7]. Genetic deficiency of Sestrin2 in mice or of its homologue in Drosophila results in diverse age- and obesity-associated metabolic pathologies such as accumulation of lipid droplets and protein aggregates, mitochondrial dysfunction, muscle degeneration and insulin resistance [8, 9]. Some of these pathologies are found to be caused by defective autophagy, which can happen due to misregulation of AMP-activated protein kinase (AMPK) and mechanistic target of rapamycin complex 1 (mTORC1) signaling. Sestrin2 associates with AMPK and promotes its activation by an upstream kinase LKB1 [10, 11]. This activation of AMPK can result in mTORC1 inhibition and subsequent autophagy activation [12]. Consistent with its identified biological functions, Sestrin2-deficienct cells are unable to promote autophagy under certain stress conditions [12].
In addition to regulating autophagy through the AMPK-mTORC1 axis, Sestrin2 can have a direct control over autophagy. A recent study showed that Sestrin2 physically associates with p62 and promotes degradation of its substrate Keap1 [13]. During oxidative stress, Sestrin2/p62-mediated Keap1 degradation is important for boosting Nrf2-dependent antioxidant gene transcription to ensure cell survival [13]. However, the molecular mechanism of why p62-Sestrin2 association is important for autophagic degradation of Keap1 and other p62 targets has been obscure. In this study, we demonstrate that Sestrin2 and p62 physically associate with Unc-51-like protein kinase 1 (ULK1), an autophagy-initiating protein kinase. ULK1 phosphorylates p62, and it is Sestrin2’s role to promote the phosphorylation of p62 by ULK1. Because p62 phosphorylation is known to facilitate its own degradation [14-16], our results explain how Sestrin2 can promote autophagic degradation of p62 and its substrates.
Results
Physical association among Sestrin2, ULK1 and p62
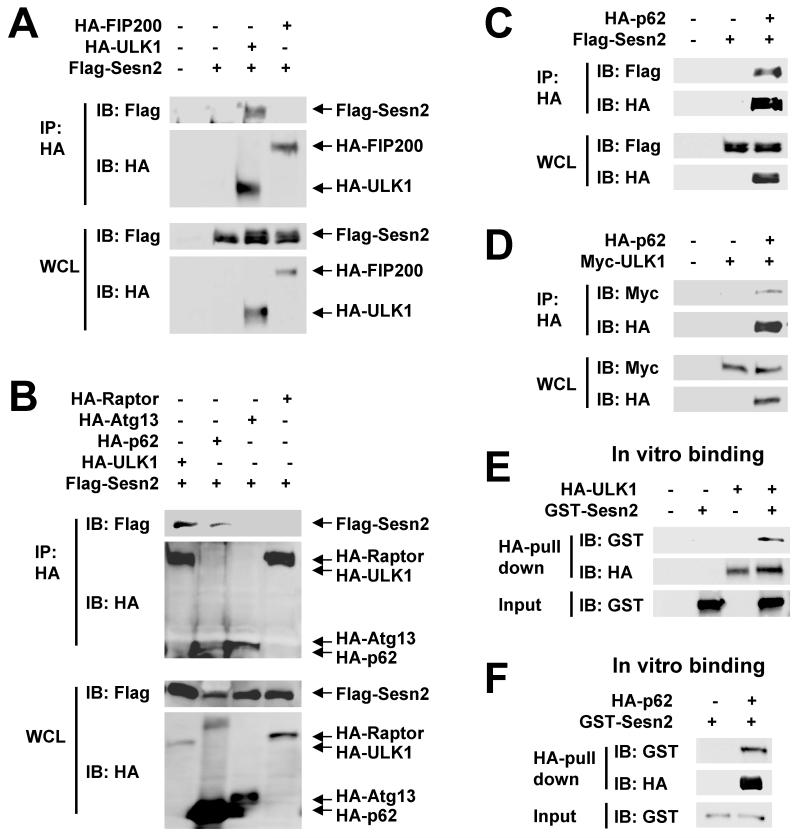
To understand how Sestrin2 regulates autophagy, we tested for physical interaction between Sestrin2 and known autophagy-regulating proteins. As a result, we identified ULK1 as a new binding partner for Sestrin2; ULK1 and p62, but not ULK1’s regulatory subunits Atg13 and FIP200, were shown to physically bind to Sestrin2 in co-immunoprecipitation assays (Fig. 1A-C). We found that p62 also interacted with ULK1 in cells (Fig. 1D). The associations between Sestrin2 and ULK1 and between Sestrin2 and p62 were also observed in vitro; recombinant Sestrin2 proteins expressed in and purified from E. coli were able to bind to ULK1 and p62 in mammalian cell lysates (Fig. 1E,F).
Fig. 1. Sestrin2 physically associates with ULK1 and p62.
(A-D) ULK1 and p62 are associated with Sestrin2 in cells. HEK293 cells were transfected with plasmids expressing indicated proteins. HA-tagged proteins were immunopurified (IP) from whole cell lysates (WCL) and subjected to immunoblotting (IB). FIP200 and Atg13 are regulatory subunits of ULK1, and Raptor was used as a negative control. (E,F) ULK1 and p62 bind to Sestrin2 in vitro. HEK293 cells were transfected with plasmids expressing HA-tagged ULK1 (E) or p62 (F). Cell lysates were incubated with GST-Sestrin2 recombinant protein expressed and purified from E. coli, and immunoprecipitated using anti-HA antibody. Pulled-down GST-Sestrin2 proteins were assayed by immunoblotting.
Each of the three Sestrin2 subdomains was sufficient to bind to ULK1 or p62
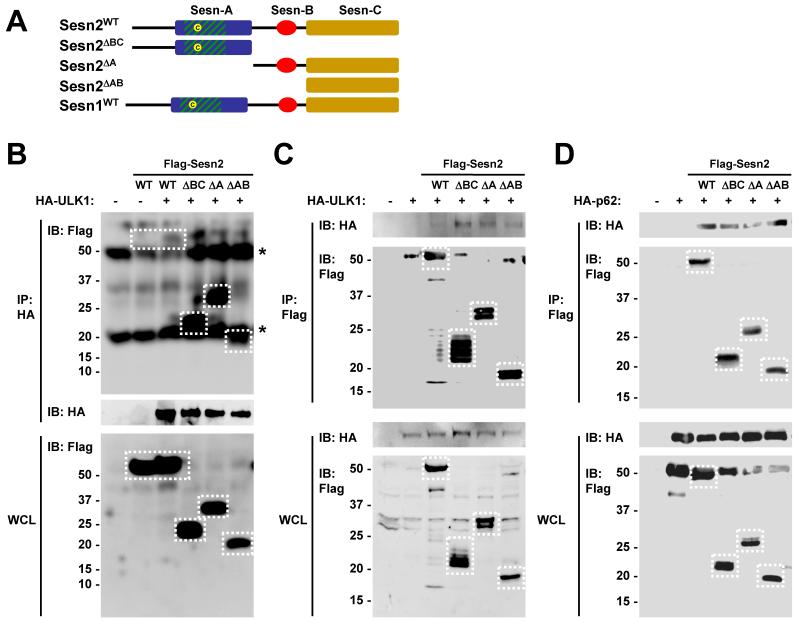
As previously determined through phylogenic analyses [17], Sestrin proteins contain three subdomains designated as Sesn-A, Sesn-B and Sesn-C (Fig. 2A). Thus, we determined which subdomain of Sestrin2 is required for association with ULK1 and p62. Truncated Sestrin2 mutants lacking Sesn-B and Sesn-C domains (Sesn2ΔBC), Sesn-A domain (Sesn2ΔA) or Sesn-A and Sesn-B domains (Sesn2 ΔAB) were used in the protein binding assays. Sesn2ΔBC, Sesn2 ΔA and Sesn2 ΔAB, as well as the full-length form of Sestrin2 (Sesn2WT), were able to physically associate with ULK1 (Fig. 2B,C) and with p62 (Fig. 2D), suggesting that each of the subdomains of Sestrin2 can independently interact with ULK1 and p62. Interestingly, truncated Sestrin2 proteins associated with ULK1 more strongly than the full-length form (Fig. 2B,C), introducing the possibility that each subdomain may have an inhibitory effect on the others when interacting with ULK1.
Fig. 2. Mapping the ULK1- and p62-interacting domains of Sestrin2.
(A) Diagram depicting domain structure of wild type and truncated mutants of Sestrin2. (B-D) HEK293 cells were transfected with indicated proteins, subjected to immunoprecipitation (IP) using indicated antibodies. Whole cell lysates and purified immunocomplexes were analyzed by immunoblotting (IB). Stars denote non-specific bands corresponding to immunoglobulin heavy or light chain in the immunocomplexes.
C-terminal domain of ULK1 is necessary and sufficient to bind to Sestrin2
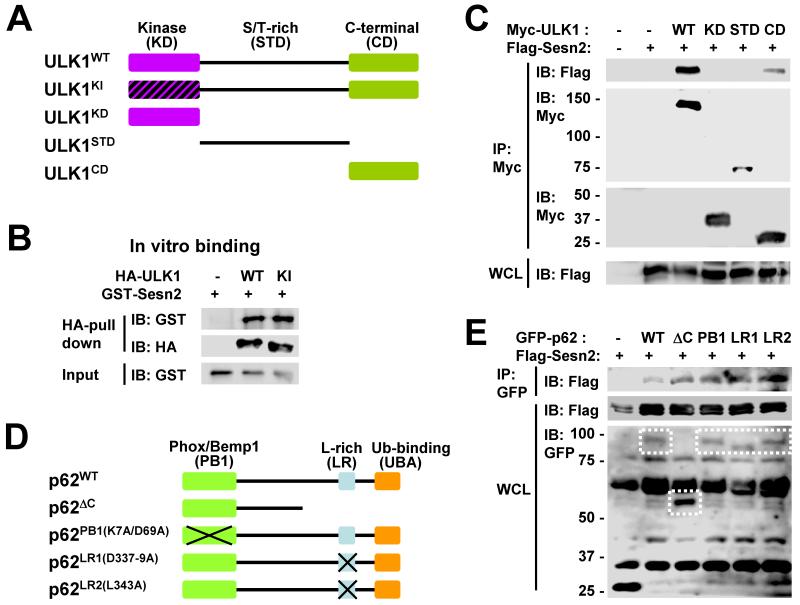
It was then determined which subdomains of ULK1 and p62 were required for association with Sestrin2. ULK1 consists of three subdomains: N-terminal kinase domain (KD), central serine/threonine-rich domain (STD) and C-terminal domain (CD) [18]. We utilized truncated ULK1 mutants containing each of these domains and a kinase-inactive (KI) ULK1 mutant [18] (Fig. 3A). The catalytic activity of ULK1 was not required for interaction with Sestrin2, as ULK1KI was able to bind to Sestrin2 in vitro as efficiently as wild-type (WT) ULK1 (Fig. 3B). Although CD of ULK1 was able to bind to Sestrin2, KD and STD were unable to associate with Sestrin2 (Fig. 3C), suggesting that Sestrin2 specifically binds to CD of ULK1. Because CD interacts with many proteins that regulate ULK1 and is important for ULK1 function and localization [19], it can be speculated that Sestrin2 may control ULK1 activity through physical association.
Fig. 3. Mapping the Sestrin2-interacting domains of ULK1 and p62.
(A) Diagram depicting domain structure of ULK1 and its mutant proteins. Kinase-inactive (KI) mutants as well as truncated proteins with kinase domain (KD), Ser/Thr-rich domain (STD) and C-terminal domain (CD) were illustrated. (B) Both ULK1WT and ULK1KI were able to pull down GST-Sestrin2 in vitro. HEK293 cells were transfected with plasmids expressing HA-tagged ULK1WT or ULKKI. Cell lysates were incubated with GST-Sestrin2 recombinant protein and immunoprecipitated using anti-HA antibody. Pulled-down GST-Sestrin2 proteins were assayed by immunoblotting (IB). (C) HEK293 cells were transfected with plasmids expressing indicated proteins. Myc-tagged proteins were immunopurified (IP) from whole cell lysates (WCL) and subjected to immunoblotting. (D) Diagram depicting domain structure of p62 and its mutant proteins. PB1 (Phox/Bemp1), LR (leucine-rich) and UBA (ubiquitin-binding) domains were truncated or mutated. (E) HEK293 cells were transfected with plasmids expressing indicated proteins. GFP-tagged proteins were immunopurified from whole cell lysates and subjected to immunoblotting.
Functionality of PB1, LR and UBA domains is dispensable for p62 binding to Sestrin2
p62 contains three notable protein-protein interaction domains: N-terminal Phox/Bemp1 domain (PB1) which associates with diverse signaling molecules, central leucine-rich domain (LR) which associates with an autophagosome marker LC3 and C-terminal ubiquitin-binding-associated domain (UBA) [3] (Fig. 3D). Deletion or mutation of each p62 domain (Fig. 3D) did not abolish its ability to interact with Sestrin2 (Fig. 3E), suggesting that the functionality of these domains is generally dispensable for association with Sestrin2.
Sestrin2 interacts with ULK1-Atg13-FIP200 complex
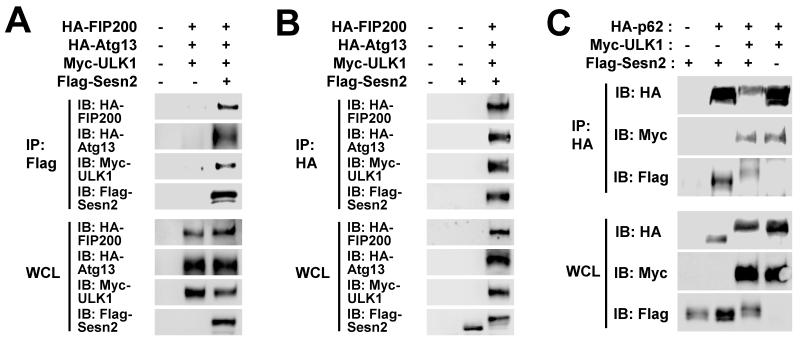
In mammalian cells, ULK1 is present in an autophagy-initiating protein kinase complex containing Atg13 and FIP200 [20-22]. Although Sestrin2 did not show direct physical interaction with Atg13 and FIP200 (Fig. 1A,B), we wondered if these subunits could associate with Sestrin2 through ULK1. Indeed, ULK1 co-transfection enabled Atg13 and FIP200 to interact with Sestrin2 (Fig. 4A,B), indicating that Sestrin2 can bind to the ULK1-Atg13-FIP200 complex as a whole.
Fig. 4. Association between Sestrin2, ULK1-Atg13-FIP200 and p62.
(A-C) Sestrin2 associates with Atg13-FIP200 and p62 when ULK1 is co-expressed. HEK293 cells were transfected with plasmids expressing indicated proteins. Immunoprecipitated proteins (IP) and whole cell lysates (WCL) were subjected to immunoblotting (IB) with indicated antibodies. Note that ULK1 co-transfection induces electromobility retardation (band shift) of Sestrin2 (B,C) and p62 (C).
ULK1 phosphorylates p62 at the Ser403 site
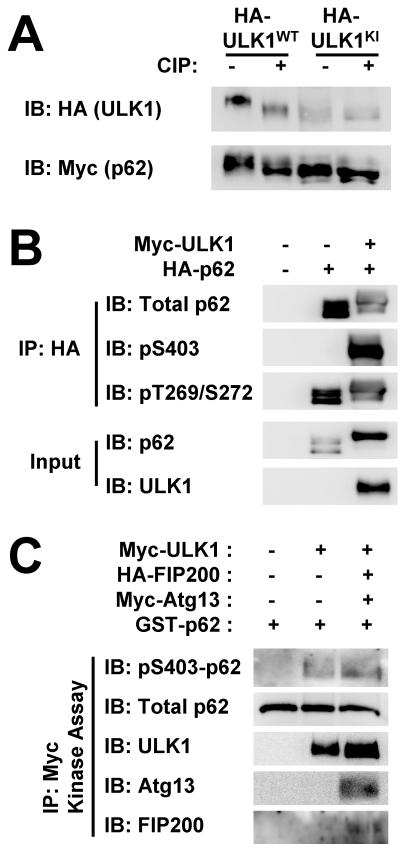
After transfection of Sestrin2, ULK1 and p62, we noticed that co-transfection of ULK1 induced an electromobility retardation (band shift) of p62 and Sestrin2 (Fig. 4B,C), indicating post-translational modification of the proteins. The band shift of p62 became slightly more pronounced when both ULK1 and Sestrin2 were expressed, compared to ULK1 presence alone (Fig. 4C; note that the bottom half of the p62 bands have been shifted up by Sestrin2 co-expression). As ULK1 is a protein kinase, it was investigated further whether the band shift of p62 was caused by ULK1-mediated phosphorylation. Indeed, ULK1KI was unable to produce the band shift of p62 (Fig. 5A), suggesting that it is dependent on the catalytic activity of ULK1. The band shift of p62 was reverted by CIP treatment (Fig. 5A), confirming that phosphorylation took place and caused the electromobility retardation.
Fig. 5. ULK1 phosphorylates p62 at Ser403.
(A) ULK1-induced band shift of p62 is phosphatase-sensitive. HEK293 cells were transfected with HA-tagged ULK1WT or ULK1KI. After treatment with calf intestinal phosphatase (CIP), cell lysates were analyzed by immunoblotting. (B) ULK1 induces Ser403 phosphorylation of p62. HEK293 cells were transfected with plasmids expressing indicated proteins. Anti-HA immunocomplexes (IP) and whole cell lysates (WCL) were analyzed by immunoblotting. (C) HEK293 cells were transfected with Myc-tagged ULK1 and its regulatory subunits FIP200 and Atg13. ULK1 kinase activity of anti-Myc immunocomplexes was assayed using recombinant GST-p62 protein. Amounts of phosphorylated and total proteins were assayed through immunoblotting.
It has been shown that Ser403 site on p62 is phosphorylated by casein kinase 2 (CK2) and TBK1 [14, 15], while Thr269 and Ser272 sites are phosphorylated by cyclin-dependent kinase 1 (CDK1) [23]. We tested if ULK1 could induce p62 phosphorylation at these known sites. ULK1 co-transfection with p62 prominently increased phosphorylation of p62 at Ser403 site, whereas no such effect was observed at Thr269 and Ser272 sites (Fig. 5B), demonstrating that ULK1 promoted phosphorylation of p62 at Ser403 site.
We then performed in vitro kinase assays to confirm that ULK1-mediated p62 phosphorylation was the result of a direct biochemical reaction. Immunopurified ULK1 was able to induce Ser403 phosphorylation of p62 in vitro (Fig. 5C). When ULK1 was co-purified with its regulatory subunits Atg13 and FIP200, the level of phosphorylation was increased (Fig. 5C), reflecting the enhanced catalytic activity of ULK1 by these subunits [20-22].
Sestrin2 promotes ULK1-mediated phosphorylation of p62
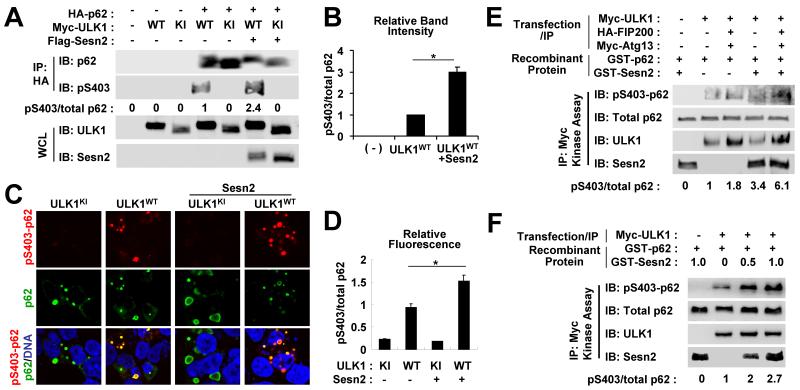
Because the ULK1-mediated band shift of p62 was slightly enhanced upon Sestrin2 co-transfection (Fig. 4C), we tested if Sestrin2 could promote this phosphorylation. Ser403 phosphorylation of p62, which was induced by ULK1WT but not ULK1KI, became much stronger upon co-expression with Sestrin2 (Fig. 6A,B). We also confirmed this finding using immunocytochemistry where total p62 and phospho-Ser403 p62 were stained green and red respectively. Red phospho-p62 signal was hardly detectable in ULK1KI-transfected cells while cells with ULK1WT transfection showed considerable red signals (Fig. 6C,D). Merged images of ULK1WT-transfected cells revealed many yellow spots that represented phosphorylated p62 aggregates in addition to some remaining green spots that implied that not all p62 aggregates were phosphorylated by ULK1WT (Fig. 6C,D). However, when Sestrin2 was co-transfected with ULK1, most p62 aggregates were either yellow or even reddish yellow (Fig. 6C,D), suggesting stronger ULK1-mediated p62 phosphorylation. These data demonstrate that Sestrin2 can promote ULK1-mediated phosphorylation of p62 in mammalian cells.
Fig. 6. Sestrin2 promotes ULK1-mediated phosphorylation of p62.
(A,B) HEK293 cells were transfected with plasmids expressing indicated proteins. Anti-HA immunocomplexes (IP) and whole cell lysates (WCL) were subjected to immunoblotting (IB). Relative phosphorylation of p62 at Ser403 (pS403/total p62) was quantified by densitometry and presented as numbers below each lane (A) and as a graph (B, n = 3). (C,D) HEK293 cells were transfected with plasmids expressing indicated proteins and immunostained with anti-phospho Ser403 p62 (red) and anti-total p62 (green) antibodies and DAPI (DNA, blue). Relative fluorescence of phosphorylated and total p62 in each aggregate was quantified and presented as a graph (n ≥ 20). (E,F) HEK293 cells were transfected with Myc-tagged ULK1 and its regulatory subunits FIP200 and Atg13. ULK1 kinase activity of anti-Myc immunocomplexes was assayed using recombinant GST-p62 protein. GST-Sesn2 protein (0.5 μg or as indicated) was directly added to the kinase reaction mixture. Amounts of phosphorylated and total proteins were assayed through immunoblotting. Relative phosphorylation of p62 at Ser403 (pS403/total p62) was quantified by densitometry and presented as numbers below each lane. *P < 0.05.
Because Sestrin2 can activate AMPK and inhibit mTORC1 [10], which can together increase ULK1 activity, it was not clear whether the direct physical association among Sestrin2, ULK1 and p62 was what contributed to the enhanced p62 phosphorylation. To rule out any involvement of indirect signaling pathways, we immunopurified ULK1 from transfected HEK293 cells and performed a kinase reaction with the addition of recombinant Sestrin2 and p62. Through this assay, we tested if Sestrin2 could directly promote ULK1-mediated p62 phosphorylation in vitro. ULK1-mediated Ser403 phosphorylation of p62 was strongly enhanced upon addition of recombinant Sestrin2 (Fig. 6E), suggesting that Sestrin2 can directly facilitate p62 phosphorylation by ULK1 without the involvement of indirect signaling pathways such as AMPK and mTORC1. The in vitro promotion of ULK1-mediated p62 phosphorylation by Sestrin2 was dose-dependent (Fig. 6F).
Endogenous Sestrin2 and ULK1 control p62 phosphorylation
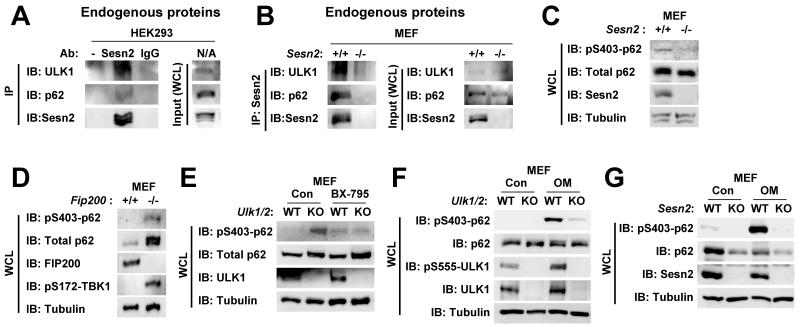
Through a co-immunoprecipitation assay, we found that endogenous Sestrin2 also associated with endogenous ULK1 and p62 in mammalian cells (Fig. 7A,B). Thus, we asked if endogenous Sestrin2 and ULK1 could regulate p62 phosphorylation as well. Consistent with our findings, Sestrin2-deficient mouse embryonic fibroblasts (MEF) exhibited a substantial decrease in Ser403 phosphorylation of p62 when compared to the control MEF (Fig. 7C). However, very unexpectedly, genetic deletion of FIP200, which completely nullifies activities of both ULK1 and its close homolog ULK2 in regulating autophagy [24], or concomitant deficiency of ULK1 and ULK2 modestly increased Ser403 phosphorylation of p62 when compared to the WT control MEF (Fig. 7D,E). Because Ser403 residue of p62 can be phosphorylated by other kinases such as CK2 and TBK1 [14, 15], it is possible that absence of ULK1 activity may have induced compensatory upregulation of these kinases. A recent report supported this idea by demonstrating that ULK1 inhibits TBK1 [25], and consistent with this report, we found that TBK1 is prominently activated in FIP200-deficient MEF (Fig. 7D). Furthermore, treatment of ULK1/2-deficient MEF with a TBK1 inhibitor BX-795 dramatically suppressed the p62 phosphorylation increased by the ULK1/2 loss (Fig. 7E). Therefore, even though ULK1 can directly phosphorylate p62, complete absence of ULK1 provokes slightly more p62 phosphorylation through indirect feedback activation of TBK1.
Fig. 7. Effect of Sestrin2 and FIP200 deficiencies on p62 phosphorylation levels.
(A,B) Physical association among endogenous Sestrin2, p62 and ULK1. HEK293 (A) and Sesn2+/+ and Sesn2−/− MEF (B) were treated with 100 μM etoposide for 16 hr to maximize endogenous Sestrin2 expression. Sestrin2 was immunopurified using protein G/A-conjugated anti-Sestrin2 antibody. Protein G/A-conjugated pre-immune IgG and empty protein G/A beads (−) were used as negative controls. Both anti-Sestrin2 immunocomplexes (IP) and whole cell lysates (WCL) were assayed through immunoblotting (IB). (C) Reduction of p62 Ser403 phosphorylation in Sestrin2-deficienct mouse embryonic fibroblasts (MEF). Whole cell lysates from wild-type (Sesn2+/+) and Sesn2−/− MEF were subjected to immunoblotting with indicated antibodies. (D) Induction of p62 Ser403 phosphorylation in FIP200-deficient MEF. Whole cell lysates from wild-type (Fip200+/+) and Fip200−/− MEF were subjected to immunoblotting with indicated antibodies. (E) Induction of basal p62 Ser403 phosphorylation in ULK1/2-deficient MEF was suppressed by BX-795, an inhibitor of TBK1. Wild-type (WT, Ulk1+/+/Ulk2+/+) and Ulk1−/−/Ulk2−/− (KO) MEF were treated with PBS (Con) or 10 μM BX-795 for 6 h, and subjected to immunoblotting with indicated antibodies. (F,G) Oligomycin-mediated energy depletion strongly induced Ser403 phosphorylation of p62 in WT MEF, but not as robustly in ULK1/2- or Sestrin2-deficient MEF (KO). WT (Ulk1+/+/Ulk2+/+ and Sesn2+/+ controls), Ulk1−/−/Ulk2−/− and Sesn2−/− MEF were treated with PBS (Con) or 10 μg/mL oligomycin for 6 h, and subjected to immunoblotting with indicated antibodies.
Sestrin2 and ULK1/2 mediate p62 phosphorylation under energetic stress
Cellular energy deprivation leads to activation of AMPK, which phosphorylates and subsequently activates ULK1 [26]. Consistent with previous reports, oligomycin, which depletes cellular ATP, elevated AMPK-mediated activatory phosphorylation of ULK1 at Ser555 (Fig. 7F). Intriguingly, Ser403 phosphorylation of p62 was very robustly increased by oligomycin treatment in WT MEF (Fig. 7F,G). The oligomycin-induced Ser403 phosphorylation of p62 was dramatically attenuated in ULK1/2-deficient MEF and Sestrin2-deficient MEF, indicating that endogenous Sestrin2-ULK1/2 mediates p62 phosphorylation during energetic stress.
ULK1 phosphorylates Sestrin2 at multiple sites
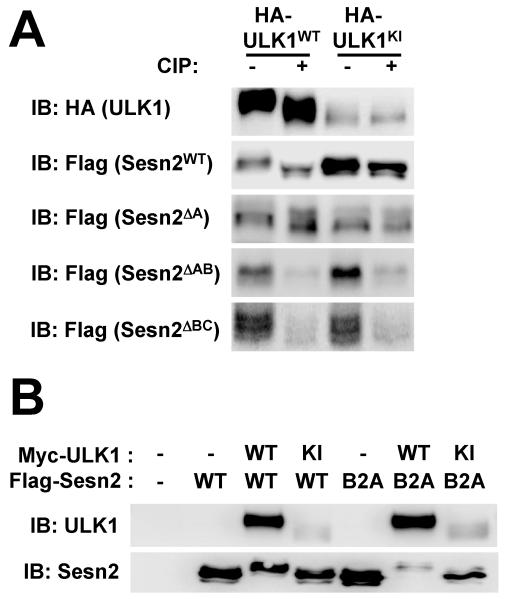
Finally, we examined if the ULK1-induced electromobility retardation of Sestrin2 (Fig. 4B,C) was caused by ULK1-mediated phosphorylation as well. The band shift of Sestrin2 was induced only by ULK1WT but not by ULK1KI (Fig. 6A and Fig. 8A,B), showing that the shift was dependent on the catalytic activity of ULK1. CIP treatment reverted the band shift (Fig. 8A), further demonstrating that the shift was caused by ULK1-mediated phosphorylation. We then attempted to identify the ULK1-mediated phosphorylation site(s) on Sestrin2. Because no post-translational modification has been previously reported for Sestrin2, we subjected truncated mutants of Sestin2 (Sesn2 ΔA, Sesn2 ΔAB and Sesn2ΔBC) to the band shift assays to identify possible domain(s) of phosphorylation. Sesn2ΔA exhibited a band shift upon ULK1WT expression while Sesn2ΔAB did not (Fig. 8A), suggesting that Sesn-B domain contains ULK1-dependent phosphorylation sites. However, Sesn2ΔBC also exhibited slight band shift upon ULK1WT expression, which is diminished by either CIP treatment or ULK1KI mutation (Fig. 8A), showing that there may be additional phosphorylation sites in Sesn-A domain. Mutating all Ser/Thr residues in Sesn-B domain (Sesn2B2A) did not abolish the band shift of Sestrin2 (Fig. 8B), confirming that ULK1 phosphorylates Sestrin2 at multiple sites.
Fig. 8. ULK1 phosphorylates Sestrin2 at multiple sites.
(A) HEK293 cells were transfected with HA-tagged ULK1WT or ULKKI and Flag-tagged wild-type or truncated Sestrin2 proteins. Cell lysates were treated with calf intestinal phosphatase (CIP) and assayed through immunoblotting (IB). (B) HEK293 cells were transfected with plasmids expressing indicated proteins. Cell lysates were assayed through immunoblotting. Sesn2B2A is a Sestrin2 mutant protein whose Ser/Thr residues in Sesn2-B domain were all substituted with Ala.
Discussion
In this study, we revealed a new function of Sestrin2 in promoting ULK1-dependent phosphorylation of p62. In addition to previously known functions of Sestrin2 in regulating AMPK and mTORC1 signaling pathways (Fig. 9A), this new function provides an explanation of how Sestrin2 specifically contributes to selective autophagy of ubiquitinated proteins, dysfunctional mitochondria and a signaling molecule Keap1, which are all mediated by ULK1 and p62 [4-6, 27]. Thus we can now explain why Sestrin2-deficient cells and tissues can accumulate large amount of ubiquitinated protein aggregates and dysfunctional mitochondria [8] and exhibit defective Nrf2 signaling that exacerbates oxidative damage during environmental stresses [13]. By controlling ULK1-mediated Ser403 phosphorylation of p62 that is required for efficient removal of p62 targets [14-16], Sestrin2 can promote selective autophagy (Fig. 9B). The Sestrin2/p62-mediated direct activation of selective autophagy can be further enhanced upon indirect upstream modulation of AMPK and mTORC1 signaling pathways, which is already known to promote both general and selective autophagy processes [18].
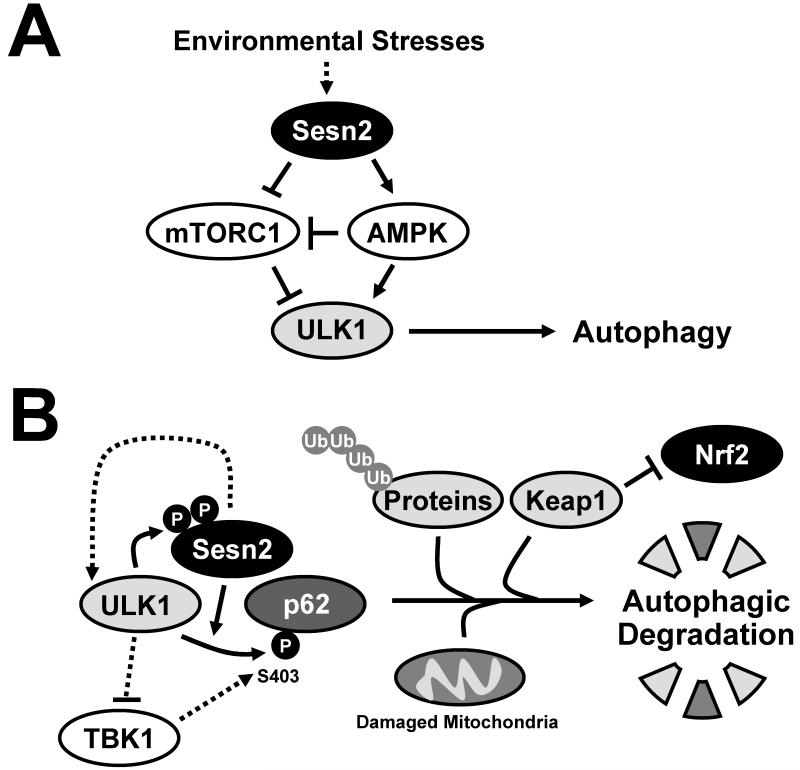
Fig. 9. Proposed roles of Sestrin2 and ULK1 in regulation of p62-mediated selective autophagy.
(A) From previous studies, Sestrin2 was shown to be a mediator of stress-induced autophagy. Stress-inducible Sestrin2 can provoke ULK1 activation through activating AMPK and inhibiting mTORC1. (B) In this study, Sestrin2 was shown to associate with ULK1 and p62 and directly promote ULK1-dependent phosphorylation of p62 at Ser403. Because phosphorylation of this site is known to facilitate autophagic degradation of p62 targets, our study provides an explanation on how Sestrin2 is required for proper elimination of ubiquitinated protein aggregates and dysfunctional mitochondria and for appropriate regulation of the Keap1-Nrf2 signaling pathway.
Although ULK1 is one of the first molecules to be identified as an autophagy mediator [27, 28], finding its biochemical substrates had been extremely difficult [29]. It is only recently discovered that ULK1 phosphorylates several autophagy regulators, such as AMBRA1 [30], Beclin-1 [31], ZIPK [32] and Atg9 [33]. In addition to these known substrates, we showed that p62 can be another important substrate of ULK1 mediating its selective autophagy-regulating function. Because ULK1 can associate with p62 even when autophagosome formation is blocked by Vps34 inhibition [34], ULK1-mediated p62 regulation may be one of the early steps of selective autophagy prior to ULK1-mediated Beclin-1/Vps34 [30, 31] or Atg9/myosin II [32, 33] regulation. Therefore, in addition to contributing to carrying out autophagy through autophagosome membrane formation and growth [30-33], ULK1 can initiate the selective autophagy process by phosphorylating p62 and bringing both p62 and its substrates to the autophagosome formation sites.
p62 is a signaling adaptor protein that has many cellular functions [3]. Recent reports suggest that phosphorylation of p62 can alter its function in regulating autophagy [14-16] and cell cycle [23]. Specifically, phosphorylation of p62 at Ser403 site is important for degradation of p62 and its target molecules [14, 15]. Since ULK1 is a well-known autophagy initiating kinase [27], and Sestrin2 is a recently characterized promoter of autophagy [8, 12, 13], our study provides a mechanism of how ULK1 and Sestrin2 can come together to facilitate selective autophagy by inducing Ser403 phosphorylation of p62. This mechanism seems to have strong physiological significance in the context of energy deprivation where Ser403 phosphorylaion of p62 is dramatically increased in a Sestrin2- and ULK1/2-dependent manner.
Materials and Methods
Plasmids, antibodies and chemicals
Flag-Sesn2WT, Flag-Sesn2ΔBC, Flag-Sesn2ΔA and Flag-Sesn2ΔAB were subcloned into pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA, USA). HA-ULK1WT, HA-ULK1KI, Myc-ULK1WT, Myc-ULK1KD, Myc-ULK1STD and Myc-ULK1CD were from K.L. Guan (UC San Diego) and M. Aghajan (Isis Pharm. Inc.) [18]. GFP-p62WT, GFP-p62ΔC, GFP-p62PB1(K7A/D69A), GFP-p62LR1(D337-9A) and GFP-p62LR2 (L343A) were from Addgene, Cambridge, MA, USA [34]. Plasmids expressing Myc-ULK1WT, Myc-ULK1KI, Myc-Atg13 and HA-Atg13 were from D.H. Kim (Univ. of Minnesota) [22]. Plasmids expressing HA-FIP200 and HA-p62 were from J.L. Guan (Univ. of Cincinnati) [24, 35]. Antibodies to detect ULK1 (4773S), p62 (5114S), phospho-Thr269/Ser272 p62 (13121S), phospho-Ser555 ULK1 (5869), phospho-Ser172 TBK1 (5483P) and TBK1 (3013S) were from Cell Signaling Technology, Danvers, MA, USA. Anti-Sestrin2 (10795-1-AP) antibody was from Proteintech, Chicago, IL, USA. For immunoprecipitation experiments, we generated additional anti-Sestrin2 antibodies from guinea pigs (PRF&L, Canadensis, PA, USA) using recombinant GST-Sestrin2 proteins purified from E. coli. Control IgG was prepared from pre-immune serum of the same animals. Anti-phospho-Ser403 p62 antibody (MABC186) was from Milipore, Billerica, MA, USA. Anti-FIP200 antibody was from J.L.Guan (Univ. of Cincinnati) [36]. Anti-hemagglutinin (HA) antibody (3F10) was from Roche, Penzberg, Upper Bavaria, Germany. Anti-Myc (9E10) and anti-GFP (12A6) antibodies were from Developmental Studies Hybridoma Bank, Iowa City, IA, USA. Anti-GST antibody (554805) was from BD Biosciences, Franklin Lakes, NJ, USA. Anti-Flag (M2) and anti-Tubulin (T5168) antibodies, as well as etoposide and oligomycin, were from Sigma, St. Louis, MO, USA. BX-795 was obtained from Selleckchem, Houston, TX, USA.
Cell culture and Transfection
HEK293 and MEF cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) containing 10% fetal bovine serum (FBS) and penicillin/streptomycin at 37°C in 5% CO2. Sesn2−/−, Fip200−/− and Ulk1−/−/Ulk2−/− MEF cells were previously described [9, 24, 37]. For transient expression of proteins, HEK293 cells were transfected with purified plasmid constructs using Lipofectamin 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were harvested two days after transfection for immunoblotting, immunoprecipitation, immunocytochemistry or other biochemical assays.
Immunocytochemistry
Two days after transfection, HEK293 cells were seeded onto glass coverslips. On the following day, cells were washed with PBS and fixed with 4% paraformaldehyde. After permeabilizing the cells with 0.3% Triton X-100, they were incubated overnight with primary antibodies. The cells were then washed with PBS and were incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 30 min and counterstained with DAPI (Invitrogen) [38]. Samples were analyzed under a FluoView 500 laser confocal microscope (Olympus, Tokyo, Japan).
Immunoprecipitation
Cell lysates were prepared in a lysis buffer (20 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM NaPPi, 1 mM β-glycerophosphate and 1 mM Na3VO4) containing 0.3% CHAPS and protease inhibitor cocktail (Roche), and immunoprecipitated with anti-HA (A2095, Sigma) or anti-Flag (A2220, Sigma) agarose bead or other antibodies conjugated to a protein G/A bead (Calbiochem, Darmstadt, Germany). The immunocomplexes were then washed four times with the lysis buffer and analyzed through immunoblotting [39].
In vitro pull-down assay
Full-length mouse Sestrin2 cDNA was cloned in pGEX-4T-1 (Amersham) and expressed in BL21 (DE3) E. Coli. The recombinant proteins were purified using a Glutathione-Sepharose 4B column (Amersham, Amersham, UK). Cell lysates prepared from HEK293 cells transfected with HA-ULK1 or HA-p62 were incubated with GST-Sestrin2 at 37°C for 3 hr. After the incubation, HA-ULK1 or HA-p62 was immunoprecipitated, and the presence of GST-Sestrin2 in the immunocomplexes was examined through immunoblotting (see below).
In vitro kinase assay
Myc-ULK1WT was purified from transfected HEK293 cells through immunoprecipitation using a protein G/A bead-conjugated anti-myc antibody. The kinase reaction was performed in a solution of 25 mM MOPS, pH 7.5, 1 mM EGTA, 0.1 mM Na3VO4, 15 mM MgCl2 and 100 μM ATP. GST-p62 (BML-UW1035; Enzo Life Sciences, Farmingdale, NY, USA) was used as a substrate. GST-Sestrin2 was directly added to the reaction. The reaction mixtures were incubated at 37°C for 1 hr.
Calf intestinal alkaline phosphatase (CIP) assay
Cell lysates were prepared in the lysis buffer described above and incubated with 1 unit/μL CIP (M0290S, New England Biolabs, Ipswich, MA, USA) at 30°C overnight [39].
Immunoblotting
Cell lysates, immunocomplexes and enzymatic reaction mixtures were boiled in SDS sample buffer for 5 min, separated by SDS-PAGE, transferred to PVDF membranes and probed with primary antibodies. After incubation with secondary antibodies conjugated with HRP, chemiluminescence was detected using LAS4000 (GE, Fairfield, CT, USA) systems [9]. All of the contrast adjustments were linear and uniformly applied to the whole image in each panel. Quantification of relative band intensities was done by densitometry. P values were calculated using Student’s t test.
Acknowledgments
We thank Drs. M. Karin, K.L. Guan (UCSD), A.V. Budanov (VCU), S. Tooze (Cancer Research UK), A. Saltiel, R.A. Miller, L. Rui, D. Lombard, S. Pletcher (UM), D.H. Kim (UMN), J.L. Guan (Univ. of Cincinnati), M. Aghajan (Isis Pharm. Inc.) and Santa Cruz Biotech Inc. for sharing cell lines, reagents and access to lab equipment. We also thank Dr. S. Hwang (Univ. of Chicago) for valuable discussions. This work was supported by grants from American Association for the Study of Liver Diseases/American Liver Foundation, Ellison Medical Foundation (AG-NS-0932-12) and NIH (P30AG024824, P30AG013283, P30DK034933, P30DK089503, P30CA046592 and R21AG045432).
Abbreviations
- AMPK
AMP-activated protein kinase
- mTORC1
mechanistic target of rapamycin complex 1
- ULK1
Unc-51-like protein kinase 1
- MEF
mouse embryonic fibroblasts
- TBK1
Tank-binding protein kinase 1
- CK2
casein kinase 2
- CDK1
cyclin-dependent kinase 1
- CIP
Calf intestinal alkaline phosphatase
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 6.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Budanov AV, Karin M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013;18:792–801. doi: 10.1016/j.cmet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Budanov AV, Talukdar S, Park EJ, Park H, Park H-W, Bandyopadhyay G, Li N, Aghajan M, Jang I, et al. Maintenance of metabolic homeostasis by Sestrin2 and Sestrin3. Cell Metab. 2012;16:311–321. doi: 10.1016/j.cmet.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanli T, Linher-Melville K, Tsakiridis T, Singh G. Sestrin2 modulates AMPK subunit expression and its response to ionizing radiation in breast cancer cells. PLoS One. 2012;7:e32035. doi: 10.1371/journal.pone.0032035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 13.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, Rhee SG. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Budanov AV, Lee JH, Karin M. Stressin’ Sestrins take an aging fight. EMBO Mol Med. 2010;2:388–400. doi: 10.1002/emmm.201000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linares JF, Amanchy R, Greis K, Diaz-Meco MT, Moscat J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol Cell Biol. 2011;31:105–117. doi: 10.1128/MCB.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alers S, Loffler AS, Wesselborg S, Stork B. The incredible ULKs. Cell Commun Signal. 2012;10:7. doi: 10.1186/1478-811X-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 29.Deminoff SJ, Herman PK. Identifying Atg1 substrates: four means to an end. Autophagy. 2007;3:667–673. doi: 10.4161/auto.4771. [DOI] [PubMed] [Google Scholar]
- 30.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011;30:636–651. doi: 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of atg9 by the atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbi S, Ueda H, Zheng C, Cooper LA, Zhao J, Christopher R, Guan JL. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol Biol Cell. 2002;13:3178–3191. doi: 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda H, Abbi S, Zheng C, Guan JL. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol. 2000;149:423–430. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAlpine F, Williamson LE, Tooze SA, Chan EY. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013;9:361–373. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ro SH, Jung CH, Hahn WS, Xu X, Kim YM, Yun YS, Park JM, Kim KH, Seo M, Ha TY, et al. Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy. 2013;9:2103–2114. doi: 10.4161/auto.26563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim M, Park HL, Park HW, Ro SH, Nam SG, Reed JM, Guan JL, Lee JH. Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy. 2013;9:1201–1213. doi: 10.4161/auto.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]